Abstract
Natural killer (NK) cells, which can exert early and powerful anti-tumour and anti-viral responses, are important components of the innate immune system. DNAX accessory molecule-1 (DNAM-1) is an activating receptor molecule expressed on the surface of NK cells. Recent findings suggest that DNAM-1 is a critical regulator of NK cell biology. DNAM-1 is involved in NK cell education and differentiation, and also plays a pivotal role in the development of cancer, viral infections and immune-related diseases. However, tumours and viruses have developed multiple mechanisms to evade the immune system. They are able to impair DNAM-1 activity by targeting the DNAM-1 receptor–ligand system. We have reviewed the roles of DNAM-1, and its biological functions, with respect to NK cell biology and DNAM-1 chimeric antigen receptor-based immunotherapy.
Keywords: DNAM-1, DNAM-1 ligands, infections, natural killer cells, tumours
Introduction
The biological characteristics and functions of natural killer (NK) cells, such as their education and differentiation, recognition and activation, killing and immunoregulatory functions have been thoroughly investigated over the decades.1–5 Natural killer cells possess the ability to eliminate malignant transformed cells, virus-infected cells and some stressed cells, without causing damage to healthy cells.6 Upon their activation, NK cells secrete numerous pro-inflammatory cytokines and chemokines, such as interferon-γ (IFN-γ), tumour necrosis factor-α, interleukin-6 (IL-6), granulocyte–macrophage colony-stimulating factor, and macrophage inflammatory proteins 1α and 1β, to modulate immune functions.3 With dual roles in innate and adaptive immunity, NK cells are critical components of the immune system, and contribute to establishing defence mechanisms against infections and tumours, and maintaining immune system homeostasis.7
Natural killer cells express a number of germline-encoded activating and inhibitory receptors in humans. Inhibitory receptors include killer immunoglobulin-like receptors, immunoglobulin-like transcript 2 receptors and the CD94/NKG2A receptor.8 The NK cell activation receptors can be classified into two different categories, based on the specificity of receptor–ligand recognition: MHC-dependent receptors, such as activating killer immunoglobulin-like receptors and CD94/NKG2C, and non-MHC-dependent receptors, such as NK cell group 2 member D (NKG2D), natural cytotoxicity receptors, NKp65, NKp80, T-cell immunoglobulin and mucin-containing domain 3, and DNAX accessory molecule-1 (DNAM-1, which is also known as CD226).9
DNAM-1, which binds to the nectin molecule CD112 (also known as nectin-2 or PRR2) and the nectin-like molecule CD155 (also known as poliovirus receptor PVR or Necl5), has recently emerged as a pivotal regulator of NK cell-mediated functions against cancer, viral infections and immune-related pathologies.10–13 It is involved in intercellular adhesion, lymphocyte signalling, lymphokine secretion and cytotoxicity induced by cytotoxic T lymphocytes and NK cells.11 In recent years, results from a series of studies have highlighted the involvement of DNAM-1 in NK cell education, differentiation, immune synapse formation and cytokine production in NK cell biology.14–18 In addition, DNAM-1 exerts its synergistic roles in regulating NK cell activation and functions with three molecules: T-cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domains (TIGIT); CD96, which is also known as T-cell activation increased late expression (or TACTILE); and class I restricted T-cell-associated molecule (CRTAM).19
In this article, we have reviewed recent advances in DNAM-1 research, and how the expression of DNAM-1 ligands (DNAM-1Ls) is regulated. We have also briefly outlined the synergistic combinations of NK cell activation receptors. We discuss the involvement of DNAM-1 in cancer, infections and autoimmune diseases. In addition, we explored immune evasion mechanisms employed by malignant transformed and virus-infected cells, in an attempt to further understand the role of DNAM-1 in novel anti-tumour and anti-virus immunotherapeutic strategies.
Expression and modulation of DNAM-1 on NK cells
DNAM-1 is a 65 000 molecular weight immunoglobulin-like transmembrane glycoprotein that is expressed on the surface of NK cells, T lymphocytes, platelets, monocytes and a subset of B cells.11 The gene CD226, which encodes DNAM-1, is conserved between mice and humans, and is located on chromosomes 18E4 and 18q22.3, respectively. DNAM-1 was first discovered in T lymphocytes and was associated with the differentiation of cytotoxic T lymphocytes. It was originally designated T-cell lineage-specific activation antigen-1.10 It is also expressed on platelets, and participates in platelet activation and aggregation; therefore, it is also known as platelet and T-cell activation antigen-1.11
DNAM-1 contains three domains: an extracellular domain of 230 amino acids, comprising two immunoglobuiln-like domains and eight N-linked glycosylation sites; a transmembrane domain of 28 amino acids; and a cytosolic domain of 60 amino acids, with four putative tyrosine residues and one serine residue for phosphorylation, which recruits signal proteins. The structural basis of the interactions between DNAM-1 and its ligands is unclear. Liu and colleagues showed that membrane-bound and soluble DNAM-1 bound to immunoglobulin V-set domains of nectin-2 (nectin-2v). DNAM-1 was found to bind to nectin-2 through the same interface as that used by the nectin-2v dimer.20 In comparison with CD226-extracellular domain 1 and full-length CD226, the effector functions of DNAM-1, such as recognition, adhesion, immune synapse formation and cytotoxicity, are dependent on the structure of DNAM-1 extracellular domain 1.14
MicroRNAs (miRNAs) are a class of non-coding endogenous small RNAs that post-transcriptionally regulate gene expression by binding to the 3′ untranslated regions of target genes. There is an increasing body of evidence to indicate that miRNAs could regulate the expression of NK cell activation receptor and function-related genes (KLRK1, PRF1, GZMB and IFNG).21 We found that miR-422a targets the 3′-untranslated region of CD226, and that miR-422a down-regulation results in a significant increase in DNAM-1 expression in NK cells. Consequently, this results in the up-regulation of the abilities of NK cells to recognize and kill tumour cells (unpublished data).
Expression of DNAM-1 on NK cells can also be regulated by soluble factors and cell–cell interactions.22–26 In humans, IL-2 or IL-15 alone or in combination with heat-shock protein 70-derived 14-mer peptide, granulocyte–macrophage colony-stimulating factor/IL-2 fusion protein, and glucocorticoid up-regulates DNAM-1 expression,23–25 whereas indoleamine 2,3-dioxygenase, transforming growth factor-β and chronic CD155 exposure by some tumour cells can down-regulate DNAM-1 expression on NK cells.22,26
DNAM-1 ligands
CD112 and CD155 are DNAM-1Ls that belong to the nectin and nectin-like (Necl) protein families, comprising nectin 1-4 and Necl 1-5, respectively.12,27 These two molecules are broadly distributed on normal neuronal, epithelial, endothelial, fibroblastic cells, and on transformed and pathogen-infected cells.12 They are also expressed at the cell surface of immune cells such as monocytes, dendritic cells (DCs), and activated T cells. DNAM-1 receptor–ligand interactions mediate the cross-talk between NK cells and other immune cells, to maintain homeostasis.28
CD155 is a transmembrane glycoprotein whose external domain mediates cell attachment to the extracellular matrix molecule vitronectin, while its intracellular domain interacts with dynein. CD155 serves as a cellular receptor for poliovirus. CD112 is a plasma membrane component of adherens junctions. It is involved in the cell–cell spread of viruses. Results from a previous study demonstrated that blocking CD155 signalling blunted NK cell-mediated destruction of tumour cells. However, blocking CD112 signals failed to inhibit NK cell-mediated killing, suggesting that CD155 is the most important ligand in DNAM-1-mediated cytotoxicity.29 Therefore, it can be seen that DNAM-1Ls also mediate the recognition and killing of target cells through their interactions with the DNAM-1 activating receptor.
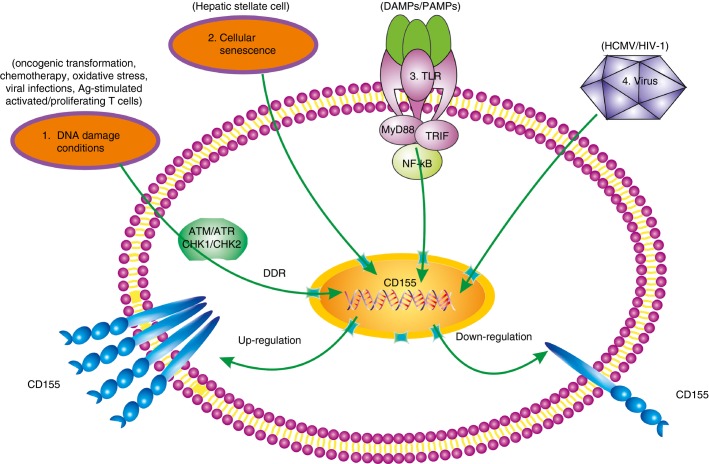
Similar to the induction of NKG2D ligands, target cells initiate an intrinsic response to cellular stress, which results in the aberrant expression of DNAM-1Ls.30 Atypical expression of DNAM-1Ls, especially CD155, is affected by multiple pathological conditions, such as tumorigenesis, inflammation-associated diseases, virus infections and other certain stressors (Fig.1).
Figure 1.
Dual regulation of CD155 expression. 1, DNA damage conditions, including oncogenic transformation, chemotherapy, oxidative stress, viral infections and antigen-stimulated activated/proliferating T cells (referred to as dysregulated proliferation), stimulate CD155 gene transcription through the DNA damage sensors ataxia-telangiectasia mutated (ATM), ATM and Rad3-related (ATR), and check-point kinase 1/2 (CHK1/2). 2, Cellular senescence such as hepatic stellate cells can also induce up-regulation of CD155 expression. 3, Toll-like receptor (TLR) interactions with pathogen- and damage-associated molecular patterns (PAMPs and DAMPs) also increase CD155 expression in nuclear factor-κB (NF-κB)-dependent way (the former three factors in green line) 4, whereas both human cytomegalovirus (HCMV) and HIV-1 can induce down-regulation of CD155 expression (the final factor in black line). DDR, DNA damage response; MYD88, myeloid differentiation factor 88; TRIF, Toll/interleukin-1 receptor domain-containing adaptor inducing interferon-β.
The DNA damage response is a complex signalling network that could be employed in limiting tumorigenesis.31 Results from a number of studies have demonstrated that DNA damage induced by oncogenic transformation, chemotherapy, oxidative stress, viral infections and antigen-stimulated activated/proliferating T cells activates the expression of certain genes, including ATM, ATR, CHEK1 and CHEK2, which subsequently results in the expression of the genes encoding CD155 and CD112.32–35 It was recently shown that nitric oxide induced the expression of CD155 at very high levels. CD155 was then able to exert its anti-cancer activity through activation of the DNA damage response network.36
Krizhanovsky and colleagues showed that the up-regulation of CD155 expression, induced by cellular senescence, could mediate the killing of hepatic stellate cells by NK cells, so inhibiting fibrosis progression.37 Hepatic stellate cells are special cells that contribute to the development of liver fibrosis, while senescence limits the fibrogenic response to tissue damage. It was recently shown that CD155 expression correlated with toll-like receptor engagement of viral or bacterial components on antigen-presenting cells. Upon activation, toll-like receptors recruit myeloid differentiation factor 88 (MyD88), Toll/IL-1 receptor domain-containing adaptor inducing IFN-β and nuclear factor-κB, the expression of CD155 is up-regulated.38
Several viruses, including human cytomegalovirus (HCMV), Epstein–Barr virus and HIV-1, can induce the aberrant expression of DNAM-1Ls.39–42 For example, HCMV induced low-level expression of CD155 and CD112 on target cells through proteasome-mediated degradation during infection to combat NK cell-mediated destruction.39,40 B cells infected with Epstein–Barr virus exhibited up-regulated expression of CD112. As a result, these B cells were more susceptible to NK cell-mediated killing.41 In addition, HIV-1, through its Nef and Vpu proteins, is able to down-regulate the expression of DNAM-1 in cytotoxic cells, and CD155 expression in HIV-infected cells. This helps HIV-infected cells avoid NK cell-dependent cytotoxicity.43 It has since been demonstrated that Vpr could over-ride the effects induced by Nef, contributing to the up-regulation of CD155 in the DNA damage response network.35
Critical roles of DNAM-1 in NK cell functions
DNAM-1 is a co-activation receptor involved in the regulation of NK cell adhesion, lymphocyte signalling, lymphokine secretion and cytotoxicity. However, there is an increasing body of evidence that indicates greater involvement of DNAM-1 with respect to NK cell biology.14–19 DNAM-1 has now been shown to be involved in NK cell education and differentiation, immune synapse formation, cytokine production and cross-talk with DCs and T cells. DNAM-1 also acts synergistically with CD96, TIGIT and CRTAM to regulate NK cell functions.14–16,18,19,28,44
At present, there is no known marker that can be used to distinguish educated from non-educated NK cells. Therefore, elucidating the cellular and molecular mechanisms governing NK cell education would be a critical step in understanding NK cell regulation. Anfossi et al.45 were the first to recognize that DNAM-1 was associated with NK cell education, courtesy of its differing expression patterns in educated and non-educated NK cells. Subsequently, Enqvist et al. examined a number of activation receptors and adhesion molecules on the surface of NK cells, and discovered that only DNAM-1 expression was involved in the expression of educating self-specific inhibitory receptors. These findings were an indicator that DNAM-1 was an intrinsic marker of educated NK cells, and was potentially associated with cytotoxicity.17
Two research groups have provided evidence of the pivotal role of DNAM-1 during NK cell differentiation.15,18 Nabekura et al. showed that DNAM-1−/− Ly49H+ NK cells could not be expanded in culture. In addition, their differentiation into memory NK cells was inhibited when they were infected with a murine CMV, implying that DNAM-1 was essential for their differentiation into memory NK cells.15 Martinet et al. found that DNAM-1+ NK cells were able to differentiate into DNAM-1− NK cells, regardless of the stage of maturation. Moreover, the development was independent of CD11b phenotype. In contrast, DNAM-1− NK cells retained their DNAM-1− phenotype and were unable to re-express DNAM-1. DNAM-1+ NK cells exhibited functional differences compared with their DNAM-1− counterparts, such as regulated cytokine secretion and anti-tumour functions.18 Interestingly, these functional differences are likely to be a consequence of distinct gene expression profiles exhibited by DNAM-1 per se, but not the co-stimulatory signals provided by the engagement of DNAM-1–CD155 interactions. These results suggest that DNAM-1 might serve as an adjuvant differentiation marker, complementary to the traditional CD11b molecule in mice.18
The stimulation of DNAM-1 induces its involvement in the formation of immunological synapse. In contrast, blocking DNAM-1 impairs the formation of immunological synapse, resulting in decreased adhesion between NK cells and target cells, and a reduction in the level of DNAM-1-mediated cytolysis.14,46
The proportion of IFN-γ+ NK cells is reduced in DNAM-1-deficient mice compared with that in wild-type controls, in response to stimulation with bacterial lipopolysaccharide.16 CD96 is known to inhibit the production of IFN-γ in NK cells. With regard to the opposite role of CD96 and DNAM-1 in regulating the production of IFN-γ of NK cells, DNAM-1 appears to play a crucial role in the up-regulation of NK cell-mediated IFN-γ secretion during inflammation.
Based on DNAM-1 receptor–ligand interactions in vitro, DNAM-1 appears to play a vital role in the management of DCs and T cells. Interactions between DNAM-1 and CD155, and/or CD112 contribute to the NK-mediated killing of immature and mature DCs.28 Furthermore, DNAM-1 cooperates with NKp30, inducing NK cells to lyse immature DCs and promoting the proliferation of mature DCs.47 The DNAM-1/DNAM-1L axis is involved in the cross-talk between NK and T cells. NK cells can recognize and lyse antigen-stimulated activated/proliferating T cells in graft-versus-host disease (GVHD) or other autoimmune diseases. They can also promote the differentiation of T helper cells.44,48
TIGIT, CD96 and CRTAM are able to bind to nectins and nectin-like molecules to modulate the functions of NK cells, which brought about the complexity of DNAM-1 involvement in NK cell biology. The role of DNAM-1 in collaborating with these three molecules, and in balancing the activation of NK cells, was recently reported.19
DNAM-1 signalling (pathway) and the synergistic activation of NK cells
The mechanisms by which NK cells are activated by an array of innate activating and inhibitory receptors have been widely studied.5 Other researchers have reviewed the synergistic activation of NK cells,5 and the signalling pathways involved for activating receptors such as natural cytotoxicity receptors, NKG2D, 2B4 and DNAM-1.49,50,13
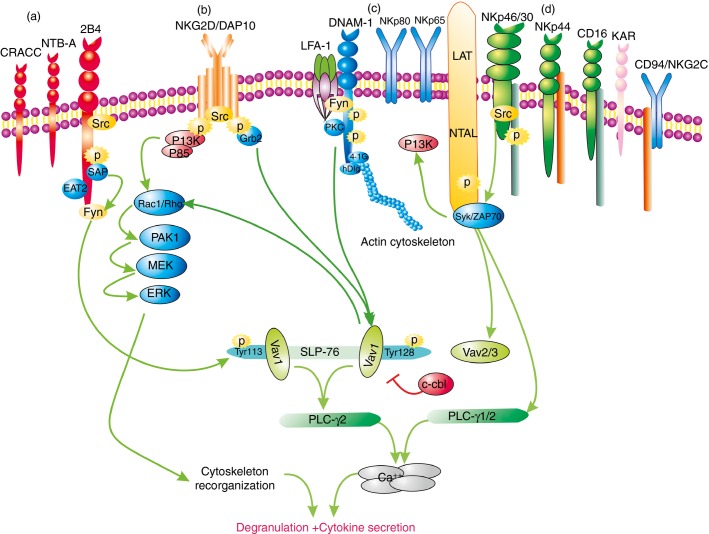
The involvement of multiple diverse signalling pathways increases the complexity, and the diversity of regulation, for NK cell responsiveness.6 There are several different activating signalling pathways involved in human NK cells: (i) the ITAM–Syk/ZAP70 pathway, as observed for natural cytotoxicity receptors, CD94/NKG2C, KAR and CD16(FcγRIIIA); (ii) the DAP10 (YINM)-PI3K/Grb2 pathway, for NKG2D; (iii) the ITSM-SAP(SLAM-associated protein)/EAT-2 pathway, for the signalling lymphocytic-activation molecule (SLAM) family receptors 2B4, NTB-A (CD84) and CRACC (CD319); and (iv) other tyrosine-based signalling motif activating pathways, for DNAM-1, NKp80 and NKp65 (Fig.2).
Figure 2.
Multiple activating human natural killer (NK) cell receptors and representative signalling pathways of 2B4, NKG2D, DNAM-1 and NKp46/30. (a) SLAM family receptor CRACC (CD319), NTB-A (CD84), and 2B4; e.g. 2B4 transduces signal in ITSM-SAP/EAT-2-Fyn-dependent pathway. (b) NKG2D; e.g. NKG2D/DAP10 transduces signal in DAP10-PI3K-MAPK/Grb2-Vav1-dependent pathway. (c) DNAM-1, NKp80, and NKp65; e.g.DNAM-1 signalling cascade is described in text. (d) NKp46/NKp30, NKp44, CD16 (FcγRIIIA), KAR and CD94/NKG2C. e.g.NKp46/NKp30 transduces signal in ITAM-Syk/ZAP70-dependent pathway, and results in subsequently activation of phosphatidylinositol-3 kinase (PI3K), Vav2/3 and phospholipase C (PLC)-γ1/2. The first step to transduce intracellular signal of all activation receptors is to be phosphorylated by Src family kinases. 2B4 can induce Src homology 2(SH2) domain-containing leucocyte phosphoprotein of 76 000 MW (SLP-76) phosphorylation at Tyr113, whereas NKG2D and DNAM-1 can only induce SLP-76 phosphorylation at Tyr128. Two Vav1, which could inhibit degradation by the E3 ubiquitin ligase c-cbl, bind to two phosphotyrosines of SLP-76 and result in synergistic activation of NK cells. All activation receptors induce ultimately PLC-γ1/2 activation, Ca2+ flux, cytoskeletal reorganization, and result in degranulation and cytokine secretion. SLAM, signalling lymphocytic-activation molecule; ITSM, immunoreceptor tyrosine-based switch motif; SAP, SLAM-associated protein; EAT2, Ewing-sarcoma-associated transcripts 2; MAPK, mitogen-activated protein kinase; Grb2, grow factor binding protein 2; Syk, spleen tyrosine kinase; ZAP, ζ chain-associated protein kinase; LFA-1, lymphocyte function-associated antigen 1; LAT, linker for the activation of T cells; NTAL, non-T-cell activation linker.
It is clear that DNAM-1, 2B4, NKG2D,or NKp46, acting individually are not sufficient to trigger the activation of NK cells.6 At least two of these receptors must act in synergy to induce NK cell cytotoxicity and the production of cytokines. The redundant signalling pathways that exist allow NK cells to respond rapidly to changes in the cellular environment. It has been demonstrated that among these activation receptors, there is no preferred receptor, nor is there a hierarchy.6 Therefore, it can be seen that DNAM-1, as a co-activation receptor, plays a pivotal role in regulating NK cell activation.
There are a number of key events that occur in the DNAM-1 signalling pathway. Upon engagement, DNAM-1 Ser329 is phosphorylated by protein kinase C. Leucocyte function-associated antigen-1 (LFA-1) then cross-links with DNAM-1, resulting in the recruitment of DNAM-1 to lipid rafts and its engagement with the cytoskeleton.17,51 Fyn src kinase is essential for DNAM-1 Tyr322 phosphorylation, which is induced by LFA-1.52 DNAM-1 works synergistically with 2B4 to phosphorylate Tyr128 and Tyr113 of Src homology 2(SH2) domain-containing leucocyte phosphoprotein of 76 000 MW (SLP-76), resulting in two Vav1 molecules binding to one SLP-76 molecule, and subsequent phospholipase Cγ2 activation, Ca2+ flux, cytoskeletal reorganization, degranulation and secretion.53 When target cells are recognized, LFA-1 co-localizes with DNAM-1 at the immune synapse, which is required for the transduction of downstream signals. At present, it is not clear how DNAM-1 coordinates with LFA-1, spatially and temporally, in the immune synapse.
Two key molecules, SLP-76 and Vav, are essential for regulating the synergistic activation of NK cells (Fig.2). It is necessary that Tyr113 and Tyr128 of SLP-76 are phosphorylated. It has been shown that 2B4 phosphorylates SLP-76 at Tyr113, and NKG2D and DNAM-1 phosphorylate Tyr128. These observations suggest synergistic activation among 2B4, NKG2D and DNAM-1.53 In addition, two Vav molecules are recruited to SLP-76, thereby inhibiting the degradation of Vav by the E3 ubiquitin ligase c-cbl.54 It remains to be determined how NKp46 cooperates with DNAM-1, 2B4 and NKG2D, and whether there are additional synergistic activation mechanisms that are independent of SLP-76.
DNAM-1 in the control of cancer
The functional interaction of DNAM-1 with its ligands is involved in NK cell-mediated recognition, controlling and killing of tumour cells in vivo and in vitro, highlighting the link between extrinsic and intrinsic cellular mechanisms.55–63 In RMA primary lymphoma mice, Tahara-Hanaoka et al. described the role of DNAM-1 in controlling the development of tumours in vivo. The over-expression of CD155 and CD112 in cancer cells resulted in tumour rejection, and involved a DNAM-1-dependent mechanism that was mediated by NK cells.55 Experimental evidence for the in vivo relevance of DNAM-1 in the control of tumour metastasis was recently shown in mice lacking DNAM-1. These DNAM-1−/− mice contained more lung metastases than wild-type mice. It was possible for NK cells to inhibit the metastasis of melanoma lesions in this model, with CD155 seemingly a key ligand in the NK cell-mediated suppression of metastases.16,56
Iguchi-Manaka et al. demonstrated low levels of cytotoxicity in DNAM-1−/− mice with DNAM-1L-expressing tumours. They also showed that DNAM-1−/− mice were more susceptible to tumour development than wild-type mice, suggesting a critical role for DNAM-1 in tumour immunosurveillance.57 Similar to the results seen for membrane-bound DNAM-1 with respect to anti-tumour responses, the interaction of soluble DNAM-1 with CD155 and CD112 also resulted in the inhibition of tumour growth in a dose-dependent manner. It is also possible that soluble DNAM-1 might inhibit the metastatic potential of cancer cells, although further work is required to confirm this.11,64
In coordination with extrinsic cellular mechanisms that drive the anti-tumour response, intrinsic cellular mechanisms ultimately enforce the over-expression of DNAM-1Ls on targets so that they are recognized and attacked by NK cells. DNAM-1Ls are expressed on the surface of many tumour cells, such as lung carcinoma, primary human leukaemia, myeloma, melanoma, neuroblastoma, ovarian cancer, colorectal carcinoma and Ewing sarcoma cells.29,58,62,65–69 In myeloid leukaemia, CD155 and CD112 are expressed on the surface of leukaemia cells, with NK cell-mediated destruction of leukaemia cells dependent on DNAM-1 interactions.65 In addition, CD155-expressing neuroblastoma cells are efficiently killed by NK cells. Blocking either DNAM-1 or CD155 results in the significant inhibition of tumour cell lysis.29
However, cancer cells have established several mechanisms to evade DNAM-1-mediated killing. The first and most obvious strategy is the down-regulation of DNAM-1 on the surface of cytotoxic lymphocytes, which can be induced by cytokines, or by chronic exposure to DNAM-1Ls.26,70 In non-small cell lung carcinoma, DNAM-1 down-regulation is induced by interactions between NK cells and tumour cells, or through the secretion of the immunoregulatory factors transforming growth factor-β and indoleamine 2,3-dioxygenase. This results in the impairment of NK cell effector functions and immune surveillance, and the promotion of tumour progression.26 Soluble DNAM-1 levels are significantly higher in the sera of patients with cancer than in healthy controls.71 Soluble CD155 is also present in serum and can block DNAM-1 recognition mediated by cytotoxic cells, so helping tumour cells to evade the immune attack. In addition, tumours are able to reduce the expression levels of DNAM-1Ls on the surface of cells. Qu et al. examined CD155 expression in a series of hepatocellular carcinoma tissue samples. They saw that patients with hepatocellular carcinoma and low CD155 expression levels had a poor prognosis. These findings suggest that the down-regulation of CD155 expression is an immune system evasion strategy employed by tumours.72
Some tumours manipulate DNAM-1-mediated signalling and induce high expression levels of DNAM-1Ls. High-level expression of CD155 in the tumour vasculature was shown to mediate the invasion and migration of fibrosarcoma and glioblastoma tumour cells, implying that CD155 possibly contributes to tumorigenesis.73 In non-small cell lung carcinoma, CD155 was found to be expressed on lung carcinoma cells, and mediated tumour migration during the early stage of tumour development. The down-regulation of DNAM-1 was then induced during tumour progression.69 Similar to findings regarding NKG2D ligands, over-expression of DNAM-1Ls is considered a critical danger signal that alerts cytotoxic cells to combat tumour cells. Therefore, CD155 appears to play dual roles in eliminating and promoting tumours.74
The induction of DNAM-1L expression on tumour cells is a promising therapeutic strategy for cancer. The DNAM-1 receptor system has already been used as a target for anti-cancer therapies. Multimodal treatments involving surgery, radiotherapy, chemotherapy and palliative treatment, can help to improve the prognosis of a variety of malignancies.75 Cell immunotherapies, especially those involving NK and T cells, have the potential to be very specific, and have already been applied to the clinical treatment of certain malignancies and haematological diseases.76,77
A major breakthrough was recently made, involving T lymphocytes equipped with a chimeric antigen receptor (CAR) for the treatment of cancer. Unfortunately, NK cells have not receive similar attention.78 A recent study reported that DNAM-1 CAR T cells equipped with different signalling molecules, such as CD3ζ, CD28, CD40 and 4-1BB, exhibited high levels of cytotoxicity and produced low levels of IFN-γ against DNAM-1L-expressing tumour cells.79 NK cells have been engineered to contain CARs; and they have been shown to be therapeutically beneficial in the treatment of cancer. However, their effects are weak compared with those seen for CAR T cells, and anti-tumour immunity is inconsistent.78 Chu et al. reported that CS1 CAR NK cells (CS1, referred to as a surface protein that is highly expressed on multiple myeloma cells) exhibited enhanced cellular cytotoxicity and increased IFN-γ production, and showed a specific CS1-dependent recognition and suppression of multiple myeloma in vitro and ex vivo.80
A specific therapy that combines extracellular with intracellular responses shows some potential for the future treatment of malignancies. Given the similarities and importance of the activation receptors NKG2D and DNAM-1 in the immunosurveillance of malignancies, the NKG2D/DNAM-1 receptor system could be employed as a diagnostic tool and a therapeutic target for cancer.81
DNAM-1 in the control of infections
7Natural killer cells have a crucial role in controlling and limiting viral and bacterial infections. It has been reported that DNAM-1 is involved in controlling lymphocytic choriomeningitis virus, HCMV, hepatitis C virus (HCV) and HIV-1 infections.82–85 The loss of DNAM-1 was previously shown to lead to a delay in the clearance of lymphocytic choriomeningitis virus, and higher titres of virus.82 Results from several studies have shown that DNAM-1 plays some role in the NK cell-mediated antiviral immune response during the early stages of infection. As an example, NK cells can recognize and kill HCMV-infected myeloid DCs, so overcoming immune system evasion strategies of the virus.83 The binding of viral or bacterial components with toll-like receptors on DCs can up-regulate the expression of DNAM-1Ls, such that they are recognized by the DNAM-1 activating receptor.38 Interferon-α-activated NK cells efficiently recognize and kill HCV-infected hepatic cells in patients with acute HCV infections, and this was found to be dependent on the DNAM-1 activating receptor.84 The importance of DNAM-1 with respect to the ways that NK cells control viral infections is underscored by the fact that several viruses have developed mechanisms to evade the immune system, and can impair DNAM-1-mediated recognition and killing.39,40,42,43
Interactions between DNAM-1/DNAM-1L have been found to mediate the migration of leucocytes, including NK cells, from the blood to secondary lymphoid organs or inflamed tissues during bacterial infections.86,87 Bottino et al. found that CD155 and CD112 are present at the cell junctions on primary vascular endothelial cells. Monoclonal antibodies against DNAM-1 or CD155 inhibited transendothelial NK cell migration, suggesting that DNAM-1 is involved in transendothelial migration in some way.12 It has subsequently been shown that NK cells rapidly migrate from the blood into injured tissues during inflammation.87 Observations from other studies indicate that NK cells might be involved in sepsis; however, it remains unclear whether DNAM-1 is involved in this process.88
DNAM-1 in immune-related diseases
Allogeneic bone marrow transplantation is an effective therapy for many malignancies. However, GVHD is a serious complication following the transplantation of bone marrow, and is caused by an alloreactive donor T cell-mediated cellular immune response against host tissues.89 NK cells can kill host antigen-presenting cells and exert anti-GVHD effects; therefore, it is believed that they can control the development of GVHD.90 Allogeneic antigen-presenting cells and activated T cells can be destroyed through mechanisms mediated by DNAM-1.48 Using mouse models, the role of DNAM-1 on the surface of CD8+ T cells in GVHD was determined. Co-stimulation of DNAM-1 induced the exacerbation of GVHD, while blocking DNAM-1 attenuated the effects of GVHD.91,92 It remains to be seen whether DNAM-1 drives the anti-GVHD effects of NK cells in humans.
Defects in the functional responses of NK cells are frequently observed in patients with autoimmune disorders. A single nucleotide polymorphism in CD226 can result in the amino acid substitution Gly307Ser (rs763361). This correlates with the development of a wide range of immune-related diseases, such as systemic lupus erythematosus, systemic sclerosis (SSc), type I diabetes and rheumatoid arthritis (RA).93–96
Systemic sclerosis is a chronic autoimmune disease that affects connective tissue, and is characterized by dermal fibrosis and thickening of the skin. In a mouse model of SSc, features of fibrosis were suppressed in DNAM-1−/− mice, whereas wild-type DNAM-1+/+ mice exhibited profibrotic characteristics. Therefore, it was postulated that the expression of DNAM-1 contributes to SSc pathogenesis, while blocking DNAM-1 could be a promising treatment for SSc and associated fibrotic diseases.94
Rheumatoid arthritis is a chronic and systemic inflammatory disorder, in which inflammation of the joints and associated tissues results in the progressive destruction of bone and cartilage. In an in vitro model of RA, DNAM-1 and DNAM-1L were expressed on the surface of NK cells and RA fibroblast-like synoviocytes (RA-FLS), respectively.97 The RA-FLS were recognized and cleared by NK cells in a DNAM-1-dependent manner. Hence, the involvement of DNAM-1 in NK cell-mediated cytotoxicity against RA-FLS could be one mechanism influencing local joint inflammation in RA.97
Many studies have linked dysfunctional NK cells to autoimmune disorders. The Gly307Ser mutation in DNAM-1 appears to lead to the dysfunction of NK cells.98 Results from one study, involving an experimental autoimmune encephalomyelitis (EAE) model, have provided some novel insights into the role of NK cells that limit the progress of autoimmune diseases.99 The depletion of NK cells resulted in the exacerbation of EAE in this particular model. This result supported the hypothesis that the absence of NK cell regulatory roles is associated with a severe EAE phenotype in mice. NK cells appear to exert a key role in the initiation of autoimmune diseases, but their role(s) appear to be diminished once a disorder is established.98
Concluding remarks
It has been clearly shown that interactions with DNAM-1 and its ligands, in particular CD155, play a key role in controlling various pathological conditions, such as cancer, infectious diseases and autoimmunity. The potential role of DNAM-1 in the treatment of tumours has attracted much attention as DNAM-1Ls are widely expressed on the surface of tumour cells. In addition, a significant breakthrough has been made in understanding the role of DNAM-1 with respect to NK cell education, differentiation and memory.
There remain many features of NK cells that require further investigation. Following on from the discovery of DNAM-1+ and DNAM-1− NK cells, the relationship between different NK cell subtypes and their genotypes requires attention, as does the formation of specialized NK cell repertoires. The crystal structure of the human DNAM-1 receptor and the CD155 ligand are yet to be clarified. The mechanism of regulation for CD155 expression remains partially understood, with little known concerning the mechanism of regulation for CD112 expression. A better understanding of the anomalous expression of DNAM-1Ls in different pathologies might provide clues as to how to enhance the immune response and immune surveillance, and in the development of therapeutic targets. It is unclear at the moment whether additional cellular pathways induce the expression of DNAM-1Ls.
It is necessary to determine whether DNAM-1Ls can be shed or secreted from the surface of tumour cells. New methods that effectively recognize and kill tumour cells and virus-infected cells need to be developed, especially given their ability to develop mechanisms to evade recognition and clearance by the host immune system. It is not clear how the numerous innate receptors and intracellular signalling molecules regulate NK cell responsiveness spatially and temporally; therefore, this should be investigated. Whether the Gly307Ser mutation affects the downstream signalling of DNAM-1, and the functions of cytotoxic cells, also requires further investigation. Finally, the feasibility and availability of DNAM-1 CAR-based cell immunotherapy should be studied in a clinical setting. Results from all of these proposed studies will enhance our understanding of DNAM-1 with respect to NK cell biology, and hopefully pave the way for more effective DNAM-1-based therapies.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (Grant no. 81172786) and Youth Chenguang project of Science and Technology of Wuhan City (Grant no. 201050231077).
Glossary
- Aa
amino acid
- ATM
Ataxia-telangiectasia, mutated
- ATR
ATM and Rad3-related
- CAR
chimeric antigen receptor
- CHEK1/2
Check-point kinase 1/2
- CRTAM
class I restricted T cell-associated molecule
- DC
dendritic cell
- DNAM-1
DNAX accessory molecule-1
- DNAM-1L
DNAM-1 ligand
- EAE
experimental autoimmune encephalomyelitis
- EAT2
Ewing-sarcoma associated transcripts 2
- Grb2
grow factor binding protein 2
- GVHD
graft-versus-host disease
- HCMV
human cytomegalovirus
- HCV
hepatitis C virus
- HIV-1
human immunodeficiency virus type 1
- IFN-γ
interferon-gamma
- IL
interleukin
- ITSM
immunoreceptor tyrosine-based switch motif
- LAT
linker for the activation of T cells
- LFA-1
lymphocyte function-associated antigen 1
- MAPK
mitogen activated protein kinase
- MHC
major histocompatibility complex
- MiRNA
microRNA
- MYD88
myeloid differentiation factor 88
- NKG2D
natural killer cell group 2 member D
- NK
natural killer
- NTAL
non-T cell activation linker
- PAMPs/DAMPs
pathogen- and damage-associated molecular patterns
- RA-FLS
rheumatoid arthritis Fibroblast-like synoviocytes
- SAP
SLAM-associated protein
- SLAM
signalling lymphocytic-activation molecule
- SLP-76
Src homology 2(SH2) domain-containing leukocyte phosphoprotein of 76 kD
- SSc
systemic sclerosis
- Syk
spleen tyrosine kinase
- TIGIT
T cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibitory motif (ITIM) domains
- ZAP
zeta chain-associated protein kinase
Disclosures
All authors declare no competing financial interests.
References
- Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer. 1975;16:216–29. doi: 10.1002/ijc.2910160204. [DOI] [PubMed] [Google Scholar]
- Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5:112–17. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- Colucci F, Caligiuri MA, Di Santo JP. What does it take to make a natural killer? Nat Rev Immunol. 2003;3:413–25. doi: 10.1038/nri1088. [DOI] [PubMed] [Google Scholar]
- Watzl C, Urlaub D, Fasbender F, Claus M. Natural killer cell regulation – beyond the receptors. F1000Prime Rep. 2014;6:87. doi: 10.12703/P6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzl C. How to trigger a killer: modulation of natural killer cell reactivity on many levels. Adv Immunol. 2014;124:137–70. doi: 10.1016/B978-0-12-800147-9.00005-4. [DOI] [PubMed] [Google Scholar]
- Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–58. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–9. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 2004;23:255–9. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellora F, Castriconi R, Dondero A, Carrega P, Mantovani A, Ferlazzo G, Moretta A, Bottino C. Human NK cells and NK receptors. Immunol Lett. 2014;161:168–73. doi: 10.1016/j.imlet.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Burns GF, Triglia T, Werkmeister JA, Begley CG, Boyd AW. TLiSA1, a human T lineage-specific activation antigen involved in the differentiation of cytotoxic T lymphocytes and anomalous killer cells from their precursors. J Exp Med. 1985;161:1063–78. doi: 10.1084/jem.161.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya A, Campbell D, Hannum C, Yssel H, Franz-Bacon K, McClanahan T, et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 1996;4:573–81. doi: 10.1016/s1074-7613(00)70060-4. [DOI] [PubMed] [Google Scholar]
- Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198:557–67. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andrade LF, Smyth MJ, Martinet L. DNAM-1 control of natural killer cells functions through nectin and nectin-like proteins. Immunol Cell Biol. 2014;92:237–44. doi: 10.1038/icb.2013.95. [DOI] [PubMed] [Google Scholar]
- Hou S, Ge K, Zheng X, Wei H, Sun R, Tian Z. CD226 protein is involved in immune synapse formation and triggers Natural Killer (NK) cell activation via its first extracellular domain. J Biol Chem. 2014;289:6969–77. doi: 10.1074/jbc.M113.498253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabekura T, Kanaya M, Shibuya A, Fu G, Gascoigne NRJ, Lanier LL. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity. 2014;40:225–34. doi: 10.1016/j.immuni.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CJ, Martinet L, Gilfillan S, Souza-Fonseca-Guimaraes F, Chow MT, Town L, et al. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat Immunol. 2014;15:431–8. doi: 10.1038/ni.2850. [DOI] [PubMed] [Google Scholar]
- Enqvist M, Ask EH, Forslund E, Carlsten M, Abrahamsen G, Beziat V, et al. Coordinated expression of DNAM-1 and LFA-1 in educated NK cells. J Immunol. 2015;194:4518–27. doi: 10.4049/jimmunol.1401972. [DOI] [PubMed] [Google Scholar]
- Martinet L, Ferrari De Andrade L, Guillerey C, Lee JS, Liu J, Souza-Fonseca-Guimaraes F, et al. DNAM-1 expression marks an alternative program of NK Cell maturation. Cell Rep. 2015;11:85–97. doi: 10.1016/j.celrep.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15:243–54. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- Liu J, Qian X, Chen Z, Xu X, Gao F, Zhang S, et al. Crystal structure of cell adhesion molecule nectin-2/CD112 and its binding to immune receptor DNAM-1/CD226. J Immunol. 2012;188:5511–20. doi: 10.4049/jimmunol.1200324. [DOI] [PubMed] [Google Scholar]
- Beaulieu AM, Bezman NA, Lee JE, Matloubian M, Sun JC, Lanier LL. MicroRNA function in NK-cell biology. Immunol Rev. 2013;253:40–52. doi: 10.1111/imr.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB, El-Jawhari JJ, Neilson AL, Hall GD, Melcher AA, Meade JL, Cook GP. Human tumour immune evasion via TGF-beta blocks NK cell activation but not survival allowing therapeutic restoration of anti-tumour activity. PLoS ONE. 2011;6:e22842. doi: 10.1371/journal.pone.0022842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromadnikova I, Pirkova P, Sedlackova L. Influence of in vitro IL-2 or IL-15 alone or in combination with Hsp-70-derived 14-mer peptide (TKD) on the expression of NK cell activatory and inhibitory receptors. Mediators Inflamm. 2013;2013:405295. doi: 10.1155/2013/405295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penafuerte C, Bautista-Lopez N, Boulassel MR, Routy JP, Galipeau J. The human ortholog of granulocyte macrophage colony-stimulating factor and interleukin-2 fusion protein induces potent ex vivo natural killer cell activation and maturation. Cancer Res. 2009;69:9020–8. doi: 10.1158/0008-5472.CAN-09-2322. [DOI] [PubMed] [Google Scholar]
- Moustaki A, Argyropoulos KV, Baxevanis CN, Papamichail M, Perez SA. Effect of the simultaneous administration of glucocorticoids and IL-15 on human NK cell phenotype, proliferation and function. Cancer Immunol Immunother. 2011;60:1683–95. doi: 10.1007/s00262-011-1067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer I, Fridman WH, Sautes-Fridman C. Tumor microenvironment in NSCLC suppresses NK cells function. Oncoimmunology. 2012;1:244–6. doi: 10.4161/onci.1.2.18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang W, Wu DA, Chen C, Xu QZ, Zhao B, et al. Molecular cloning, characterization and three-dimensional modeling of porcine nectin-2/CD112. Vet Immunol Immunopathol. 2009;132:257–63. doi: 10.1016/j.vetimm.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Pende D, Castriconi R, Romagnani P, Spaggiari GM, Marcenaro S, Dondero A, et al. Expression of the DNAM-1 ligands, Nectin-2 (CD112) and poliovirus receptor (CD155), on dendritic cells: relevance for natural killer-dendritic cell interaction. Blood. 2006;107:2030–6. doi: 10.1182/blood-2005-07-2696. [DOI] [PubMed] [Google Scholar]
- Castriconi R, Dondero A, Corrias MV, Lanino E, Pende D, Moretta L, Bottino C, Moretta A. Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: critical role of DNAX accessory molecule-1-poliovirus receptor interaction. Cancer Res. 2004;64:9180–4. doi: 10.1158/0008-5472.CAN-04-2682. [DOI] [PubMed] [Google Scholar]
- Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol. 2013;31:413–41. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat Rev Immunol. 2009;9:568–80. doi: 10.1038/nri2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113:3503–11. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- Soriani A, Fionda C, Ricci B, Iannitto ML, Cippitelli M, Santoni A. Chemotherapy-elicited upregulation of NKG2D and DNAM-1 ligands as a therapeutic target in multiple myeloma. Oncoimmunology. 2013;2:e26663. doi: 10.4161/onci.26663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxford JL, Tang MLF, Pan MF, Huang CW, Kamran N, Phua CML, et al. ATM-dependent spontaneous regression of early E mu-myc-induced murine B-cell leukemia depends on natural killer and T cells. Blood. 2013;121:2512–21. doi: 10.1182/blood-2012-08-449025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena L, Giuliani E, Matusali G, Cohen EA, Doria M. The human immunodeficiency virus type 1 Vpr protein upregulates PVR via activation of the AIR-mediated DNA damage response pathway. J Gen Virol. 2013;94:2664–9. doi: 10.1099/vir.0.055541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fionda C, Abruzzese MP, Zingoni A, Soriani A, Ricci B, Molfetta R, et al. Nitric oxide donors increase PVR/CD155 DNAM-1 ligand expression in multiple myeloma cells: role of DNA damage response activation. BMC Cancer. 2015;15:17. doi: 10.1186/s12885-015-1023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–67. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamran N, Takai Y, Miyoshi J, Biswas SK, Wong JS, Gasser S. Toll-like receptor ligands induce expression of the costimulatory molecule CD155 on antigen-presenting cells. PLoS ONE. 2013;8:e54406. doi: 10.1371/journal.pone.0054406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasec P, Wang EC, Davison AJ, Vojtesek B, Armstrong M, Griffin C, et al. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol. 2005;6:181–8. doi: 10.1038/ni1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prod'homme V, Sugrue DM, Stanton RJ, Nomoto A, Davies J, Rickards CR, et al. Human cytomegalovirus UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112. J Gen Virol. 2010;91:2034–9. doi: 10.1099/vir.0.021931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappworth IY, Wang EC, Rowe M. The switch from latent to productive infection in Epstein–Barr virus-infected B cells is associated with sensitization to NK cell killing. J Virol. 2007;81:474–82. doi: 10.1128/JVI.01777-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduan S, Reif T, Schindler M, Schubert U. HIV-1 Vpu mediated downregulation of CD155 requires alanine residues 10, 14 and 18 of the transmembrane domain. Virology. 2014;464:375–84. doi: 10.1016/j.virol.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusali G, Potesta M, Santoni A, Cerboni C, Doria M. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol. 2012;86:4496–504. doi: 10.1128/JVI.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano E, Joller N, Cao Y, Kuchroo VK, Hafler DA. The CD226/CD155 interaction regulates the proinflammatory (Th1/Th17)/anti-inflammatory (Th2) balance in humans. J Immunol. 2013;191:3673–80. doi: 10.4049/jimmunol.1300945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Ramsbottom KM, Hawkins ED, Shimoni R, McGrath M, Chan CJ, Russell SM, Smyth MJ, Oliaro J. Cutting edge: DNAX accessory molecule 1-deficient CD8+ T cells display immunological synapse defects that impair antitumor immunity. J Immunol. 2014;192:553–7. doi: 10.4049/jimmunol.1302197. [DOI] [PubMed] [Google Scholar]
- Balsamo M, Zambello R, Teramo A, Pedrazzi M, Sparatore B, Scordamaglia F, et al. Analysis of NK cell/DC interaction in NK-type lymphoproliferative disease of granular lymphocytes (LDGL): role of DNAM-1 and NKp30. Exp Hematol. 2009;37:1167–75. doi: 10.1016/j.exphem.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Ardolino M, Zingoni A, Cerboni C, Cecere F, Soriani A, Iannitto ML, Santoni A. DNAM-1 ligand expression on Ag-stimulated T lymphocytes is mediated by ROS-dependent activation of DNA-damage response: relevance for NK-T cell interaction. Blood. 2011;117:4778–86. doi: 10.1182/blood-2010-08-300954. [DOI] [PubMed] [Google Scholar]
- Koch J, Steinle A, Watzl C, Mandelboim O. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 2013;34:182–91. doi: 10.1016/j.it.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Upshaw JL, Leibson PJ. NKG2D-mediated activation of cytotoxic lymphocytes: unique signaling pathways and distinct functional outcomes. Semin Immunol. 2006;18:167–75. doi: 10.1016/j.smim.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Ralston KJ, Hird SL, Zhang X, Scott JL, Jin B, Thorne RF, et al. The LFA-1-associated molecule PTA-1 (CD226) on T cells forms a dynamic molecular complex with protein 4.1G and human discs large. J Biol Chem. 2004;279:33816–28. doi: 10.1074/jbc.M401040200. [DOI] [PubMed] [Google Scholar]
- Shibuya K, Shirakawa J, Kameyama T, Honda S, Tahara-Hanaoka S, Miyamoto A, et al. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J Exp Med. 2003;198:1829–39. doi: 10.1084/jem.20030958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Long EO. Complementary phosphorylation sites in the adaptor protein SLP-76 promote synergistic activation of natural killer cells. Sci Signal. 2012;5:ra49. doi: 10.1126/scisignal.2002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Das A, Gross CC, Bryceson YT, Long EO. Synergistic signals for natural cytotoxicity are required to overcome inhibition by c-Cbl ubiquitin ligase. Immunity. 2010;32:175–86. doi: 10.1016/j.immuni.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara-Hanaoka S, Shibuya K, Kai H, Miyamoto A, Morikawa Y, Ohkochi N, Honda S, Shibuya A. Tumor rejection by the poliovirus receptor family ligands of the DNAM-1 (CD226) receptor. Blood. 2006;107:1491–6. doi: 10.1182/blood-2005-04-1684. [DOI] [PubMed] [Google Scholar]
- Chan CJ, Andrews DM, McLaughlin NM, Yagita H, Gilfillan S, Colonna M, Smyth MJ. DNAM-1/CD155 interactions promote cytokine and NK cell-mediated suppression of poorly immunogenic melanoma metastases. J Immunol. 2010;184:902–11. doi: 10.4049/jimmunol.0903225. [DOI] [PubMed] [Google Scholar]
- Iguchi-Manaka A, Kai H, Yamashita Y, Shibata K, Tahara-Hanaoka S, Honda S, et al. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med. 2008;205:2959–64. doi: 10.1084/jem.20081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven DH, de Hooge AS, Mooiman EC, Santos SJ, ten Dam MM, Gelderblom H, et al. NK cells recognize and lyse Ewing sarcoma cells through NKG2D and DNAM-1 receptor dependent pathways. Mol Immunol. 2008;45:3917–25. doi: 10.1016/j.molimm.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Fregni G, Perier A, Pittari G, Jacobelli S, Sastre X, Gervois N, et al. Unique functional status of natural killer cells in metastatic stage IV melanoma patients and its modulation by chemotherapy. Clin Cancer Res. 2011;17:2628–37. doi: 10.1158/1078-0432.CCR-10-2084. [DOI] [PubMed] [Google Scholar]
- Morgado S, Sanchez-Correa B, Casado JG, Duran E, Gayoso I, Labella F, Solana R, Tarazona R. NK cell recognition and killing of melanoma cells is controlled by multiple activating receptor-ligand interactions. J Innate Immun. 2011;3:365–73. doi: 10.1159/000328505. [DOI] [PubMed] [Google Scholar]
- Pende D, Bottino C, Castriconi R, Cantoni C, Marcenaro S, Rivera P, et al. PVR (CD155) and Nectin-2 (CD112) as ligands of the human DNAM-1 (CD226) activating receptor: involvement in tumor cell lysis. Mol Immunol. 2005;42:463–9. doi: 10.1016/j.molimm.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Lakshmikanth T, Burke S, Ali TH, Kimpfler S, Ursini F, Ruggeri L, et al. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J Clin Investig. 2009;119:1251–63. doi: 10.1172/JCI36022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor S, Durst M, Jansen L, Accardi R, Tommasino M, Trunk MJ, et al. Activating NK cell receptor ligands are differentially expressed during progression to cervical cancer. Int J Cancer. 2008;123:2343–53. doi: 10.1002/ijc.23733. [DOI] [PubMed] [Google Scholar]
- Hou S, Zheng X, Wei H, Tian Z, Sun R. Recombinant soluble CD226 protein directly inhibits cancer cell proliferation in vitro. Int Immunopharmacol. 2014;19:119–26. doi: 10.1016/j.intimp.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Pende D, Spaggiari GM, Marcenaro S, Martini S, Rivera P, Capobianco A, et al. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112) Blood. 2005;105:2066–73. doi: 10.1182/blood-2004-09-3548. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Su T, He L, Wang H, Ji G, Liu X, Zhang Y, Dong G. Identification and functional analysis of ligands for natural killer cell activating receptors in colon carcinoma. Tohoku J Exp Med. 2012;226:59–68. doi: 10.6120/tjem.226.59. [DOI] [PubMed] [Google Scholar]
- Carlsten M, Bjorkstrom NK, Norell H, Bryceson Y, van Hall T, Baumann BC, et al. DNAX accessory molecule-1 mediated recognition of freshly isolated ovarian carcinoma by resting natural killer cells. Cancer Res. 2007;67:1317–25. doi: 10.1158/0008-5472.CAN-06-2264. [DOI] [PubMed] [Google Scholar]
- Pfeiffer M, Seitz G, Ruck P, Mueller C, Steinle A, Lang P, et al. CD155 is involved in NK-cell mediated lysis of human hepatoblastoma in vitro. Front Biosci (Elite Ed) 2011;3:1456–66. doi: 10.2741/e346. [DOI] [PubMed] [Google Scholar]
- Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71:5412–22. doi: 10.1158/0008-5472.CAN-10-4179. [DOI] [PubMed] [Google Scholar]
- Martinovic KMM, Babovic NL, Dzodic RR, Jurisic VB, Tanic NT, Konjevic GM. Decreased expression of NKG2D, NKp46, DNAM-1 receptors, and intracellular perforin and STAT-1 effector molecules in NK cells and their dim and bright subsets in metastatic melanoma patients. Melanoma Res. 2014;24:295–304. doi: 10.1097/CMR.0000000000000072. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhang T, Zhuang R, Zhang Y, Jia W, Song C, et al. Increased levels of soluble CD226 in sera accompanied by decreased membrane CD226 expression on peripheral blood mononuclear cells from cancer patients. BMC Immunol. 2009;10:34. doi: 10.1186/1471-2172-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu P, Huang X, Zhou X, Lu Z, Liu F, Shi Z, et al. Loss of CD155 expression predicts poor prognosis in hepatocellular carcinoma. Histopathology. 2015;66:706–14. doi: 10.1111/his.12584. [DOI] [PubMed] [Google Scholar]
- Sloan KE, Eustace BK, Stewart JK, Zehetmeier C, Torella C, Simeone M, et al. CD155/PVR plays a key role in cell motility during tumor cell invasion and migration. BMC Cancer. 2004;4:73. doi: 10.1186/1471-2407-4-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagumo Y, Iguchi-Manaka A, Yamashita-Kanemaru Y, Abe F, Bernhardt G, Shibuya A, Shibuya K. Increased CD112 expression in methylcholanthrene-induced tumors in CD155-deficient mice. PLoS ONE. 2014;9:e112415. doi: 10.1371/journal.pone.0112415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W. Infusions of allogeneic natural killer cells as cancer therapy. Clin Cancer Res. 2014;20:3390–400. doi: 10.1158/1078-0432.CCR-13-1766. [DOI] [PubMed] [Google Scholar]
- Boissel L, Betancur-Boissel M, Lu W, Krause DS, Van Etten RA, Wels WS, Klingemann H. Retargeting NK-92 cells by means of CD19- and CD20-specific chimeric antigen receptors compares favorably with antibody-dependent cellular cytotoxicity. Oncoimmunology. 2013;2:e26527. doi: 10.4161/onci.26527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Wang X, Jin H. Cell transfer therapy for cancer: past, present, and future. J Immunol Res. 2014;2014:525913. doi: 10.1155/2014/525913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingemann H. Are natural killer cells superior CAR drivers? Oncoimmunology. 2014;3:e28147. doi: 10.4161/onci.28147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MR, Zhang T, Alcon A, Sentman CL. DNAM-1-based chimeric antigen receptors enhance T cell effector function and exhibit in vivo efficacy against melanoma. Cancer Immunol Immunother. 2015;64:409–18. doi: 10.1007/s00262-014-1648-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Deng Y, Benson DM, He S, Hughes T, Zhang J, et al. CS1-specific chimeric antigen receptor (CAR)-engineered natural killer cells enhance in vitro and in vivo antitumor activity against human multiple myeloma. Leukemia. 2014;28:917–27. doi: 10.1038/leu.2013.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T, Onishi H, Katano M. Cancer immunotherapy using NKG2D and DNAM-1 systems. Anticancer Res. 2012;32:2241–7. [PubMed] [Google Scholar]
- Welch MJ, Teijaro JR, Lewicki HA, Colonna M, Oldstone MBA. CD8 T cell defect of TNF-α, and IL-2 in DNAM-1 deficient mice delays clearance in vivo of a persistent virus infection. Virology. 2012;429:163–70. doi: 10.1016/j.virol.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri G, Muntasell A, Romo N, Saez-Borderias A, Pende D, Geraghty DE, et al. NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood. 2011;117:848–56. doi: 10.1182/blood-2010-08-301374. [DOI] [PubMed] [Google Scholar]
- Stegmann KA, Bjorkstrom NK, Ciesek S, Lunemann S, Jaroszewicz J, Wiegand J, et al. Interferon-α-stimulated natural killer cells from patients with acute hepatitis C virus (HCV) infection recognize HCV-infected and uninfected hepatoma cells via DNAX accessory molecule-1. J Infect Dis. 2012;205:1351–62. doi: 10.1093/infdis/jis210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Presti R, Vermi W, Lavender K, Turnbull E, Ochsenbauer-Jambor C, et al. Loss of DNAM-1 contributes to CD8+ T-cell exhaustion in chronic HIV-1 infection. Eur J Immunol. 2010;40:949–54. doi: 10.1002/eji.200940234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N, Imbert AM, Devilard E, Fabre S, Chabannon C, Xerri L, et al. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J Exp Med. 2004;199:1331–41. doi: 10.1084/jem.20032206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural-killer cells and dendritic cells: “l'union fait la force”. Blood. 2005;106:2252–8. doi: 10.1182/blood-2005-03-1154. [DOI] [PubMed] [Google Scholar]
- Chiche L, Forel JM, Thomas G, Farnarier C, Vely F, Blery M, Papazian L, Vivier E. The role of natural killer cells in sepsis. J Biomed Biotechnol. 2011;2011:986491. doi: 10.1155/2011/986491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomchik WD, Couzens MS, Tang CB, McNiff J, Robert ME, Liu J, Shlomchik MJ, Emerson SG. Prevention of graft versus host disease by inactivation of host antigen-presenting cells. Science. 1999;285:412–5. doi: 10.1126/science.285.5426.412. [DOI] [PubMed] [Google Scholar]
- Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- Nabekura T, Shibuya K, Shibuya A. DNAX accessory molecule-1 (DNAM-1) plays an important role in alloreactive CD8+ T cells responsible for the exacerbation of acute graft-versus-host disease. Proc Natl Acad Sci USA. 2011;108:E34–E. [Google Scholar]
- Koyama M, Kuns RD, Olver SD, Lineburg KE, Lor M, Teal BE, et al. Promoting regulation via the inhibition of DNAM-1 after transplantation. Blood. 2013;121:3511–20. doi: 10.1182/blood-2012-07-444026. [DOI] [PubMed] [Google Scholar]
- Du Y, Tian L, Shen LX, Wang F, Yu LK, Song Y, Zhu JF, Du R. Association of the CD226 single nucleotide polymorphism with systemic lupus erythematosus in the Chinese Han population. Tissue Antigens. 2011;77:65–7. doi: 10.1111/j.1399-0039.2010.01568.x. [DOI] [PubMed] [Google Scholar]
- Avouac J, Elhai M, Tomcik M, Ruiz B, Friese M, Piedavent M, et al. Critical role of the adhesion receptor DNAX accessory molecule-1 (DNAM-1) in the development of inflammation-driven dermal fibrosis in a mouse model of systemic sclerosis. Ann Rheum Dis. 2013;72:1089–98. doi: 10.1136/annrheumdis-2012-201759. [DOI] [PubMed] [Google Scholar]
- Elhai M, Chiocchia G, Marchiol C, Lager F, Renault G, Colonna M, et al. Targeting CD226/DNAX accessory molecule-1 (DNAM-1) in collagen-induced arthritis mouse models. J Inflamm. 2015;12:9. doi: 10.1186/s12950-015-0056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattana TC, Santos AS, Fukui RT, Mainardi-Novo DT, Costa VS, Santos RF, Matioli SR, da Silva ME. CD226 rs763361 is associated with the susceptibility to type 1 diabetes and greater frequency of GAD65 autoantibody in a Brazilian cohort. Mediators Inflamm. 2014;2014:694948. doi: 10.1155/2014/694948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen N, Pascal V, Fasth AE, Sundstrom Y, Galsgaard ED, Ahern D, et al. Balance between activating NKG2D, DNAM-1, NKp44 and NKp46 and inhibitory CD94/NKG2A receptors determine natural killer degranulation towards rheumatoid arthritis synovial fibroblasts. Immunology. 2014;142:581–93. doi: 10.1111/imm.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel LA, Yokoyama WM, French AR. Natural killer cells in human autoimmune disorders. Arthritis Res Ther. 2013;15:216. doi: 10.1186/ar4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Shi FD, Jung S, Pien GC, Wang J, Salazar-Mather TP, et al. The neuronal chemokine CX3CL1/fractalkine selectively recruits NK cells that modify experimental autoimmune encephalomyelitis within the central nervous system. FASEB J. 2006;20:896–905. doi: 10.1096/fj.05-5465com. [DOI] [PubMed] [Google Scholar]