Abstract
Galectins are carbohydrate-binding proteins that act as extracellular signaling molecules in various normal and pathological processes. Galectin bioactivity is mediated by specific non-covalent interactions with cell-surface and extracellular matrix (ECM) glycoproteins, which can enhance or inhibit signaling events that influence various cellular behaviors, including adhesion, proliferation, differentiation, and apoptosis. Here, we developed a materials approach to modulate galectin bioactivity by mimicking natural galectin-glycoprotein interactions. Specifically, we created a variant of a peptide that self-assembles into β-sheet nanofibers under aqueous conditions, QQKFQFQFEQQ (Q11), which has an asparagine residue modified with the monosaccharide N-acetylglucosamine (GlcNAc) at its N-terminus (GlcNAc-Q11). GlcNAc-Q11 self-assembled into β-sheet nanofibers under similar conditions as Q11. Nanofibrillar GlcNAc moieties were efficiently converted to the galectin-binding disaccharide N-acetyllactosamine (LacNAc) via the enzyme β-1,4-galactosyltransferase and the sugar donor UDP-galactose, while retaining β-sheet structure and nanofiber morphology. LacNAc-Q11 nanofibers bound galectin-1 and -3 in a LacNAc concentration-dependent manner, although nanofibers bound galectin-1 with higher affinity than galectin-3. In contrast, galectin-1 bound weakly to GlcNAc-Q11 nanofibers, while no galectin-3 binding to these nanofibers was observed. Galectin-1 binding to LacNAc-Q11 nanofibers was specific because it could be inhibited by excess soluble β-lactose, a galectin-binding carbohydrate. LacNAc-Q11 nanofibers inhibited galectin-1-mediated apoptosis of Jurkat T cells in a LacNAc concentration-dependent manner, but were unable to inhibit galectin-3 activity, consistent with galectin-binding affinity of the nanofibers. We envision that glycopeptide nanofibers capable of modulating galectin-1 bioactivity will be broadly useful as biomaterials for various medical applications, including cancer therapeutics, immunotherapy, tissue regeneration, and viral prophylaxis.
Introduction
Galectins are receiving increasing attention as therapeutic targets due to their important role as extracellular signaling molecules in various normal and pathological processes, including immunological tolerance and inflammation,1,2 antigen-specific T cell activation,3 cancer progression,4-6 platelet activation,7 angiogenesis,8 and viral infection.9 Galectins are a 15-member protein family characterized by a highly conserved carbohydrate-recognition domain (CRD) that mediates non-covalent interactions with β-galactosides, such as β-lactose and Ν-acetyllactosamine (LacNAc).10 Galectins can be sub-divided into 3 general families: proto-type galectins, which non-covalently associate into homodimers having two identical carbohydrate-recognition domains; tandem-repeat galectins with distinct N- and C-terminal CRDs that are covalently linked; and galectin-3, which assembles into a pentamer via an N-terminal domain.11 Within extracellular microenvironments, galectins bind to various cell surface and ECM glycoproteins bearing β-galactosides via specific carbohydrate-dependent interactions. Due to their multivalent CRDs, galectins can simultaneously bind multiple different cell surface and ECM glycoproteins, essentially acting as non-covalent cross-linkers to organize or cluster different proteins into “bioactive” signaling synapses or sequester receptors in an inactive state.12 Galectin-mediated glycoprotein crosslinking can enhance or inhibit “outside-in” cellular signaling events that govern various cellular behaviors, including adhesion,13-19 proliferation,20,21 apoptosis,22-25 and differentiation.26-29 Despite their highly conserved CRDs, however, different galectins can have similar, synergistic, or antagonistic activity within a given physiological or pathological setting. For example, whereas galectin-1 and galectin-8 co-stimulate antigen-specific T cell responses, galectin-3 antagonizes these responses,3 suggesting that therapeutics that selectively inhibit galectin-1 activity could suppress antigen-specific immune responses, whereas those that selectively inhibit galectin-3 activity could boost antigen-specific immunity. In addition, galectin-1 and galectin-3 induce different responses in neutrophils and T cells, in particular with galectin-1 up-regulating T cell production of IL-10, an anti-inflammatory cytokine, and down-regulating expression of interferon-γ.25 Furthermore, galectin-1 suppresses Thelper (Th)-1-type cytokines in models of autoimmune disease,30 whereas galectin-3 suppresses Th-2-type cytokines.31 In experimental autoimmune encephalomyelitis, an experimental model for multiple sclerosis, galectin-1 deficiency increased disease severity,32 whereas galectin-3 deficiency decreased disease severity,33 suggesting that selective inhibition of galectin-3 may provide therapeutic benefit. Alternatively, galectin-1 enhances human immunodeficiency virus infection by mediating association of the virus with host cells, whereas galectin-3 does not enable HIV-host association.34 As a result, developing carbohydrate-based therapeutics that can modulate galectin bioactivity by interfering with specific galectin-glycoprotein interactions is a rapidly growing area of research.
To date, a wide variety of small molecules related to or inspired by β-galactosides have been investigated as galectin inhibitors.35-37 However, achieving robust therapeutic efficacy has been challenged by the relatively low affinity of monovalent carbohydrates for galectins, which results from the high entropic penalty associated with small molecule binding to a protein and the limited enthalpy afforded by weak non-covalent bonds formed between proteins and carbohydrates. Throughout natural systems, binding between galectins and carbohydrates is often enhanced via high-density carbohydrate display by glycoproteins, typically referred to as the “glycocluster effect”.38 As a result, synthetic glycoclusters are gaining interest as alternatives to small molecule inhibitors of galectin-glycoprotein interactions. For example, dendrimers, cyclodextrins, glycopeptides and other supramolecular architectures can provide glycoclusters with enhanced lectin-binding affinity, despite often having a relatively low carbohydrate valency.39-42 Glycopolymers can provide an even greater degree of carbohydrate multivalency, and in turn lectin-binding affinity, than low-molecular weight glycoclusters, and many different types of glycopolymers have been synthesized based on natural and synthetic monomers.43 Living polymerization enables synthesis of glycopolymers with well-defined carbohydrate density and valency, because it can eliminate the compositional drift associated with conventional co-polymer synthesis. Such compositional precision is important because carbohydrate density and valency are key determinants of lectin-glycan cross-linking.44 Recent work demonstrated that chemically-defined poly(LacNAc) polymers immobilized onto solid-phase substrates can provide biomaterials capable of harnessing galectin bioactivity, in particular for mediating reversible immobilization of galectin-binding ECM proteins onto biomaterial surfaces.45,46 Here, we sought to create water-soluble biomaterials having LacNAc concentrations comparable to conventional glycopolymers, in which LacNAc content can be easily and precisely varied to optimize nanofiber efficacy for modulating galectin bioactivity.
Non-covalent self-assembly of peptides into supramolecular nanofibers can provide biomaterials with polymer-like structural features and well-defined composition of integrated functional ligands.47-54 In particular, self-assembling peptide “monomers” with different pendant functional groups, such as peptides, proteins, or small molecules, can be readily synthesized in high yield and purity using standard methods. Self-assembly of any single monomer can provide β-sheet nanofibers having activity related to the pendant ligand. In addition, different monomers mixed together in the pre-assembled state can co-assemble into multi-functional nanofibers, and the composition of each functional ligand integrated into the nanofibers often correlates with monomer mole fraction in solution during assembly.55,56 Toward the development of therapeutic biomaterials, peptide monomers are advantageous because they are made from natural, biodegradable molecules, while the resulting nanofibers are non-inflammatory and minimally immunogenic, unless modified with a non-self epitope.57,58 Finally, since carbohydrates are covalently linked to the side-chain of asparagine (i.e. “n-linked”) or serine/threonine (i.e. “mucin-like) amino acids in natural glycoproteins,59 a carbohydrate conjugated to the side chain of an asparagine, serine, or threonine amino acid appended onto the terminus of a self-assembling peptide domain will provide biomaterials that more closely approximate n-linked or mucin-like carbohydrate presentation by natural glycoproteins. Thus, we proposed that nanofibers fabricated via self-assembly of carbohydrate-modified peptides, or “glycopeptides”, would provide a useful alternative to conventional glycopolymers for creating water-soluble, galectin-binding biomaterials.
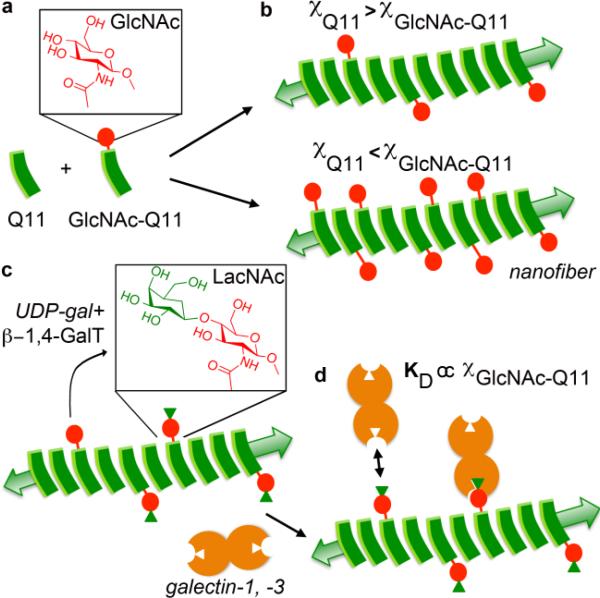
In this study, we created a variant of a peptide that self-assembles into β-sheet nanofibers under aqueous conditions, QQKFQFQFEQQ (Q11),60 which has an asparagine residue modified with the monosaccharide N-acetylglucosamine (GlcNAc) at its N-terminus (GlcNAc-Q11) (Fig 1a). We proposed that nanofibers with tunable carbohydrate concentration could be fabricated via simple mixing of GlcNAc-Q11 and Q11 at various molar ratios in the pre-assembled state (Fig. 1b), based on previous observations with multi-component peptide co-assemblies and peptide-protein composite assemblies.55,56 In turn, we proposed that GlcNAc moieties displayed by nanofibers could be converted to the galectin-binding β-galactoside, LacNAc, via the enzyme β-1,4-galactosyltransferase (β-1,4-GalT) and a galactose donor (Fig. 1c), based on previous demonstrations of the synthetic utility of this mechanism to fabricate LacNAc modified surfaces.61 Finally, we proposed that nanofibers displaying LacNAc would bind galectin-1 and galectin-3 in a LacNAc-dependent manner (Fig. 1d), and that nanofiber LacNAc content could be optimized to create biomaterials with improved efficacy for modulating galectin-1 bioactivity as an extracellular signaling molecule.
Figure 1. Schematic of self-assembled glycopeptide nanofibers that bind galectins.
a) β-sheet fibrillizing peptide Q11, and a Q11 variant having an asparagine residue with an N-linked n-acetylglucosamine (GlcNAc) moiety. b) Q11 and GlcNAc-Q11 co-assemble into nanofibers in which GlcNAc content of the nanofibers is dependent on the molar ratio of Q11 to GlcNAc-Q11 (χ). c) Pendant GlcNAc moieties along the nanofiber are converted to the β-galactoside, n-acetyllactosamine (LacNAc) by the enzyme β-1,4-galactosyltransferase (β-1,4-GalT) and the sugar donor UDP-galactose (UDP-gal). d) LacNAc-Q11 nanofibers bind galectin-1 and -3, and nanofiber galectin binding affinity can be tailored by varying the molar ratio of Q11 to GlcNAc-Q11 present during assembly.
Methods
Peptide Synthesis and Purification
The β-sheet fibrillizing peptide, Ac-QQKFQFQFEQQ-Am (Q11),60 and its glycosylated analog GlcNAc-Q11, were synthesized using standard Fmoc protocols for solid-phase peptide synthesis on a CS336X automated peptide synthesizer (CS Bio). Reagents for peptide synthesis were purchased from Novabiochem, unless stated otherwise. Q11 was synthesized on a 0.25 mmol scale using a Rink amide resin. Fmoc deprotection was accomplished with 20% piperidine (Sigma) in dimethylformamide (DMF) (Fisher). Peptide was cleaved and deprotected by incubating in a cocktail of 9.5:0.25:0.25 trifluoroacetic acid (TFA) (Fisher):triisopropylsilane (TIS) (Sigma):water for 2.5 hours. Peptide was precipitated and washed with cold diethyl ether (Fisher). Finally, peptide was dried, dissolved in water, frozen and lyophilized.
GlcNAc-Q11 (Asn(Ac3NH-β-Glc)-SGSG-QQKFQFQFEQQ-Am) was synthesized and cleaved/deprotected using the same methods as for Q11 above. Fmoc-Asn(Ac3NH-β-Glc)-OH was purchased from Novabiochem. Ac3NH-β-Glc was deacetylated to give Asn(GlcNAc)-SGSG-Q11 by incubating the peptide in a sodium methoxide solution (0.5 M in methanol, pH 10) (Acros Organics) for 30 minutes. Peptide precipitate was sedimented by centrifugation, and sodium methoxide supernatant was decanted. Peptide pellet was then washed with methanol, and sedimented by centrifugation. Centrifugation and methanol washing steps were repeated twice. Peptide was dried in vacuo, dissolved in water, frozen, and lyophilized.
Peptides were purified via reversed phase high performance liquid chromatography (RP-HPLC) using a Varian ProStar HPLC system equipped with a Grace-Vydac C-18 column, or a DionexTM Ultimate 3000TM System (Thermo Scientific) equipped with a C-18 column (Thermo Scientific). The mobile phase consisted of (A) water and (B) acetonitrile, both with 0.1% TFA. Elution was achieved with a linear gradient varying (B) from 25 to 35% over 10 min. Peptide was detected by absorbance at 215 nm.
MALDI-Time-of-Flight Mass Spectrometry (MALDI-TOF-MS)
For MALDI-TOF-MS analysis, 1 μL of peptide purified by RP-HPLC was mixed with 1 μL of α-cyano-4-hydroxycinnamic acid (10 mg/mL in 70% acetonitrile and 0.1% TFA), spotted onto a MALDI target, and dried. Samples were analyzed using reflectron, positive ion mode on an AB SCIEX TOF/TOFTM 5800 equipped with a 1 KHz N2 OptiBeamTM on-axis laser. Laser power was used at the threshold level required to generate signal.
Nanofiber Preparation
GlcNAc-Q11 and Q11 nanofibers were prepared using established methods for self-assembly of Q11 and other Q11 derivatives.60 In particular, Q11 nanofibers were prepared by first dissolving lyophilized peptide powder in water at specified concentrations. Similarly, nanofibers with variable content of GlcNAc were prepared by first mixing GlcNAc-Q11 and Q11 peptides at different molar ratios in the powder form, and then dissolving them in water at the desired concentration to favor their co-assembly.62 Peptides were dissolved in water by alternating cycles of vortexing and sonication, and then incubated at room temperature for 30 minutes, during which time the peptides begin to self-assemble. Aqueous peptide solutions were then diluted 10-fold in neutral aqueous buffers, such as 1x phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4), or 20 mM HEPES with 10 mM MnCl2, based on prior observations that neutral aqueous buffers accelerate Q11 self-assembly.60
We used an RP-HPLC assay to determine the correlation between GlcNAc-Q11 mole fraction in solution during nanofiber assembly and GlcNAc content of the resultant nanofibers, similar to previous methods.56 GlcNAc-Q11 and Q11 were mixed as dry powders at different molar ratios (χGlcNAc-Q11 = 0.01, 0.05, 0.10, 0.25 and 0.50), dissolved in water as described above, diluted 10-fold in 1x PBS to a final concentration of 3.2 mM, and incubated for 30 minutes. Nanofibers were then sedimented by centrifugation at 12000xg for 5 min, similar to previous methods.55 Greater than 90% of peptide in solution was sedimented by centrifugation, independent of carbohydrate content (data not shown), consistent with previous reports.55 Following centrifugation, supernatant above the nanofiber pellet was carefully removed via pipet, nanofibers were resuspended in TFA, and nanofibers were disassembled via vigorous pipetting. GlcNAc-Q11 and Q11 content in each TFA solution was then independently analyzed via RP-HPLC, using methods described above. Specifically, an RP-HPLC chromatogram was collected from each sample containing a mixture of GlcNAc-Q11 and Q11 in TFA. The area under the GlcNAc-Q11 peak (~18.5 min; confirmed via MALDI-TOF-MS) and Q11 peak (~19.5 min; confirmed via MALDI-TOF-MS) was calculated. RP-HPLC chromatograms of serial dilutions of TFA solutions with known concentrations of GlcNAc-Q11 or Q11 were then collected and the area under the GlcNAc-Q11 or Q11 peak was used to create standard curves of absorbance versus GlcNAc-Q11 concentration or Q11 concentration, respectively. The Q11 standard curve was used to convert the area under the Q11 peak in each sample RP-HPLC chromatogram to Q11 concentration in the sample. Similarly, the GlcNAc-Q11 standard curve was used to convert the area under the GlcNAc-Q11 peak in each sample RP-HPLC chromatogram to GlcNAc-Q11 concentration in the sample. Mole fraction of GlcNAc-Q11 in each sample was then calculated by dividing GlcNAc-Q11 concentration in the sample by the total peptide concentration in the sample (i.e. concentration of Q11 + concentration of GlcNAc-Q11) in that sample. Chromatograms from 3 samples at each GlcNAc-Q11 feed mole fraction (χGlcNAc-Q11 = 0.01, 0.05, 0.10, 0.25 and 0.50) were collected and analyzed, and all standards were run in duplicate.
Enzymatic Synthesis of LacNAc-Q11 Nanofibers
Enzymatic conversion of nanofibrillar GlcNAc to LacNAc was performed in a total volume of 20 μL for all MALDI-TOF and transmission electron microscopy experiments. For all enzymatic conversion experiments, we used a basis of 2 nmol GlcNAc-Q11 incubated with 2.4 mUnits (mU) of β-1,4-galactosyltransferase (β-1,4-GalT) from bovine milk (Sigma) and 10 nmol sugar donor, uridine diphosphate galactose (UDP-gal) (Carbosynth and Sigma). 1 U corresponds to the amount of enzyme that transfers 1 μmole galactose to GlcNAc per min at 37°C, pH 7. Here, the reaction mixture contained 0.0024 U of β-1,4-GalT, which is expected to transfer ~2.4 nmol galactose to GlcNAc per minute. Thus, enzymatic conversion of 2 nmol GlcNAc-Q11 to LacNAc was expected to approach completion within ~1 minute under these reaction conditions. However, we assumed that tethering GlcNAc to nanofibers would decrease enzymatic reaction efficiency due to greatly reduced substrate diffusivity in the reaction medium, and thus we allowed the reaction to proceed at 37 °C for 18 hours in a humid environment in 20 mM HEPES and 10 mM MnCl2 buffer (pH 7.5).
To determine enzyme reaction kinetics, we quantified the decrease in GlcNAc-Q11 MALDI-TOF-MS peak intensity as a function of time. Specifically, nanofibers containing 0.25 mM GlcNAc-Q11/0.75 mM Q11 (5 nmol GlcNAc) were incubated with 12 mU β-1,4-GalT and 50 nmol UDP-gal in 20 mM HEPES and 10 mM MnCl2 buffer (pH 7.5) to maintain the basis of 2.4 mU β-1,4-GalT/2 nmol GlcNAc-Q11 and 10 nmol UDP-gal. 1 μL aliquots were taken from the reaction mixture at different time points (0, 1, 5, 10, 60, 300, 600, and 1080 min) and mixed with 5 μL of pre-chilled 10% aqueous acetonitrile solution to quench enzyme activity. Samples were then analyzed using MALDI-TOF-MS as described above. To account for differences in ionization efficiencies of different molecules, the GlcNac-Q11 peak intensity in MALDI-TOF-MS spectra at each time point was normalized to the Q11 peak intensity within the same spectrum. Conversion efficiency was then calculated from: [GlcNAc-Q11/Q11 peak intensity at time = t]/[GlcNAc-Q11/Q11 peak intensity at time = 0]. Conversion efficiency versus time was plotted to establish the kinetic profile of the reaction. Reactions were run in triplicate to assess variability.
Enzymatic synthesis was scaled up to maintain the basis of 2.4 mU β-1,4-GalT/2 nmol GlcNAc-Q11 and 10 nmol UDP-gal for co-precipitation experiments, circular dichroism experiments, and Jurkat T cell apoptosis experiments (described below). For these experiments, total Q11 concentration was 1 mM, with χGlcNAc-Q11 = 0.01, 0.05, 0.10, 0.25 0.50, or 1. After enzymatic treatment, nanofibers were sedimented by centrifugation and washed with 1x PBS three times prior to further studies, including co-precipitation experiments and Jurkat T cell apoptosis experiments.
Transmission Electron Microscopy
Nanofiber morphology before and after enzymatic conversion from GlcNAc-Q11 to LacNAc-Q11 was investigated using transmission electron microscopy (TEM). Nanofibers formed as described above at a total Q11 concentration of 1 mM were diluted in 1x PBS to a final concentration of 0.125 mM. Nanofibers were then adsorbed on a formvar-carbon coated 400 mesh copper grid (FCF400-Cu-UB, EMS) by floating the grids on 5 μL drops on parafilm for 1 min. Samples were negatively stained with 1% uranyl acetate in water, and analyzed using a FEI Tecnai F30 TEM.
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectroscopy was used to study β-sheet structure of glycosylated nanofibers. 0.5, 1.25, and 2.5 mM GlcNAc-Q11 or GlcNAc-Q11:Q11 (1:9 molar ratio) were dissolved in water, diluted 10-fold in 1x phosphate buffer with potassium fluoride (137 mM KF, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4), and incubated overnight at room temperature. Additionally, 1 mM GlcNAc-Q11 was dissolved in water, diluted 10-fold in 20 mM HEPES + 10 mM MnCl2 with or without UDP-gal and β-1,4-GalT as described above, and incubated overnight. After overnight incubation, nanofibers were sedimented by centrifugation at 12000xg for 5 min, supernatant containing β-1,4-GalT was removed via pipet, and nanofibers were resuspended in 1x phosphate buffer with potassium fluoride. CD spectra were collected at room temperature in the 200-260 nm range using an AVIV 202 CD spectrometer with quartz cells of 1 mm pathlengths. Each sample was analyzed three times and averaged spectra were reported.
Expression and purification of recombinant human galectin-1 and galectin-3
Genes encoding human galectin-1 or galectin-3 (Origene) were inserted into pET-21d vectors between the NcoI site and XhoI site. For the galectin-1 gene, the XhoI site was mutated to a stop codon to remove the His6 tag using the QuikChange II site-directed mutagenesis kit (Agilent), according to manufacturer's instructions. Origami B (DE3) E.coli were transformed with pET-21d-galectin-1 or pET-21d-galectin-3 vectors and selected on 100 μg/mL ampicillin- and 50 μg/mL kanamycin A-doped LB/agar plates overnight at 37°C. Positive clones were picked and used to inoculate 10 mL of LB broth containing 100 μg/mL ampicillin- and 50 μg/mL kanamycin A. Cultures were grown overnight at 37° C, 220 rpm on an orbital shaker. Plasmids were isolated from cultures via a plasmid mini-prep kit (Qiagen), according to manufacturer's instructions, and sequenced in the University of Chicago gene sequencing core facility. Cultures of positive clones were then sub-cultured into 1 L 100 μg/mL ampicillin- and 50 μg/mL kanamycin A-containing LB broth and grown at 37 °C, 220 rpm on an orbital shaker until an O.D. at 600 nm = 0.6-0.8 was reached. Cultures were then spiked with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and incubated for 4 h at 37 °C, 100 rpm in an orbital shaker. Bacteria were pelleted by centrifugation, culture media was decanted, bacteria were resuspended in 1x PBS (with 8 mM dithiothreitol (DTT) in the case of galectin-1, which was included in all buffers to maintain galectin-1 in the “active” reduced state63). Bacteria were lysed with 1x BugBuster protein extraction reagent (Novagen), 1 eComplete protease inhibitor tablet (Santa Cruz Biotechnology), 300 units DNAse I from bovine pancreas (Sigma), and 100 μg lysozyme (Sigma) for 20 min. Lysed bacteria were cleared by centrifugation, and supernatant containing recombinant galectin-1 or galectin-3 was loaded onto columns containing α-lactose:agarose (for galectin-1) or HisPur cobalt resin (for galectin-3) equilibrated with 1x PBS. Columns were washed with 20-30 column volumes of 1x PBS to remove loosely bound bacterial proteins. Galectin-1 was eluted with increasing concentrations of β-lactose in 1x PBS with 8 mM DTT. Galectin-3 was eluted with increasing concentrations of imidazole in 1x PBS. β-lactose or imidazole were removed by 5 rounds of concentrating sample 10-fold via centrifugation in centrifugal filter tubes (MWCO 3kDa) (Millipore), followed by 10-fold dilution with 1x PBS plus 8 mM DTT (galectin-1) or 1x PBS (galectin-3). Protein molecular weight and purity were analyzed with SDS-PAGE. Protein purity was ≥ 90%, as determined by densitometry of SDS-PAGE gels. Endotoxin contaminants in fractions containing galectin-1 or galectin-3 were removed by Triton X-114 cloud-point precipitation, according to established methods.55,64 Endotoxin content was analyzed with the limulus amoebocyte lysate assay (Lonza), according to manufacturer's instructions, and was below 0.1 EU/mL in all stocks.
Co-precipitation assays with galectin-1, galectin-3, wheat germ agglutinin, and β-lactose
GlcNAc-Q11 nanofibers for binding studies (1 mM Q11 containing χGlcNAc-Q11 = 0.01, 0.05, 0.10, 0.25, or 0.50) were prepared as described above. LacNAc-Q11 nanofibers for binding studies (1 mM Q11 containing χGlcNAc-Q11 = 0.01, 0.05, 0.10, 0.25, or 0.50) were prepared as described above and then treated with β-1,4-GalT and UDP-gal to convert nanofibrillar GlcNAc to LacNAc. Nanofibers were then mixed with (a) 4 μM recombinant human galelctin-1 with or without 100 mM β-lactose (Sigma) in 1x PBS plus 8 mM DTT (b) 3 μM recombinant galectin-3 in 1x PBS, or (c) 12.5 μM wheat germ agglutinin in 1x PBS. Solutions of nanofibers and lectins were incubated at room temperature for 1 h. Nanofibers were then sedimented by centrifugation at 12000xg for 5 min and supernatant was collected. 20 μL supernatant was mixed 1:1 with 20 μL 2x Laemmli buffer and analyzed for galectin-1 or galectin-3 using SDS-PAGE electrophoresis (galectin-1 14.2 kDa; galectin-3 27.2 kDa) on precast “Any kD gels” (Bio-Rad), followed by Coomassie Blue staining. Galectin-1 and galectin-3 supernatant samples were also diluted 1:2 with 1x PBS and analyzed for tryptophan fluorescence (excitation 280 nm, emission 345 nm) using a SpectraMax M5 plate-reader in glass-bottom black 96-well plates. Tryptophan fluorescence intensity was converted to galectin-1 or galectin-3 concentration using a standard curve of tryptophan fluorescence of samples with known galectin-1 and galectin-3 concentrations. Wheat germ agglutinin concentration in supernatant was determined with the BCA assay (Fisher), according to manufacturer's instructions.
Serum protein binding to LacNAc-Q11 nanofibers
LacNAc-Q11 nanofibers for serum binding studies (1 mM Q11 containing χGlcNAc-Q11 = 0.01, 0.05, 0.10, 0.25, or 0.50) were prepared as described above and then treated with β-1,4-GalT and UDP-gal to convert nanofibrillar GlcNAc to LacNAc. Nanofibers were mixed with 10% FBS, and incubated for one hour at room temperature. Nanofibers were sedimented by centrifugation at 12000xg for 5 min. Supernatant was then collected via pipet and analyzed for total protein content using the BCA Protein Assay kit (Pierce) according to the manufacturer's instructions.
Tryptophan fluorescence quenching of galectin-1
Galectin-1 tryptophan fluorescence quenching by LacNAc-Q11 nanofibers or β-lactose was analyzed using a SpectraMax M5 plate-reader (excitation 280 nm, emission 345 nm) in glass-bottom black 96-well plates. LacNAc-Q11 nanofibers for tryptophan fluorescence quenching studies (1 mM Q11 containing χGlcNAc-Q11 = 0.01, 0.05, 0.10, 0.25, or 0.50) were prepared as described above and then treated with β-1,4-GalT and UDP-gal to convert nanofibrillar GlcNAc to LacNAc. Fluorescence intensity of unbound protein was determined from solutions of 5 μM galectin-1 in 1x PBS plus 8 mM DTT. Fluorescence intensity of bound protein was determined from solutions containing 5 μM galectin-1 in 1x PBS plus 8 mM DTT plus (a) LacNAc-Q11 nanofibers or (b) 0.01-100 mM β-lactose. Due to the 3 phenylalanine residues, Q11 also emits fluorescence upon excitation at 280 nm. Thus, background fluorescence intensity was determined from solutions of 1x PBS containing (a) LacNAc-Q11 nanofibers alone, or (b) 0.01-10 mM β-lactose alone. Tryptophan fluorescence quenching was reported as: fluorescence of unbound protein – fluorescence of bound protein, where bound protein = (fluorescence of galectin-1 plus nanofibers or β-lactose) – (fluorescence of nanofibers or β-lactose alone) (i.e. background signal), and unbound protein = (fluorescence intensity of galectin-1 alone). Dissociation constants were calculated by non-linear regression using GraphPad Prism software.
Analysis of Jurkat T cell apoptosis by galectin-1 and galectin-3 in the presence or absence of LacNAc-Q11 nanofibers
Jurkat T cells were obtained from the laboratory of Dr. Anita S. Chong at the University of Chicago, and were grown in suspension culture in RPMI 1640 with 10% heat-inactivated fetal bovine serum, which we referred to as “culture media”. For all experiments, culture media was supplemented with 100 μg/mL polymyxin b (Invivogen) to inhibit any endotoxin in galectin-1 or galectin-3 stocks that was not removed via Triton X-114 cloud-point precipitation. For cytotoxicity experiments, 20,000 Jurkat T cells per sample were incubated for 4 h in culture media supplemented with 0-2 mM GlcNAc-Q11 in 1x PBS, or LacNAc-Q11 nanofibers (1 mM Q11 containing χGlcNAc-Q11 = 0.01, 0.05, 0.10, 0.25, 0.50, or 1, prepared as described above and then treated with β-1,4-GalT and UDP-gal to convert nanofibrillar GlcNAc to LacNAc). Untreated controls were incubated in culture media supplemented with 1x PBS. For all galectin-1 apoptosis experiments, 20,000 Jurkat T cells per sample were incubated in culture media supplemented with 10 μM galectin-1 and 8 mM DTT, and untreated controls were incubated in culture media supplemented with 8 mM DTT. For all galectin-3 apoptosis experiments, 20,000 Jurkat T cells per sample were incubated in culture media supplemented with 5 μM galectin-3, and untreated controls were incubated in culture media. Culture media for experimental and untreated “control” groups was additionally supplemented with 1 mM Q11 nanofibers with varying LacNAc concentrations, prepared as described above, 250 μM β-lactose (Sigma), or 500 μM thiodigalactoside (TDG) (Santa Cruz Biotechnology). There was no discernible difference in cell agglutination or metabolic activity between control groups supplemented with Q11 nanofibers, β-lactose, or TDG, suggesting that they had no effect on Jurkat T cell viability in the absence of galectin-1 or galectin-3.
For agglutination experiments, cells were incubated for 1 h at 37 °C, and then imaged at 100x magnification with a Zeiss Axioscope inverted microscope. All images were scaled down to the same size using ImageJ software (NIH).
Phosphatidylserine exposure on the outer leaflet of the cell membrane was visualized with an Annexin V-eGFP staining kit (Biovision), according to manufacturer's instructions and an established protocol.63 Briefly, Jurkat T cells were maintained in culture media supplemented with 10 μM galectin-1 and 8 mM DTT, with or without Q11 nanofibers, for 1 h. Samples were then cooled on ice for 10 min, 0.2 M β-lactose was added to the sample at a 1:1 (v/v) ratio, and the samples were manually inverted for 30 s. 0.2 M β-lactose was then added at a 5-fold volumetric excess, each sample was centrifuged for 4 min at 1500 rpm on an Eppendorf mini-spin, and the supernatant was carefully removed to avoid disturbing the cell pellet. Cells were resuspended in 500 μL “binding buffer”. 5 μL stock Annexin V-eGFP was then added to the binding buffer solution, and samples were incubated for 5 min in the dark. Samples were then mounted on microscope slides and imaged at 100× magnification using a Zeiss Axioscope inverted epifluorescent microscope under bright-field and with a GFP filter cube. Fluorescent images were pseudo-colored and scaled down to the same size with ImageJ software (NIH).
Jurkat T cell metabolic activity was assessed after 4 h of culture using the CellTiter-Fluor metabolic activity assay kit (Promega), according to manufacturer's instructions. Briefly, 2× CellTiter-Fluor reagent was added to each sample at a 1:1 (v/v) ratio and incubated for 2 h at 37 °C. Sample fluorescence was then measured using a SpectraMax M5 plate-reader (excitation 370 nm/emission 495 nm). CellTiter-Fluor fluorescence in culture media lacking Jurkat T cells was subtracted from sample fluorescence. Metabolic activity in samples was reported as the sample fluorescence normalized to fluorescence of untreated cells.
Statistical analysis
All experimental and control groups had n = 3 for co-precipitation, tryptophan fluorescence quenching, and Jurkat T cell apoptosis experiments. Statistical differences between groups were analyzed using student's t-test (two groups) or ANOVA with Tukey's post-hoc (multiple groups) using GraphPad Prism software.
Results
GlcNAc-Q11 synthesis, self-assembly, and co-assembly with Q11
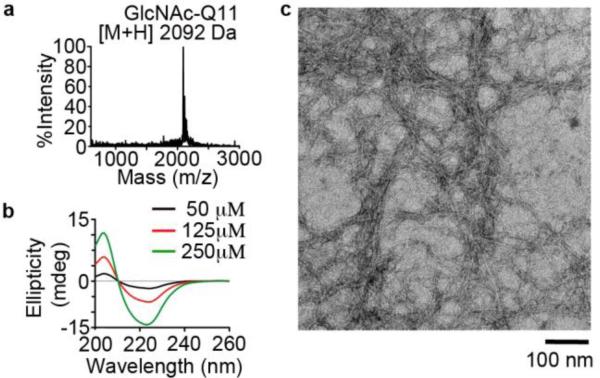
First we characterized synthesis and self-assembly of a glycosylated Q11 variant, GlcNAc-Q11 (Fig. 2). Following purification, MALDI-TOF-MS identified a peak at a mass to charge ratio (m/z) of [M+H]+ = 2092 Da (Figure 2a), which was consistent with the expected mass of GlcNAc-Q11. CD indicated that neutral aqueous buffered solutions of GlcNAc-Q11 were rich in β-sheet structures over a range of concentrations (50-250 μM), based on the minimum at 224 nm and maximum at 205 nm (Figure 2b). TEM identified high-aspect ratio nanofibers with a morphology similar to Q11 and other Q11 derivatives (Fig. 2c).56 Taken together, these results demonstrated that a Q11 variant having an appended GlcNAc moiety self-assembled into β-sheet nanofibers.
Figure 2. GlcNAc-Q11 self-assembled into β-sheet nanofibers.
a) MALDI-TOF mass spectrum of GlcNAc-Q11 (expected [M+H] = 2092 Da). b) CD spectra of GlcNAc-Q11 (χ = 1.0) at various concentrations in aqueous buffer (pH 7.4). c) TEM of GlcNAc-Q11 (χ = 1.0) after assembly in aqueous buffer (pH 7.4).
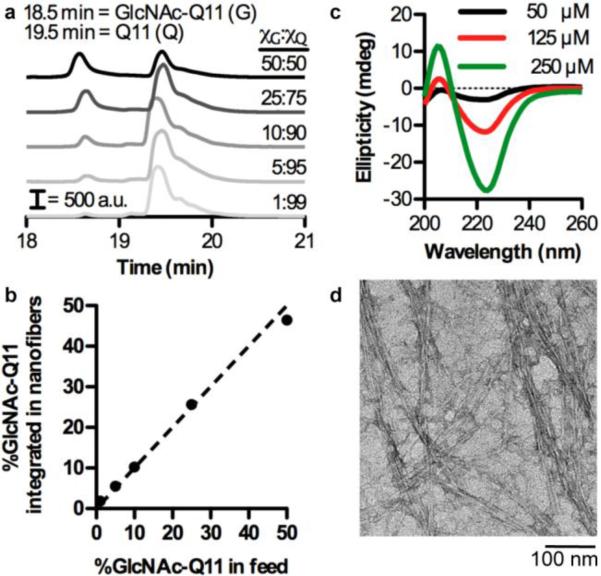
Next, we characterized co-assembly of GlcNAc-Q11 and Q11 into multi-component nanofibers using RP-HPLC, CD, and TEM (Fig. 3). Q11 eluted at ~19.5 min and GlcNAc-Q11 eluted at ~18.5 min when passed over a C18 column and eluted with a linear gradient of water and acetonitrile. As the molar ratio of GlcNAc-Q11 increased relative to Q11, the size of the GlcNAc-Q11 peak at 18.5 min increased relative to the Q11 peak at 19.5 min (Fig. 3a). Analysis of the area under the GlcNAc-Q11 and Q11 peaks in RP-HPLC chromatograms demonstrated that GlcNAc-Q11 mole fraction in solution during nanofiber assembly was predictive of nanofiber carbohydrate content over the range of χGlcNAc-Q11 = 0.01-0.50 (Fig. 3b). CD demonstrated that solutions of GlcNAc-Q11 and Q11 (molar ratio 1:9) were rich in β-sheet structures over a range of concentrations (50-250 μM), based on the minimum at 224 nm and maximum at 205 nm (Fig. 3c), similar to solutions of GlcNAc-Q11 alone (Fig. 2b). In addition, TEM identified high-aspect ratio nanofibers in solutions of GlcNAc-Q11 and Q11 (molar ratio 1:9) (Fig. 3d), with a morphology similar to GlcNAc-Q11 (Fig. 2c). Thus, simple mixing of Q11 and its glycosylated analog in the pre-assembled state enabled tailoring of nanofiber carbohydrate content, consistent with established methods to fabricate Q11 nanofibers with tunable composition of integrated peptide or protein ligands.55,56
Figure 3. Carbohydrate content of Q11 nanofibers can be tailored by varying the molar ratio of GlcNAc-Q11 to Q11 during assembly.
a) HPLC traces of GlcNAc-Q11 and Q11 mixed at various molar ratios, co-assembled in aqueous buffer, sedimented by centrifugation, and disassembled with trifluoroacetic acid. b) GlcNAc content of nanofibers determined from the area under GlcNAc-Q11 and Q11 HPLC trace peaks in (a) (n = 3, mean ± s.d.). c) CD spectra of 10% GlcNAc-Q11/90% Q11 at various concentrations in neutral aqueous buffer. d) TEM of 10% GlcNAc-Q11/90% Q11.
Enzymatic conversion of nanofibrillar GlcNAc monosaccharides to LacNAc disaccharides
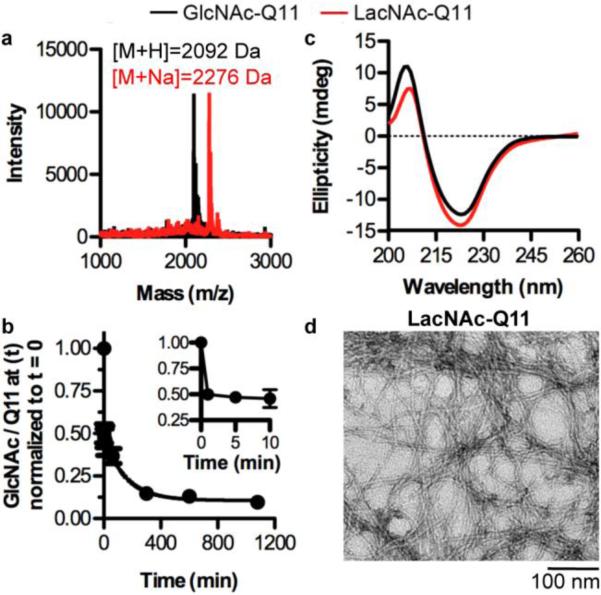
Here, we characterized an enzymatic route to convert nanofibrillar GlcNAc monosaccharides to LacNAc disaccharides (Fig 4), the latter being expected to bind galectins. An enzymatic approach was taken due to the potential for carbohydrate degradation under conditions required for solid-phase peptide synthesis and purification.65 MALDI-TOF-MS demonstrated that the molecular weight of GlcNAc-Q11 incubated with β-1,4-GalT and UDP-gal in MnCl2-containing neutral buffer increased by 162 Da ([M+Na]+ = 2276 Da], as expected for the addition of galactose via condensation of 4-OH on GlcNAc and 1-OH on galactose (Fig. 4a). In addition, MALDI-TOF-MS peak intensity for GlcNAc-Q11 (m/z = 2092 Da) was very low, suggesting the GlcNAc conversion reaction was efficient even at a very high GlcNAc density on the nanofibers. The +162 Da increase in mass required nanofibers to be incubated with both UDP-gal and β-1,4-GalT (data not shown), and the extent of reaction was greatly reduced when magnesium was added as a co-factor in place of manganese (data not shown), consistent with previous reports.66 Based on analysis of MALDI-TOF-MS peak intensity, nearly 50% of GlcNAc in the reaction medium was enzymatically converted to LacNAc within 1 min (Fig. 4b, inset), which increased to ~90% conversion by 6 h (Fig. 4b). Based on the reaction conditions, it was expected that enzymatic conversion of GlcNAc to LacNAc would reach completion within ~1 minute. The slowed reaction kinetics observed here may be due to the decreased diffusion of the GlcNAc substrate in the reaction medium when conjugated to Q11 nanofibers. Additionally, the remaining 10% of unreacted GlcNAc may be due to loss of enzymatic activity via incubation at 37°C, hydrolysis of UDP-gal, or steric shielding of GlcNAc moieties by the nanofibers. Nonetheless, CD demonstrated that the β-sheet content was similar before and after enzymatic treatment of nanofibers to convert GlcNAc to LacNAc (Fig. 4c). In addition, TEM identified high-aspect ratio nanofibers after enzymatic treatment (Fig. 4d), which have a similar morphology to untreated GlcNAc-Q11 (Fig. 2c). Thus, these results demonstrated that an enzymatic synthesis route can be used to fabricate β-sheet nanofibers decorated with the galectin-binding disaccharide, LacNAc.
Figure 4. Pendant GlcNAc moieties along Q11 nanofibers were efficiently converted to LacNAc.
a) MALDI-TOF mass spectrum of GlcNAc-Q11 (expected [M+H] = 2092 Da) (black) and LacNAc-Q11 (expected [M+Na] = 2275 Da) (red) prepared via treatment of GlcNAc-Q11 (χ = 1.0) with β-1,4-GalT and UDP-gal. b) Kinetic analysis of nanofibrillar GlcNAc conversion to LacNAc, as determined by the ratio of the GlcNAc-Q11 MALDI intensity to the Q11 MALDI intensity at time t divided by the ratio of the GlcNAc-Q11 MALDI intensity to the Q11 MALDI intensity at t = 0 (χGlcNAc-Q11:χQ11 = 25:75, n = 3, mean ± s.d.). c) CD spectra of GlcNAc-Q11 (χ = 1.0) before (black) and after (red) treatment with β-1,4-GalT and UDP-gal. d) TEM of GlcNAc-Q11 (χ = 1.0) after assembly in aqueous buffer (pH 7.4) and treatment with β-1,4-GalT and UDP-gal.
Lectin binding to self-assembled glycopeptide nanofibers
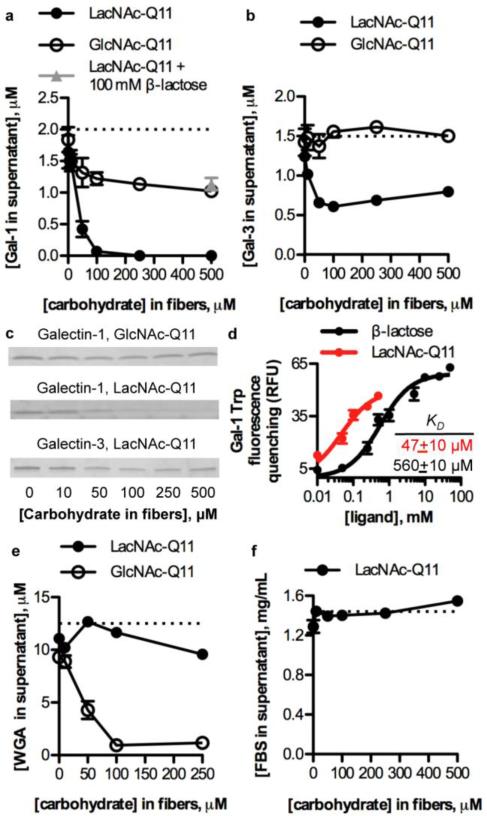
We characterized binding of galectin-1 and galectin-3 to Q11 nanofibers with varied GlcNAc or LacNAc concentration using co-precipitation and tryptophan fluorescence quenching assays (Fig. 5a-d). The extent of galectin-1 binding to GlcNAc-Q11 nanofibers before and after enzymatic treatment increased with increasing carbohydrate concentration in the nanofibers (Fig. 5a). However, converting GlcNAc to LacNAc increased nanofiber galectin-1 binding affinity, as indicated by a significantly lower concentration of galectin-1 in supernatants above LacNAc-Q11 nanofibers when compared to supernatants above GlcNAc-Q11 nanofibers, which was consistent with previous reports demonstrating increased binding affinity of galectin-1 for β-galactosides when compared to GlcNAc.67 Binding of galectin-1 to LacNAc-Q11 nanofibers was mediated by specific protein-carbohydrate interactions, since it was inhibited by a molar excess of β-lactose (Fig. 5a, gray triangle). Galectin-3 also bound to LacNAc-Q11 nanofibers in a LacNAc concentration-dependent manner, however, galectin-3 failed to bind to GlcNAc-Q11 nanofibers (Fig. 5b), consistent with previous reports demonstrating minimal binding of galectin-3 to GlcNAc-Q11.67 SDS-PAGE analysis of supernatants above GlcNAc-Q11 or LacNAc-Q11 nanofibers incubated with galectin-1 or galectin-3 qualitatively demonstrated that LacNAc-Q11 nanofibers have higher affinity for galectin-1 than galectin-3, since there was no detectable galectin-1 present in supernatants above nanofibers with [LacNAc] ≥ 100 μM, whereas galectin-1 or galectin-3 were detected in supernatants above nanofibers with [GlcNAc] ≥ 100 μM or [LacNAc] ≥ 100 μM, respectively (Fig. 5c). Thus, tailoring nanofiber LacNAc concentration via simple mixing of molecular variants in the pre-assembled state can give rise to synthetic glycoclusters with increased affinity for galectin-1 when compared to galectin-3.
Figure 5. Lectin binding to LacNAc-Q11 nanofibers.
Binding of 2 μM galectin-1 to Q11 nanofibers with 0-500 μM LacNAc or GlcNAc. b) Binding of 1.5 μM galectin-3 to Q11 nanofibers with 0-500 μM LacNAc or GlcNAc. c) PAGE gels of supernatants above nanofibers sedimented by centrifugation, qualitatively demonstrating increased binding of galectin-1 to LacNAc-Q11 nanofibers. d) Tryptophan fluorescence quenching of galectin-1 by LacNAc-Q11 nanofibers or soluble β-lactose. e) Binding of 12.5 μM wheat germ agglutinin (WGA), a GlcNAc-binding lectin, to Q11 nanofibers with 0-250 μM LacNAc or GlcNAc, demonstrating correlation between carbohydrate type and lectin-binding specificity. f) Non-specific binding of fetal bovine serum to Q11 nanofibers with 0-500 μM LacNAc.
Based on the observation that LacNAc-Q11 nanofibers had higher affinity for galectin-1 than galectin-3, we compared the affinity of galectin-1 for nanofibrillar LacNAc and soluble β-lactose using tryptophan fluorescence quenching. The dissociation constant of galectin-1 for LacNAc-Q11 was nearly an order of magnitude lower than its dissociation constant for soluble β-lactose (Fig. 5d), as demonstrated by an increased extent of tryptophan fluorescence quenching by LacNAc-Q11 nanofibers when compared to quenching by an identical concentration of β-lactose. Thus, this data suggests that self-assembly of glycopeptides into β-sheet nanofibers can provide synthetic glycoclusters having increased galectin-binding affinity when compared to monomeric carbohydrates.
Different lectins often have binding specificity for different carbohydrates, and here we used co-precipitation experiments to investigate correlations between the type of carbohydrate conjugated to Q11 nanofibers and their lectin-binding specificity. Specifically, we analyzed GlcNAc-Q11 and LacNAc-Q11 nanofiber co-precipitation with wheat germ agglutinin (WGA), a lectin that preferentially binds GlcNAc.68 Extent of WGA binding to GlcNAc-Q11 nanofibers increased as a function of GlcNAc concentration, whereas WGA bound only weakly to LacNAc-Q11 nanofibers over the range of carbohydrate concentrations tested (Fig. 5e). These observations indicated that conjugating galactose onto GlcNAc-Q11 nanofibers via β-1,4-GalT inhibited WGA recognition of GlcNAc. When considered alongside data demonstrating that galectin-1 and galectin-3 preferentially bound to LacNAc-Q11 nanofibers (Fig. 5a-c), these results suggest that nanofiber lectin-binding specificity can also be tailored by changing the type of carbohydrate conjugated to Q11 nanofibers.
Finally, in light of previous demonstrations that LacNAc can be used to reduce non-specific protein adsorption onto nanomaterials,70 here we investigated fetal bovine serum binding onto LacNAc-Q11 nanofibers. The concentration of fetal bovine serum proteins bound non-specifically to Q11 nanofibers was low and independent of nanofiber LacNAc concentration (Fig. 5f). In addition, SDS-PAGE analysis of nanofibers after co-precipitation with fetal bovine serum suggested that the profile of non-specifically bound proteins did not vary significantly over the range of LacNAc concentrations tested (data not shown). Together, these results indicate that pendant LacNAc moieties neither increase nor decrease non-specific serum protein binding to self-assembled β-sheet peptide nanofibers, and suggest that LacNAc-Q11 nanofibers may be useful as biomaterials to modulate galectin bioactivity within extracellular microenvironments.
Efficacy of glycopeptide nanofibers as inhibitors of galectin-mediated T cell apoptosis
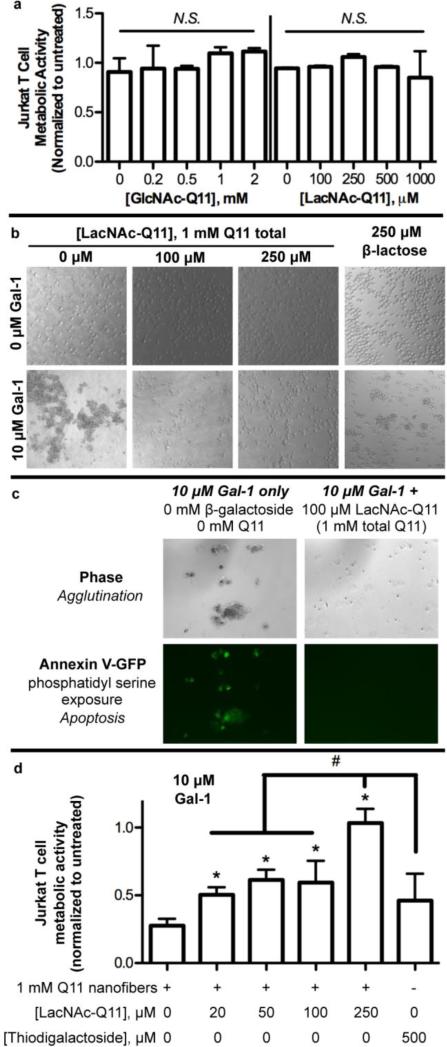
Galectin-1-mediated T cell apoptosis is a key mediator of tumor immune privilege,71,72 and therapeutics capable of inhibiting galectin-mediated T cell apoptosis are of interest for enhancing anti-tumor immunity. Based on observations that nanofibrillar LacNAc bound galectin-1 with higher affinity than soluble β-lactose, we compared the efficacy of LacNAc-Q11 nanofibers and β-lactose for inhibiting galectin-1-mediated Jurkat T cell apoptosis, an established model of galectin-1 bioactivity (Fig. 6).22 Neither GlcNAc-Q11, nor LacNAc-Q11, were cytotoxic to Jurkat T cells over the range of concentrations used in these studies (Fig. 6a), indicating that Q11 nanofibers were suitable for characterizing the efficacy of nanofibrillar LacNAc for modulating galectin-1 bioactivity.
Figure 6. LacNAc-Q11 nanofibers inhibited galectin-1-mediated Jurkat T cell agglutination and apoptosis.
a) Cytotoxicity of GlcNAc-Q11 and LacNAc-Q11 nanofibers. b) Bright-field photomicrographs of Jurkat T cell agglutination in culture media with or without Gal-1 in the presence of 1 mM Q11 nanofibers with 0, 100, or 250 μM LacNAc, or 250 μM β-lactose. c) Bright-field and fluorescent photomicrographs of Jurkat T cells stained with Annexin V-eGFP to assess phosphatidylserine exposure, an early marker of apoptosis, in media supplemented with 10 μM Gal-1 alone or with 1 mM Q11 nanofibers having 100 μM LacNAc-Q11. d) Metabolic activity of Jurkat T cells in media supplemented with 10 μM Gal-1 plus 1 mM Q11 nanofibers with 0-250 μM LacNAc or 500 μM thiodigalactoside, a known galectin inhibitor and stable analog of β-lactose. * represents p < 0.05 compared to 0 μM LacNAc-Q11 or, # represents p < 0.05 compared to 250 μM LacNAc group, ANOVA with Tukey's post-hoc.
First, we characterized Jurkat T cell agglutination, an early marker of apoptosis, via galectin-1 in the presence of LacNAc-Q11 nanofiber formulations that co-precipitated >90% of galectin-1 in solution (i.e. [carbohydrate] ≥ 100 μM, Fig. 5a) or soluble β-lactose. Galectin-1 protein caused agglutination of Jurkat T cells, as noted by the predominance of large cell aggregates rather than single suspended cells (Fig 6b). In contrast, galectin-mediated T cell agglutination was inhibited by LacNAc-Q11 nanofibers or soluble β-lactose, albeit with differing effectiveness (Fig. 6b). In particular, single cells were predominantly observed in cultures treated with LacNAc-Q11 nanofibers, whereas both single cells and cell aggregates were observed in cultures treated with soluble β-lactose. Qualitatively, this suggested that LacNAc-Q11 nanofibers are more effective galectin-1 inhibitors than soluble β-lactose, which was not surprising in light of our observations that galectin-1 affinity for LacNAc-Q11 nanofibers was higher than its affinity for soluble β-lactose (Fig. 5d).
Next, we analyzed phosphatidylserine exposure on the outer leaflet of Jurkat T cell membranes, which is a marker of apoptosis known to be up-regulated by galectin-1.22,63 Based on observations that LacNAc-Q11 nanofibers having [carbohydrate] ≥ 100 μM inhibited Jurkat T cell agglutination (Fig. 6b), we limited qualitative analysis of phosphatidylserine exposure to a single LacNAc-Q11 formulation. Jurkat T cells agglutinated via galectin-1 stained positively for phosphatidylserine on the outer leaflet of the cell membrane, as visualized by Annexin V-eGFP (Fig 6c), consistent with previous reports.22,63 In contrast, Jurkat T cells in cultures containing galectin-1 and a LacNAc-Q11 formulation that inhibited agglutination were negative for phosphatidylserine exposure (Fig 6b). Because LacNAc-Q11 nanofibers were not cytotoxic (Fig. 6a), we inferred that cells treated with LacNAc-Q11 nanofibers were viable and not necrotic in these qualitative studies.
To quantify the efficacy of LacNAc nanofibers as inhibitors of galectin-1 bioactivity, we analyzed loss of Jurkat T cell metabolic activity, a later marker of apoptosis. In particular, Jurkat T cells were cultured in media supplemented with galectin-1 and nanofibers with varying LacNAc mole fractions, or thiodigalactoside (TDG), a stable β-lactose analog and known galectin-1 inhibitor.73 Here, we investigated a broader range of LacNAc-Q11 nanofiber formulations to identify correlations between nanofiber galectin-1-binding affinity and efficacy for modulating galectin-1 bioactivity. Metabolic activity of Jurkat T cells was significantly reduced when cultured in media supplemented with galectin-1 and Q11 nanofibers lacking LacNAc (Fig. 6d). In contrast, 1 mM Q11 nanofibers with pendant LacNAc moieties maintained Jurkat T cell metabolic activity in the presence of galectin-1 to an extent that was dependent on LacNAc concentration, such that nanofibers fabricated with χGlcNAc-Q11 = 0.25 (i.e. 250 μM carbohydrate) nearly completely inhibited galectin-1 induced loss of Jurkat T cell metabolic activity (Fig. 6d). Notably, 500 μM TDG (a 2-fold molar excess of the highest LacNAc-Q11 concentration) was not effective at maintaining Jurkat T cell metabolic activity (Fig. 6d), which was consistent with qualitative results suggesting that a soluble β-galactoside was unable to inhibit galectin-1-mediated Jurkat T cell apoptosis to the same extent as LacNAc-Q11 nanofibers (Fig. 6b).
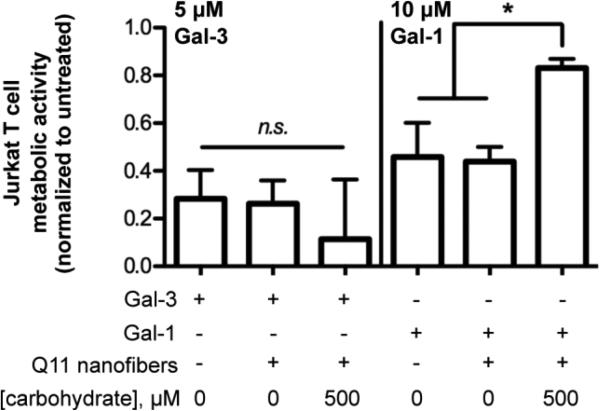
T cells also undergo apoptosis in response to galectin-3, although galectin-1 preferentially induces apoptosis of double-negative and double-positive CD4/CD8 T cells, whereas galectin-3 preferentially induces apoptosis of CD4/CD8 double-negative T cells, via differential binding of each galectin to T cell surface receptors.22 Jurkat T cells undergo apoptosis via both galectin-1 and galectin-3,22 and here we used Jurkat T cells as a model to compare the efficacy of LacNAc-Q11 nanofibers as inhibitors of galectin-1 or galectin-3 apoptosis. Q11 nanofibers with pendant LacNAc moieties did not inhibit galectin-3-mediated Jurkat T cell apoptosis to a measurable extent when compared to nanofibers lacking LacNAc or cultures lacking nanofibers (Fig. 7, left). In contrast, an identical nanofiber formulation robustly inhibited galectin-1-mediated apoptosis when compared to nanofibers lacking LacNAc or cultures lacking nanofibers (Fig. 7, right). These results were consistent with our observations that LacNAc-Q11 nanofibers have higher affinity for galectin-1 than galectin-3 (Fig. 5a-c).
Figure 7. Efficacy of LacNAc-Q11 nanofibers for inhibiting galectin-3 or galectin-1-mediated Jurkat T cell apoptosis.
d) Metabolic activity of Jurkat T cells in culture media supplemented with 5 μM Gal-3 or 10 μM Gal-1 in the absence of Q11 nanofibers, in the presence of 1 mM Q11 nanofibers without LacNAc, or in the presence of 1 mM Q11 with LacNAc (χGlcNAc-Q11 = 0.5). * represents p < 0.05, ANOVA with Tukey's post-hoc.
Taken together, these results demonstrated that self-assembled glycopeptide nanofibers can inhibit galectin-1-mediated T cell apoptosis more effectively than soluble β-galactosides, likely due to the formation of glycoclusters with enhanced galectin-binding affinity. Notably, nanofiber galectin-binding affinity, as determined via co-precipitation assays, was predictive of nanofiber efficacy for inhibiting galectin bioactivity.
Discussion
Here, we demonstrated that a glycosylated variant of a β-sheet fibrillizing peptide can self-assemble into high-aspect ratio nanofibers. These nanofibers can bind to galectins in a carbohydrate-dependent manner, and nanofiber lectin-binding specificity and affinity can be tailored by varying the type and concentration of carbohydrate integrated into the nanofibers. The ease of tuning carbohydrate concentration within these nanofibers via simple mixing of glycosylated and non-glycosylated peptides in the pre-assembled state highlights the flexibility of this platform for creating synthetic glycoclusters with tailored carbohydrate content. Self-assembly of LacNAc into supramolecular nanofibers increased its affinity for galectin-1 by nearly an order of magnitude when compared to soluble β-lactose. As with other polyvalent carbohydrates, this increased binding affinity is likely due in part to increased carbohydrate avidity, or the “glycocluster effect”, provided by self-assembly. In addition, binding affinity was likely enhanced by the reduced entropic penalty resulting from galectin-1 association with LacNAc immobilized on a supramolecular nanofiber having a relatively high molecular weight. As a result, galectin-binding nanofibers are more effective than conventional small molecules for inhibiting galectin-1-mediated T cell apoptosis, suggesting their potential as therapeutic biomaterials.
The ability to append a carbohydrate ligand onto a self-assembling domain to create functional nanofibers adds significant new biomaterial design opportunities to a platform that already provides robust design flexibility via integration of peptide or protein ligands.49-52,55,56 Although other self-assembling carbohydrate variants are receiving attention for fabricating functional nanomaterials,53,74-78 one inherent advantage of the β-sheet fibrillizing platform used herein is that the concentration of functional ligand integrated into the nanofiber can be precisely varied via simple mixing of different self-assembling molecules in the pre-assembled state.55,56 Importantly, this allows for nanofiber functional properties to be tailored according to application specific needs. In this case, nanofiber affinity for galectin-1 and -3, and efficacy for inhibiting galectin-1 bioactivity, could be tailored by varying the molar ratio of glycosylated and non-glycosylated peptides present during nanofiber assembly. Notably, we observed that binding affinity of galectin-1 for LacNAc-Q11 nanofibers was greater than that of galectin-3. The observed differences in galectin-binding affinity of LacNAc-Q11 nanofibers may be the result of steric clashing due to the larger size of galectin-3 when compared to galectin-1 (MW ~ 27 kDa versus 14 kDa, respectively), or the self-assembly of galectin-3 into pentamers and galectin-1 into dimers. Although the exact mechanism governing these differences in binding are beyond the scope of this report, they are nonetheless consistent with previous work demonstrating that galectin-1 binds strongly to the human immunodeficiency virus coat glycoprotein GP120 having high LacNAc density, whereas galectin-3 is unable to bind.34 More importantly, different galectins can have contrasting activities as inhibitory or stimulatory signaling molecules within distinct physiological or pathological settings, such as antigen-specific T cell responses,3 T cell apoptosis,22 autoimmune disease severity,32,33 viral infection,9 and angiogenesis.79 Thus, the potential to tailor galectin binding affinity by modifying nanofiber composition may lead to biomaterials with optimal efficacy for modulating galectin bioactivity in different therapeutic settings.
In addition to biophysical constraints, lectin binding to glycopeptide nanofibers is also governed by carbohydrate chemistry. In particular, galectins preferentially bind to β-galactosides, such as LacNAc and β-lactose, with different galectins having fine specificity for sialylated, fucosylated, or repeated variants.80-82 Efforts to create synthetic galectin-binding glycoclusters are often challenged by the hydrolytic instability of the glycosidic bonds linking different carbohydrates.65 Within natural systems, GlcNAc is enzymatically converted to LacNAc by β-1,4-GalT in the presence of an activated galactose donor, such as UDP-galactose.83 This bio-inspired route has been used previously to fabricate LacNAc-modified substrates in vitro,61 and here we used this same synthesis route to fabricate LacNAc-modified nanofibers. We chose a synthetic route in which GlcNAc moieties are modified after Q11 self-assembly into nanofibers, rather than in the pre-assembled state, because Q11 nanofibers can be sedimented by centrifugation to separate them from low molecular weight proteins, unused reactants, or side products.55,64 Thus, self-assembled nanofibers essentially provided a pseudo-solid-phase that greatly simplifies purification via cycles of centrifugation and washing steps. Nanofibrillar GlcNAc moieties were converted to LacNAc in high yield via β-1,4-GalT, and the reaction was dependent on the enzyme, sugar donor, and metal co-factor. These results were consistent with previous work demonstrating that an enzyme can efficiently react with a fibrillized ligand to mediate protein immobilization onto nanofibers.64 Taken together, these observations demonstrate that enzyme-catalyzed reactions are generally useful for post-assembly modification of self-assembled peptide nanofibers displaying appropriate substrates. Here, this was particularly important for enhancing nanofiber galectin-binding affinity, since nanofibers bearing GlcNAc moieties had low affinity for galectin-1 and no observable affinity for galectin-3. Moving forward, sequential enzymatic reactions inspired by natural carbohydrate synthesis may be broadly useful for fabricating nanofibers with more complex carbohydrates. In turn, precisely tailoring carbohydrate chemistry may provide more finely tuned control of nanofiber lectin-binding affinity or specificity than what is afforded by biophysical constraints alone. Thus, combining the ability to vary carbohydrate concentration via simple mixing of Q11 variants in the pre-assembled state with appropriate enzymatic reactions to vary carbohydrate type post-assembly is likely to provide significant biomaterial design flexibility. In turn, this flexibility is likely to facilitate fabrication of synthetic glycoclusters with tailored lectin-binding properties as therapeutics for various applications, including viral prophylaxis, immunotherapy, and cancer. In addition, co-assembly of carbohydrate-modified Q11 variants with Q11 variants bearing peptide or protein ligands may provide a simple route for fabricating nanofibers that can replicate glycoprotein structure and function, ultimately leading to new classes of therapeutic biomaterials or next-generation mimics of the extracellular matrix.
Efficacy of LacNAc-Q11 nanofibers as inhibitors of galectin-mediated T cell apoptosis provided an initial demonstration of their potential as therapeutic biomaterials. Optimal LacNAc-Q11 nanofiber formulations were a more effective inhibitor of galectin-1-mediated T cell agglutination and apoptosis than conventional small molecule inhibitors, such as β-lactose and its stable variant TDG. This observation was not necessarily surprising, given that LacNAc-Q11 affinity for galectin-1 was nearly an order of magnitude higher than that of soluble β-lactose, likely due to the glycocluster effect provided by LacNAc-Q11 self-assembly. LacNAc-Q11 nanofibers were also more effective for inhibiting Jurkat T cell apoptosis by galectin-1 than galectin-3, suggesting that these materials may be effective for inhibiting galectin-1 signaling in antigen-specific T cell responses,3 or suppressing galectin-1-mediated apoptosis of CD4/CD8 double-negative and double-positive T cells,22 while having little effect on galectin-3. Thus, self-assembled nanofibers provide a promising alternative to small molecule galectin inhibitors that have prevailed to date, in part due to their increased binding affinity that can optimized by tailoring carbohydrate concentration, type, or both. However, small molecule inhibitors and low-molecular weight glycoclusters have other inherent limitations that may also be addressable with self-assembled glycopeptide nanofibers. In particular, systemic delivery of small molecules or low-molecular weight glycoclusters will likely result in off-target effects due to rapid and extensive bio-distribution, and in turn inhibition of galectin activity underlying normal physiological processes. For example, endogenous galectins inhibit entry of certain viruses,9 and therefore systemic inhibition of galectin activity may render patients susceptible to opportunistic viral infections. In contrast, localized delivery of galectin-inhibiting nanofibers having low diffusivity due to their high molecular weight may be less likely to increase susceptibility to opportunistic infection. In addition, although not explicitly investigated here, galectin inhibitors based on self-assembled peptide nanofibers are anticipated to be biocompatible, minimally inflammatory, and non-immunogenic,57,58 thereby enhancing their safety profile as therapeutic biomaterials.
Conclusions
Self-assembly of glycopeptides into high-aspect ratio nanofibers gives rise to synthetic glycoclusters that can non-covalently bind to galectins, and in turn modulate their activity as extracellular signals. The concentration of carbohydrate integrated into the nanofibers can be easily and precisely varied via simple mixing of different self-assembling peptide variants in the pre-assembled state, while carbohydrate type can be tailored via enzymatic reactions in the post-assembled state. Due to ease of fabrication, precise control over material composition, inherent ‘glycocluster effect’, and efficacy for modulating galectin bioactivity, self-assembled glycopeptide nanofibers provide a promising alternative to low-molecular weight polyvalent scaffolds or glycopolymers as synthetic glycoclusters and glycoprotein mimetics. We envision that these nanofibers will be broadly useful as biomaterials that can modulate galectin bioactivity in various normal and pathological settings, including innate and adaptive immunity, cancer, viral infection, wound healing, and tissue regeneration.
Acknowledgements
This research was supported by the National Institutes of Health (NIBIB, 1R01EB009701; NCI, U54 CA151880; NIAID, 1F32AI096769) and the National Science Foundation (DMR-1455201). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and BioEngineering, the National Institute of Allergy and Infectious Disease, the National Cancer Institute, the National Institutes of Health, or the National Science Foundation. MALDI-TOF was performed in the University of Chicago Mass Spectrometry facility and the University of Florida Mass Spectrometry facility, with support from NSF CHE MRI 1040016. CD was performed in the University of Chicago Biophysics Core. TEM was performed in the University of Chicago Materials Research Center.
Footnotes
Conflicts of Interest.
Antonietta Restuccia, Ye F. Tian, Joel H. Collier, and Gregory A. Hudalla declare that they have no conflicts of interest.
Ethical Standards.
No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
References
- 1.Cedeno-Laurent F, Dimitroff CJ. Galectin-1 research in T cell immunity: past, present and future. Clin Immunol. 2012;142:107–116. doi: 10.1016/j.clim.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabinovich GA, Toscano MA. Turning ‘sweet’ on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat Rev Immunol. 2009;9:338–352. doi: 10.1038/nri2536. [DOI] [PubMed] [Google Scholar]
- 3.Tribulatti MV, Figini MG, Carabelli J, Cattaneo V, Campetella O. Redundant and antagonistic functions of galectin-1, -3, and -8 in the elicitation of T cell responses. J Immunol. 2012;188:2991–2999. doi: 10.4049/jimmunol.1102182. [DOI] [PubMed] [Google Scholar]
- 4.Fortuna-Costa A, Gomes AM, Kozlowski EO, Stelling MP, Pavao MS. Extracellular galectin-3 in tumor progression and metastasis. Front Oncol. 2014;4:138. doi: 10.3389/fonc.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heusschen R, Griffioen AW, Thijssen VL. Galectin-9 in tumor biology: a jack of multiple trades. Biochim Biophys Acta. 2013;1836:177–185. doi: 10.1016/j.bbcan.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Ito K, Stannard K, Gabutero E, Clark AM, Neo SY, Onturk S, Blanchard H, Ralph SJ. Galectin-1 as a potent target for cancer therapy: role in the tumor microenvironment. Cancer Metastasis Rev. 2012;31:763–778. doi: 10.1007/s10555-012-9388-2. [DOI] [PubMed] [Google Scholar]
- 7.Schattner M, Rabinovich GA. Galectins: new agonists of platelet activation. Biol Chem. 2013;394:857–863. doi: 10.1515/hsz-2013-0108. [DOI] [PubMed] [Google Scholar]
- 8.Thijssen VL, Griffioen AW. Galectin-1 and -9 in angiogenesis: a sweet couple. Glycobiology. 2014;24:915–920. doi: 10.1093/glycob/cwu048. [DOI] [PubMed] [Google Scholar]
- 9.Baum LG, Garner OB, Schaefer K, Lee B. Microbe-Host Interactions are Positively and Negatively Regulated by Galectin-Glycan Interactions. Front Immunol. 2014;5:284. doi: 10.3389/fimmu.2014.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. Introduction to galectins. Glycoconj J. 2004;19:433–440. doi: 10.1023/B:GLYC.0000014072.34840.04. [DOI] [PubMed] [Google Scholar]
- 11.Yang RY, Rabinovich GA, Liu FT. Galectins: structure, function and therapeutic potential. Expert Rev Mol Med. 2008;10:e17. doi: 10.1017/S1462399408000719. [DOI] [PubMed] [Google Scholar]
- 12.Rabinovich GA, Toscano MA, Jackson SS, Vasta GR. Functions of cell surface galectin-glycoprotein lattices. Curr Opin Struct Biol. 2007;17:513–520. doi: 10.1016/j.sbi.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy Y, Arbel-Goren R, Hadari YR, Eshhar S, Ronen D, Elhanany E, Geiger B, Zick Y. Galectin-8 functions as a matricellular modulator of cell adhesion. J Biol Chem. 2001;276:31285–31295. doi: 10.1074/jbc.M100340200. [DOI] [PubMed] [Google Scholar]
- 14.Horiguchi N, Arimoto K, Mizutani A, Endo-Ichikawa Y, Nakada H, Taketani S. Galectin-1 induces cell adhesion to the extracellular matrix and apoptosis of non-adherent human colon cancer Colo201 cells. J Biochem. 2003;134:869–874. doi: 10.1093/jb/mvg213. [DOI] [PubMed] [Google Scholar]
- 15.Espelt MV, Croci DO, Bacigalupo ML, Carabias P, Manzi M, Elola MT, Munoz MC, Dominici FP, Wolfenstein-Todel C, Rabinovich GA, Troncoso MF. Novel roles of galectin-1 in hepatocellular carcinoma cell adhesion, polarization, and in vivo tumor growth. Hepatology. 2011;53:2097–2106. doi: 10.1002/hep.24294. [DOI] [PubMed] [Google Scholar]
- 16.Friedrichs J, Manninen A, Muller DJ, Helenius J. Galectin-3 regulates integrin alpha2beta1-mediated adhesion to collagen-I and -IV. J Biol Chem. 2008;283:32264–32272. doi: 10.1074/jbc.M803634200. [DOI] [PubMed] [Google Scholar]
- 17.Hughes RC. Galectins as modulators of cell adhesion. Biochimie. 2001;83:667–676. doi: 10.1016/s0300-9084(01)01289-5. [DOI] [PubMed] [Google Scholar]
- 18.Moiseeva EP, Spring EL, Baron JH, de Bono DP. Galectin 1 modulates attachment, spreading and migration of cultured vascular smooth muscle cells via interactions with cellular receptors and components of extracellular matrix. J Vasc Res. 1999;36:47–58. doi: 10.1159/000025625. [DOI] [PubMed] [Google Scholar]
- 19.Kuwabara I, Liu FT. Galectin-3 promotes adhesion of human neutrophils to laminin. J Immunol. 1996;156:3939–3944. [PubMed] [Google Scholar]
- 20.Lei CX, Zhang W, Zhou JP, Liu YK. Interactions between galectin-3 and integrinbeta3 in regulating endometrial cell proliferation and adhesion. Hum Reprod. 2009;24:2879–2889. doi: 10.1093/humrep/dep250. [DOI] [PubMed] [Google Scholar]
- 21.Wan SY, Zhang TF, Ding Y. Galectin-3 enhances proliferation and angiogenesis of endothelial cells differentiated from bone marrow mesenchymal stem cells. Transplant Proc. 2011;43:3933–3938. doi: 10.1016/j.transproceed.2011.10.050. [DOI] [PubMed] [Google Scholar]
- 22.Stillman BN, Hsu DK, Pang M, Brewer CF, Johnson P, Liu FT, Baum LG. Galectin-3 and galectin-1 bind distinct cell surface glycoprotein receptors to induce T cell death. J Immunol. 2006;176:778–789. doi: 10.4049/jimmunol.176.2.778. [DOI] [PubMed] [Google Scholar]
- 23.Sturm A, Lensch M, Andre S, Kaltner H, Wiedenmann B, Rosewicz S, Dignass AU, Gabius HJ. Human galectin-2: novel inducer of T cell apoptosis with distinct profile of caspase activation. J Immunol. 2004;173:3825–3837. doi: 10.4049/jimmunol.173.6.3825. [DOI] [PubMed] [Google Scholar]
- 24.Motran CC, Molinder KM, Liu SD, Poirier F, Miceli MC. Galectin-1 functions as a Th2 cytokine that selectively induces Th1 apoptosis and promotes Th2 function. Eur J Immunol. 2008;38:3015–3027. doi: 10.1002/eji.200838295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stowell SR, Qian Y, Karmakar S, Koyama NS, Dias-Baruffi M, Leffler H, McEver RP, Cummings RD. Differential roles of galectin-1 and galectin-3 in regulating leukocyte viability and cytokine secretion. J Immunol. 2008;180:3091–3102. doi: 10.4049/jimmunol.180.5.3091. [DOI] [PubMed] [Google Scholar]
- 26.Chan J, O'Donoghue K, Gavina M, Torrente Y, Kennea N, Mehmet H, Stewart H, Watt DJ, Morgan JE, Fisk NM. Galectin-1 induces skeletal muscle differentiation in human fetal mesenchymal stem cells and increases muscle regeneration. Stem Cells. 2006;24:1879–1891. doi: 10.1634/stemcells.2005-0564. [DOI] [PubMed] [Google Scholar]
- 27.Arikawa T, Matsukawa A, Watanabe K, Sakata KM, Seki M, Nagayama M, Takeshita K, Ito K, Niki T, Oomizu S, Shinonaga R, Saita N, Hirashima M. Galectin-9 accelerates transforming growth factor beta3-induced differentiation of human mesenchymal stem cells to chondrocytes. Bone. 2009;44:849–857. doi: 10.1016/j.bone.2009.01.365. [DOI] [PubMed] [Google Scholar]
- 28.Goldring K, Jones GE, Thiagarajah R, Watt DJ. The effect of galectin-1 on the differentiation of fibroblasts and myoblasts in vitro. J Cell Sci. 2002;115:355–366. doi: 10.1242/jcs.115.2.355. [DOI] [PubMed] [Google Scholar]
- 29.Yang RY, Hsu DK, Yu L, Chen HY, Liu FT. Galectin-12 is required for adipogenic signaling and adipocyte differentiation. J Biol Chem. 2004;279:29761–29766. doi: 10.1074/jbc.M401303200. [DOI] [PubMed] [Google Scholar]
- 30.Baum LG, Blackall DP, Arias-Magallano S, Nanigian D, Uh SY, Browne JM, Hoffmann D, Emmanouilides CE, Territo MC, Baldwin GC. Amelioration of graft versus host disease by galectin-1. Clin Immunol. 2003;109:295–307. doi: 10.1016/j.clim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Cortegano I, del Pozo V, Cardaba B, de Andres B, Gallardo S, del Amo A, Arrieta I, Jurado A, Palomino P, Liu FT, Lahoz C. Galectin-3 down-regulates IL-5 gene expression on different cell types. J Immunol. 1998;161:385–389. [PubMed] [Google Scholar]
- 32.Toscano MA, Bianco GA, Ilarregui JM, Croci DO, Correale J, Hernandez JD, Zwirner NW, Poirier F, Riley EM, Baum LG, Rabinovich GA. Differential glycosylation of TH1, TH2 and TH-17 effector cells selectively regulates susceptibility to cell death. Nat Immunol. 2007;8:825–834. doi: 10.1038/ni1482. [DOI] [PubMed] [Google Scholar]
- 33.Jiang HR, Al Rasebi Z, Mensah-Brown E, Shahin A, Xu D, Goodyear CS, Fukada SY, Liu FT, Liew FY, Lukic ML. Galectin-3 deficiency reduces the severity of experimental autoimmune encephalomyelitis. J Immunol. 2009;182:1167–1173. doi: 10.4049/jimmunol.182.2.1167. [DOI] [PubMed] [Google Scholar]
- 34.St-Pierre C, Manya H, Ouellet M, Clark GF, Endo T, Tremblay MJ, Sato S. Host-soluble galectin-1 promotes HIV-1 replication through a direct interaction with glycans of viral gp120 and host CD4. J Virol. 2011;85:11742–11751. doi: 10.1128/JVI.05351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieters RJ. Inhibition and detection of galectins. Chembiochem. 2006;7:721–728. doi: 10.1002/cbic.200600011. [DOI] [PubMed] [Google Scholar]
- 36.Ingrassia L, Camby I, Lefranc F, Mathieu V, Nshimyumukiza P, Darro F, Kiss R. Anti-galectin compounds as potential anti-cancer drugs. Curr Med Chem. 2006;13:3513–3527. doi: 10.2174/092986706779026219. [DOI] [PubMed] [Google Scholar]
- 37.Oberg CT, Leffler H, Nilsson UJ. Inhibition of galectins with small molecules. Chimia (Aarau) 2011;65:18–23. doi: 10.2533/chimia.2011.18. [DOI] [PubMed] [Google Scholar]
- 38.Lundquist JJ, Toone EJ. The cluster glycoside effect. Chem Rev. 2002;102:555–578. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- 39.Galan MC, Dumy P, Renaudet O. Multivalent glyco(cyclo)peptides. Chem Soc Rev. 2013;42:4599–4612. doi: 10.1039/c2cs35413f. [DOI] [PubMed] [Google Scholar]
- 40.Martinez A, Ortiz Mellet C, Garcia Fernandez JM. Cyclodextrin-based multivalent glycodisplays: covalent and supramolecular conjugates to assess carbohydrate-protein interactions. Chem Soc Rev. 2013;42:4746–4773. doi: 10.1039/c2cs35424a. [DOI] [PubMed] [Google Scholar]
- 41.Hatano K, Matsuoka K, Terunuma D. Carbosilane glycodendrimers. Chem Soc Rev. 2013;42:4574–4598. doi: 10.1039/c2cs35421g. [DOI] [PubMed] [Google Scholar]
- 42.Sansone F, Casnati A. Multivalent glycocalixarenes for recognition of biological macromolecules: glycocalyx mimics capable of multitasking. Chem Soc Rev. 2013;42:4623–4639. doi: 10.1039/c2cs35437c. [DOI] [PubMed] [Google Scholar]
- 43.Miura Y. Design and synthesis of well-defined glycopolymers for the control of biological functionalities. Polymer Journal. 2012;44:679–689. [Google Scholar]
- 44.Godula K, Bertozzi CR. Density variant glycan microarray for evaluating cross-linking of mucin-like glycoconjugates by lectins. J Am Chem Soc. 2012;134:15732–15742. doi: 10.1021/ja302193u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauerzapfe B, Krenek K, Schmiedel J, Wakarchuk WW, Pelantova H, Kren V, Elling L. Chemo-enzymatic synthesis of poly-N-acetyllactosamine (poly-LacNAc) structures and their characterization for CGL2-galectin-mediated binding of ECM glycoproteins to biomaterial surfaces. Glycoconj J. 2009;26:141–159. doi: 10.1007/s10719-008-9172-2. [DOI] [PubMed] [Google Scholar]
- 46.Beer MV, Rech C, Gasteier P, Sauerzapfe B, Salber J, Ewald A, Moller M, Elling L, Groll J. The next step in biomimetic material design: poly-LacNAc-mediated reversible exposure of extra cellular matrix components. Adv Healthc Mater. 2013;2:306–311. doi: 10.1002/adhm.201200080. [DOI] [PubMed] [Google Scholar]
- 47.Collier JH, Rudra JS, Gasiorowski JZ, Jung JP. Multi-component extracellular matrices based on peptide self-assembly. Chem Soc Rev. 2010;39:3413–3424. doi: 10.1039/b914337h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aida T, Meijer EW, Stupp SI. Functional supramolecular polymers. Science. 2012;335:813–817. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galler KM, Aulisa L, Regan KR, D'Souza RN, Hartgerink JD. Self-assembling multidomain peptide hydrogels: designed susceptibility to enzymatic cleavage allows enhanced cell migration and spreading. J Am Chem Soc. 2010;132:3217–3223. doi: 10.1021/ja910481t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou M, Smith AM, Das AK, Hodson NW, Collins RF, Ulijn RV, Gough JE. Self-assembled peptide-based hydrogels as scaffolds for anchorage-dependent cells. Biomaterials. 2009;30:2523–2530. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Loo Y, Zhang S, Hauser CA. From short peptides to nanofibers to macromolecular assemblies in biomedicine. Biotechnol Adv. 2012;30:593–603. doi: 10.1016/j.biotechadv.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Woolfson DN, Mahmoud ZN. More than just bare scaffolds: towards multi-component and decorated fibrous biomaterials. Chem Soc Rev. 2010;39:3464–3479. doi: 10.1039/c0cs00032a. [DOI] [PubMed] [Google Scholar]
- 53.Li XM, Kuang Y, Xu B. “Molecular trinity” for soft nanomaterials: integrating nucleobases, amino acids, and glycosides to construct multifunctional hydrogelators. Soft Matter. 2012;8:2801–2806. doi: 10.1039/C2SM06920B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giano MC, Pochan DJ, Schneider JP. Controlled biodegradation of self-assembling beta-hairpin peptide hydrogels by proteolysis with matrix metalloproteinase-13. Biomaterials. 2011;32:6471–6477. doi: 10.1016/j.biomaterials.2011.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudalla GA, Sun T, Gasiorowski JZ, Han H, Tian YF, Chong AS, Collier JH. Gradated assembly of multiple proteins into supramolecular nanomaterials. Nat Mater. 2014 doi: 10.1038/nmat3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung JP, Nagaraj AK, Fox EK, Rudra JS, Devgun JM, Collier JH. Co-assembling peptides as defined matrices for endothelial cells. Biomaterials. 2009;30:2400–2410. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, Pompano RR, Santiago FW, Maillat L, Sciammas R, Sun T, Han H, Topham DJ, Chong AS, Collier JH. The use of self-adjuvanting nanofiber vaccines to elicit high-affinity B cell responses to peptide antigens without inflammation. Biomaterials. 2013;34:8776–8785. doi: 10.1016/j.biomaterials.2013.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rudra JS, Tian YF, Jung JP, Collier JH. A self-assembling peptide acting as an immune adjuvant. Proc Natl Acad Sci U S A. 2010;107:622–627. doi: 10.1073/pnas.0912124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shylaja M, Seshadri HS. Glycoproteins - an Overview. Biochem Educ. 1989;17:170–178. [Google Scholar]
- 60.Collier JH, Messersmith PB. Enzymatic modification of self-assembled peptide structures with tissue transglutaminase. Bioconjug Chem. 2003;14:748–755. doi: 10.1021/bc034017t. [DOI] [PubMed] [Google Scholar]
- 61.Ban L, Mrksich M. On-chip synthesis and label-free assays of oligosaccharide arrays. Angew Chem Int Edit. 2008;47:3396–3399. doi: 10.1002/anie.200704998. [DOI] [PubMed] [Google Scholar]
- 62.Gasiorowski JZ, Collier JH. Directed intermixing in multicomponent self-assembling biomaterials. Biomacromolecules. 2011;12:3549–3558. doi: 10.1021/bm200763y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pace KE, Hahn HP, Baum LG. Preparation of recombinant human galectin-1 and use in T-cell death assays. Method Enzymol. 2003;363:499–518. doi: 10.1016/S0076-6879(03)01075-9. [DOI] [PubMed] [Google Scholar]
- 64.Hudalla GA, Modica JA, Tian YF, Rudra JS, Chong AS, Sun T, Mrksich M, Collier JH. Self-Adjuvanting Supramolecular Vaccine Carrying a Folded Protein Antigen. Advanced Healthcare Materials. 2013;2:1114–1119. doi: 10.1002/adhm.201200435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buskas T, Ingale S, Boons GJ. Glycopeptides as versatile tools for glycobiology. Glycobiology. 2006;16:113R–136R. doi: 10.1093/glycob/cwj125. [DOI] [PubMed] [Google Scholar]
- 66.Yu H, Chokhawala H, Karpel R, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- 67.Horlacher T, Oberli MA, Werz DB, Krock L, Bufali S, Mishra R, Sobek J, Simons K, Hirashima M, Niki T, Seeberger PH. Determination of carbohydrate-binding preferences of human galectins with carbohydrate microarrays. Chembiochem. 2010;11:1563–1573. doi: 10.1002/cbic.201000020. [DOI] [PubMed] [Google Scholar]
- 68.Nagata Y, Burger MB. Wheat Germ Agglutinin: Molecular Characteristics and Specificity for Sugar Binding. J Biol Chem. 1974;249:3116–3122. [PubMed] [Google Scholar]
- 69.Goldstein IJ, Poretz RD. Isolation, Physicochemical Characterization, and Carbohydrate-binding Specificity of Lectins. In: Liener IE, Sharon N, Goldstein IJ, editors. The Lectins: Properties, Functions, and Applications in Biology and Medicine, eds. Academic Press; 1986. pp. 103–115. [Google Scholar]
- 70.Garcia I, Sanchez-Iglesias A, Henriksen-Lacey M, Grzelczak M, Penades S, Liz-Marzan LM. Glycans as Biofunctional Ligands for Gold Nanorods: Stability and Targeting in Protein-Rich Media. J Am Chem Soc. 2015;137:3686–3692. doi: 10.1021/jacs.5b01001. [DOI] [PubMed] [Google Scholar]
- 71.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16:137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 72.Banh A, Zhang J, Cao H, Bouley DM, Kwok S, Kong C, Giaccia AJ, Koong AC, Le QT. Tumor galectin-1 mediates tumor growth and metastasis through regulation of T-cell apoptosis. Cancer Res. 2011;71:4423–4431. doi: 10.1158/0008-5472.CAN-10-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stannard KA, Collins PM, Ito K, Sullivan EM, Scott SA, Gabutero E, Darren Grice I, Low P, Nilsson UJ, Leffler H, Blanchard H, Ralph SJ. Galectin inhibitory disaccharides promote tumour immunity in a breast cancer model. Cancer Lett. 2010;299:95–110. doi: 10.1016/j.canlet.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 74.Du X, Zhou J, Guvench O, Sangiorgi FO, Li X, Zhou N, Xu B. Supramolecular assemblies of a conjugate of nucleobase, amino acids, and saccharide act as agonists for proliferation of embryonic stem cells and development of zygotes. Bioconjug Chem. 2014;25:1031–1035. doi: 10.1021/bc500187m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kiyonaka S, Sada K, Yoshimura I, Shinkai S, Kato N, Hamachi I. Semi-wet peptide/protein array using supramolecular hydrogel. Nat Mater. 2004;3:58–64. doi: 10.1038/nmat1034. [DOI] [PubMed] [Google Scholar]
- 76.Komatsu H, Matsumoto S, Tamaru S, Kaneko K, Ikeda M, Hamachi I. Supramolecular hydrogel exhibiting four basic logic gate functions to fine-tune substance release. J Am Chem Soc. 2009;131:5580–5585. doi: 10.1021/ja8098239. [DOI] [PubMed] [Google Scholar]
- 77.Li X, Kuang Y, Shi J, Gao Y, Lin HC, Xu B. Multifunctional, biocompatible supramolecular hydrogelators consist only of nucleobase, amino acid, and glycoside. J Am Chem Soc. 2011;133:17513–17518. doi: 10.1021/ja208456k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jung JH, Amaike M, Shinkai S. Sol-gel transcription of novel sugar-based superstructures composed of sugar-integrated gelators into silica: creation of a lotus-shaped silica structure. Chem Commun. 2000:2343–2344. [Google Scholar]
- 79.Etulain J, Negrotto S, Tribulatti MV, Croci DO, Carabelli J, Campetella O, Rabinovich GA, Schattner M. Control of angiogenesis by galectins involves the release of platelet-derived proangiogenic factors. PLoS One. 2014;9:e96402. doi: 10.1371/journal.pone.0096402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, Leffler H, Smith DF, Cummings RD. Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem. 2008;283:10109–10123. doi: 10.1074/jbc.M709545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Solis D, Mate MJ, Lohr M, Ribeiro JP, Lopez-Merino L, Andre S, Buzamet E, Canada FJ, Kaltner H, Lensch M, Ruiz FM, Haroske G, Wollina U, Kloor M, Kopitz J, Saiz JL, Menendez M, Jimenez-Barbero J, Romero A, Gabius HJ. N-domain of human adhesion/growth-regulatory galectin-9: preference for distinct conformers and non sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution. Int J Biochem Cell Biol. 2010;42:1019–1029. doi: 10.1016/j.biocel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 82.Nagae M, Nishi N, Nakamura-Tsuruta S, Hirabayashi J, Wakatsuki S, Kato R. Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue. J Mol Biol. 2008;375:119–135. doi: 10.1016/j.jmb.2007.09.060. [DOI] [PubMed] [Google Scholar]
- 83.Ramakrishnan B, Shah PS, Qasba PK. alpha-Lactalbumin (LA) stimulates milk beta-1,4-galactosyltransferase I (beta 4Gal-T1) to transfer glucose from UDP-glucose to N-acetylglucosamine. Crystal structure of beta 4Gal-T1 × LA complex with UDP-Glc. J Biol Chem. 2001;276:37665–37671. doi: 10.1074/jbc.M102458200. [DOI] [PubMed] [Google Scholar]