Abstract
The eukaryotic genome is organized in the three-dimensional nuclear space in a specific manner that is both a cause and a consequence of its function. This organization is in part established by a special class of architectural proteins of which CTCF is the best characterized. Although CTCF has been assigned a variety of often contradictory roles, new results help draw a unifying model to explain the many functions of this protein. CTCF creates boundaries between topologically associating domains in chromosomes and, within these domains, CTCF facilitates interactions between transcription regulatory sequences. Thus, CTCF links the architecture of the genome to its function.
Keywords: epigenetics, histones, chromatin, transcription
Introduction
Eukaryotic genomes are dynamically packaged into multiple levels of organization, from nucleosomes to chromatin fibers to large-scale chromosomal domains occupying defined territories of the nucleus. The three dimensional interplay of protein–DNA complexes facilitates timely realization of intricate nuclear functions such as transcription, replication, DNA repair and mitosis1. A combination of microscopy and chromosome conformation capture (3C)-related approaches2 revealed that CCCTC-binding factor (CTCF) is in large part responsible for bridging the gap between nuclear organization and gene expression. CTCF is the main insulator protein described in vertebrates. Initially characterized as a transcription factor capable of activating or repressing gene expression in heterologous reporter assays3, 4, CTCF was later found to display properties characteristic of insulators i.e. the ability to interfere with enhancer-promoter communication or buffer transgenes from chromosomal position effects caused by heterochromatin spreading. These properties, observed using transgene assays, were interpreted to suggest a role for insulators in restricting enhancer-promoter interactions and establishing functional domains of gene expression.
In this Review, we discuss recent evidence, arising from the use of 3C-related techniques, indicating that the diverse properties of CTCF and other insulator proteins are based on their broader role in mediating inter- and intra-chromosomal interactions between distant sites in the genome. As a result of these interactions, CTCF elicits specific functional outcomes that are context-dependent, determined by the nature of the two sequences brought together and the proteins with which they interact. Consequently, CTCF contributes to the establishment of a three-dimensional (3D) structure of the chromatin fiber in the nucleus that is both an effector and a consequence of genome function. Because the role of CTCF extends well beyond that originally attributed to insulator proteins, and its functional effects are based on its ability to mediate interactions between distant sequences, we propose the term “architectural” rather than “insulator” to describe this type of protein.
CTCF interactions with DNA, proteins and ncRNA
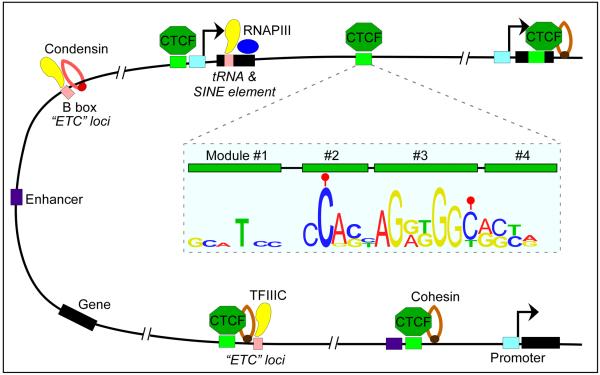
CTCF is conserved in most bilaterian phyla but it is absent from yeast, C. elegans and plants5. It contains a highly conserved 11 zinc finger DNA-binding domain6 and it is present at 55,000–65,000 sites in mammalian genomes7, normally located in linker regions surrounded by well-positioned nucleosomes8. Of these, approximately 5,000 sites are ultraconserved between mammalian species and tissues, and correspond to high affinity sites9, whereas 30–60% of CTCF sites show cell-type-specific distribution8, 10–12. The location of CTCF sites with respect to genomic features provides insights into the possible roles of this protein. Approximately 50% of CTCF binding sites reside within intergenic regions, ~15% are located near promoters and ~40% are intragenic (exons and introns)7, 12 (Fig.1). Surprisingly, and in view of the original role attributed to CTCF as an enhancer blocker, enhancer elements are enriched for this protein13, 14, suggesting that a subset of CTCF sites may be important in regulating transcription in order to establish cell lineage-specific programs. Experiments using the ChIP-exo technique uncovered a 52 bp CTCF binding motif containing four CTCF binding modules15, 16 (Fig.1).
Figure 1. Features of CTCF binding sites in the genome.
CTCF binding sites are associated with different genetic elements. The majority of sites are intergenic and co-localize with cohesin. In addition, a fraction of CTCF binding sites are located near RNA polymerase III (RNAPIII) type II genes (e.g. tRNA and SINE elements) and ETC loci, suggesting that TFIIIC and CTCF may cooperate in some aspects of the function of this protein. The 12 bp consensus sequence of CTCF sites is embedded within binding modules 2 and 3 as determined by the ChIP-exo technique. DNA methylation (filled red ball) of cytosine residues occurs at positions 2 and 12 of the consensus sequence in a subset of CTCF sites.
The presence of CpG in the DNA consensus sequence of the CTCF binding site supports the notion that methylation of cytosine residues at carbon atom 5 of the base (5mC) in those sites containing CpG may underlie, at least in part, CTCF target selectivity in different cell-types17. Recent studies indicate that DNA methylation plays a widespread role in regulating CTCF occupancy at many genes, including CDKN2A (which encodes INK4A and ARF)18, BCL619 and BDNF20. Stamatoyannopoulos and colleagues have mapped the occupancy of CTCF in 19 human cell types and, by comparing this information with DNA methylation data from parallel reduced representation bisulfite sequencing, they found that 41% of cell type-specific CTCF sites are linked to differential DNA methylation21 (Fig.2). Conversely, at 67% of sites that showed DNA methylation variability, DNA methylation was associated with concomitant down-regulation of cell type-specific CTCF occupancy. CTCF can also affect the methylation status of DNA by forming a complex with poly(ADP-ribose) polymerase 1 (PARP1) and DNA (cytosine-5)-methyltransferase 1 (DNMT1). CTCF activates PARP1, which can then inactivate DNMT1 by poly(ADP-ribosyl)ation, and thus maintains methyl-free CpGs in the DNA22, 23. An additional level of complexity in the interaction between CTCF and its target sequence can arise from the oxidation of 5mC to 5-hydroxymethylcytosine (5hmC)24, 25, 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC)26 by TET enzymes. Genome-wide profiling analysis of 5hmC has shown that this modification, and to a lesser extent 5fC, is enriched at sites in the genome containing CTCF27, 28. Furthermore, identification of proteins that bind to different oxidized derivatives of 5mC identified CTCF as a 5caC-specific binder29. These results underscore the complexity and possible importance of the relationship between DNA methylation status and plasticity of CTCF occupancy. However, the presence of cell-type specific CTCF binding sites that are not differentially methylated suggests the existence of other mechanisms to regulate DNA occupancy by this protein (Fig.2).
Figure 2. Regulation of CTCF binding to DNA.

Constitutive CTCF sites present in cells from different tissues are present in non-methylated and nucleosome-free regions. Cell-type specific CTCF binding is partly regulated by differential DNA methylation and nucleosome occupancy across different cell-types. This suggests that cells may use ATP-dependent chromatin remodeling complexes to regulate nucleosome occupancy at specific CTCF sites and control the interaction of this protein with DNA. In addition, the methylation status of cell-type-specific CTCF binding sites may be determined by a combination of activities of de novo methyltransferases and TET enzymes that regulate the presence and levels of 5mC at specific sites. Immortalized cancer cell lines contain high levels of 5mC at CTCF sites, which correlates with the low CTCF occupancy in these cells (Filled red circle: methylated DNA; open circle: unmethylated DNA).
One such mechanism is covalent modification of CTCF, which undergoes post-translational modifications that include sumolyation30 and poly(ADP-ribosy)lation31. In breast cancer cells, defective poly(ADP-ribosyl)ation of CTCF leads to its dissociation from the CDKN2A locus, resulting in aberrant silencing of this tumor suppressor gene32. In Drosophila melanogaster, poly(ADP-ribosy)lation of CP190 and CTCF facilitates their interaction, tethering to the nuclear matrix, and intra-chromosomal contacts33.
Interaction between CTCF and other proteins may represent an additional strategy by which the function of this protein can be regulated at various genomic locations during cell differentiation. Although different proteins have been implicated in CTCF function at specific loci34, including Yin-Yang 1 (YY1), Kaiso, CHD8, PARP1, Maz, JunD, ZNF143, Prdm5, and Nucleophosmin, only cohesin has been shown to be required to stabilize most CTCF-mediated chromosomal contacts and to be essential for CTCF function at most sites in the genome35–41. Interaction between CTCF and the cohesin complex takes place through the carboxy-terminal region of CTCF and the SA2 subunit of cohesin35. Like CTCF, cohesin is present in intergenic regulatory regions, promoters, introns and 5'UTRs of genes during interphase. Depending on the cell line, 50–80% of CTCF sites in the genome are also occupied by cohesin, and downregulation of cohesin using RNAi results in disruption of CTCF-mediated intra-chromosomal interactions42–44. A second protein that may cooperate with CTCF at a subset of sites in the genome is TFIIIC, a factor required for the transcription of tRNAs, 5S rRNA, SINE B2 elements, and other non-coding RNAs by RNA polymerase III (RNAPIII)45. TFIIIC also binds to many genomic sites devoid of RNAPIII that are called `extra-TFIIIC' (ETC) loci (Fig.1). In yeast, both the tRNA genes and ETC loci have been shown to cluster and tether DNA sequences to the nuclear periphery45, 46. Furthermore, SINE B2 elements and human tRNA genes, both of which contain binding sites for TFIIIC, can act as enhancer-blocking insulators in transgene assays47, 48, and genome-wide analyses revealed that CTCF and its binding partner cohesin are found in the vicinity of many tRNA genes and ETC loci in mouse49 and human cells50, 51 (Fig.1).
In addition to proteins, several observations suggest that RNAs may also cooperate with CTCF to stabilize interactions mediated by this protein. D. melanogaster architectural proteins such as CP190 require the Rm62 RNA helicase for proper function, and interactions between Rm62 and CP190 depend on the presence of RNA52. Similar observations have been made in mammalian cells, where CTCF has been shown to interact with the DEAD-box RNA helicase p68 and its associated ncRNA, both of which are required for proper CTCF function53. These observations, together with new findings indicating that CTCF can itself bind Jpx RNA54, offer support to the idea that ncRNAs may play an important role in stabilizing interactions mediated by CTCF and its protein partners.
Mechanisms of CTCF function
A large body of evidence strongly supports the idea that the mechanism underlying the diverse functions of CTCF in genome biology is based on its ability to mediate long-range interactions between two or more genomic sequences. This evidence first came from chromosome conformation capture (3C) analyses of the H19/Igf2 and murine β-globin loci. At the imprinted maternal H19/Igf2 locus, work using circular chromosome conformation capture (4C) indicated that the H19 imprinting control region (ICR) forms extensive inter- and intra-chromosomal interactions across the genome, many of which require the presence of CTCF binding sites within the ICR55. Binding of CTCF at multiple DNase I hypersensitive sites (DHSs) is also required to maintain a specific chromatin architecture at the murine β-globin locus56. Similarly, D. melanogaster architectural proteins BEAF-32 and ZW5 were shown to mediate long-range interactions at the hsp70 locus and between copies of the gypsy retrotransposon57, 58.
Taken together, these observations suggest that the effect of CTCF and other architectural proteins on gene expression is a result of their ability to bring in close proximity sequences located far apart in the linear genome. Assuming that this mechanism underlies all functions of CTCF, results of locus-specific and genome-wide studies can be interpreted to suggest that CTCF-mediated contacts regulate aspects of genome function in a manner that is context-dependent i. e. the functional outcomes of these interactions depend on the nature of the sequences adjacent to CTCF binding sites and perhaps the presence of other specific chromatin proteins. Below we critically analyze existing evidence for these various roles in an attempt to generate a model that reflects the function of this protein in its endogenous genomic context.
The classical roles of CTCF
CTCF as a chromatin barrier
The function of CTCF and other architectural proteins was initially analyzed using transgene assays, where results were often interpreted to suggest that these proteins act as barriers to the processive spread of heterochromatin59.. Indeed, sequences with bona fide barrier activity in yeast have been shown to recruit histone acetyltransferases that antagonize the spreading of silencing histone modifications60. However, although this interpretation of experimental results is often used to explain results obtained in higher eukaryotes, recent studies offer a different view of how architectural proteins may regulate gene expression. For example, Felsenfeld and colleagues have demonstrated the presence of CTCF-dependent enhancer blocking but CTCF-independent barrier function in the chicken HS4 insulator. In this case, the barrier function depends on USF1, which recruits histone acetyltransferases61.
Results from genome-wide studies of the localization of CTCF in relation to various histone modifications also do not offer strong support for a role of CTCF as a barrier. Human CD4+ T cells and HeLa cells contain around 30,000 domains of H3K27me3 (a histone modification characteristic of silenced chromatin), of which ~1,600 and ~800 contain CTCF at one of the domain borders, respectively8. This represents only 2–4% of the domain borders, a relatively low number if CTCF was primarily involved in the establishment or maintenance of these silenced domains. Similarly, it has been suggested that CTCF may contribute to the formation of chromosome domains associated with the nuclear lamina. Human lung fibroblasts contain 2,688 borders flanking lamin-associated domains (LADs). These domains are enriched in oriented promoters, CpG islands and CTCF binding sites. Approximately 9% of LAD borders contain CTCF (245 out of 2,688; 120 additional borders contain a combination of promoters, CpGs and/or CTCF) within 10 kb of the boundary62. Although the correlation is tantalizing, it is unlikely that CTCF is the main contributor to the formation of these borders or that this is one of the primary functions of this protein.
Despite the lack of strong evidence from genome-wide localization analyses, the role of CTCF has been interpreted as domain barrier in several studies. Recent mapping of CTCF-mediated intra- and inter-chromosomal interactions in mouse embryonic stem cells (ESCs) using ChIA-PET, a technique that combines ChIP with 3C analyses, seems to lend support to the notion that CTCF may be, at least in part, responsible for the establishment of functional expression domains83. A total of 1480 CTCF-containing cis-interacting loci were identified by this strategy. Cluster analyses of intra-chromosomal interactions with seven histone modification signatures and RNAPII profiles uncovered four distinct categories of CTCF-mediated loops. One class (155 out of 1295 loops smaller than 1 Mb, 12%) contains active H3K4me1, H3K4me2 and H3K36me3 histone modifications inside but repressive H3K9me3, H3K20me3 and H3K27me3 modifications outside of the loops. A second class (142, 11%) of loops has the reverse pattern of histone modifications. Although the evidence is only correlative, and the number of CTCF-mediated interactions is very small compared to the total number of sites of this protein in the genome, the existence of these two types of loops in which CTCF flanks histone modifications with opposite effects on transcription is suggestive, but not conclusive proof, of a role for this protein in separating functional domains of gene expression.
Some examples of locus-specific analyses seem to also support this view of CTCF-mediated barrier activity. For instance, the Wilms' tumor 1 (Wt1) transcription factor can activate or repress expression of the mouse Wnt4 gene in a cell-type specific manner by controlling the state of chromatin in a domain whose boundaries are defined by CTCF. Mutation of CTCF leads to spreading of histone modifications outside of the delimited genomic domain, causing aberrant expression of neighboring genes and suggesting a role for CTCF in the establishment or maintenance of the Wnt4 domain by creating a functional barrier63.
Although the ChIA-PET and Wnt4 results are best explained by assuming that CTCF is cable of barrier activity, a similar explanation of results obtained from studies at other loci may appear less straightforward. For example, groups of androgen responsive genes demarcated by CTCF binding sites tend to have similar epigenetic and expression profiles, suggesting that CTCF establishes domains were these genes are co-regulated64. Downregulation of CTCF results in a decrease in the expression of genes within the domain while genes outside of the domain are unaffected, a result that can be explained if CTCF is involved in targeting regulatory sequences to androgen-responsive promoters and, in its absence, transcription of these genes decreases. Similarly, the mouse HOXA locus forms two distinct chromatin loops around CTCF binding site 5 (CBS5) as ESCs differentiate into neural progenitor cells. The loop containing the HOXA1-7 gene cluster upstream of CBS5 is marked by active H3K4me3 modifications, whereas the loop containing the downstream HOXA9-13 gene cluster is enriched for repressive H3K27me3 marks65. Knockdown of CTCF results in the loss of 3D conformation and the concomitant spread of H3K27me3 modifications across the locus. These results can be explained based on a barrier function for CTCF but it is equally possible that CTCF binding sites in the HOXA locus may participate in bringing together regulatory sequences for gene activation or repression. This explanation is supported by results obtained in D. melanogaster, where the role of CTCF in the maintenance of H3K27me3-enriched domains delimited by CTCF and other architectural proteins was analyzed in detail. When CTCF was knocked down, H3K27me3 domains showed significant reduction in the level of this histone modification within the domain. However, little or no spreading of H3K27me3 was observed outside of the demarcated domains66, 67. This suggests that Drosophila CTCF may help maintain the level of silencing within domains, but not its spreading, presumably by clustering of H3K27me3 loci and Polycomb group (PcG) proteins into Polycomb bodies66, 68. Based on these results, we suggest that there is little causal evidence to support a generalized functional role for CTCF in separating domains with different epigenetic marks. Instead, alternative mechanistically different processes, such as those involving looping between regulatory sequences, may better explain some of the observations previously interpreted in this context.
CTCF as an enhancer blocker
Although CTCF has been extensively characterized for its ability to block enhancer activity in transgene assays, there has been little evidence to support such a role for this protein in its normal genomic context. However, some recent studies suggest that CTCF can indeed act as enhancer blocker at specific loci. For example, induction of the Eip75B gene by treatment of D. melanogaster Kc cells with the steroid hormone ecdysone results in the downregulation of one of the Eip75B transcripts encoded by this gene that is expressed from an alternative upstream promoter. This is caused by activation of a poised CTCF site by recruitment of CP190, increasing its interaction with a distant CTCF site and topologically separating the downregulated promoter of the Eip75B gene from its enhancer69. Some genome-wide studies also suggest an enhancer blocking function for CTCF. A search for conserved regulatory motifs in the human genome led to the finding of 15,000 CTCF sites that separate adjacent genes that show markedly reduced correlation in gene expression when compared to genes in a similar arrangement but not separated by CTCF sites70. A similar observation has been made for the BEAF-32 protein of D. melanogaster71, suggesting that CTCF and other architectural proteins may allow neighboring gene pairs to be differentially regulated. However, the classical enhancer-blocker function of CTCF appears to contradict more recent results supporting a function for CTCF as a facilitator of enhancer function. Below we describe in detail some of this new information in order to underscore the widespread, but not widely acknowledged, role for CTCF as a positive regulator of various transcription processes. In the end, models to explain how CTCF controls gene expression need to account for these two apparently contradictory functions, enhancer blocker and enhancer facilitator, of this protein.
An updated view of CTCF function
CTCF helps tether distant enhancers to their promoters
Recent observations seem to contradict the idea of enhancer blocking as a predominant role for CTCF. For example, Dekker and colleagues examined interactions between promoters and their regulatory sequences using the Chromosome Conformation Capture Carbon Copy (5C) technique and found that 79% of long-range interactions between distal elements and promoters are not blocked by the presence of one or more intervening CTCF-bound sites72. Instead, a fraction of these interacting distal elements is significantly enriched for CTCF and/or histone modifications characteristic of active enhancers (H3K4me1,2 and H3K4me3), lending strong support to the concept that one of the main roles of CTCF in genome function may be to facilitate the interaction between regulatory sequences. Activation of transcription requires the assembly of specific activators, the Mediator complex and the basal transcription machinery in a process that involves long-range chromosomal interactions between distal enhancers and proximal promoter elements. The enrichment of CTCF sites at promoter and intergenic regions observed in ChIP-seq studies also suggests that one of the main functions of CTCF may be to target regulatory elements to their cognate promoters. This conclusion is supported by the finding of a significant overlap between cell-type-specific CTCF binding sites and enhancer elements73, as well as studies at several individual loci. For example, CTCF-mediated topological organization of the major histocompatibility complex class II (MHCII) locus precedes transcriptional activation74. Activation of MHC-II gene expression by interferon-γ (IFNγ) treatment requires the looping of the XL9 enhancer element and its cognate promoters that is mediated by CTCF, class II transactivator (CIITA) and specific transcription factors75.
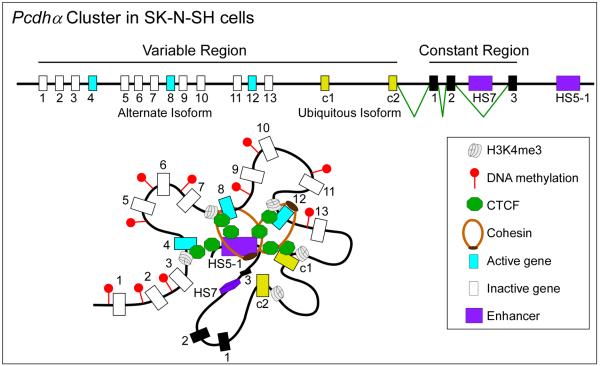
CTCF has also been shown to be important in regulating the expression of complex gene clusters in which regulatory sequences are far from some of their target genes. For instance, in human islets, CTCF maintains long-range interactions between the insulin (INS) and synaptotagmin 8 (SYT8) genes necessary for SYT8 transcription76. In the mammalian brain, neuronal diversity is attained through a combination of stochastic promoter choice and alternative pre-mRNA processing of the protocadherin (Pcdh) genes. Each Pcdh mRNA contains a variable 5' exon followed by a common region. The Pcdh gene cluster is comprised of more than 50 different 5' exons, each preceded by its own promoter (Fig.3). CTCF and cohesin bind to most of these promoters77 and the distant enhancer element HS5-178. Alternative isoform expression requires CTCF-mediated DNA looping between the HS5-1 enhancer and active Pcdhα promoters79, 80 (Fig.3). Conditional knockout of CTCF in mouse postmitotic projection neurons leads to reduced expression of Pcdh genes, neuronal defects and abnormal behavior, suggesting that CTCF is required to tether the HS5-1 enhancers to the various promoters81.
Figure 3. CTCF regulates enhancer-promoter interactions in a multi-gene cluster.
The human Pcdhα gene cluster contains 13 similar, tandemly arranged, variable first exons (1 to 13, shown in blue if they are transcribed or in white if they are not) and two related c-type ubiquitous first exons (c1 and c2, shown in yellow). Each of these 15 variable first exons is adjacent to its own promoter and is spliced to three downstream constant exons (1 to 3, shown in black). Pcdhα alternate isoforms are expressed stochastically, whereas all the c-type isoforms are expressed ubiquitously in all cells. The SK-N-SH cells depicted here express isoforms 4, 8 and 12. Promoter choice and the formation of an active chromatin hub is mediated by CTCF-cohesin DNA looping between the distal HS5-1 enhancer and distinct promoters at the Pcdhα gene cluster. Individual variable exons (blue and white rectangles) or ubiquitous exons (yellow rectangles) may be expressed and joined to the three exons from the constant region (black rectangles) by pre-mRNA splicing. Binding of CTCF to the promoter preceding individual exons is correlated with the level of gene activity. The active promoters are distinguished from the inactive promoters by an enrichment for H3K4me3 and a depletion of DNA methylation, which leads to expression of the downstream genes (blue rectangles).
A third recent example underscoring the role of CTCF in promoting enhancer-promoter interactions comes from studies in mouse ESCs, where the TATA-binding protein associated factor 3 (TAF3), a component of the core promoter-recognition complex TFIID, is required for endodermal differentiation. In addition to promoters, TAF3 also localizes to distal sites containing CTCF and cohesin and the two sequences form a loop in a TAF3-dependent manner82 (Fig.4). Given the role of TAF3 in regulating lineage commitment in ESCs, it is possible that the distal elements containing both CTCF and TAF3 binding sites may have acquired H3K4me1/2 pre-patterning in ESCs to become endodermal enhancers, thus supporting the idea that CTCF can tether distal regulatory sequences to their target promoters.
Figure 4. CTCF facilitates endodermal enhancer-promoter interactions in ESCs.
Recruitment of TAF3 at endodermal enhancers by CTCF and chromatin looping activates Mapk3 in ESCs. Apart from being a component of TFIID at core promoters, TAF3 may also associate with other transcription factors across the genome in ESCs. For instance, TAF3 represses the activity of pluripotency-associated transcription factors (OCT4, SOX2 and NANOG).
Observations made using genome-wide analyses of intra-chromosomal interactions also support a role of CTCF in facilitating contacts between transcription regulatory sequences. Analysis of CTCF-mediated interactions using ChIA-PET in mouse ESCs suggests that this protein may be involved in clustering promoters of different genes, perhaps to establish transcription factories83. Interestingly, 28% of genes whose promoters are brought into close proximity (<10 kb) to p300 sites by CTCF-mediated contacts are upregulated in mouse ESCs, and knockdown of CTCF results in downregulation of some of these genes, supporting the notion that CTCF may be involved in mediating enhancer-promoter interactions during transcription initiation83.
CTCF regulates recombination at the antigen receptor loci
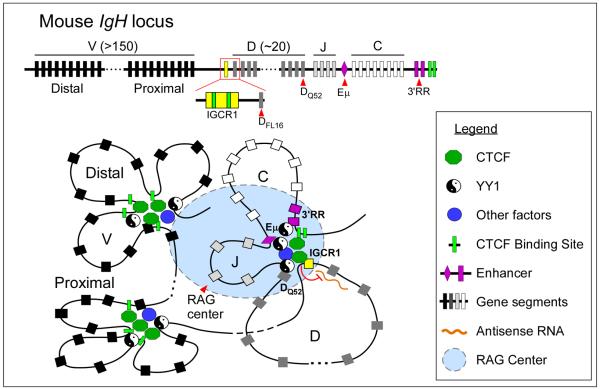
The role of CTCF in mediating enhancer-promoter communication may also contribute to the regulation of other nuclear processes like V(D)J recombination. The B cell immunoglobulin (Ig) and T cell receptor (Tcr) loci comprise multiple copies of variable (V), diversity (D), joining (J) and constant (C) gene segments that span across large genomic regions (Fig.5). During the adaptive response, unique epigenetic features and 3D chromatin architecture at these loci provide the framework for recombinase-activating gene (RAG)-mediated DNA recombination of the gene segments to generate antigen receptor diversity84. While not essential for progression of V(D)J recombination, CTCF-mediated long-range chromatin interactions may influence lineage/stage-specificity and proper segment choice during recombination.
Figure 5. CTCF regulates V(D)J recombination.
V(D)J recombination at antigen receptor loci is regulated by chromatin accessibility, which correlates with active histone modifications and transcription. CTCF may influence the outcome of V(D)J recombination by regulating enhancer-promoter interactions and locus compaction. At the IgH locus, CTCF-mediated looping of DH-JH-CH segments imposes ordered recombination (DH-to-JH) by controlling the communication of enhancers (Eμ and 3'RR) with distinct gene segments. Binding of CTCF at IGCR1 blocks the influence of the Eμ enhancer on proximal VH segments and prevents the spread of active histone modification from DH into the proximal VH region. In addition, it inhibits the level of antisense transcription within the DH region and modulates locus compaction in collaboration with other factors (e.g. YY1, Ikaros, Pax5, E2A). As a consequence, CTCF within IGCR1 may bias the rearrangement of distal (over proximal) VH segments with DJH joins.
Looping between distant CTCF binding sites may bring together distant gene segments. In pro-B cells, chromatin looping of CTCF sites at the IgH locus occurs independently of the Eμ enhancer and contributes to the compaction of the locus85, 86 (Fig.5). In double-positive thymocytes, CTCF-mediated looping between the Eα enhancer and specific promoters within the Tcrα/δ locus facilitates Vα-Jα over Vδ-Dδ-Jδ rearrangement87. By establishing interactions between specific sequences, CTCF may also impede other sequences from contacting each other. This, in fact, may be the basis for the enhancer-blocking function of CTCF. In the IgH locus, two CTCF binding sites within the Intergenic Control Region 1 (IGCR1) mediate ordered and lineage-specific VH-to-DJH recombination as well as biasing distal over proximal VH rearrangements88. Positioned between the VH and DH clusters, IGCR1 suppresses the transcriptional activity and the rearrangement of proximal VH segments by forming a CTCF-mediated loop that presumably isolates the proximal VH promoter from the influence of the downstream Eμ enhancer (Fig.5). Similarly, in pre-pro-B cells, CTCF promotes distal over proximal Vκ rearrangement by blocking the communication between specific enhancer and promoter elements in the Igκ locus89.
CTCF regulates transcription pausing and alternative mRNA splicing
The existence of a fraction of CTCF binding sites in the 5' UTR and introns of genes suggests a role of CTCF in regulating transcriptional events downstream of the initiation step. Indeed, recent studies indicate that CTCF can control both pausing of RNAPII and alternative mRNA splicing. For example, CTCF binds to both the first intron and upstream regulatory elements in the mouse Myb locus. During erythroid differentiation, looping between the first intron, promoter and upstream enhancer elements mediated by CTCF and key erythroid transcription and elongation factors is required for RNAPII-mediated transcriptional elongation and high expression of the Myb gene90. This 3D architecture is lost upon differentiation, when CTCF interferes with RNAPII elongation at the first intron, leading to low expression of the Myb gene. Here, the dual functions of CTCF in transcription initiation and pausing appear to rely on its ability to stabilize long-range interactions with regulatory sequences and to impede the elongation of RNAPII. The effect of CTCF on RNAPII elongation may be widespread, given that the genome-wide presence of CTCF at promoter-proximal regions in 5'UTRs correlates strongly with high pausing indexes91.
In other cases, hindering elongation of RNAPII by CTCF may result in the inclusion or exclusion of specific exons in the mature mRNA transcript. One example of this phenomenon occurs at the CD45 gene in humans, which encodes alternatively spliced transcripts during lymphocyte differentiation. Binding of CTCF to exon 5 of the gene promotes its inclusion in the CD45 mRNA whereas disruption of CTCF binding results in exclusion of this exon. Interestingly, it appears that DNA methylation of CTCF recognition sequences in exon 5 determines whether this protein binds to exon sequences, since knockdown of DNMT1 during late stages of lymphocyte differentiation leads to CTCF binding and inclusion of exon 5 in CD45 transcripts92 (Fig.6).
Figure 6. CTCF promotes alternative mRNA splicing.

Mutually exclusive DNA methylation and CTCF binding may regulate alternative splicing. At the CD45 gene, DNA methylation at exon 5 inhibits CTCF binding, which leads to fairly unimpeded transcriptional elongation by RNA polymerase II (RNAPII) and subsequent exclusion of exon 5 during splicing of the resultant mRNA (upper panel). By contrast, hypomethylation of exon 5 leads to CTCF binding and RNAPII stalling, which promotes the inclusion of exon 5 (lower panel).
Genome topology may rationalize CTCF roles
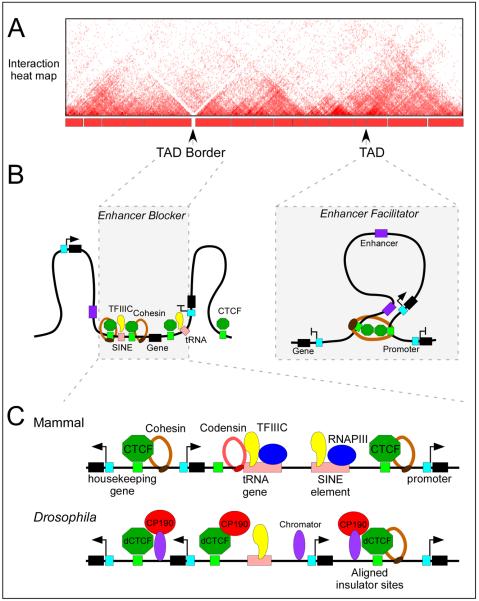
Results from experiments aimed at mapping all interactions in the genome using Hi-C suggest that genomes of higher eukaryotes are organized into topologically associating domains (TADs), defined by a high frequency of interactions within domains and a low frequency of interactions between adjacent domains (Fig.7). In D. melanogaster, TAD boundaries are gene-dense regions enriched for highly transcribed genes and clusters of architectural protein sites, including CTCF, BEAF-32, Su(Hw), Mod(mdg4), Chromator, and CP19093, 94. Similarly, TAD borders in mammals are enriched for CTCF and Rad21 binding sites, housekeeping and tRNA genes, and SINE elements95 (Fig.7). The enrichment of CTCF and Rad21 at TAD borders may have a causal role in determining their establishment. This conclusion is supported by results from experiments in which a 58 kb region located at the border between the Tsix and Xist TADs in the mouse X-chromosome was deleted. Elimination of these sequences, which include a CTCF site and the Xist, Tsix and Xite genes, leads to increased interactions in the previous inter-TAD border region and to the formation of a new TAD border at an adjacent location96.
Figure 7. CTCF regulates three-dimensional genome architecture.
A) Cartoon of an interaction heat map of a chromosome segment around 2.5 Mb in length depicting data generated by Hi-C in mammalian cells. The TADs and their borders are indicated.
B) The presence of multiple CTCF and TFIIIC binding sites at TAD borders may contribute to the establishment of the border. This arrangement may explain the observed function of CTCF as an enhancer blocker. On the other hand, CTCF binding sites within TADs may facilitate enhancer-promoter looping through the recruitment of cohesin. The blue box denotes the promoter of the gene.
C) Chromatin features of TAD borders in mammals and Drosophila melanogaster. The TAD borders in mammals are enriched for housekeeping and tRNA genes, SINE elements and CTCF binding sites. In D. melanogaster, they are enriched for highly transcribed genes and clusters of binding sites for various architectural proteins. The role of TFIIIC, cohesin and condensin proteins in mediating TAD border formation remains to be determined.
Recent results using Hi-C to explore the role of CTCF and cohesin in the 3D organization of the mammalian genome support a similar conclusion of CTCF acting as a boundary protein between TADs, although the details vary between the different studies97–99. Depletion of cohesin in HEK293 human embryonic kidney cells results in a general loss of intra-chromosomal interactions without affecting the TAD organization, whereas depletion of CTCF causes a similar decrease of intra-domain interactions concomitant with an increase in interactions between adjacent TADs97. Cohesin-deficient post-mitotic mouse astrocytes also exhibit a decrease in long-range interactions mediated by CTCF and cohesin, but additionally display a relaxation of TAD organization98. This TAD relaxation could be a consequence of a reduction in TAD border strength due to the lack of cohesin binding or to an increase in inter-TAD interactions as observed in CTCF-depleted HEK293 cells. A similar decrease in cohesin-mediated interactions was observed in cohesin-depleted developing mouse thymocytes arrested in G1, with an increase in alternative interactions resulting in changes to gene expression99.
The presence of only 15% of genomic CTCF binding sites at TAD borders while the other 85% are present inside TADs95 indicates that CTCF and cohesin alone are insufficient to separate different TADs, a conclusion supported by the relatively mild effects on TAD organization observed in cells depleted of CTCF or cohesin. In D. melanogaster, CTCF forms clusters with other architectural proteins at TAD borders, and it is possible that vertebrate CTCF may adopt a similar strategy. Several lines of evidence suggest that TFIIIC may be a candidate architectural protein that cooperates with CTCF at TAD borders in vertebrates. As discussed above, TFIIIC co-localizes with CTCF near many tRNA genes and ETC loci in mammalian cells49–51, and binds SINE elements and tRNA genes, both of which functionally behave as enhancer-blocking insulators in humans47, 48 and are enriched at TAD borders95. Since CTCF has been shown to recruit cohesin in mammalian cells, and TFIIIC interacts with cohesin and condensin in yeast, CTCF and TFIIIC may possibly act as docking sites for these proteins to stabilize interactions required for the formation of TAD borders. TAD borders do not allow interactions between sequences located in the two adjacent TADs. Thus, it is possible that sequences present at these borders represent the enhancer-blocking insulators previously characterized in transgene assays (Fig.7). Additional studies will be needed to clarify whether clustering of CTCF, TFIIIC, cohesin and condensin occur at TAD boundaries of mammalian cells and if their presence is required for border formation.
The majority of CTCF binding sites (~85%) reside within TADs and, by definition, are unable to form a border. What is the role of CTCF at these sites? Studies in pre-pro-B cells using Hi-C suggest that CTCF located within TADs is primarily involved in mediating short-range intra-TAD interactions100. As we discussed above, the function of these CTCF-mediated interactions may be to direct enhancers located within the TAD to the appropriate gene promoter. In large mammalian genomes, the resolution of Hi-C data is limited by the number of sequencing reads, and this restricts the amount of structural information that can be obtained at the sub-TAD level. The use of 5C over large (1–2 Mb) genomic regions has made possible the mapping of finer topologies at the sub-megabase scale101. These topologies originate from interactions mediated by CTCF, cohesin and Mediator, either alone or in various combinations. Many of these interactions change during cell differentiation and occur between genomic regions containing epigenetic signatures characteristic of enhancers and promoters. Furthermore, it appears that different combinations of these three architectural proteins mediate interactions at different length scales, whereas CTCF in combination with cohesin is enriched in constitutive interactions that are present in mouse ESCs and do not change when these cells differentiate into neural progenitor cells. These results are suggestive of functional specialization of CTCF-mediated contacts as a consequence of interactions between this protein and various partners. The formation of different complexes with other proteins inside TADs and at TAD borders may underlie the different functions of CTCF in genome organization and explain its apparently contradictory properties as an enhancer facilitator and enhancer blocker (Fig.7).
Conclusions and Perspectives
The emerging theme from recent studies is that CTCF functions as an architectural protein that contributes to the establishment of genome topology. This is attained at two levels that are likely to be inter-related and account for most previous observations. At a global level, interactions mediated by CTCF and other architectural proteins result in the formation of TADs. At a more local sub-megabase scale, CTCF may be involved in fine-tuning intra-chromosomal interactions within TADs to regulate various aspects of gene expression.
Cooperation of CTCF with other protein partners, perhaps regulated by covalent modifications, may determine its functional specificity. First, association with other proteins such as TFIIIC, cohesins and condensins at specific genomic locations may result in the formation of TAD borders by precluding interactions across these sites, thus explaining the observed enhancer blocking properties of this protein. Second, association with other proteins such as cohesin and Mediator, may define the range and stability of chromosomal interactions within TADs, explaining other roles in transcription. The ability of CTCF to bind RNA opens the possibility of a role for ncRNAs in helping stabilize these contacts and perhaps regulate their function. Finally, covalent modifications of CTCF and its partners are likely to also influence its regulatory potential. The principal outcome of CTCF-mediated contacts is to regulate transcription at various levels, including initiation, promoter selection, promoter-proximal pausing and splicing. To date, the role of CTCF in determining 3D genome organization has been considered mostly in the context of its effect on transcription during G1 but both the protein and the architectural properties of the genome it controls are likely to be also important at other stages of the cell cycle, including DNA replication during S phase and chromosome condensation during mitosis102. In particular, how the TAD organization relates to the structure of metaphase chromosomes and how this affects gene expression at the beginning of G1 are important issues for future studies.
Acknowledgments
Work in the authors' lab is supported by U.S. Public Health Service Award R01 GM035463 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Glossary terms
- Chromosome conformation capture (3C)
A ligation-based technique used to map interactions between two specific genomic regions.
- ChIP-exo
An extension of ChIP–seq that includes exonuclease trimming after immunoprecipitation to increase the resolution of the mapped transcription-factor-bound sites.
- Circular chromosome conformation capture (4C)
The combination of inverse PCR and high-throughput sequencing with the chromosome conformation capture (3C) technique that allows the profiling of chromatin interactions between a known specific locus and multiple unknown sites.
- DNase I hypersensitive site
Chromosomal region that is readily degraded by deoxyribonuclease I (DNase I) owing to decreased nucleosome occupancy. These sites are associated with open chromatin conformation and the binding of transcription factors.
- Nuclear lamina
A scaffold of proteins comprised mainly of lamin A/C and B that is predominantly found in the nuclear periphery associated with the inner surface of the nuclear membrane.
- Chromosome Conformation Capture Carbon Copy (5C)
A technique used to profile all chromatin interactions in specific regions of the genome by the hybridization of a mixture of DNA primers to chromosome conformation capture (3C) templates followed by high-throughput sequencing.
- Mediator complex
The ~30-subunit co-activator complex required for successful transcription of RNA polymerase II promoters of metazoans genes. Its interaction with RNA polymerase II and site-specific factors facilitates the enhancer-promoter communication.
- ChIA-PET
A technique used to determine the chromosomal interactions that are mediated by a specific chromatin binding protein by combining chromatin immunoprecipitation (ChIP) with chromosome conformation capture (3C)-type analysis.
- Adaptive response
The acquired immune response to the specific antigen presented on a pathogen that typically triggers immunological memory.
- Pro-B Cell
Earliest developmental stage of B-cells in the bone marrow defined as the CD19+ cytoplasmic IgM− or B220+ CD43+ population that has incomplete rearrangement of the immunoglobulin heavy-chain.
- Double-positive thymocytes
Immature T cells characterized by the expression of CD4 and CD8 cell-surface markers that will differentiate into single-positive thymocytes after their T-cell receptor interact with self-peptide-MHC ligands in the thymus.
- Pre-pro-B cell
The lymphoid progenitors found in the bone marrow that contain the CLP-2s surface marker and lack heavy-chain DJ rearrangements.
- Hi-C
An extension of the chromosome conformation capture (3C) technique that incorporates a biotin-labeled nucleotide at the ligation junction to allow selective purification of chimeric DNA ligated products for high-throughput sequencing. This method generates matrices of interaction frequencies across the genome.
Biographies
CHIN-TONG ONG Chin-Tong Ong received a Ph.D. in Developmental Biology from Washington University in St. Louis, Missouri, USA. Currently a postdoctoral fellow in the Corces laboratory, his research focuses on understanding how specific post-translational modifications of architectural proteins affect their function in transcription and genome organization.
VICTOR G. CORCES Victor G. Corces received a Ph.D. in Chemistry from the Universidad Autonoma of Madrid in Spain and did postdoctoral training at Harvard University, Massachusetts, USA. He is currently an HHMI Professor at Emory University, Georgia, USA. His research aims to understand the mechanisms by which the three-dimensional organization of the genome regulates gene expression. For additional information, see http://www.biology.emory.edu/research/Corces/
References
- 1.Van Bortle K, Corces VG. Nuclear organization and genome function. Annu Rev Cell Dev Biol. 2012;28:163–87. doi: 10.1146/annurev-cellbio-101011-155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Laat W, Dekker J. 3C-based technologies to study the shape of the genome. Methods. 2012;58:189–91. doi: 10.1016/j.ymeth.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baniahmad A, Steiner C, Kohne AC, Renkawitz R. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell. 1990;61:505–14. doi: 10.1016/0092-8674(90)90532-j. [DOI] [PubMed] [Google Scholar]
- 4.Lobanenkov VV, et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–53. [PubMed] [Google Scholar]
- 5.Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci U S A. 2012;109:17507–12. doi: 10.1073/pnas.1111941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–7. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Tian Y, Shu W, Bo X, Wang S. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One. 2012;7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt D, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–48. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 13.DeMare LE, et al. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 2013;23:1224–34. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song L, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–67. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–19. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakahashi H, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–89. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genome-wide mapping of different CTCF mutants reveals that combinatorial clustering of its 11 zinc fingers recognizes a wide array of DNA modules.
- 17.Engel N, West AG, Felsenfeld G, Bartolomei MS. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat Genet. 2004;36:883–8. doi: 10.1038/ng1399. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez C, et al. CTCF is a DNA methylation-sensitive positive regulator of the INK/ARF locus. Biochem Biophys Res Commun. 2010;392:129–34. doi: 10.1016/j.bbrc.2009.12.159. [DOI] [PubMed] [Google Scholar]
- 19.Lai AY, et al. DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J Exp Med. 2010;207:1939–50. doi: 10.1084/jem.20100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang J, et al. Nicotinamide adenine dinucleotide (NAD)-regulated DNA methylation alters CCCTC-binding factor (CTCF)/cohesin binding and transcription at the BDNF locus. Proc Natl Acad Sci U S A. 2010;107:21836–41. doi: 10.1073/pnas.1002130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–8. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates that cell-type specific CTCF binding sites across 19 diverse human cell types are linked to the differential DNA methylation patterns within the cells.
- 22.Zampieri M, et al. ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem J. 2012;441:645–52. doi: 10.1042/BJ20111417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guastafierro T, et al. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem. 2008;283:21873–80. doi: 10.1074/jbc.M801170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song CX, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–91. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 30.MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29:714–25. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu W, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–10. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 32.Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell. 2009;34:271–84. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong CT, Van Bortle K, Ramos E, Corces VG. Poly(ADP-ribosyl)ation regulates insulator function and intrachromosomal interactions in Drosophila. Cell. 2013;155:148–59. doi: 10.1016/j.cell.2013.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work shows that poly(ADP-ribosyl)ation regulates CTCF-mediated intrachromosomal interactions and its association with other architectural protein in Drosophila.
- 34.Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J Cell Sci. 2009;122:1275–84. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]
- 35.Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol. 2011;31:2174–83. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 37.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–14. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–66. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galli GG, et al. Genomic and proteomic analyses of Prdm5 reveal interactions with insulator binding proteins in embryonic stem cells. Mol Cell Biol. 2013;33:4504–16. doi: 10.1128/MCB.00545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie D, et al. Dynamic trans-acting factor colocalization in human cells. Cell. 2013;155:713–24. doi: 10.1016/j.cell.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–3. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–6. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nativio R, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirkland JG, Raab JR, Kamakaka RT. TFIIIC bound DNA elements in nuclear organization and insulation. Biochim Biophys Acta. 2013;1829:418–24. doi: 10.1016/j.bbagrm.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–72. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 47.Lunyak VV, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–51. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 48.Raab JR, et al. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31:330–50. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carriere L, et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 2012;40:270–83. doi: 10.1093/nar/gkr737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oler AJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–8. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moqtaderi Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–40. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genome-wide mapping of distinct components of RNA polymerase III complex in human cells reveals the association of TFIIIC with many ETC loci that are located near CTCF binding sites.
- 52.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–41. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 53.Yao H, et al. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–55. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun S, et al. Jpx RNA Activates Xist by Evicting CTCF. Cell. 2013;153:1537–51. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work shows that the interaction of Jpx ncRNA with CTCF is necessary for the eviction of CTCF from the promoter to facilitate Xist transcription.
- 55.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 56.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–75. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–74. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–50. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 60.Oki M, Kamakaka RT. Barrier function at HMR. Mol Cell. 2005;19:707–16. doi: 10.1016/j.molcel.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 61.Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol Cell Biol. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 63.Essafi A, et al. A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev Cell. 2011;21:559–74. doi: 10.1016/j.devcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taslim C, et al. Integrated analysis identifies a class of androgen-responsive genes regulated by short combinatorial long-range mechanism facilitated by CTCF. Nucleic Acids Res. 2012;40:4754–64. doi: 10.1093/nar/gks139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci U S A. 2011;108:7391–6. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz YB, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012;22:2188–98. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Bortle K, et al. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012;22:2176–87. doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li HB, et al. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol. 2011;31:616–25. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wood AM, et al. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie X, et al. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A. 2007;104:7145–50. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Ramos E, Corces VG. The BEAF-32 insulator coordinates genome organization and function during the evolution of Drosophila species. Genome Res. 2012;22:2199–207. doi: 10.1101/gr.142125.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mapping of long-range interactions between distal elements and TSSs indicates that CTCF binding sites are enriched at many enhancer elements and enhancer-promoter interaction may not be disrupted by intervening CTCF sites.
- 73.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–20. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Majumder P, Boss JM. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol Cell Biol. 2010;30:4211–23. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–98. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat Struct Mol Biol. 2011;18:372–8. doi: 10.1038/nsmb.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Golan-Mashiach M, et al. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res. 2012;40:3378–91. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2011;108:17195–200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo Y, et al. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A. 2012;109:21081–6. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monahan K, et al. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc Natl Acad Sci U S A. 2012;109:9125–30. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–57. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 82.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–31. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that CTCF directly recruits TAF3 to promoter distal sites that act as enhancers for endoderm lineage differentiation in ES cells.
- 83.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–8. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that CTCF-mediated chromatin interactions may separate chromatin domains and facilitate enhancer-promoter contacts.
- 84.Bossen C, Mansson R, Murre C. Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol. 2012;30:337–56. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 85.Guo C, et al. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147:332–43. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Degner SC, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A. 2011;108:9566–71. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shih HY, et al. Tcra gene recombination is supported by a Tcra enhancer- and CTCF-dependent chromatin hub. Proc Natl Acad Sci U S A. 2012;109:E3493–502. doi: 10.1073/pnas.1214131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–30. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that binding of CTCF to a key V(D)J recombination regulatory region mediates chromatin looping to regulate ordered and lineage-specific rearrangement of the IgH locus.
- 89.Ribeiro de Almeida C, et al. The DNA-binding protein CTCF limits proximal Vkappa recombination and restricts kappa enhancer interactions to the immunoglobulin kappa light chain locus. Immunity. 2011;35:501–13. doi: 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 90.Stadhouders R, et al. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 2012;31:986–99. doi: 10.1038/emboj.2011.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paredes SH, Melgar MF, Sethupathy P. Promoter-proximal CCCTC-factor binding is associated with an increase in the transcriptional pausing index. Bioinformatics. 2013;29:1485–7. doi: 10.1093/bioinformatics/bts596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that DNA methylation and binding of CTCF at exons regulate alternative splicing by controlling the elongation rate of RNA polymerase II.
- 93.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–84. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 95.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript shows that the mammalian genome is organized into topollogically associating domains separated by boundaries enriched for CTCF and other architectural proteins.
- 96.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]; This and the following three papers show that CTCF and cohesin are required to maintain the organization of topollogically associating domains.
- 97.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–29. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seitan VC, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–77. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Phillips-Cremins JE, et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013;153:1281–95. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–53. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]