Abstract
miRNAs are a conserved class of approximately 22 nucleotide (nt) short non-coding RNAs that normally silence gene expression via translational repression and/or degradation of targeted mRNAs in plants and animals. Identifying the whereabouts of miRNAs potentially informs miRNA functions, some of which are perhaps specialized to specific cellular compartments. In this review, I discuss the significance of miRNA localizations in the cytoplasm, including those at RNA granules and endomembranes, and the export of miRNAs to extracellular space. I explore how miRNA localizations and functions are regulated by protein modifications on the core miRNA-binding protein Argonaute (AGO) during normal and stress conditions, and conclude by discussing new AGO partners, non-AGO miRNA-binding proteins, and the emergent understanding of miRNAs found in the nucleoplasm, nucleoli, and mitochondria.
miRNA: A Moving Target
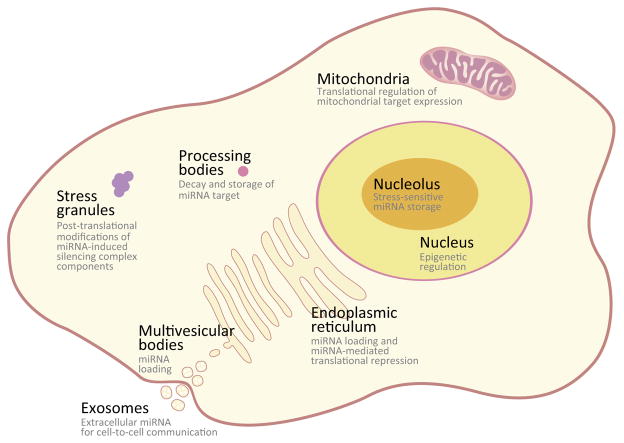
In animals, most miRNA genes are transcribed by RNA polymerase II in the nucleus (reviewed in [1]). The primary miRNA transcript is processed by Drosha in the nucleus to become one or more precursor miRNAs (pre-miRNAs), which bear a hairpin structure with a 2-nt overhang at the 3′ end. The overhang structure is recognized by Exportin 5 for export to the cytoplasm, where pre-miRNAs are processed by Dicer to become an approximately 22-nt duplex. The duplex is then loaded onto one of the AGO family members (e.g., AGO1–4 in humans), which selectively retains one strand as the mature miRNA. Emergent data indicate that mature miRNAs then localize in subcellular compartments in the cytoplasm and, surprisingly, the nucleus (Figure 1, Key Figure). In this review, I discuss the potential functions of miRNAs in these compartments.
Figure 1. Key Figure.
Proposed Location Specific Functions of Mature miRNAs.
To effect gene silencing, miRNAs require AGOs and other silencing factors to form differently sized miRNA-induced silencing complexes (miRISC), ranging from approximately 100 kDa to 3 MDa [2–4]. Depending on the degree of sequence complementarity between miRNAs and their RNA targets, as well as the protein composition of miRISCs, the outcomes of miRNA binding are different. Perfect complementarity between miRNAs and RNA targets allows AGO2 (the only AGO that has slicer activity) to cleave RNA targets. Nearly all animal miRNAs identify RNA targets with perfect complementarity at the so-called ‘seed sequence’ (2ndn–7th nucleotide), where the recruitment of the AGO-binding protein GW182 forms a canonical miRISC to repress translation and/or accelerate deadenylation and degradation of mRNAs. Thus, when considering the whereabouts of miRNA functions, it is important to consider the localizations of miRNAs, the core miRNA-binding AGO family members, and other miRISC components.
The Possible Roles of P-bodies in miRNA Silencing
Given that AGO is required for functional miRISCs, its localization has been used as the proxy for identifying sites of miRNA functions ([5,6]; reviewed in [7]). Fluorescent microscopy studies indicate that mammalian AGO members are localized prominently in cytoplasmic bodies known as P-bodies (PBs) [8], which are enriched with RNA decay factors, including deadenylases. Besides AGO, PBs are also enriched with GW182, which bridges AGO proteins with deadenylases. Importantly, both miRNAs and their targets are recruited to PBs [5]. Such localizations suggest that PBs are a major site for miRNA-directed deadenylation and subsequent decay of mRNA targets. However, quantitation of the cytoplasmic localization of AGO2 indicates that, even though it has >tenfold higher concentration at PBs than the neighboring cytoplasmic space, <1% of cytoplasmic AGO2 is localized in PBs [9]. Moreover, cells depleted of microscopically visible PBs have normal miRNA-mediated translational repression and degradation of mRNAs [7,10]. Although PBs could be sites for decay of miRNA targets, these data argue that the major actions of miRNAs occur elsewhere in the cytoplasm, likely as submicroscopic complexes.
PBs are also proposed to be storage sites of miRNA-targeted mRNAs destined for future translation [11]. For example, a subpopulation of CAT-1 mRNAs repressed by miR-122 is localized at PBs, but upon amino acid starvation, these mRNAs are delocalized from PBs and translation of this transcript resumes [11]. Given that such starvation-induced translation occurs even with treatment of a transcriptional inhibitor, it is likely that formerly repressed CAT-1 mRNAs are relocated to polysomes for translation. A possible source of these repressed mRNAs is from PBs. Photokinetic experiments further indicate that a subpopulation of mRNAs is immobilized in PBs upon amino acid starvation, which is released upon the relief of the stress [12]. However, a dilemma for this model is that neither poly(A)+ mRNAs nor the poly(A)-binding protein are detected in PBs [8,12]; hence, it is difficult to envisage that those PB-localized miRNA-targeted mRNAs still have poly(A) tails, which are required for efficient translation. The role of PBs as an mRNA storage for future translation was first demonstrated in yeast studies using two reporter genes [13]. However, a recent yeast genome-wide survey indicated that such behavior is restricted to a subpopulation of genes and that mRNAs switching from translational silence to active translation also tend to have strong associations with poly(A)-binding protein and have long poly(A) tails [14]. Therefore, either these transcripts at PBs would have to be adenylated by a cytoplasmic poly(A) polymerase before translation or the observed starvation-induced translation is mainly due to the relocation of cytosolic, non-PB-localized transcripts to polysomes.
A potential clue of AGO function at PBs may come from its dynamic behavior there: AGO exhibits slow exchange kinetics between PBs and the cytoplasm, indicating that most AGO stays at PBs after entry [9,15,16]. Given that other protein components at PBs can rapidly exchange with the neighboring cytoplasm, such slow kinetics specific to AGO indicate that this core miRNA-binding protein is somehow anchored to one or more immobile components within PBs. Such AGO anchorage can be altered upon physiological stimulation or disrupted by expression of pathological repeats of the Huntingtin (HTT) gene from Huntington’s disease [15,16], suggesting that the immobilization of AGO at PBs is a regulated process. Future photokinetic experiments coupled with a systematic knockdown or overexpression screen could identify the immobile components and delineate the mechanism of how AGOs anchor at PBs, which in turn may shed light on the possible roles of PBs in miRNA silencing.
The Underappreciated Roles of Endomembranes in miRNA Silencing
Should AGO localization represent all functional miRISCs, where do 99% of these reside in the cytoplasm if not in PBs? Biochemical experiments indicate that a portion of AGO and Dicer co-fractionates with endoplasmic reticulum (ER) and Golgi (reviewed in [17]). In fact, the first antibody to detect AGO in mammalian cells was derived by using intracellular membranes as antigens [18]. Genetic screens in plants and nematodes further identified the requirement of a lipid synthesis pathway component in RNA silencing [19,20], consistent with possible roles of endomembrane in miRNA silencing. Here, I discuss two particular fractions within the endomembrane system: ER and multivesicular bodies.
ER: A Possible Site of miRNA-Mediated Translational Repression
Three recent studies from human, plant, and fly cells indicated possible regulatory roles of ER in miRNA-mediated repression [21–23]. In Drosophila S2 cells, a class of polysome-bound miRISC (P-miRISC) is formed upon serum starvation and some of these P-miRISCs associate and co-sediment with ER components [23]. Under serum starvation, P-miRISC is responsible for augmenting miRNA-mediated repression by five- to tenfold. Such repression occurs only at the translational level and is unaffected by the knockdown of the canonical miRISC component GW182. Consistent with these findings, P-miRISC lacks GW182, but AGO binds to another partner, Loqs-PB, instead. In humans, the homolog of Loqs-PB, TAR RNA binding protein (TRBP), is also identified in rough ER (i.e., ribosome-associated ER) [21]; TRBP is required for loading miRNAs to AGO and anchoring the resulting miRISCs to ER membranes [21]. In Arabidopsis, miRNA-mediated repression is regulated by an integral membrane protein ALTERED MERISTEM PROGRAM 1 (AMP1) on rough ER. As in the Drosophila case, the repression is mediated at the translational level only [22]. Given that AMP1 homologs have been identified in both mammals and nematodes, such a translation-specific miRNA-mediated repression is likely to be conserved.
A genome-wide survey indicated that ER-associated mRNAs encode both cytoplasmic and nuclear proteins, in addition to membrane-bound proteins, suggesting that ER is a regulatory hub for all types of transcript [24]. Given that the density of ribosomes on ER-associated transcripts is twofold higher than those transcripts bound by cytosolic ribosomes, such local enrichment of both ribosomes and miRISCs at rough ER potentially allows for higher efficiency to sample and silence translating mRNAs that have miRNA-binding sites [21–24]. One way to examine the mechanism of how miRNAs regulate ER-bound transcripts is to use a modified ribosome profiling technique that targets only ER-bound ribosomes [25], and specifically ask whether P-miRISCs cause a reduction in the density of ribosomes on mRNAs targeted by specific miRNAs. The ribosome occupancy patterns will signify whether ribosomes are paused at particular sites during initiation or elongation, or dropped off prematurely, potentially explaining mechanistically how miRNAs repress translation at ER.
Multivesicular Body-Bound miRNAs Trafficking Inside and Outside of Cells
Multivesicular bodies (MVBs) are membrane-bound compartments that represent the late stage of endosomes and are responsible for sorting molecules to lysosomes for degradation, either to the plasma membrane for secretion or back to the Golgi/ER for reuse. MVB formation requires the endosomal sorting complex required for transport (ESCRT) and depletion of certain ESCRT components inhibits miRNA silencing. Moreover, blocking the pathway for MVB turnover increases the amount of miRNA-loaded AGOs and stimulates miRNA silencing [26,27]. Taken together, these data suggest that MVBs act as centers for miRNA loading or recycling.
Regulated fusion of MVBs with plasma membrane results in the secretion of exosomes to the extracellular space. These exosomes encompass select miRNAs and miRISC components, including AGO2 and GW182 [28]. Notably, these exosomal AGO2/miRNAs can modulate the gene expression of recipient cells within the cancer microenvironment, between immune cells, and between neuronal synapses (e.g., [29–31]). The burning question is how miRISCs and specific miRNAs are sorted into exosomes for secretion. Recently, such exosomal sorting was found to be inversely correlated to the degree of miRISC association with polysomes in high cell density conditions when growth is restricted, where an increased association of miRISC with polysomes results in an impaired miRNA export via exosomes [32]. Such polysome sequestration results in an increased intracellular miRNA levels, but without an increase in miRNA repression. Although it is unclear whether the sequestration could be miRNA specific, these data suggest that the subcellular localizations of miRISCs are tied to one another and are differentially regulated based on growth conditions.
New AGO Partners and Non-AGO miRNA-Binding Proteins
Although AGO binds to GW182 and the binding is important for miRNA silencing [33], not all AGOs are bound to GW182. As illustrated earlier for P-miRISC in Drosophila, the binding of AGO to Loqs-PB and GW182 is mutually exclusive [34], suggesting that altered compositions of miRISCs result in distinct functional consequences [4,11,35,36]. For example, AGO was recently shown to bind to the human prion protein PrPC [37]; this is the protein that, when misfolded, causes neurological pathologies, including variant Creutzfeldt–Jakob disease. PrPC co-fractionates with miRISC components on endosomes, including MVBs [27], and its binding to AGO promotes the formation or stability of GW182-containing miRISC [37]. Alternatively, most AGO-bound miRNAs in adult tissues exist as low-molecular-weight complexes that are not associated with mRNA and GW182 [2]; these miRNAs are not actively engaged in target repression, but may act as miRNA reservoirs instead [2]. However, these low-molecular-weight miRNA reservoirs can re-incorporate back to a canonical GW182-containing miRISC upon physiological stimulation (e.g., mitogenic responses for quiescent cells [38] or T cell activation [2]). It is currently unclear whether these miRNA reservoirs concentrate in particular cellular compartments.
Another conundrum recently observed is that the presence of miRNAs does not necessitate the presence of AGO. Rather than correlating with the absolute amount present in cells, the silencing potential of a miRNA is correlated with the degree of its binding to AGO [39] as well as its association with GW182 [2]. The correlation difference could be because only a small fraction (<10%) of miRNAs are AGO bound [21,39,40], and miRNAs may bind to mRNA targets independent of AGO binding [40]. Interestingly, certain miRNAs can bind directly to proteins, such as Heterogeneous nuclear ribonucleoprotein E2 (hnRNP E2; miR-328) [41], TAR DNA-binding protein 43 (TDP-43; miR-1, miR-206) [42], and the toll-like receptor TLR7/8 (miR-21, miR29a) [43], or to the HIV Gag protein nonspecifically [44]. However, it is unclear how AGOs and miRNA can be stably present in cells without one another, because their stabilities are interdependent [45–48]. The physical presence of AGOs is critical for miRNA stability [46] where mature miRNAs are depleted in cells lacking AGOs [45]. Alternatively, unloaded AGOs are degraded in cells through the proteasome or lysosome [47,48]. Therefore, the degree and prevalence of such phenomena (miRNAs not binding to AGOs and/or miRNA binding to proteins other than AGO) remains unclear. To make the AGO–miRNA relation more complicated, recent crosslink immunoprecipitation (CLIP)-RNA seq data using four different antibodies suggest that AGO can bind to mRNA independent of miRNAs [49,50]. Notably, such miRNA-independent binding can be lost during stress [50]. As a result of this newly discovered biology, the localization of miRNA-free AGO, AGO-free miRNA, GW182-free miRISC, and other noncanonical forms of miRISCs, should be explored more rigorously at the cellular and biochemical levels.
Localization and Assembly of miRISCs Regulated by Protein Modifications
How are these various miRISCs assembled and distributed in a regulatory manner? One possible mechanism is regulated through reversible post-translational modifications on AGO/miRISC complexes [51]. For example, phosphorylation at Ser-387 of AGO2 by mitogen-activated protein kinase (MAPK) or Akt3 is important for its localization to PBs and favors miRISC to repress translation [51,52]. AGO1–4 are increasingly poly(ADP-ribosyl)ated upon various stressors when miRNA activities are reduced [53,54]. Under some stress conditions, a sizable fraction of AGOs and poly(ADP-ribosylation) polymerases relocalize to cytoplasmic structures called ‘stress granules’ [9,53], suggesting that these structures provide local sites for AGO modification to reduce miRNA activities. In addition, other protein modifications, including prolyl 4-hydroxylation [51,55], ubiquitination [47,51,56], and SUMOylation [57], regulate AGO stability, whereas phosphorylation at specific sites impairs miRNA binding or loading [51,58,59]. Given such indispensable roles in miRNA silencing, AGOs can globally alter miRNA activities in specific context [51], such as upon hypoxia [55,59] or immune stimulation [2,56]. Yet, selective regulation of a subset of miRNAs is possible, and recent reports showed that hypoxia-induced phosphorylation of AGO2 at Tyr-393 inhibits the maturation of pre-miRNAs with long loops, a subclass of miRNAs enriched for tumor-suppressive functions [59].
Although diverse protein modifications of AGOs are increasingly identified, we are still at an early stage in the mapping of how the upstream signaling pathway regulates the assembly of miRISCs. Upon T cell activation, phosphoinositide 3-kinase (PI3K) –Akt–mammalian target of rapamycin (mTOR) signaling promotes the conversion of low-molecular-weight AGO-bound miRNA reservoirs to high-molecular-weight complexes that include target mRNAs and GW182 [2]. In Drosophila S2 cells, insulin inhibits the formation of membrane-associated Ago1/miRNA complexes, but not P-miRISC, while the protein kinase C chemical activator PMA inhibits the formation of both [23]. Systematic understanding of protein modifications on miRISC components and their upstream signaling will not only allow us to understand how the miRNA regulatory network responds to external stimuli, but may also open new avenues for therapy. For example, although some miRNAs could be increasingly expressed in malignant cells, overall miRNA levels are reduced in patients’ tumors [60]. Therefore, to alter the course of tumorigenesis due to global loss of miRNAs, one possible therapeutic avenue is to increase the activity of the remaining miRNAs in tumors by selectively blocking a specific AGO modification that impairs miRNA activities.
The Unexpected Localizations of miRNAs
Nucleus
Although the final processing step and the loading of mature miRNA onto AGO occur in the cytoplasm, localization of mature miRNA in the nucleus was noted early on, for example, 20% of miR-21 was found in the nuclear extract, accounting for approximately 500 copies per cell [61]. Subsequently, miR-29b has been observed to be preferentially enriched in the nucleus with a hexanucleotide localization signal at the 3′ end, which is sufficient to direct the nuclear localization of an unrelated chemically synthesized 21-nt short interfering RNA (siRNA) [62]. Recent genome-wide studies have indicated that the phenomenon seems to be more widespread, in that most miRNAs are present in both nucleus and cytoplasm [63–67]. Some of these miRNAs, such as miR-21 and let-7, have been shown to cleave the RNA targets that have perfect sequence complementarity with the corresponding miRNAs in nuclear extracts, indicating that these miRNAs are bound with AGO2 [61,68]. Consistently, siRNAs can silence nuclear-localized RNAs, including 7SK, U6, Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), and Nuclear paraspeckle assembly transcript 1 (NEAT1) [3,66,68], supporting the notion that short RNA-loaded AGO2 can be present in the nucleus. However, one caveat of these knockdown experiments is that they were performed in dividing cells, where cytoplasmic factors may have access to nuclear species during mitosis.
Using a method independent of biochemical fractionation, fluorescence correlation spectroscopy on GFP-tagged AGO2 stably expressed in human cells revealed that AGO2 exists as a complex of 158 kDa in the nucleus in contrast to a large approximately 3 MDa complex in the cytoplasm [3]. This small nuclear complex was thought to comprise only AGO2 and a short RNA [3]. However, recent biochemical data argue for the nuclear presence of other proteins involved in biogenesis and the silencing of mature miRNAs, such as Dicer, TRBP, and GW182 [66]. Yet, unlike the cytoplasmic counterpart, these nuclear extracts lack the ability to load duplex short RNAs onto AGO2 [66], indicating that short RNA-loaded AGO2 originates from the cytoplasm [3] where the nuclear import of AGO2 is partly mediated by importin 8 [69]. The nuclear retention of these loaded AGO2 is dependent on the mode of miRNA binding to RNA targets, where an miRNA remains in the nucleus longer if it binds to the target with a central bulge [3]. Several reports have indicated that some nuclear-localized miRNAs are involved in gene silencing and/or activation at promoters (reviewed in [67]); however, more experiments are needed to ascertain whether such effects are direct. For example, gene-editing technologies, such as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) [70], can be used to test whether single point mutations on potential miRNA binding sites on DNA can abrogate the epigenetic regulation and whether the effect can be rescued by a miRNA with compensatory mutations.
Nucleolus
The nucleolus is the major subcompartment within the nucleus, which is responsible for ribosome biogenesis, cell cycle control, and cell signaling [71]. Using fluorescent in situ hybridization, miR-206 was first identified to be localized in the nucleolus in rat myoblast cells [72]. To date, there are approximately 40 miRNAs identified to be nucleolus associated in multiple cell lines [73–75]. These include miR-191, miR-484, miR-193b, miR-93, and miR-574, which have been identified in isolated nucleoli using Taqman assays, northern blots, and high-throughput sequencing [75,76]. Preliminary characterization indicates that these nucleolar miRNAs have several interesting properties in terms of their cytoplasmic transport and AGO association.
Treatment with Leptomycin B results in depletion of the cytoplasmic fraction of nucleolar-localized miRNAs (e.g., miR-484, miR-21, and miR-31), suggesting these miRNAs are actively trafficking to the cytoplasm in a manner dependent on Exportin 1 [75,76]. Viral infection or transfection of exogenous nucleic acids into cells result in redistribution of miR-484 from the nucleolus to the nucleoplasm and then to the cytoplasm [75]. Whether Exportin 1 is involved in this redistribution in response to viral infection remains to be tested. Notably, Exportin 1 associates with AGO and is responsible for the shuttling of mature miR-16 and miR-29b between the nucleus and the cytoplasm [77]. Therefore, it will be of interest to test whether this Exportin 1-mediated transport of nuclear and/or nucleolar miRNAs is dependent on AGO binding, for example, using cells knocked out of all AGOs [45]. Intriguingly, compared with miR-20a, which is exclusively localized to the cytoplasm, miR-484 binds less to AGO2, suggesting that miR-484 localization in the nucleolus is independent of AGO2. It remains unclear whether this nucleolar miRNA can bind to other proteins or bind directly to its targets without AGO (as proposed by [40]). Interestingly, a fraction of spliced IGF2 mRNAs, which have predicted binding sites for miR-206 and four other nucleolus-localized miRNAs at the 3′ untranslated regions (UTR), were also found in the nucleolus [78]. Given that the protocol to isolate highly purified nucleoli from mammalian cells is well established, future experiments can rigorously test whether insulin growth factor 2 (IGF2) or other mRNA targets are specifically bound by AGO2 and/or miRNAs.
Mitochondria
Mitochondria are cytoplasmic, double membrane-bound organelles responsible for cellular energy production; each mitochondrion contains its own circular DNA that encodes 13 proteins, 22 tRNAs, and two rRNAs [79]. Several studies have identified, surprisingly, a broad range of miRNAs (up to hundreds) from mitochondria isolated from rat, mouse, and human cells and/or tissues [80–87]. The mitochondrial miRNA localizations were further supported by treatment with RNAases to remove cytosolic RNAs associated with outer mitochondrial membranes [80,82–84,87]. In terms of miRNA machinery, AGO2, but not Dicer and TRBP, is found in isolated mitochondria [80,85,87] and, intriguingly only AGO2, but not AGO1 and AGO3, was selectively imported to mitochondria [87]. It was estimated that 13% of the total AGO2 localizes in mitochondria in the undifferentiated myoblast cell line C2C12, and that the level increases to 33% after differentiation [87]. Taken together, these data suggest that a sizeable fraction of AGO2 and miRNAs is present in mitochondria.
As shown by both immunoprecipitation and CLIP, AGO2 is associated with mitochondrially encoded mRNA targets [85,87], but two reports suggest opposite roles of these miRNAs identified in mitochondria in regulating the expression of mitochondrial mRNAs. Overexpression of miR-181c in primary cultures and rat models results in reduced protein level of mitochondrially encoded cytochrome c oxidase subunit 1 (COX1), which contains a highly conserved binding site for the 3rd–8th nucleotide of miR-181c at the 3′ UTR [85,88]. Although miR-181c was shown to repress the expression of a luciferase reporter mRNA containing the COX1 3′ UTR that is expressed in the cytosol, the study did not definitively show that miR-181c represses COX1 mRNA in the mitochondria. Alternatively, miR-1 increases the translation of COX1 and another mitochondrially encoded NADH dehydrogenase 1 (ND1) [87], contrary to the canonical repressive role of a miRNA. This translational activation is sensitive to the mitochondrial translation inhibitor chloramphenicol and is abrogated in AGO2-knockout cells. Upon adding back AGO2 to these knockout cells, miR-1 represses the expression of cytosolic miR-1 mRNA targets, but increases Cyclooxygenase 1 (COX1) and NADH-ubiquinone oxidoreductase chain 1 (ND1) expression. More significantly, adding back an AGO2 that is fused with a mitochondrial-targeting signal peptide results in an increase in COX1 and ND1 expression but no change in the expression of cytosolic targets. Although the molecular mechanism of the translational activation remains unknown, the mitochondria do not have GW182 (the miRISC component required for miRNA-mediated repression) and GW182 knockdown does not affect COX1 and ND1 expression. Furthermore, the activation is dependent on both the 5′ seed of miR-1 and its 3′ end sequence. It will be of interest to examine which protein partners bound to AGO2 are required for translational activation in mitochondria and to test whether the 3′ end sequence could be a transferrable mitochondrial localization signal.
Concluding Remarks
Future characterization of these varied location-specific miRNA functions will benefit from deciphering the local composition of these individual miRISCs and the types of target they are associated with (see Outstanding Questions). Recent developments in molecular tools, such as the use of promiscuous biotinylation, should allow investigation of the organelle-specific proteomes and transcriptomes associated with miRNAs [25,89–91]. Special attention should be paid to the possible distinct functions of individual AGO members at specific organelles (e.g., AGO2 in mitochondria). Conversely, could there be proteins, other than hnRNP E2, TLR7/8, TDP-43, and HIV Gag proteins, associated with functionally active miRNAs independent of AGOs? Such biochemical understandings may shed light on noncanonical roles of miRNAs and miRNA-dependent translational activation as proposed in mitochondria. In vitro biochemical assays for miRNA functions should also be tailored to include nonprotein components, such as the type of lipids within the membranes, or to use highly purified, intact organelles. Although mature miRNA species are known to be the effectors of miRNA silencing, recent studies suggest that such roles could be expanded to pre-miRNAs [92], which originate from the nucleus [1] and could be identified in nucleoli and mitochondria [74,84]. Given that the miRNA-processing factors Drosha and its partner DiGeorge syndrome chromosomal region 8 (DGCR8) are enriched in the nucleolus [93,94], it will be of interest to see whether there are specific functions for pre-miRNAs locally made there.
Most importantly, a quantitative view of miRISC composition and miRNA–target interaction should be established to appreciate whether such localized function is of physiological significance. Considerable efforts have established strategies to determine the copy number of AGOs, miRNAs, and their targets required for global miRNA functions [39,40,95,96]. Such numbers will need to be re-evaluated in the context of local concentration within these varied subcompartments. As discussed above, the recent arrival of the gene-editing technique CRISPR opens the possibility to probe defined location-specific miRNA–target interactions. It is now possible to introduce single-point mutations to test whether mammalian miRNAs in the nucleus can act like siRNAs in yeast or Piwi-interacting (pi)RNAs in Drosophila in generating epigenetic chromatin marks [67]. Similarly, these gene-editing technologies can be applied to mitochondrial genomes to examine whether specific miRNA–mitochondrial mRNA target interaction is of physiological significance.
Lastly, it is paramount to delineate the mechanisms of how miRNAs and/or miRISCs are distributed in intracellular locations and exported from cells. The intracellular distribution is likely dependent on cell types; for example, heart muscle cells have at least 5000 mitochondria per cell and mitochondrially localized miRNAs will likely have a larger role there in modulating the expression of proteins responsible for energy production. Other mechanisms in play are protein modifications on miRISC components and localization signals within miRNAs, particularly those at the 3′ end, similar to the signal identified for the nuclear miR-29b [62]. Just as a small-molecule drug can be used to target protein-modification enzymes in modulating miRISC distribution, a defined nucleotide motif will be instrumental for designing next-generation siRNAs for location-specific miRNA targets. This new biological knowledge could then be applied in therapies when the location-specific miRNA functions become apparent. However, the key question remains; are these location-specific miRNA functions physiologically relevant?
Trends.
Mature miRNAs localize in multiple subcellular locations in the cytoplasm, such as RNA granules, endomembranes, and mitochondria, and secrete outside cells via exosomes.
Recent studies have revealed that mature miRNAs can also localize to the nucleus, where they could function in epigenetic regulation.
The distributions of canonical and noncanonical forms of miRNA induced silencing complexes suggest that different subcellular locations are required for the processing and degradation of miRNA itself or for silencing or activation of miRNA targets.
These subcellular distributions are differentially regulated by post translational modifications as a function of cellular conditions, but one major question is whether such location specific miRNAs are physiologically relevant.
Outstanding Questions.
What is the regulatory mechanism accounting for the distribution of miRNAs in different intracellular locations and export outside cells?
What are the targeting signals for miRNAs and their associated proteins to distinct subcellular locations?
If AGO only binds 10% of miRNAs, what other non AGO proteins are bound by miRNAs? Alternatively, are they associated with their targets independent of protein binding? Do these complexes have specific functions?
Do individual AGOs have specific functions in different organelles?
How many copies of miRNAs are sufficient for their localized functions to have a physiological effect? Are these location specific miRNA functions physiologically relevant?
Acknowledgments
The author would like to thank Phillip Sharp, Yoshinari Ando, Marriki Laiho, Sam Das, Jesse Zamudio, and Joe Fischer for critical reading of the manuscript, and Yun Wah Lam for discussing the current state of the field of nucleolar miRNAs. The miRNA research in the Leung lab is funded by the National Institutes of Health (NIH) grant R01 GM104135.
References
- 1.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 2.La Rocca G, et al. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci USA. 2015;112:767–772. doi: 10.1073/pnas.1424217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohrt T, et al. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Höck J, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 7.Leung AKL, Sharp PA. Quantifying Argonaute proteins in and out of GW/P-bodies: implications in microRNA activities. Adv Exp Med Biol. 2013;768:165–182. doi: 10.1007/978-1-4614-5107-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cougot N, et al. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung AKL, et al. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eulalio A, et al. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharyya SN, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Aizer A, et al. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J Cell Sci. 2014;127:4443–4456. doi: 10.1242/jcs.152975. [DOI] [PubMed] [Google Scholar]
- 13.Brengues M, et al. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arribere JA, et al. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol Cell. 2011;44:745–758. doi: 10.1016/j.molcel.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cougot N, et al. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci. 2008;28:13793–13804. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savas JN, et al. A role for huntington disease protein in dendritic RNA granules. J Biol Chem. 2010;285:13142–13153. doi: 10.1074/jbc.M110.114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YJ, et al. Traffic into silence: endomembranes and post-transcriptional RNA silencing. EMBO J. 2014;33:968–980. doi: 10.1002/embj.201387262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cikaluk DE, et al. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodersen P, et al. Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGO-NAUTE 1 in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:1778–1783. doi: 10.1073/pnas.1112500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi Z, Ruvkun G. The mevalonate pathway regulates microRNA activity in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:4568–4573. doi: 10.1073/pnas.1202421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stalder L, et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013;32:1115–1127. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu PH, et al. Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol Cell. 2013;52:113–123. doi: 10.1016/j.molcel.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jan CH, et al. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:716. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbings DJ, et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 28.Cortez MA, et al. MicroRNAs in body fluids: the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Squadrito ML, et al. Endogenous RNAs modulate micro-RNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 30.Ono M, et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in meta-static breast cancer cells. Sci Signal. 2014;7:ra63. doi: 10.1126/scisignal.2005231. [DOI] [PubMed] [Google Scholar]
- 31.Morel L, et al. Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J Biol Chem. 2013;288:7105–7116. doi: 10.1074/jbc.M112.410944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh S, et al. Polysome arrest restricts miRNA turnover by preventing their exosomal export in growth retarded mammalian cells. Mol Biol Cell. 2015;26:1072–1083. doi: 10.1091/mbc.E14-11-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun JE, et al. The role of GW182 proteins in miRNA-mediated gene silencing. Adv Exp Med Biol. 2013;768:147–163. doi: 10.1007/978-1-4614-5107-5_9. [DOI] [PubMed] [Google Scholar]
- 34.Miyoshi K, et al. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA. 2009;15:1282–1291. doi: 10.1261/rna.1541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frohn A, et al. Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol Cell Proteomics. 2012;11:1442–1456. doi: 10.1074/mcp.M112.017756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbings D, et al. Human prion protein binds Argonaute and promotes accumulation of microRNA effector complexes. Nat Struct Mol Biol. 2012;19:517–524. doi: 10.1038/nsmb.2273. [DOI] [PubMed] [Google Scholar]
- 38.Olejniczak SH, et al. Long-lived microRNA-Argonaute complexes in quiescent cells can be activated to regulate mitogenic responses. Proc Natl Acad Sci USA. 2013;110:157–162. doi: 10.1073/pnas.1219958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flores O, et al. Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res. 2014;42:4629–4639. doi: 10.1093/nar/gkt1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janas MM, et al. Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA. 2012;18:2041–2055. doi: 10.1261/rna.035675.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eiring AM, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King IN, et al. The RNA-binding protein TDP-43 selectively disrupts microRNA-1/206 incorporation into the RNA-induced silencing complex. J Biol Chem. 2014;289:14263–14271. doi: 10.1074/jbc.M114.561902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen AK, et al. MicroRNA binding to the HIV-1 Gag protein inhibits Gag assembly and virus production. Proc Natl Acad Sci USA. 2014;111:E2676–E2683. doi: 10.1073/pnas.1408037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamudio JR, et al. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156:920–934. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149–1157. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 47.Smibert P, et al. Homeostatic control of Argonaute stability by microRNA availability. Nat Struct Mol Biol. 2013;20:789–795. doi: 10.1038/nsmb.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez NJ, Gregory RI. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA. 2013;19:605–612. doi: 10.1261/rna.036434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung AKL, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karginov FV, Hannon GJ. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013;27:1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jee D, Lai EC. Alteration of miRNA activity via context-specific modifications of Argonaute proteins. Trends Cell Biol. 2014;24:546–553. doi: 10.1016/j.tcb.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horman SR, et al. Akt-mediated phosphorylation of Argonaute 2 downregulates cleavage and upregulates translational repression of microRNA targets. Mol Cell. 2013;50:356–3567. doi: 10.1016/j.molcel.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leung AKL, et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seo GJ, et al. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu C, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol. 2011;31:4760–4774. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bronevetsky Y, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sahin U, et al. Sumoylation of human argonaute 2 at lysine-402 regulates its stability. PLoS ONE. 2014;9:e102957. doi: 10.1371/journal.pone.0102957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazumder A, et al. A transient reversal of miRNA-mediated repression controls macrophage activation. EMBO Rep. 2013;14:1008–1016. doi: 10.1038/embor.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen J, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams BD, et al. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 62.Hwang HW, et al. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 63.Liao JY, et al. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park CW, et al. Mature microRNAs identified in highly purified nuclei from HCT116 colon cancer cells. RNA Biol. 2010;7:606–614. doi: 10.4161/rna.7.5.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeffries CD, et al. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA. 2011;17:675–686. doi: 10.1261/rna.2006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gagnon KT, et al. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roberts TC. The microRNA biology of the mammalian nucleus. Mol Ther Nucleic Acids. 2014;3:e188. doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robb GB, et al. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 69.Weinmann L, et al. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 70.Hsu PD, et al. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boulon S, et al. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Politz JCR, et al. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci USA. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcon E, et al. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008;16:243–260. doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- 74.Politz JCR, et al. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li ZF, et al. Dynamic localisation of mature microRNAs in Human nucleoli is influenced by exogenous genetic materials. PLoS ONE. 2013;8:e70869. doi: 10.1371/journal.pone.0070869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bai B, et al. Small RNA expression and deep sequencing analyses of the nucleolus reveal the presence of nucleolus-associated microRNAs. FEBS Open Bio. 2014;4:441–449. doi: 10.1016/j.fob.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castanotto D, et al. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci USA. 2009;106:21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reyes-Gutierrez P, et al. A mRNA and cognate microRNAs localize in the nucleolus. Nucleus. 2014;5:636–642. doi: 10.4161/19491034.2014.990864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Temperley RJ, et al. Human mitochondrial mRNAs: like members of all families, similar but different. Biochim Biophys Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bandiera S, et al. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS ONE. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lung B, et al. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kren BT, et al. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bian Z, et al. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010;20:1076–1078. doi: 10.1038/cr.2010.119. [DOI] [PubMed] [Google Scholar]
- 84.Barrey E, et al. Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Das S, et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sripada L, et al. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS ONE. 2012;7:e44873. doi: 10.1371/journal.pone.0044873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Das S, et al. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS ONE. 2014;9:e96820. doi: 10.1371/journal.pone.0096820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roux KJ, et al. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rhee HW, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williams CC, et al. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roy-Chaudhuri B, et al. Regulation of microRNA-mediated gene silencing by microRNA precursors. Nat Struct Mol Biol. 2014;21:825–832. doi: 10.1038/nsmb.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu H, et al. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 94.Shiohama A, et al. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 95.Bosson AD, et al. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Denzler R, et al. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]