Abstract
The composition of the microbiome in health and disease has only recently become a major research focus. Although it is clear that an imbalance or dysbiosis in the microbiota is associated with disease, its interrelatedness to disease penetrance is largely unknown. Inflammatory bowel disease (IBD) is an excellent disease in which to explore these questions because of the extensive genetic studies identifying disease susceptibility loci and the ability to easily sample the intestinal microbiota in IBD patients due to the accessibility of stool samples. In addition, mouse models of IBD have contributed to our understanding of the interrelatedness of the gut microbiota and genes associated with IBD. The power of the mouse studies is that multiple colitis models exist that can be used in combination with genetically modified mice that harbour deficiencies in IBD susceptibility genes. Collectively, these studies revealed that bacterial dysbiosis does occur in human IBD and in mouse colitis models. In addition, with an emphasis on immune genes, the mouse studies provided evidence that specific immune regulatory proteins associated with IBD influence the gut microbiota in a manner consistent with disease penetrance. In this review, we will discuss studies in both humans and mice that demonstrate the impact of immunodeficiences in interleukin-10, interleukin-17, nucleotide-binding oligomerization domain (NOD) 2, NOD-like receptor proteins 3 and 6, Toll-like receptor or IgA have on the interrelatedness between the composition of the gut microbiota and disease penetrance of IBD and its mouse models.
Keywords: colitis, dybiosis, immunodeficiency, inflammatory bowel disease, microbiota
Introduction
It has long been appreciated that the host microbiota can influence disease with an emphasis on infectious disease due to either the outgrowth of a pathogenic commensal or the introduction of a new bacterial species. More recently, science has embraced the concept that the host and its microbiota are in balance, existing in a symbiotic relationship. Although all mucosal surfaces and the skin harbour commensals, the largest bacterial load exists in the intestinal tract. Indeed, the intestinal tract harbours up to 1014 total bacteria, with the total human microbiota making up at least 90% of the total cellular content of the human body.1,2 Imbalance or dysbiosis in the microbiota between the abundance of beneficial and harmful organisms can either contribute to disease or are a consequence of disease. The cause and effect of dysbiosis in disease pathogenesis has been difficult to discern. This question has perhaps been best studied in inflammatory bowel disease (IBD) in humans, which is manifested in two forms: Crohn's disease (CD) and ulcerative colitis (UC).3 The environment, through its impact on the gut microbiota, is thought to be a major driver of IBD in genetically susceptible individuals.3,4 The analysis of alterations in the microbiota in IBD has largely been accomplished by measuring the bacterial load and composition in faeces using high throughput 16S rRNA sequencing. 16S rRNA is exclusively expressed in prokaryotes and is evolutionarily conserved allowing for PCR expansion of all species of bacterial DNA without background from the mammalian genome.5,6 In IBD, increases in the Gram-negative anaerobes Enterobacteriaceae and Bacteroides among others have been observed.4,7–10 In addition, IBD has been associated with decreased abundance of the presumably beneficial bacteria Bifidobacteria.9,11 Hence, it is not clear whether the gain of potentially pathogenic bacteria or the loss of protective species contributes more to susceptibility and penetrance of IBD.
It is clear that the environment influences the gut microbiota, but genetic susceptibility is an important contributor determining whether gut inflammation driven by bacteria will lead to IBD. More than 160 IBD susceptibility loci have been identified, with pathways associated with microbial sensing and immune regulatory function being highly represented.3,12 IBD is a highly variable disease that can effect any area of the intestinal tract from the mouth to the anus.3 This difference in disease penetrance is evident from studies in monozygotic twins, whereby concordance is only 10–20% for UC and 50–75% for CD.3,13–15 In this review, we discuss the impact of immunodeficiencies in genes associated with IBD on the composition of the microbiome and their collective impact on colitis disease penetrance in both humans and mice.
Interleukin-10 is essential for gut homeostasis and the prevention of colitis
Interleukin-10 (IL-10) is a potent anti-inflammatory cytokine that is essential for intestinal homeostasis.16 This is evidenced by the development of spontaneous enterocolitis in IL-10-deficient mice.17 In the gut, IL-10 is produced by a number of cells including B cells, T cells and macrophages as well as intestinal epithelial cells (IEC).16 The cellular sources deemed of most importance in suppression of intestinal inflammation by IL-10 are CD4+ T regulatory (Treg) cells and IEC (Fig.1).18–23 In the gut, two distinct populations of IL-10-producing Treg cells exist that accumulate at sites of inflammation.16 The first expresses Foxp3 and conditional deletion of IL-10 in Foxp3+ cells resulted in spontaneous enterocolitis.24 In inflammation associated with colitis IL-10 plays an additional role whereby IL-10 receptor (IL-10R) signalling in Treg cells is essential for the maintenance of Foxp3 expression.25 The second population are termed Tr1 cells, which do not express Foxp3 and are induced via chronic T-cell activation at sites of inflammation in the presence of IL-27 and IL-10.26–28 However, unlike Foxp3+ Treg cells, a specific role for Tr1 cells in either preventing or treating colitis has not been firmly established using conditional ablation.
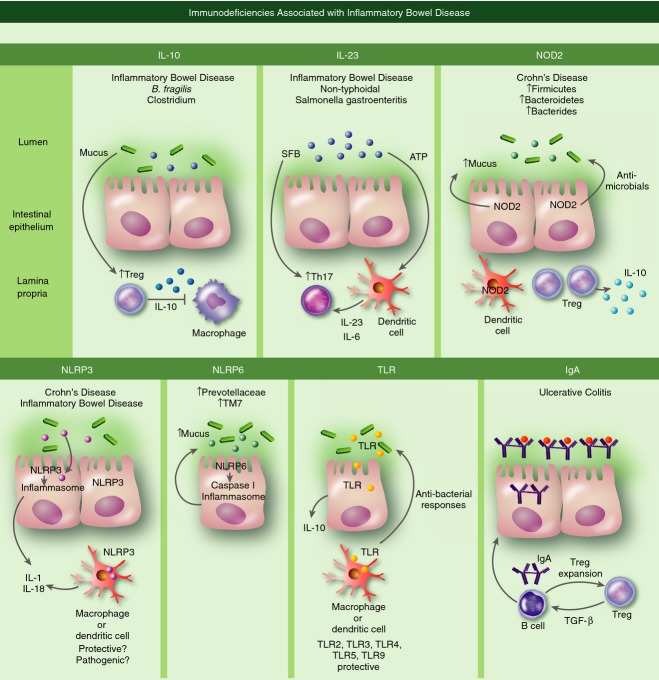
Figure 1.
Immune mechanisms controlling gut homeostasis. Interleukin-10 (IL-10): Humans with immune deficiencies in IL-10 signalling have increased susceptibility to inflammatory bowel disease (IBD). Gut bacteria can promote the homeostasis of regulatory T (Treg) cells that generate anti-inflammatory IL-10 important in dampening macrophage activation. IL-23: Humans with immune deficiencies in IL-23 signalling have increased susceptibility to IBD. IL-23 and IL-6 produced by dendritic cells in response to ATP generated by gut bacteria are important cytokines in maintaining the stability of T helper type 17 (Th17) cells. Th17 cells can be induced by segmented filamentous bacterium (SFB) present in the gut microbiota. Nucleotide-binding oligomerization domain 2 (NOD2): Loss of function mutations in NOD2 increase susceptibility to Crohn's disease in humans. NOD2 present in intestinal epithelial cells promotes their production of mucus and anti-microbials. NOD2 is also associated with the expansion of dendritic cells and Treg cells that presumably generate protective IL-10. NOD-like receptor protein 3 (NLRP3): Polymorphisms in the regulatory region of the NLRP3 gene and in genes encoding the IL-10R and IL-18R are linked to increased susceptibility to IBD. Microbial signalling activates the NLRP3 inflammasome that cleaves pro-IL-1 and IL-18. Experimental data in mouse models of IBD are conflicting regarding whether NLRP3 deficiency is pathogenic or protective. NLRP6: NLRP6 activates the caspase 1 inflammasome and is thought to regulate mucus production. Toll-like receptor (TLR): TLR signalling can lead to the generation of IL-10. Studies in mice deficient in TLR2, TLR3, TLR4, TLR5 or TLR9 indicate that they are protective in mouse models of IBD. IgA: Humans immunodeficient in IgA production can have increased risk for ulcerative colitis (UC). IgA is produced by B cells in the lamina propria in response to transforming growth factor-β (TGF-β) production by Treg cells. B cells in turn promote Treg expansion. Secretory IgA crosses the endothelial cell layer where it binds bacteria in the lumen.
The IL-10R is a heterodimer composed of the IL-10R1 and IL-10RB subunits and downstream signalling occurs via activation of the Janus kinase-signal transducer and activator of transcription (STAT) signalling pathway, with STAT3 being of importance in anti-inflammatory signalling in macrophages.29 Deletion of either IL-10RB or STAT3 in myeloid cells rendered mice susceptible to spontaneous enterocolitis.30–33 In humans, although immune deficiencies in IL-10 or its receptor are rare, genome-wide association studies revealed a linkage between susceptibility to IBD and polymorphisms in the IL10 gene (Fig.1).34,35 In addition, loss of function mutations in IL10RA and IL10RB led to early-onset colitis in humans.36–38
In mice immunodeficient in IL-10, a link between the microbiome and colitis disease penetrance is clear. This is evident by the finding that spontaneous enterocolitis occurs in a microbiome-dependent manner and is colony dependent.17,39–41 In particular, the Gram-negative opportunistic pathogen Helicobacter hepaticus has been associated with colitis. Although Helicobacter hepaticus-free Il10−/− mice do not succumb to spontaneous enterocolitis, its introduction is sufficient to drive disease.39,42,43 However, the penetrance of H. hepaticus-induced colitis is dependent upon the genetic background of the mice and the composition of the enteric microbiome.39,41–44 The finding that monoassociation of germ-free Il10−/− mice with H. hepaticus did not lead to colitis indicates that co-colonization with multiple bacteria is required to drive pathogenic inflammatory responses.45,46 Why H. hepaticus monoassociation is not sufficient to drive colitis is not completely understood, but it has been hypothesized to be the result of an immune system defect in the germ-free mice that cannot mount a full inflammatory response because of underdeveloped lymphoid tissues.16,47–51 The immune system defects in germ-free mice are reversed upon colonization with commensal bacteria introduced by co-housing with specific pathogen-free mice.50 In Il10−/− germ-free mice, co-colonization with H. hepaticus and Lactobacillus reuteri resulted in severe typhlocolitis.45 Bilophila wadsworthia, a Gram-negative anaerobic bacteria, has also been associated with colitis induction in Il10−/− mice when fed a diet high in saturated fat due to induction of taurocholic acid.52 Interestingly, monoassociation with Bilophila wadsworthia in Il10−/− germ-free mice was sufficient to induce colitis.52
Susceptibility to colitis is clearly dependent upon the composition of the microbiome in Il10−/− mice, but specific bacterial species also provide protection by altering the composition of specific immune cells in the gut mucosa. Of particular interest are IL-10-producing Foxp3+ Treg cells (Fig.1). In this regard, the daily administration of a probiotic mixture of Bifidobacteria, Lactobacilli and Streptococcus salivarius to mice in the remission period of colitis induced by 2,4,6-trinitrobenzene sulphonic acid (TNBS) resulted in less severe disease following the second course of treatment and was associated with an increase in IL-10 production by lamina propria mononuclear cells.53 However, the cellular source of the IL-10 was not investigated. In a later study, with the capacity to study Foxp3+ cells, it was shown that monoassociation of Bacteroides fragilis in germ-free mice increased the numbers of IL-10-producing CD4+ Foxp3+ Treg cells in the colon, but not in the mesenteric lymph nodes (Fig.1).54 The increase in Treg cells was shown to be dependent upon polysaccharide A expression by B. fragilis and Toll-like receptor (TLR) 2 signalling.54 It was also shown that polysaccharide A was sufficient to increase both Treg numbers and IL-10 and to protect from TNBS-induced colitis.54 The accumulation of Treg cells in the colon is consistent with a study showing that of the various gut-associated lymphoid tissues, Treg numbers were only decreased in the colonic lamina propria in germ-free mice.55 This study also showed that colonization of germ-free mice with Clostridium clusters IV and XIVa led to an accumulation of colonic IL-10-producing CD4+ Foxp3+ Treg cells (Fig.1).55 However, no increase in Treg cells was noted after monoassociation with Lactobacillus or B. fragilis.55 Clostridium-abundant mice also exhibited attenuated disease severity in dextran sodium sulphate (DSS)-induced colitis.55 CD4+ Foxp3+ Treg cells are composed of natural Treg cells that develop in the thymus and inducible Treg cells that differentiate at sites of inflammation in a transforming growth factor(TGF)-β-dependent manner.56–58 The above studies concluded that the increase in Treg cells following colonization with either B. fragilis or Clostridium was due to inducible Treg differentiation.54,55 However, it should be noted that both natural and inducible Treg populations are required to suppress colitis in the CD45RBhigh CD4+ T-cell transfer model.59
The above cumulative studies in Il10−/− and germ-free mice clearly demonstrate the essential role for Treg cells and IL-10 in the maintenance of tolerance in the intestinal tract keeping immune responses in check and thereby preventing colitis. They also highlight the potential for probiotic use of specific bacterial strains via induction of IL-10-producing Treg cells for the suppression of gut inflammation and attenuation of IBD in humans.
IL-17-producing T cells are induced by the microbiota
Interleukin-17 is a pro-inflammatory cytokine important for protection against extracellular pathogens and is produced by T helper type 17 (Th17) CD4+ cells and by additional immune cell types.60 Interleukin-17 immunodeficiencies in humans are associated with recurrent or persistent infection associated with chronic mucocutaneous candidiasis and staphylococcal dermatitis.61 Individuals who are immunodeficient for STAT3 also succumb to similar infections, which is thought to be due to its induction of IL-17 production.62 The Th17 cells are plastic and a key cytokine in their maintenance is IL-23.63–65 In models of colitis, IL-23 has emerged as a pathogenic factor, including in H. hepaticus-induced T-cell-dependent colitis.66–68 Interleukin-23 has been linked to human IBD in genome-wide association studies that identified polymorphisms in the IL-23R gene locus associated with disease susceptibility (Fig.1).69 Humans with primary immunodeficiencies in IL-23-dependent pathways are susceptible to non-typhoidal Salmonella gastroenteritis (Fig.1).70 Complicating the investigation into the exact role of Th17 cells in colitis is their production of multiple cytokines in addition to IL-17 (IL-17A) including IL-17F, IL-21 and IL-22 and granulocyte–macrophage colony-stimulating factor.71,72 In mouse models of colitis, Th17-associated cytokines play both pathogenic and protective roles.71 Given that IL-23 has multiple effects on immune function, it is likely that it also contributes to intestinal inflammation in an IL-17-independent manner.
Although the exact role of Th17 cells in IBD is not clear, their development in the gut is regulated by the commensal segmented filamentous bacterium (SFB) (Fig.1).73 Experimental evidence suggested that the increase in Th17 cells was due to SFB induction of serum amyloid A production in the terminal ileum epithelial cells, which was probably through IL-23 production by dendritic cells promoting Th17 generation (Fig.1).73 In addition, the inclusion of SFB in a bacterial cocktail used to colonize germ-free mice that were subsequently infected with the pathogen Citrobacter rodentium, attenuated colonic inflammation.73. Similarly, another study reported that ATP derived from commensal bacteria promoted the differentiation of Th17 cells through the induction of Th17-promoting cytokines, such as IL-6 and IL-23, by a subset of lamina propria CD11c+ cells (Fig.1).74 Of importance to colitis is the observation that ATP administration exacerbated T-cell-mediated colitis in SCID mice that was associated with an increase in IL-17-producing CD4+ T cells (Fig.1).74 Interestingly, we recently showed that CD11c+ dendritic cells are also implicated in the homeostasis of CD4+ Foxp3+ Treg cells.75 Hence dendritic cells are an important link between Treg and Th17 cells. In addition, IL-17 is very important in preventing skin infections and a similar mechanism whereby the local microbiota promotes IL-17 production has been speculated to promote anti-bacterial functions of keratinocytes.76 This is consistent with skin infections in humans with altered IL-17 immunity.
NLR deficiency alters the composition of the intestinal microbiota
Nucleotide-binding oligomerization domain (NOD)-like receptors (NLR) play an important role in mediating host–microbe interactions in the intestine. NLR are expressed by IEC and immune cells and are important for the recognition of pathogen-associated molecular patterns expressed by microorganisms. Deficiencies in NLR are associated with intestinal inflammation in humans and mice.77 Interestingly, some NLR proteins have been shown to contribute to the homeostatic maintenance of the gut microbiota.
Nucleotide-binding oligomerization domain containing 2 (NOD2), a member of the NLR family, plays an important role in controlling intestinal inflammation. Loss of function of NOD2 results in uncontrolled inflammatory responses and is associated with CD in humans (Fig.1).78–80 In particular, gene variants leading to the inability of NOD2 to sense the bacterial peptidoglycan structure muramyl dipeptide have been implicated in susceptibility to CD.81–83 Interestingly, CD patients with NOD2 mutations were shown to have an altered microbiota composition in the ileum.84 A higher prevalence of Bacteroides, Bacteroidetes and Firmicutes was observed compared with healthy controls (Fig.1).84 In addition, altered frequencies of Faecalibacterium and Escherichia have also been associated with NOD2 risk alleles in CD patients with NOD2 mutations.85
Given the association of NOD2 with CD it was surprising that Nod2−/− mice did not develop spontaneous intestinal inflammation nor exhibit enhanced susceptibility to DSS-colitis86,87. However, in a subsequent study Nod2−/− mice were shown to exhibit exacerbated disease in an antigen-specific model of colitis, which was linked to TLR2 responsiveness.87,88 In addition, Nod2 deficiency resulted in microbial dysbiosis consisting of increased bacterial load in the faeces and ileum along with a significant increase in the abundance of Bacteroidetes and Firmicutes compared with wild-type (WT) mice (Fig.1).84,89,90 The absence of Helicobacter in the mouse colony examined in the Petnicki-Ocwieja study afforded the opportunity to determine that NOD2 plays a role in the clearance of this opportunistic pathogen after colonization by oral gavage.89 Consistent with that finding is the increased susceptibility to H. hepaticus induction of granulomatous inflammation in the ileum of Nod2−/− mice.91 Following a similar theme as with other immunodeficiencies associated with colitis, NOD2 was associated with the production of protective IL-10 and the expansion of dendritic cells and Treg cells (Fig.1).92
As NOD1 shares similar structure and downstream signalling molecules as NOD2,93 and is crucial for controlling Helicobacter infection,94 a compensatory gain of function or increased expression of NOD1 in the absence of NOD2 can be speculated. Other than Helicobacter, NOD1 is also required for the clearance of Clostridium and prevention of bacterial translocation.95 Hence, NOD1-mediated regulation of the microbiota is a possibility. Indeed, the total bacterial burden was increased 100-fold in Nod1−/− mice with significant over-representation of Bacteroides, Clostridiales and Enterobacteriaceae.51 However, it should be noted that NOD1 has not been linked to increased susceptibility to IBD.77
NLRP3 is a member of the NLR family of proteins that contains a pyrin domain and functions to assemble the inflammasome, which is a multimeric complex composed of caspase 1, caspase-5, Pycard and the adaptor protein NALP1/Apoptosisassociated speck-like protein containing CARD adaptor (ASC).96–98 Inflammasomes act as sensors of damage as well as microbe-associated molecular patterns and are responsible for the proteolytic cleavage and maturation of pro-IL-1 and pro-IL-18 (Fig.1).97,98 A single nucleotide polymorphism analysis revealed a set of single nucleotide polymorphisms in the regulatory region of the NLRP3 gene in patients with CD (Fig.1).99 In addition, genetic studies linked polymorphisms in genes encoding both the IL-10R and IL-18R with susceptibility to IBD (Fig.1).77 The role that NLRP3 plays in colitis susceptibility has been examined in mice with contradictory results (Fig.1). In one study, Nlrp3−/− mice were shown to exhibit more severe DSS-induced and TNBS-induced colitis.100 This study also demonstrated that NLRP3 plays a role in controlling commensal overgrowth and bacteraemia as evidenced by an increase in bacterial numbers in the stool, colon, mesenteric lymph node and liver in Nlrp3−/− mice.100 However, the extent to which NLRP3 regulates the microbial composition during the steady state is not clear. This issue was addressed comparing NLlrp3+/+ and NLlrp3−/− littermates with Nlrp3−/− mice harbouring a unique bacterial composition with members of the family Enterobacteriaceae including the species Citrobacter, Proteus and Shigella.101 In addition, increases in the genera Mycobacterium, Collinsella, Subdoligranulum and Clostridium were detected.101 With the large difference in composition of the intestinal microbiota it was not surprising that Nlrp3−/− mice also exhibited increased susceptibility to DSS-induced and TNBS-induced colitis.101 Although the above reports indicate an anti-inflammatory protective role for NLRP3 in colitis, the opposite result was also obtained whereby Nlrp3−/− mice exhibited less severe disease using a similar model of DSS-induced colitis.102 The reason for the difference in disease penetrance in Nlrp3−/− mice is not clear, but could be due to differences in percentage of DSS used in the colitis induction protocol or to differences in the composition of the intestinal microbiota. In particular, the presence of Citrobacter, as noted in the Hirota et al.,101 study, is of interest given that Citrobacter rodentium-induced colitis was more severe in mice lacking NLRP3.103
Similarly to NLRP3, NLRP6 uses the ASC to activate a Caspase-1 inflammasome and Nlrp6−/− mice exhibited more severe DSS-induced colitis.104–108 To determine whether the microbiota influenced disease penetrance in the Nlrp6−/− mice they were co-housed with WT mice, which was sufficient to transfer the severe colitis phenotype.106 To evaluate the potential colitogenic microbiota present in Nlrp6−/− mice, 16S rRNA analysis was performed, demonstrating an increase in abundance of Prevotellaceae and TM7 (Fig.1).106 This same study evaluated Nlrp3−/− mice that upon co-housing with WT mice exhibited attenuated colitis.106 Hence NLRP3- and NLRP6-deficient mice serve as a clear example of how immunodeficiency influences the composition of the microbiota that in turn alters disease penetrance.
Altered microbiota in NLR-deficient mice could potentially be attributed to the anti-bacterial functions of these sentinel proteins that play an important role in host–pathogen interactions. NOD2 signalling, leads to the generation of anti-microbials like β-defensin-2, Human neutrophil peptide 1 (HNP-1) and malignant brain tumors 1 (DMBT1) (Fig.1).109–111 It is thought that NOD2 regulates mucus production, an important anti-microbial mechanism in the gut, via its interaction with Polypeptide N-Acetylgalactosaminyltransferase 2 (GALNT2) (Fig.1).112 Similarly, a recent study reported a role for NLRP6 in regulating mucus production (Fig.1).113 Goblet cell autophagy was impaired in Nlrp6−/− mice, resulting in abrogated mucus secretion.113 Hence, impaired host defence in the absence of NLR proteins might lead to an expansion of pathogenic bacteria and their systemic dissemination that could trigger an inflammatory response via signalling through TLR.
TLR are protective in colitis
TLR are a family of pattern recognition receptors each recognizing distinct pathogen-associated molecular patterns.114 Genome-wide association studies studies have not specifically identified polymorphisms in TLR genes as susceptibility loci for IBD.115 However, the TLR2-R753Q variant alters disease penetrance, conferring increased risk to develop pancolitis.116 A dominant-negative TLR5 variant was shown to confer resistance to development of CD.117 In addition, a single nucleotide polymorphism in TLR9 has been linked to CD-associated variants in other genes.118 Although the Tlr4 gene is localized to a CD susceptibility locus, strong evidence does not exist for its role in IBD penetrance.119 When the expression of TLR was assessed in IEC it was shown that non-IBD samples expressed high levels of TLR3 and TLR5 and low levels of TLR2 and TLR4.119,120 In IBD patients, TLR2 and TRL5 levels were unchanged and TLR3 either remained the same or was down-regulated depending on the IBD disease state, whereas TLR4 was greatly increased.120 Hence, although TLR are important in sensing of microorganisms in the gut the extent to which they modulate disease penetrance in IBD remains unresolved.
TLR2, TLR4 and TLR5 all signal through the adaptor protein Myd88, whereas TLR3 does not and TLR4 also signals in a Myd88-independent manner.119 To determine whether Myd88 is required for colitis induction, DSS-colitis was induced in Myd88−/− mice with the finding that they exhibited more severe disease.121 These data suggest that TLR are protective. When individual TLR were examined for DSS-colitis penetrance; mice deficient in TLR2, TLR3, TLR4, TLR5 or TLR9 all exhibited more severe disease (Fig.1).122–127 A role for multiple TLR in protection against DSS-colitis is consistent with each TLR sensing different bacterial pathogen-associated molecular patterns.
Although it is not clear how individual TLR confer protection, bacterial up-regulation of IL-10 production via TLR signalling in IEC as well as in innate and adaptive immune cells is a likely mechanism (Fig.1).16,128,129 Another possible mechanism is that immunodeficiencies in specific TLR that alter sensing of particular bacterial species could lead to an alteration in the composition of the intestinal microbiota. Indeed, Tlr5−/− mice were shown to have a phylum-level shift in which 116 bacterial phylotypes were either enriched or reduced, with no amplification or loss of a particular species.130 In addition, Tlr5−/− mice with transiently unstable microbiotas containing increases in proteobacteria exhibited colitis as compared to their non-colitic siblings, which stabilized their microbiotas resembling those of WT mice.131 In particular, increased levels of the enterobacteria species Escherichia coli in close proximity to the gut epithelium was a feature of colitis.131 Extensive studies have linked adherent-invasive E. coli (AIEC) with CD, but not in the context of a human immunodeficiency.132 In germ-free Tlr5−/− mice, monoassociation with the CD-associated AIEC LF82 strain resulted in moderate colitis.131 To determine whether commensal bacteria contribute to the virulence of AIEC LF82 germ-free mice were moved to specific pathogen-free housing before introduction of AIEC LF82 to the gut microbiome, after which they developed spontaneous colitis with weight loss and diarrhoea as early as 3 days and was associated with decreased clearance of the bacteria.131 Spontaneous disease did not occur in WT mice or following introduction of the commensal flagellate E. coli strain F-18 to Tlr5−/− mice.131 Overall, the above study demonstrated that one likely protective mechanism of TLR5 in Crohn's disease is through its recognition of AIEC flagellin promoting its clearance.131
IgA deficiency alters the gut microbiota
An important adaptive immune mechanism in gut homeostasis at the mucosal surface is B-cell secretion of IgA that functions to neutralize pathogens and microbial toxins and by confining commensals within the intestinal lumen.133,134 Secretory IgA produced in the lamina propria can cross the epithelial layer and be deposited in the gut lumen where it either diffuses or binds to the IEC mucus layer (Fig.1).133 IgA deficiency is the most prevalent primary immunodeficiency and is characterized by low or absent serum IgA.133 While up to 85–90% of IgA-deficient individuals remain asymptomatic, a number of gastrointestinal infections and disorders, including UC, have been observed (Fig.1).133 In mice, IgA deficiency resulted in more severe DSS-induced colitis.135 IgA production is dependent upon TGF-β, which we and others showed could be provided by CD4+ Foxp3+ Treg cells (Fig.1).136–138 In addition, we recently showed that mice deficient in B cells exhibit more severe DSS-induced colitis.138 In this same study, consistent with our previous observations, we found that B cells confer protection by inducing proliferation of Treg cells that in turn promote IgA isotype class switching likely through their production of TGF-β (Fig.1).138–141 In addition, we found no role for B-cell production of IL-10 in our study, consistent with Treg cells as the primary source of IL-10.138
Although IgA is crucial for regulating microbes in the gut, conversely, the microbiota promotes IgA production.142,143 Interestingly, IgA deficiency led to an aberrant microbial composition in the gut. In activation-induced cytidine deaminase (Aid)-deficient mice, B cells failed to undergo class switch recombination, resulting in a complete loss of IgA production.144 Aid−/− mice harboured 100-fold more anaerobic flora in the small intestine compared with WT mice.145 A later study further evaluated dysbiosis in Aid−/− mice and found increased prevalence of SFB and Clostridium species in the small intestine, which was reversed with IgA.146 Although dysbiosis in the absence of IgA is evident from these studies, one outstanding question is whether IgA preferentially binds to and limits certain microbes in the gut (Fig.1). Recently, specificity of IgA towards Enterobacteriaceae in the microbiota of mice was reported.135 It was shown that the microbiota in newborn mice was dominated by Enterobacteriaceae, which was replaced by Bacteroidetes and Firmicutes in the adult.135 In the absence of IgA, the microbiota transition from the immature to mature stage did not occur, resulting in a higher prevalence of Enterobacteriaceae, which may contribute to more severe DSS-induced colitis.135 Interestingly, changes in the IgA repertoire due to deficiency in PD-1 also resulted in an increase in Enterobacteriaceae.141 Similarly, a reduction in high-affinity IgA production due to a deficiency in Myd88 specifically in T cells led to an altered microbial composition in the gut.147 From these studies it is apparent that IgA plays an important role in shaping the microbiota by restraining certain bacteria. It is tempting to speculate that IgA specifically neutralizes potential host–microbe equilibrium-altering and inflammation-promoting bacteria. Indeed, in a recent study it was found that colitogenic intestinal bacteria were highly coated with IgA in mice (Fig.1).148 Furthermore, faecal bacteria with high IgA coating isolated from IBD patients upon transfer to germ-free mice conferred susceptibility to DSS-induced colitis.148
Concluding remarks
The above collective studies support the concept of co-evolution of the host immune system and the microbiota for maintaining homeostasis at the mucosal surface. In addition, it is clear that the immunological state of the intestinal tract is influenced by both host immune factors and the composition of the intestinal microbiota. Alterations in a variety of different immune regulatory loops are able to alter the gut homeostasis balance leading to dysbiosis and colitis-associated inflammation. Further insight into how and which species of the microbiota are affected by immunodeficiencies associated with IBD are still needed to determine which bacteria are pathogenic and which are protective. Although probiotics are considered a viable therapy for IBD, it still not clear how or whether they can reset the correct bacterial balance thereby alleviating inflammation in ongoing disease. This is particularly challenging because of differences in genetic susceptibility in each IBD patient that have the potential to uniquely alter the gut microbiota.
Acknowledgments
This work was supported by National Institutes of Health grants R01 AI069358-01A2, R56 AI106672-01 and the Blood Center Research Foundation.
Disclosures
The authors have no conflicts of interest.
References
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–95. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–70. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol. 2015;50:495–507. doi: 10.1007/s00535-015-1064-1. [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–90. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Vandamme P. Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol Lett. 2003;228:45–9. doi: 10.1016/S0378-1097(03)00717-1. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenbergen S, Samsom JN. Maintenance of small intestinal and colonic tolerance by IL-10-producing regulatory T cell subsets. Curr Opin Immunol. 2012;24:269–76. doi: 10.1016/j.coi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004;53:1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–92. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier C, Neut C, Mizon C, Cortot A, Colombel JF, Mizon J. Fecal βd-galactosidase production and Bifidobacteria are decreased in Crohn's disease. Dig Dis Sci. 1997;42:817–22. doi: 10.1023/a:1018876400528. [DOI] [PubMed] [Google Scholar]
- Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–10. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant SR. Update on the heritability of inflammatory bowel disease: the importance of twin studies. Inflamm Bowel Dis. 2011;17:1–5. doi: 10.1002/ibd.21385. [DOI] [PubMed] [Google Scholar]
- Halfvarson J, Bodin L, Tysk C, Lindberg E, Jarnerot G. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124:1767–73. doi: 10.1016/s0016-5085(03)00385-8. [DOI] [PubMed] [Google Scholar]
- Spehlmann ME, Begun AZ, Burghardt J, Lepage P, Raedler A, Schreiber S. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14:968–76. doi: 10.1002/ibd.20380. [DOI] [PubMed] [Google Scholar]
- Kole A, Maloy KJ. Control of intestinal inflammation by interleukin-10. Curr Top Microbiol Immunol. 2014;380:19–38. doi: 10.1007/978-3-662-43492-5_2. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- Maloy KJ, Salaun L, Cahill R, Dougan G, Saunders NJ, Powrie F. CD4+CD25+ TR cells suppress innate immune pathology through cytokine-dependent mechanisms. J Exp Med. 2003;197:111–9. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Hu B, Xu D, Liew FY. CD4+CD25+ regulatory T cells cure murine colitis: the role of IL-10, TGF-β, and CTLA4. J Immunol. 2003;171:5012–7. doi: 10.4049/jimmunol.171.10.5012. [DOI] [PubMed] [Google Scholar]
- Uhlig HH, Coombes J, Mottet C, Izcue A, Thompson C, Fanger A, et al. Characterization of Foxp3+CD4+CD25+ and IL-10-secreting CD4+CD25+ T cells during cure of colitis. J Immunol. 2006;177:5852–60. doi: 10.4049/jimmunol.177.9.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AO, Beiting DP, Tato C, John B, Oldenhove G, Lombana CG, et al. The cytokines interleukin 27 and interferon-γ promote distinct Treg cell populations required to limit infection-induced pathology. Immunity. 2012;37:511–23. doi: 10.1016/j.immuni.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A, Siewe L, Strittmatter E, Deckert M, Schluter D, Stenzel W, et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med. 2004;200:1289–97. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, Neves JF, Dowds CM, Baker K, Glickman J, Davidson NO, et al. Protective mucosal immunity mediated by epithelial CD1d and IL-10. Nature. 2014;509:497–502. doi: 10.1038/nature13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–58. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Murai M, Turovskaya O, Kim G, Madan R, Karp CL, Cheroutre H, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–84. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groux H, O'Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–42. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol. 2007;8:1380–9. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- Wakkach A, Fournier N, Brun V, Breittmayer JP, Cottrez F, Groux H. Characterization of dendritic cells that induce tolerance and T regulatory 1 cell differentiation in vivo. Immunity. 2003;18:605–17. doi: 10.1016/s1074-7613(03)00113-4. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, et al. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187:571–8. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouval DS, Biswas A, Goettel JA, McCann K, Conaway E, Redhu NS, et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40:706–19. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim KW, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency causes severe spontaneous colitis. Immunity. 2014;40:720–33. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- Moran CJ, Walters TD, Guo CH, Kugathasan S, Klein C, Turner D, et al. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2013;19:115–23. doi: 10.1002/ibd.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, et al. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am J Gastroenterol. 2011;106:1544–55. doi: 10.1038/ajg.2011.112. [DOI] [PubMed] [Google Scholar]
- Kotlarz D, Beier R, Murugan D, Diestelhorst J, Jensen O, Boztug K, et al. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology. 2012;143:347–55. doi: 10.1053/j.gastro.2012.04.045. [DOI] [PubMed] [Google Scholar]
- Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS ONE. 2013;8:e70783. doi: 10.1371/journal.pone.0070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Hao L, Medzhitov R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25:319–29. doi: 10.1016/j.immuni.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and γ-interferon-dependent mechanism. Infect Immun. 1998;66:5157–66. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler G, Wos-Oxley ML, Smoczek A, Zschemisch NH, Neumann D, Pieper DH, et al. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota–host interactions. Inflamm Bowel Dis. 2012;18:943–54. doi: 10.1002/ibd.21895. [DOI] [PubMed] [Google Scholar]
- Dieleman LA, Arends A, Tonkonogy SL, Goerres MS, Craft DW, Grenther W, et al. Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect Immun. 2000;68:5107–13. doi: 10.1128/iai.68.9.5107-5113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whary MT, Taylor NS, Feng Y, Ge Z, Muthupalani S, Versalovic J, et al. Lactobacillus reuteri promotes Helicobacter hepaticus -associated typhlocolitis in gnotobiotic B6.129P2-IL-10(tm1Cgn) (IL-10–/–) mice. Immunology. 2011;133:165–78. doi: 10.1111/j.1365-2567.2011.03423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingam NA, Robinson CJ, Bergin IL, Eaton KA, Huffnagle GB, Young VB. The effects of intestinal microbial community structure on disease manifestation in IL-10–/– mice infected with Helicobacter hepaticus. Microbiome. 2013;1:15. doi: 10.1186/2049-2618-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol. 1963;42:471–83. [PMC free article] [PubMed] [Google Scholar]
- Benveniste J, Lespinats G, Adam C, Salomon JC. Immunoglobulins in intact, immunized, and contaminated axenic mice: study of serum IgA. J Immunol. 1971;107:1647–55. [PubMed] [Google Scholar]
- Manolios N, Geczy CL, Schrieber L. High endothelial venule morphology and function are inducible in germ-free mice: a possible role for interferon-γ. Cell Immunol. 1988;117:136–51. doi: 10.1016/0008-8749(88)90083-4. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–85. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–10. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-β-bearing regulatory cells. J Immunol. 2005;174:3237–46. doi: 10.4049/jimmunol.174.6.3237. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–6. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–61. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, et al. Conversion of peripheral CD4+CD25– naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–86. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D, Lin W, Edwards B, Ziegelbauer J, Salzman NH, Carlson MR, et al. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J Immunol. 2009;182:3461–8. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–8. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH, Jakobsen MA, Larsen CS. Identification of a novel STAT3 mutation in a patient with hyper-IgE syndrome. Scand J Infect Dis. 2013;45:235–8. doi: 10.3109/00365548.2012.715750. [DOI] [PubMed] [Google Scholar]
- Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–4. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–40. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–6. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, Stepankova R, et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–18. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med. 2006;203:2485–94. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinez I, Keestra AM, Spees A, Baumler AJ. The IL-23 axis in Salmonella gastroenteritis. Cell Microbiol. 2011;13:1639–47. doi: 10.1111/j.1462-5822.2011.01637.x. [DOI] [PubMed] [Google Scholar]
- Morrison PJ, Ballantyne SJ, Kullberg MC. Interleukin-23 and T helper 17-type responses in intestinal inflammation: from cytokines to T-cell plasticity. Immunology. 2011;133:397–408. doi: 10.1111/j.1365-2567.2011.03454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, et al. The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12:568–75. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, et al. ATP drives lamina propria TH17 cell differentiation. Nature. 2008;455:808–12. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Ray A, Basu S, Miller NM, Chan AM, Dittel BN. An increase in tolerogenic dendritic cell and natural regulatory T cell numbers during experimental autoimmune encephalomyelitis in Rras−/− mice results in attenuated disease. J Immunol. 2014;192:5109–17. doi: 10.4049/jimmunol.1302254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–9. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- Rubino SJ, Selvanantham T, Girardin SE, Philpott DJ. Nod-like receptors in the control of intestinal inflammation. Curr Opin Immunol. 2012;24:398–404. doi: 10.1016/j.coi.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–12. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- Li J, Moran T, Swanson E, Julian C, Harris J, Bonen DK, et al. Regulation of IL-8 and IL-1β expression in Crohn's disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13:1715–25. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–8. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–62. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–84. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–4. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, et al. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest. 2008;118:545–59. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kitani A, Murray PJ, Wakatsuki Y, Fuss IJ, Strober W. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity. 2006;25:473–85. doi: 10.1016/j.immuni.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci USA. 2009;106:15813–8. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondot S, Barreau F, Al Nabhani Z, Dussaillant M, Le Roux K, Dore J, et al. Altered gut microbiota composition in immune-impaired Nod2–/– mice. Gut. 2012;61:634–5. doi: 10.1136/gutjnl-2011-300478. [DOI] [PubMed] [Google Scholar]
- Biswas A, Liu YJ, Hao L, Mizoguchi A, Salzman NH, Bevins CL, et al. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc Natl Acad Sci USA. 2010;107:14739–44. doi: 10.1073/pnas.1003363107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho Fernandez E, Valenti V, Rockel C, Hermann C, Pot B, Boneca IG, et al. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60:1050–9. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–74. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Yamazaki T, Kamada N, Tawaratsumida K, Kim YG, Nunez G, et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol. 2011;186:4872–80. doi: 10.4049/jimmunol.1003761. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell. 2002;10:417–26. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–32. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012;13:325–32. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, et al. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–6. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–91. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota SA, Ng J, Lueng A, Khajah M, Parhar K, Li Y, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17:1359–72. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, et al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192–9. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zaki MH, Vogel P, Gurung P, Finlay BB, Deng W, et al. Role of inflammasomes in host defense against Citrobacter rodentium infection. J Biol Chem. 2012;287:16955–64. doi: 10.1074/jbc.M112.358705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-κB and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874–80. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- Grenier JM, Wang L, Manji GA, Huang WJ, Al-Garawi A, Kelly R, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-κB and caspase-1. FEBS Lett. 2002;530:73–8. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186:7187–94. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci USA. 2011;108:9601–6. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss E, Wehkamp J, Wehkamp K, Stange EF, Schroder JM, Harder J. NOD2/CARD15 mediates induction of the antimicrobial peptide human β-defensin-2. J Biol Chem. 2006;281:2005–11. doi: 10.1074/jbc.M511044200. [DOI] [PubMed] [Google Scholar]
- Yamamoto-Furusho JK, Barnich N, Hisamatsu T, Podolsky DK. MDP-NOD2 stimulation induces HNP-1 secretion, which contributes to NOD2 antibacterial function. Inflamm Bowel Dis. 2010;16:736–42. doi: 10.1002/ibd.21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstiel P, Sina C, End C, Renner M, Lyer S, Till A, et al. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol. 2007;178:8203–11. doi: 10.4049/jimmunol.178.12.8203. [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Stevens C, Phillips AM, Smith A, Drummond HE, Noble CL, et al. TLE1 modifies the effects of NOD2 in the pathogenesis of Crohn's disease. Gastroenterology. 2011;141:972–81. doi: 10.1053/j.gastro.2011.05.043. e1–2. [DOI] [PubMed] [Google Scholar]
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, et al. NLRP6 inflammasome orchestrates the colonic host–microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–59. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17:338–44. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Matricon J, Barnich N, Ardid D. Immunopathogenesis of inflammatory bowel disease. Self Nonself. 2010;1:299–309. doi: 10.4161/self.1.4.13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik M, Joossens S, Van Steen K, Van Schuerbeek N, Vlietinck R, Rutgeerts P, et al. Toll-like receptor-1, -2, and -6 polymorphisms influence disease extension in inflammatory bowel diseases. Inflamm Bowel Dis. 2006;12:1–8. doi: 10.1097/01.mib.0000195389.11645.ab. [DOI] [PubMed] [Google Scholar]
- Gewirtz AT, Vijay-Kumar M, Brant SR, Duerr RH, Nicolae DL, Cho JH. Dominant-negative TLR5 polymorphism reduces adaptive immune response to flagellin and negatively associates with Crohn's disease. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1157–63. doi: 10.1152/ajpgi.00544.2005. [DOI] [PubMed] [Google Scholar]
- Torok HP, Glas J, Endres I, Tonenchi L, Teshome MY, Wetzke M, et al. Epistasis between Toll-like receptor-9 polymorphisms and variants in NOD2 and IL23R modulates susceptibility to Crohn's disease. Am J Gastroenterol. 2009;104:1723–33. doi: 10.1038/ajg.2009.184. [DOI] [PubMed] [Google Scholar]
- Cario E. Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm Bowel Dis. 2010;16:1583–97. doi: 10.1002/ibd.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. 2000;68:7010–7. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki A, Kanai T, Ishikura T, Makita S, Uraushihara K, Iiyama R, et al. MyD88-deficient mice develop severe intestinal inflammation in dextran sodium sulfate colitis. J Gastroenterol. 2005;40:16–23. doi: 10.1007/s00535-004-1492-9. [DOI] [PubMed] [Google Scholar]
- Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology. 2007;132:1359–74. doi: 10.1053/j.gastro.2007.02.056. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Wu H, Aitken J, Kolachala VL, Neish AS, Sitaraman SV, et al. Activation of toll-like receptor 3 protects against DSS-induced acute colitis. Inflamm Bowel Dis. 2007;13:856–64. doi: 10.1002/ibd.20142. [DOI] [PubMed] [Google Scholar]
- Fukata M, Michelsen KS, Eri R, Thomas LS, Hu B, Lukasek K, et al. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1055–65. doi: 10.1152/ajpgi.00328.2004. [DOI] [PubMed] [Google Scholar]
- Ivison SM, Himmel ME, Hardenberg G, Wark PA, Kifayet A, Levings MK, et al. TLR5 is not required for flagellin-mediated exacerbation of DSS colitis. Inflamm Bowel Dis. 2010;16:401–9. doi: 10.1002/ibd.21097. [DOI] [PubMed] [Google Scholar]
- Koon HW, Shih DQ, Chen J, Bakirtzi K, Hing TC, Law I, et al. Cathelicidin signaling via the Toll-like receptor protects against colitis in mice. Gastroenterology. 2011;141:1852–63. doi: 10.1053/j.gastro.2011.06.079. e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilewitz D, Katakura K, Karmeli F, Hayashi T, Reinus C, Rudensky B, et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. 2004;126:520–8. doi: 10.1053/j.gastro.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Levast B, Li Z, Madrenas J. The role of IL-10 in microbiome-associated immune modulation and disease tolerance. Cytokine. 2015;75:291–301. doi: 10.1016/j.cyto.2014.11.027. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Matsuoka K, Sheikh SZ, Russo SM, Mishima Y, Collins C, et al. IL-10 regulates Il12b expression via histone deacetylation: implications for intestinal macrophage homeostasis. J Immunol. 2012;189:1792–9. doi: 10.4049/jimmunol.1200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–52. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–8. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- Cerutti A, Chen K, Chorny A. Immunoglobulin responses at the mucosal interface. Annu Rev Immunol. 2011;29:273–93. doi: 10.1146/annurev-immunol-031210-101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–6. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5:28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazac BB, Roes J. TGF-β receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13:443–51. doi: 10.1016/s1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Matsumoto R, Hitoshi Y, Ishii T, Sugimoto M, Araki S, et al. Transforming growth factor β induces IgA production and acts additively with interleukin 5 for IgA production. J Exp Med. 1989;170:1415–20. doi: 10.1084/jem.170.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ray A, Jiang X, Wang JY, Basu S, Liu X, et al. T regulatory cells and B cells cooperate to form a regulatory loop that maintains gut homeostasis and suppresses dextran sulfate sodium-induced colitis. Mucosal Immunol. 2015 doi: 10.1038/mi.2015.20. ; doi: 10.1038/mi.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Basu S, Williams CB, Salzman NH, Dittel BN. A novel IL-10-independent regulatory role for B cells in suppressing autoimmunity by maintenance of regulatory T cells via GITR ligand. J Immunol. 2012;188:3188–98. doi: 10.4049/jimmunol.1103354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. 2009;106:19256–61. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–9. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- Crabbe PA, Bazin H, Eyssen H, Heremans JF. The normal microbial flora as a major stimulus for proliferation of plasma cells synthesizing IgA in the gut. The germ-free intestinal tract. Int Arch Allergy Appl Immunol. 1968;34:362–75. doi: 10.1159/000230130. [DOI] [PubMed] [Google Scholar]
- Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–9. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–7. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA. 2004;101:1981–6. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O'Connell RM, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17:153–63. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–10. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]