ABSTRACT
Higher plant vasculature is characterized by two distinct developmental phases. Initially, a well-defined radial primary pattern is established. In eudicots, this is followed by secondary growth, which involves development of the cambium and is required for efficient water and nutrient transport and wood formation. Regulation of secondary growth involves several phytohormones, and cytokinins have been implicated as key players, particularly in the activation of cell proliferation, but the molecular mechanisms mediating this hormonal control remain unknown. Here we show that the genes encoding the transcription factor AINTEGUMENTA (ANT) and the D-type cyclin CYCD3;1 are expressed in the vascular cambium of Arabidopsis roots, respond to cytokinins and are both required for proper root secondary thickening. Cytokinin regulation of ANT and CYCD3 also occurs during secondary thickening of poplar stems, suggesting this represents a conserved regulatory mechanism.
KEY WORDS: Cytokinins, Secondary growth, Cyclin D, AINTEGUMENTA, Root development
INTRODUCTION
The phytohormone cytokinin regulates several root developmental processes, including elongation, apical meristem maintenance and vascular morphogenesis (Ferreira and Kieber, 2005; Higuchi et al., 2004; Mahonen et al., 2000; Nishimura et al., 2004). Cytokinin signalling, together with other hormonal cues (Miyashima et al., 2013), is also important for root secondary growth, which involves thickening of the root via proliferation of cambial cells (Spicer and Groover, 2010; Nieminen et al., 2004). Arabidopsis mutants defective in cytokinin biosynthesis develop thinner roots with a cambium composed of fewer cells, a phenotype rescued by exogenous cytokinin application (Matsumoto-Kitano et al., 2008). Cytokinin therefore appears to act at least in part through promoting cell division and hence activation of the mitotic cell cycle.
CYCD genes encode conserved regulatory sub-units of cyclin D-cyclin-dependent kinase (CYCD-CDK) complexes that promote cell cycle progression in animals and plants (Menges et al., 2007; Morgan, 1997). The CYCD3 subgroup of CYCDs is conserved across all higher plants (Menges et al., 2007) and has three members in Arabidopsis: CYCD3;1, CYCD3;2 and CYCD3;3. CYCD3s control progression through the G1/S transition (Menges et al., 2006), and also regulate the length of the temporal period of mitotic cell division during aerial organ development (Dewitte et al., 2007).
CYCD3 genes are induced by cytokinins (Menges et al., 2007, 2006; Riou-Khamlichi et al., 1999) and are rate-limiting for cytokinin responses in Arabidopsis shoots (Dewitte et al., 2007; Riou-Khamlichi et al., 1999). Here, we identify novel roles for CYCD3;1 and the AINTEGUMENTA transcription factor in root secondary growth. Prolonged expression of CYCD3;1 in leaves caused by ectopic expression of ANT (Mizukami and Fischer, 2000) has led to the suggestion that CYCD3;1 is a target of ANT (Anastasiou and Lenhard, 2007; Wu et al., 2011). However, we show here that CYCD3 and AINTEGUMENTA play independent roles in regulating secondary thickening in roots, but provide evidence that they are co-regulated by cytokinins.
RESULTS AND DISCUSSION
CYCD3;1 is rate-limiting for root secondary thickening
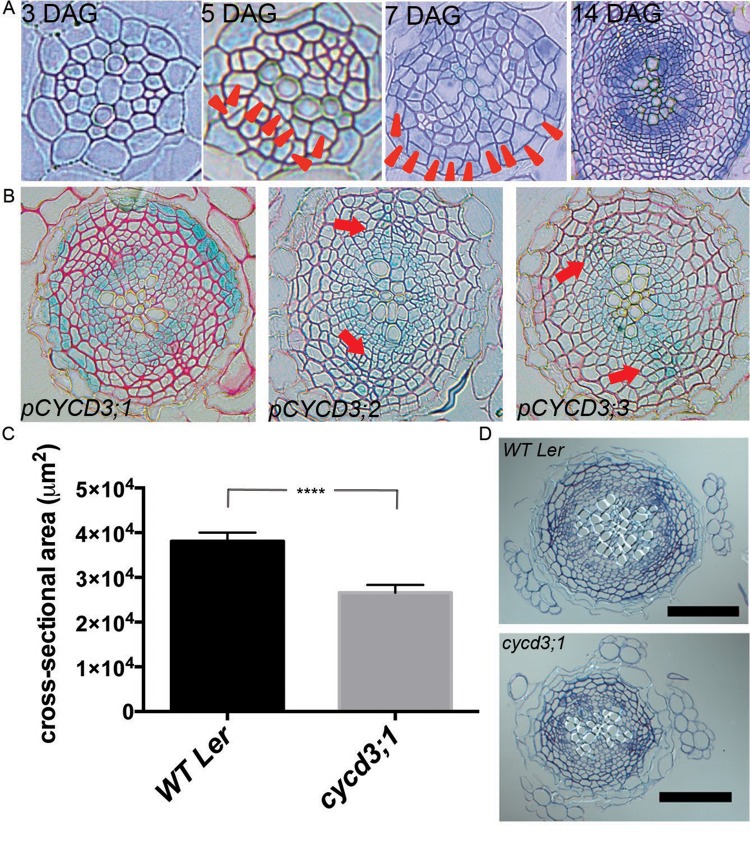
Secondary growth involves cell proliferation in the cambium (Fig. 1A). Given the known requirement for cytokinin signalling for root secondary thickening, and involvement of CYCD3s in shoot growth responses to cytokinins, we analysed the expression patterns of CYCD3s using promoter:GUS constructs. pCYCD3;1:GUS expression was observed in the innermost and outermost regions of the stele of roots undergoing secondary growth (Fig. 1A,B). These regions contain the cambium and pericycle cells respectively, both of which contribute to secondary thickening. pCYCD3;2:GUS and pCYCD3;3:GUS expression was detected in the cambium and in the phloem cells perpendicular to the primary xylem axis. CYCD3;1 expression has also been reported in whole mounts of root tissue undergoing secondary growth (Collins et al., 2015) and the vascular tissue of vegetative and flowering Arabidopsis shoot apices (Dewitte et al., 2003). Furthermore, expression of CYCD3 genes in the vascular tissue of roots undergoing primary growth was recently inferred (Collins et al., 2015) from microarray data obtained from fluorescently activated cell-sorted primary root cells (Brady et al., 2007). Collins et al. (2015) also analysed expression of CYCD3 genes in shoot cambium development with the same transcriptional fusion reporter lines used here and detected expression of all three CYCD3 genes in this tissue. These data suggest that CYCD3 genes are expressed in radially proliferating tissues and play an active role in secondary growth.
Fig. 1.
CYCD3;1 is expressed in the root cambium and regulates secondary growth. (A) Transverse sections of roots taken immediately below the hypocotyl at time points shown. Arrowheads indicate recently formed cell walls. (B) Expression patterns of CYCD3 promoter:GUS reporters. pCYCD3;1 drives GUS expression in the cambium and the pericycle (left panel), whereas pCYCD3;2 and pCYCD3;3 drive GUS expression in the cambium and phloem (middle and right panel; red arrows). (C) Stele cross-sectional area in 17 DAG Ler and cycd3;1 roots. ****P<0.0001; Error bars represent s.e.m. (D) Transverse sections of roots in C. Scale bars: 100 µm.
CYCD3 genes were recently shown to contribute to secondary growth in Arabidopsis stems with reduced hypocotyl diameter and vascular cell number in the cycd3;1-3 triple mutant generated in the Columbia background (Collins et al., 2015), although the contribution of individual CYCD3 genes was not determined. This supports a scenario in which CYCD3s are core regulators of cambial cell proliferation in both shoots and roots. Indeed, our expression data suggest that CYCD3s could play a role in root secondary growth. We compared the stele cross-sectional area in cycd3;1 to that of WT immediately under the hypocotyl during secondary growth (Fig. 1A). In order to avoid confounding effects from other polymorphisms in the Ler and Col-O backgrounds, we analysed the cycd3;1 allele in the Ler background, in which it was initially generated (Parinov et al., 1999). At 17 days after germination (DAG), cycd3;1 roots displayed a narrower stele than WT counterparts (Fig. 1C). Concomitant with reduced cell division activity, cycd3;1 roots had a reduced number of vascular cells (supplementary material Fig. S1). We conclude that CYCD3;1 promotes root secondary growth. Root diameter was reduced to a similar extent in both cycd3;1 and cycd3;1-3 triple mutants in the Ler background (supplementary material Fig. S2A,B), consistent with CYCD3;1 being primarily required among the CYCD3s genes for secondary growth in Ler roots.
ANT contributes to root secondary thickening
To identify potential regulators of CYCD3;1 expression, we performed genome-wide Pearson's correlation tests for CYCD3;1 coexpression with all known transcription factors across >500 public Affymetrix ATH1 microarray datasets annotated as conducted on root tissues. Expression of CYCD3;1 was most highly correlated with AINTEGUMENTA (ANT) (supplementary material Table S2). ANT is a member of the AP2 (APETALA2)/EREBP (Ethylene Response Element Binding Protein) family, and falls into the ANT-lineage of the AP2-like subgroup (Kim et al., 2006). Several members of the AP2-like subgroup are associated with developmental regulation during growth of young tissues (Nole-Wilson et al., 2005). ANT is involved in the control of lateral aerial organ size via the regulation of cell proliferation (Mizukami and Fischer, 2000), and is associated with fruit growth in apple (Dash and Malladi, 2012) and seasonal bud dormancy in hybrid aspen (Karlberg et al., 2011). Moreover, ectopic expression of ANT in leaves increased levels of CYCD3;1 mRNA (Mizukami and Fischer, 2000). Therefore, ANT is a candidate regulator of CYCD3;1 during secondary root growth.
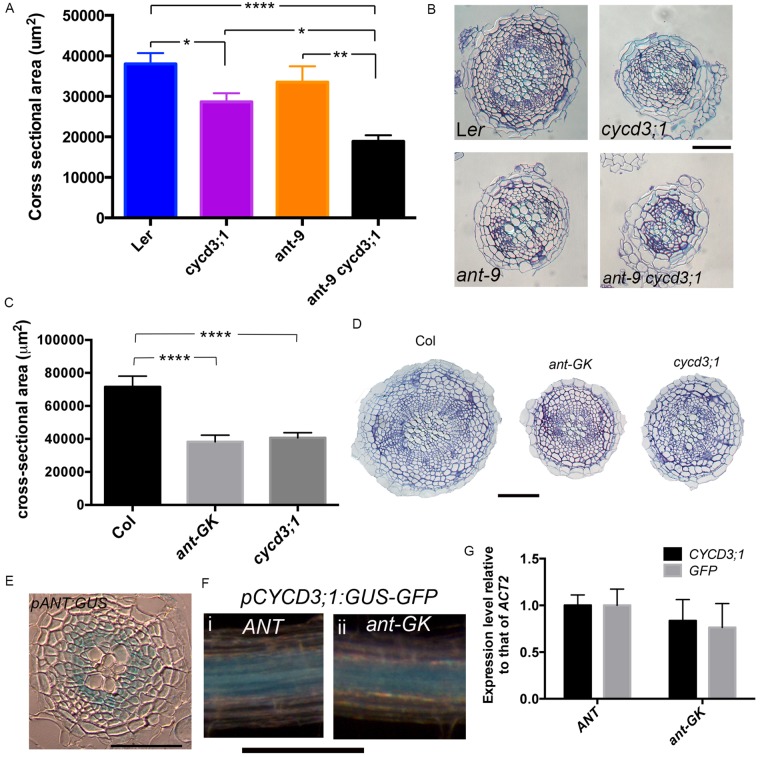
To test this hypothesis, the genetic interaction between ANT and CYCD3;1 was investigated. The original ant-9 allele isolated in the Landsberg erecta ecotype was investigated. No statistically significant reduction in root cross-sectional area was observed in ant-9 mutants (Fig. 2A,B). However, cross-sectional area was reduced to a greater extent in ant-9 cycd3;1 double mutants than in the cycd3;1 single mutant (Fig. 2A,B), suggesting a contribution of ANT to secondary thickening through a synergistic genetic interaction between ANT and CYCD3;1. To further investigate the contribution of ANT to secondary growth, we identified a new ant mutant in the Col-0 background derived from the GABI-Kat collection (supplementary material Fig. S3A). Homozygous ant-GK plants display the characteristic ant mutant phenotypes of reduced floral organ size (supplementary material Fig. S3A) and fail to produce seeds. Interestingly, root cross-sectional area and vascular cell number were reduced in ant-GK mutants to a similar extent as the reductions seen in cycd3;1 mutants (Fig. 2C,D and supplementary material Fig. S1).
Fig. 2.
Interaction between ANT and CYCD3;1 in root secondary growth. (A,C) Stele cross-sectional area in 30 DAG Ler, cycd3;1, ant-9 and ant-9 cycd3;1 roots (A) and 21 DAG Col-0, ant-GK and cycd3;1 (Col-0) roots (C). *0.01<P<0.05, **0.001<P<0.01, ****P<0.0001; Error bars represent s.e.m. (B,D) Transverse sections of roots in A (B) and C (D). Scale bars: 100 µm. (E) Transverse section from a GUS-stained 14 DAG pANT:GUS root. Scale bar: 50 μm. (F) pCYCD3;1:GUS-GFP expression in 14 DAG ANT (i) and ant-GK (ii) roots. Scale bar: 200 µm. (G) qPCR analysis of CYCD3;1 and GFP in 16 day-old ANT pCYCD3;1:GUS-GFP and ant-GK pCYCD3;1:GUS-GFP roots. Error bars: s.d. in four biological replicates, each from eight pooled roots.
To confirm that effects of loss of functional ANT and CYCD3;1 in the shoot did not influence or cause the root phenotypes described here, we grafted WT shoot scion onto ant-GK and cycd3;1 root scion. The phenotypes remained (supplementary material Fig. S4), confirming that these are independent root phenotypes. Furthermore, root elongation was not affected by ant and cycd3;1 mutations (supplementary material Fig. S6), demonstrating that the secondary growth phenotype was not caused by altered root ontogeny dynamics. The ant-GK allele is in the Col-0 background, whereas the ant-9 allele is in Ler, possibly explaining the difference in the severities of these phenotypes. We conclude that ANT also regulates root secondary thickening.
Relatively high levels of ANT mRNA have been reported in roots (Elliott et al., 1996), but the root expression pattern remains unknown. Expression of a pANT:GUS reporter was readily detected in the root cambium (Fig. 2E). Supporting this, in a transgenic line expressing a histone H2B-YFP fusion under the control of the ANT promoter (pANT:H2B-YFP), relatively strong fluorescence was observed in the stele of the more mature root (supplementary material Fig. S6).
Regulation of CYCD3;1 by ANT in shoot organs has been proposed but not demonstrated (Anastasiou and Lenhard, 2007; Horiguchi et al., 2009; Schruff et al., 2006). To test whether ANT might regulate CYCD3;1 expression in the root, a pCYCD3;1:GUS-GFP reporter was introduced into the ant-GK mutant. Visual comparison of pCYCD3;1:GUS-GFP expression in sibling F3 ant-GK and WT plants did not reveal a reduction in expression in ant-GK homozygous roots (Fig. 2F). However, qPCR analyses of root mRNA revealed a small reduction of both native CYCD3;1 and GUS-GFP transcript abundance in ant-GK pCYCD3;1:GUS-GFP mutants (Fig. 2G). This could however be explained by a relative decrease in the abundance of CYCD3;1-expressing cambial cells in the ant-GK mutant. Therefore, whilst it remains possible that ANT regulates CYCD3;1, taken together with the genetic evidence no strong regulation is indicated. Supporting this conclusion, CYCD3;1 transcript abundance was not reduced in ant-9 roots (supplementary material Fig. S7). Furthermore, an additive phenotype in the ant-9 cycd3;1 double mutant was also shown for Arabidopsis petal epidermal cell size, and CYCD3;1 expression was not reduced in young ant-9 flowers (Randall et al., 2015).
Cytokinin signalling regulates both ANT and CYCD3;1 expression
ant, cycd3;1 and cytokinin synthesis/response mutants show common phenotypes of defective root secondary thickening (Hejatko et al., 2009; Matsumoto-Kitano et al., 2008). Since cytokinins are known to regulate CYCD3;1 activity in shoot tissues (Dewitte et al., 2007), we assessed cytokinins as potential regulators of ANT and CYCD3;1 in the root cambium.
We first analysed their expression in response to exogenous cytokinin. qRT-PCR analysis of root mRNA showed that following cytokinin application to plants 14 DAG, ANT transcript levels were increased 6-fold relative to untreated plants, correlating with a smaller increase in CYCD3;1 transcript levels (supplementary material Fig. S8A). Supporting induction of CYCD3;1 by cytokinins, pCYCD3;1:GUS expression was also induced in roots after addition of the synthetic cytokinin kinetin (supplementary material Fig. S8B).
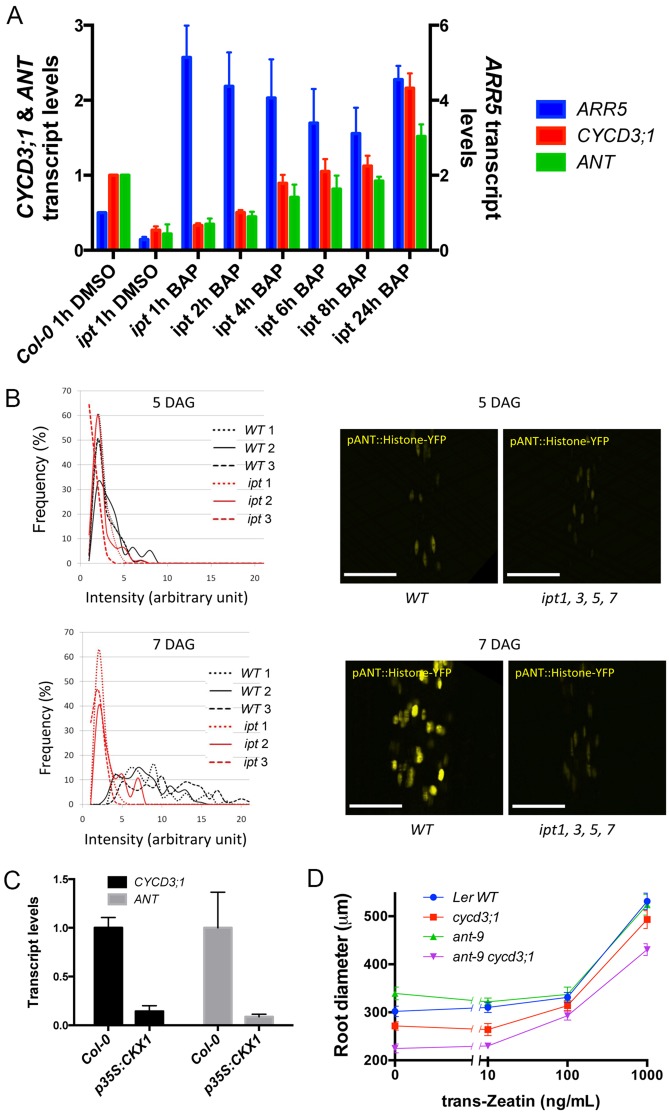
To analyse induction of ANT and CYCD3;1 with greater resolution, we measured expression at several time points following addition of cytokinins to ipt1,3,5,7 mutants. These mutants carry loss-of-function alleles for four ipt genes (ipt1,3,5,7), which encode isopentenyl transferases involved in cytokinin biosynthesis. These multiple mutants have reduced levels of isopentenyladenine as well as trans-zeatin (tZ), a cytokinin shown to have an effect on cambium proliferation (Matsumoto-Kitano et al., 2008; Miyawaki et al., 2006). Since these mutants fail to undergo proper secondary growth in roots (Matsumoto-Kitano et al., 2008), the use of this mutant should limit the amount of background ANT and CYCD3;1 expression. Consistent with cambium-expression of ANT and CYCD3;1, expression of these genes was reduced in the ipt1,3,5,7 mutant (Fig. 3A). Elevated ANT and CYCD3;1 expression was observed from 4 h after incubation with BAP (Student's t-test, P<0.05; n=3 in each case), increasing until 24 h.
Fig. 3.
ANT and CYCD3;1 respond to cytokinins in root secondary thickening. (A) qPCR analysis of ARR5, ANT and CYCD3;1 in Col-0 and ipt1;3;5;7 roots treated with DMSO and ipt1;3;5;7 roots treated with 1 µM BAP for the periods indicated. Error bars represent s.d. from 3 biological replicates. (B) Intensity of YFP signal in lines expressing pANT:H2B-YFP at 5 DAG and 7 DAG in WT plants vs ipt1,3,5,7 plants. WT1-3 and ipt1-3 each represent three independent T3 lines homozygous for pANT:H2B-YFP. ipt1-3 are also homozygous for ipt1;3;5;7 alleles. In each line, signal intensity was measured from >50 nuclei. Frequency distributions of signal intensity are shown. (C) Transcript levels of CYCD3;1 and ANT in Col-0 and p35S:CKX1 roots. s.d. in three biological replicates is shown. (D) Diameter of Ler, cycd3;1, ant-9 and ant-9 cycd3;1 roots following tZ treatments. Roots were grown for 11 days then transferred to media containing (or not for control) tZ. Error bars represent s.e.m.
We then investigated the cytokinin requirement for the increased ANT promoter activity observed during the transition from primary to secondary root growth (Fig. 3B). We analysed expression of pANT:H2B-YFP in ipt1,3,5,7 mutants. During the activation stage around 5 DAG (Fig. 1A), when the first cell division event in the procambium occurs, weak YFP signal from the pANT:H2B-YFP construct was detected in the roots of both WT and ipt1;3;5;7 plants (Fig. 3B top). Quantitation of the YFP signal from confocal images showed similar signal intensity in both WT and ipt mutant plants (Fig. 3B left). However, whereas in WT plants the signal intensified in roots during the transition stage (7 DAG), when procambial cells begin to proliferate, in ipt1;3;5;7 roots it did not (Fig. 3B). We suggest that the increase of ANT expression observed in WT roots depends on normal levels of cytokinins. Alternatively, ANT expression might be delayed due to a delay in vascular tissue development in the ipt1;3;5;7 mutant.
To further investigate the dependence of ANT and CYCD3;1 expression on cytokinin, we used p35S:CKX1 plants overexpressing cytokinin oxidase leading to lower levels of cytokinins (Werner et al., 2001). These displayed reduced abundance of ANT and CYCD3;1 transcripts in roots (Fig. 3C). Taken together, these results strongly indicate regulation of ANT and CYCD3;1 in the root vascular tissue by cytokinins.
ANT and CYCD3;1 are involved in the regulation of root secondary growth by cytokinins
To determine whether ANT and CYCD3;1 are part of the signalling mechanism by which cytokinins promote secondary thickening in roots, the root thickening response to cytokinins was analysed in mutants. Initially, the response of cycd3;1 roots to cytokinins was compared to WT. Although cycd3;1 roots were thinner than WT and remained so with low concentrations of tZ, higher concentrations of tZ restored secondary thickening to a level comparable with WT (supplementary material Fig. S9). We next compared this response in WT, ant-9, cycd3;1, and ant-9 cycd3;1 double mutants. As previously observed (Fig. 2A and supplementary material Fig. S7), without addition of cytokinins cycd3;1 and ant-9 cycd3;1 roots showed reduced diameter compared to WT Ler roots, the double mutant being thinner than the single mutant (Fig. 3D). All genotypes respond to tZ by increasing in diameter but notably the ant-9 cycd3;1 double mutant remains thinner than other genotypes at higher tZ concentrations (1000 ng/µl; Fig. 3D), supporting a synergistic interaction between ANT and CYCD3;1 and implying that these factors independently contribute to radial cell division activity in the cambium. This data also indicates that other factors can mediate the response of the cambium to cytokinins in the absence of ANT and CYCD3;1.
Conserved roles for ANT and CYCD3;1 in secondary growth
Both ANT and CYCD3 genes are widely conserved amongst angiosperms (Kim et al., 2006; Menges et al., 2007) and expression of the poplar ANT orthologue has been reported in cambial tissue (Schrader et al., 2004; Zhang et al., 2011). It was also reported that down-regulation of ANT and CYCD3 orthologues in hybrid aspen (P. tremula x tremuloides) is required for bud-growth cessation in this species (Karlberg et al., 2011). We analysed the activity of the poplar ANT orthologue AIL1 in stems undergoing secondary growth, and observed activity within the cambium (Fig. 4A), consistent with a potential role for AIL1 in the regulation of secondary growth in this species. The activity of a promoter sequence designated PttANT was also recently reported in the cambium (Etchells et al., 2015); this promoter sequence was derived independently but appears to be that of PttAIL1 according to the locus identity, supporting the expression pattern described here. As cytokinins also regulate secondary growth in poplar (Nieminen et al., 2008), we analysed the expression of PtAIL1 and the poplar CYCD3 homologue PtCYCD3;2 following cytokinin treatments. It should be noted that the CYCD3 gene number suffixes denote arbitrary order of naming in that species (Renaudin et al., 1996). qRT-PCR analyses revealed increases in relative abundances of both PtAIL1 and PtCYCD3;2 transcripts after twelve hours of cytokinin treatment (Fig. 4B). Therefore, we propose that the regulation of secondary growth by cytokinin-induced ANT and CYCD3;1 is conserved in higher plants.
Fig. 4.
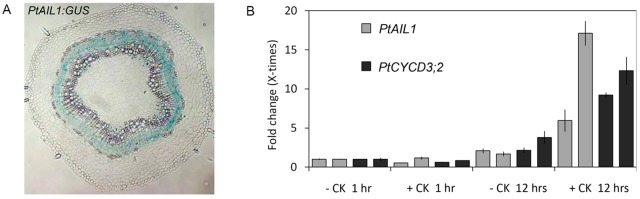
Poplar ANT is expressed in the cambium and is induced by cytokinins. Cross-section of a Poplar pAIL1:GUS stem following GUS assay (A) and qPCR analyses of PtANT and PtCYCD3;2 transcripts following treatments of cytokinin (100 nM 2iP) or mock treatments for one or twelve hours (B). Two individuals represented separately in adjacent bars were used for the experiment. Transcript levels were normalized to PttTUA2. Error bars: s.d. in four technical replicates.
Conclusions
The transcription factor ANT and the cyclin CYCD3;1 play independent roles in regulating cell division during secondary growth of Arabidopsis roots. cycd3;1, ant and cytokinin biosynthesis and receptor mutants share a common failure in cambial proliferation, and we show that these components act in conserved pathways linking cytokinins to developmentally regulated cell proliferation.
MATERIALS AND METHODS
Plant material and growth
Arabidopsis plants were grown in 16 h days at 22°C on MS medium containing 1.5% sucrose, 0.5 g/l 2-(N-morpholino)ethanesulfonic acid and 1% agar. CYCD3 promoter:GUS reporter lines, cycd3;1, ant-9, ipt1;3;5;7 and cre2;3;4 mutants have been described (Dewitte et al., 2007; Elliott et al., 1996; Higuchi et al., 2004; Miyawaki et al., 2006; Mizukami and Fischer, 2000). The cycd3;1 mutant initially isolated from a Landsberg erecta (Ler) DS element insertion library (Parinov et al., 1999) was backcrossed twice to Ler WT, and the triple cycd3;1-3 mutant in the Ler background was generated by crossing cycd3;1 with cycd3;2, and backcrossing lines hemizygous for both alleles to Ler twice before introgressing the cycd3;3 allele from the EXOTIC collection (Dewitte et al., 2007). The ant-GK mutant (GK-874H08; TAIR accession no.: 1006453905) is described in supplementary material Fig. S3. The pCYCD3;1:GUS-GFP and pANT:Histone2b-YFP (pANT:H2B-YFP) reporters were constructed in pKGWFS7 (Karimi et al., 2002). 947 bp of DNA upstream (representing sequence to the adjacent gene) of the CYCD3;1 start codon was used. 5137 bp of ANT upstream sequence was used for pANT:H2B-YFP, and 6291 bp for pANT:GUS which was isolated from a lambda EMBL3 library (David Smyth and Brian Kwan, Monash University, Melbourne, Australia). Scion of Atipt1;3;5;7 was grafted onto WT stock and used for floral dipping (Clough and Bent, 1998).
WT Populus tremula x tremuloides line T89 was used for qPCR analyses. P. tremula x tremuloides seedlings were grown for one month in long-day greenhouse conditions at 22°C in 5 litre pots. The pAIL1:GUS line has been described (Karlberg et al., 2011).
Arabidopsis micrografting
Arabidopsis plants were grafted according to a published protocol (Turnbull et al., 2002) with the following modifications. Ethanol sterilised Arabidopsis seeds were germinated on 1/2 Murashige and Skoog (MS) medium plus 1% Difco agar (pH 5.7; 1% sucrose) and grown on vertically-mounted Petri dishes under long (16 h of 80–100 µmoles light, Col-0 and ant-GK grafts) or short day conditions (8 h of 80–100 µmoles light, Ler and cycd3;1 grafts) at 20°C. Grafting was performed under sterile conditions in a laminar flow hood with a Zeiss portable dissecting microscope. 5–6 day-old seedlings were transferred to 9 cm Petri dishes that contained one layer of 2.5×4 cm sterilised Hybond N membrane (GE Healthcare) on top of two sterilised 8 cm disks of 3 mm Chr Whatman paper (Scientific Laboratory Supplies). The Whatman paper and Hybond N membrane were kept moist using a 1% sucrose solution (Col-0 and ant-GK grafts) or sterile distilled water (Ler and cycd3;1 grafts). A transverse cut was made through the hypocotyl close to the shoot using a vascular dissecting knife (Ultra Fine Micro Knife; Fine Science Tools). In addition, one cotyledon was removed to assist in aligning the grafted pieces. In the case of self-grafts, an additional 1 mm segment was cut from the hypocotyl and discarded. Grafts were assembled by butt alignment of the two cut halves with no supporting collar. After grafting, Petri dishes were sealed with parafilm, mounted vertically under long (Col-0 and ant-GK grafts) or short day conditions (Ler and cycd3;1 grafts) and monitored for 7 days at 20°C. Afterwards, successful grafts and un-grafted controls were transferred to 1/2 MS plates with 1% sucrose under long day conditions at 20°C for additional 2–3 weeks. For ant-GK related grafts and un-grafted plants, DNA was extracted and genotyping PCR was performed to confirm the genotype of each scion and stock. Roots from stock were sampled and embedded with Leica resin. 5 µm-thin plastic sections were cut at 5 mm below the hypocotyl-root junction and stained with Toluidine Blue O.
Microscopy and anatomical analyses
Transverse sectioning and GUS assays were described (Mahonen et al., 2006, 2000; Nieuwland et al., 2009). Sections were taken within 1 cm of the root-hypocotyl junction. Microscopy was performed using a Zeiss LSM 710 or a Leica SP5. Roots were stained with 100 μg/ml propidium iodide.
Cytokinin induction and quantitative PCR
BAP, kinetin and trans-zeatin were dissolved in dimethyl sulfoxide and diluted in sterile water. 2iP was dissolved in 20 mM NaPi buffer. Arabidopsis inductions were performed by transplanting onto plates containing cytokinins or equivalent control buffer. Inductions in P. tremula x tremuloides seedlings were performed by submerging stem pieces in solutions indicated. Col-0 and ipt1,3,5,7 mutants were grown on a nylon mesh (SEFAR NITEX 03-100/44) on vertical plates containing 0.8% Plant Agar (Duchefa), 1% sucrose (Duchefa), 0.5× MS medium including vitamins (Duchefa) and pH adjusted to 5.7–5.8 with 20× MES buffer (Mes monohydrate, Duchefa). 7-day-old plants were transferred with the mesh to plates containing 1 µM BAP or the respective DMSO control. Primary root samples consisting of 40 plants were harvested at 1, 2, 4, 6, 8 and 24 h after the transfer by cutting a few millimeters below the hypocotyl, primary root tips and lateral roots were discarded. RNA was isolated with Qiagen RNeasy Plant Mini Kit with an on-column DNase treatment. Additional DNase treatment with DNase I (RNase-free) (Thermo Scientific) was performed to 1 µg of RNA prior to the oligo-dT-primed cDNA synthesis with First Strand cDNA Synthesis Kit (Thermo Scientific). Expression of ANT, CYCD3;1 and ARR5 was quantified with primers listed in supplementary material Table S3with HOT FIREPol® EvaGreen® qPCR Mix Plus (no ROX) (Solis Biodyne). The Bio-Rad CFX384 was used with one cycle 95°C for 15 min, 40 cycles each consisting of 95°C for 15 s, 60°C for 30 s and 72°C for 30 s, one cycle 95°C for 10 s followed by melt curve analysis. Raw data values were normalized to the geometric mean of four control genes (Vandesompele et al., 2002) and fold changes calculated in comparison to the Col-0 expression level. The experiment was performed in triplicate.
For other experiments, RNA was isolated using TriPure (Roche Diagnostics) and cDNA was synthesized using the Ambion Retroscript kit. qRT-PCR analyses were performed as described for Arabidopsis (Menges and Murray, 2002) and P. tremula x tremuloides (Nieminen et al., 2008). Gene-specific primers are listed (supplementary material Table S1). Relative transcript levels were quantified using the ΔΔCT method (Livak and Schmittgen, 2001).
Statistics
Student's t-tests were performed in GraphPad Prism. *0.01<P<0.05 **0.001<P<0.01 ***0.0001<P<0.001 ****P<0.0001. Pearson's correlation tests were carried out in R (r-project.org).
Acknowledgements
We are most grateful to David Smyth and Brian Kwan who made the pANT:GUS line, Teva Vernoux for discussions, and Angela Marchbank, Jo Kilby, Katja Kainulainen and Mikko Herpola for excellent technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
RSR wrote the manuscript. RSR, SM & WD analysed data. RSR, SM & AA performed qPCR experiments. RSR, SM, JZ, AE & AK created and analysed tissue cross-sections. TB conducted the time-course cytokinin induction experiment in Arabidopsis. RSR & KB conducted genotyping of ant-9 cycd3;1 mutants. CWM conducted micrografting. AA, CF, MMK & JYL created transgenic lines and conducted preliminary experiments. JI conducted qPCR on Poplar-derived cDNA. TK, APH, RB, WD, YH & JAHM contributed intellectually to project planning and manuscript preparation.
Funding
Work in the J.A.H.M. lab was supported by grants BB/D011914 from the Biotechnology and Biological Sciences Research Council and a grant from the European Research Area in Plant Genomics to J.A.H.M. (BB/E024858) and Y.H., who was also supported by the Academy of Finland and Tekes.
Supplementary material
Supplementary material available online at http://bio.biologists.org/lookup/suppl/doi:10.1242/bio.013128/-/DC1
References
- Anastasiou E. and Lenhard M. (2007). Growing up to one's standard. Curr. Opin. Plant Biol. 10, 63-69. 10.1016/j.pbi.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Brady S. M., Orlando D. A., Lee J.-Y., Wang J. Y., Koch J., Dinneny J. R., Mace D., Ohler U. and Benfey P. N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801-806. 10.1126/science.1146265 [DOI] [PubMed] [Google Scholar]
- Clough S. J. and Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Collins C., Maruthi, N. M. and Jahn C. E. (2015). CYCD3 D-type cyclins regulate cambial cell proliferation and secondary growth in Arabidopsis. J. Exp. Bot. 66, 4595-4606. 10.1093/jxb/erv218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash M. and Malladi A. (2012). The AINTEGUMENTA genes, MdANT1 and MdANT2, are associated with the regulation of cell production during fruit growth in apple (Malus×domestica Borkh.). BMC Plant Biol. 12, 98 10.1186/1471-2229-12-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Riou-Khamlichi C., Scofield S., Healy J. M. S., Jacqmard A., Kilby N. J. and Murray J. A. H. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15, 79-92. 10.1105/tpc.004838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Scofield S., Alcasabas A. A., Maughan S. C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V. et al. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104, 14537-14542. 10.1073/pnas.0704166104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R. C., Betzner A. S., Huttner E., Oakes M. P., Tucker W. Q., Gerentes D., Perez P. and Smyth D. R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155-168. 10.1105/tpc.8.2.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells J. P., Mishra L. S., Kumar M., Campbell L. and Turner S. R. (2015). Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25, 1050-1055. 10.1016/j.cub.2015.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira F. J. and Kieber J. J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8, 518-525. 10.1016/j.pbi.2005.07.013 [DOI] [PubMed] [Google Scholar]
- Hejatko J., Ryu H., Kim G.-T., Dobesova R., Choi S., Choi S. M., Soucek P., Horak J., Pekarova B., Palme K. et al. (2009). The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell 21, 2008-2021. 10.1105/tpc.109.066696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Pischke M. S., Mahonen A. P., Miyawaki K., Hashimoto Y., Seki M., Kobayashi M., Shinozaki K., Kato T., Tabata S. et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 101, 8821-8826. 10.1073/pnas.0402887101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Gonzalez N., Beemster G. T. S., Inzé D. and Tsukaya H. (2009). Impact of segmental chromosomal duplications on leaf size in the grandifolia-D mutants of Arabidopsis thaliana. Plant J. 60, 122-133. 10.1111/j.1365-313X.2009.03940.x [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D. and Depicker A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193-195. 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Karlberg A., Bako L. and Bhalerao R. P. (2011). Short day-mediated cessation of growth requires the downregulation of AINTEGUMENTALIKE1 transcription factor in hybrid aspen. PLoS Genet. 7, e1002361 10.1371/journal.pgen.1002361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Soltis P. S., Wall K. and Soltis D. E. (2006). Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23, 107-120. 10.1093/molbev/msj014 [DOI] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mahonen A. P., Bonke M., Kauppinen L., Riikonen M., Benfey P. N. and Helariutta Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14, 2938-2943. 10.1101/gad.189200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen A. P., Bishopp A., Higuchi M., Nieminen K. M., Kinoshita K., Tormakangas K., Ikeda Y., Oka A., Kakimoto T. and Helariutta Y. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311, 94-98. 10.1126/science.1118875 [DOI] [PubMed] [Google Scholar]
- Matsumoto-Kitano M., Kusumoto T., Tarkowski P., Kinoshita-Tsujimura K., Vaclavikova K., Miyawaki K. and Kakimoto T. (2008). Cytokinins are central regulators of cambial activity. Proc. Natl. Acad. Sci. USA 105, 20027-20031. 10.1073/pnas.0805619105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M. and Murray J. A. H. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203-212. 10.1046/j.1365-313X.2002.01274.x [DOI] [PubMed] [Google Scholar]
- Menges M., Samland A. K., Planchais S. and Murray J. A. H. (2006). The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18, 893-906. 10.1105/tpc.105.039636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., Pavesi G., Morandini P., Bogre L. and Murray J. A. H. (2007). Genomic organization and evolutionary conservation of plant D-type cyclins. Plant Physiol. 145, 1558-1576. 10.1104/pp.107.104901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S., Sebastian J., Lee J.-Y. and Helariutta Y. (2013). Stem cell function during plant vascular development. EMBO J. 32, 178-193. 10.1038/emboj.2012.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., Tabata S., Sandberg G. and Kakimoto T. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 103, 16598-16603. 10.1073/pnas.0603522103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y. and Fischer R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 942-947. 10.1073/pnas.97.2.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. O. (1997). Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13, 261-291. 10.1146/annurev.cellbio.13.1.261 [DOI] [PubMed] [Google Scholar]
- Nieminen K. M., Kauppinen L. and Helariutta Y. (2004). A weed for wood? Arabidopsis as a genetic model for xylem development. Plant Physiol. 135, 653-659. 10.1104/pp.104.040212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen K., Immanen J., Laxell M., Kauppinen L., Tarkowski P., Dolezal K., Tahtiharju S., Elo A., Decourteix M., Ljung K. et al. (2008). Cytokinin signaling regulates cambial development in poplar. Proc. Natl. Acad. Sci. USA 105, 20032-20037. 10.1073/pnas.0805617106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland J., Maughan S., Dewitte W., Scofield S., Sanz L. and Murray J. A. H. (2009). The D-type cyclin CYCD4;1 modulates lateral root density in Arabidopsis by affecting the basal meristem region. Proc. Natl. Acad. Sci. USA 106, 22528-22533. 10.1073/pnas.0906354106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S. and Ueguchi C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16, 1365-1377. 10.1105/tpc.021477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S., Tranby T. L. and Krizek B. A. (2005). AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol. Biol. 57, 613-628. 10.1007/s11103-005-0955-6 [DOI] [PubMed] [Google Scholar]
- Parinov S., Sevugan M., Ye D., Yang W.-C., Kumaran M. and Sundaresan V. (1999). Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11, 2263-2270. 10.1105/tpc.11.12.2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R. S., Sornay E., Dewitte W. and Murray J. A. H. (2015). AINTEGUMENTA and the D-type cyclin CYCD3;1 independently contribute to petal size control in Arabidopsis: evidence for organ size compensation being an emergent rather than a determined property. J. Exp. Bot. 66, 3991-4000. 10.1093/jxb/erv200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin J.-P., Doonan J. H., Freeman D., Hashimoto J., Hirt H., Inze D., Jacobs T., Kouchi H., Rouze P., Sauter M. et al. (1996). Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol. Biol. 32, 1003-1018. 10.1007/BF00041384 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Huntley R., Jacqmard A. and Murray J. A. H. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541-1544. 10.1126/science.283.5407.1541 [DOI] [PubMed] [Google Scholar]
- Schrader J., Nilsson J., Mellerowicz E., Berglund A., Nilsson P., Hertzberg M. and Sandberg G. (2004). A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16, 2278-2292. 10.1105/tpc.104.024190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff M. C., Spielman M., Tiwari S., Adams S., Fenby N. and Scott R. J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251-261. 10.1242/dev.02194 [DOI] [PubMed] [Google Scholar]
- Spicer R. and Groover A. (2010). Evolution of development of vascular cambia and secondary growth. New Phytol. 186, 577-592. 10.1111/j.1469-8137.2010.03236.x [DOI] [PubMed] [Google Scholar]
- Werner T., Motyka V., Strnad M. and Schmülling T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98, 10487-10492. 10.1073/pnas.171304098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Li Y.-H., Wu J.-Y., Chen Q.-Z., Huang X., Chen Y.-F. and Huang X.-L. (2011). Over-expression of mango (Mangifera indica L.) MiARF2 inhibits root and hypocotyl growth of Arabidopsis. Mol. Biol. Rep. 38, 3189-3194. 10.1007/s11033-010-9990-8 [DOI] [PubMed] [Google Scholar]
- Zhang J., Gao G., Chen J.-J., Taylor G., Cui K.-M. and He X.-Q. (2011). Molecular features of secondary vascular tissue regeneration after bark girdling in Populus. New Phytol. 192, 869-884. 10.1111/j.1469-8137.2011.03855.x [DOI] [PubMed] [Google Scholar]