Abstract
Inheritance of each chromosome depends upon its centromere. A histone H3 variant, CENP-A, is essential for epigenetically marking centromere location. We find that CENP-A is quantitatively retained at the centromere upon which it is initially assembled. CENP-C binds to CENP-A nucleosomes and is a prime candidate to stabilize centromeric chromatin. Using purified components, we find that CENP-C reshapes the octameric histone core of CENP-A nucleosomes, rigidifies both surface and internal nucleosome structure, and modulates terminal DNA to match the loose wrap that is found on native CENP-A nucleosomes at functional human centromeres. Thus, CENP-C affects nucleosome shape and dynamics in a manner analogous to allosteric regulation of enzymes. CENP-C depletion leads to rapid removal of CENP-A from centromeres, indicating their collaboration in maintaining centromere identity.
Centromeres direct chromosome inheritance at cell division, and nucleosomes containing a histone H3 variant, CENP-A, are central to current models of an epigenetic program for specifying centromere location (1). The centromere inheritance model in metazoans suggests that the high local concentration of pre-existing CENP-A nucleosomes at the centromere guides the assembly of nascent CENP-A which occurs once per cell cycle following mitotic exit. This model predicts that after initial assembly into centromeric chromatin, CENP-A must be stably retained at that centromere; otherwise centromere identity would be lost before the next opportunity for new loading in the following cell cycle. Here, we use biochemical reconstitution to measure the shape and physical properties of CENP-A nucleosomes with and without its close binding partner, CENP-C, and combine these studies with functional tests that reveal the mechanisms underlying the high stability of centromeric chromatin.
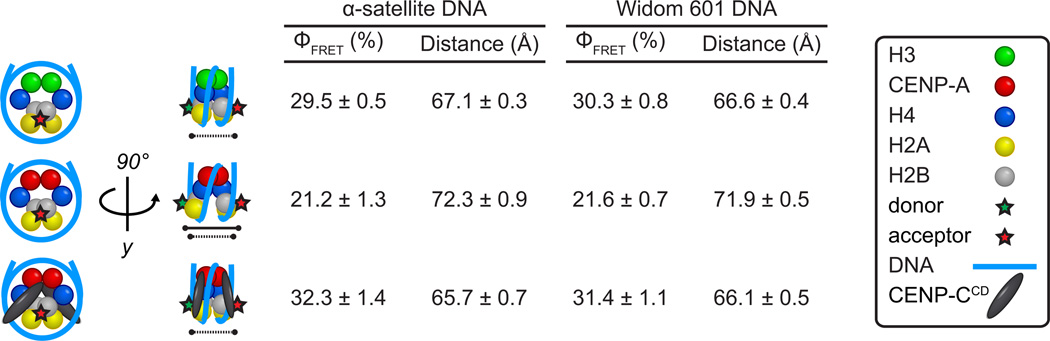
CENP-C recognizes CENP-A nucleosomes via a region termed its central domain (a.a. 426–537; CENP-CCD) (2, 3). We first considered how CENP-CCD may affect the overall shape of the CENP-A-containing nucleosome using an intranucleosomal fluorescence resonance energy transfer (FRET)-based approach. We designed an experiment to measure FRET efficiency, ΦFRET, between two fluorophores on defined positions on the H2B subunits of CENP-A nucleosomes in the absence or presence of CENP-CCD, and then used these measurements to calculate intranucleosomal distances (Figs. 1 and S1). The H2B distances for CENP-A nucleosomes in the absence of CENP-CCD are ~5 Å further apart than expected from their crystal structure (PDB ID 3AN2) (4), indicating that CENP-A-containing nucleosomes in solution prefer a histone octamer configuration not captured in the crystal structure. It is likely that CENP-A nucleosomes sample both conformations in solution, with crystal contacts stabilizing the form that was reported (4). In contrast to CENP-A nucleosomes, conventional nucleosomes have smaller H2B distances in solution (Fig. 1) that are consistent with their crystal structure (5). Separation of H2A/H2B dimers from each other is consistent with a nucleosome model based on rotation of the CENP-A/CENP-A’ interface in (CENP-A/H4)2 heterotetramers (6). Upon binding of CENP-CCD, with the known stoichiometry of two CENP-CCD molecules per nucleosome (3), the H2A/H2B distances shorten to ones that are nearly identical to conventional nucleosomes (Fig. 1). The differences we observed between H3 nucleosomes, CENP-A nucleosomes, and CENP-A nucleosomes in a complex with CENP-CCD are found using either the human α-satellite DNA sequence that corresponds to the most heavily occupied site at centromeres (7) or the completely synthetic ‘601’ nucleosome positioning sequence (8) (Fig. 1).
Figure 1. CENP-A nucleosomes have a conventional shape only upon CENP-CCD binding.
Calculated FRET efficiencies (ΦFRET) and distances between donor and acceptor fluorophores on H2B S123C for the indicated nucleosomes on either α-satellite or Widom 601 DNA. Data are shown as the mean ± s.e.m of three independent nucleosome reconstitutions.
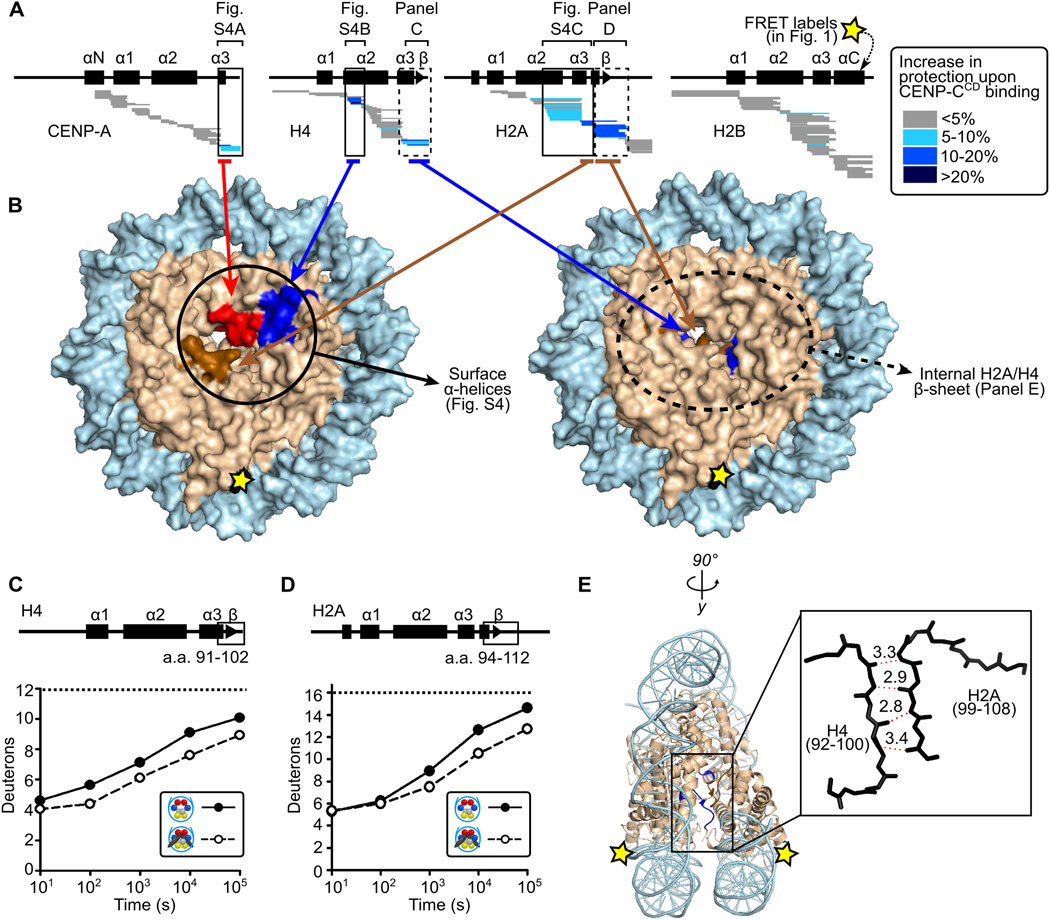
The shape change we measure within the nucleosome upon CENP-CCD binding most likely occurs through rotation at the four-helix bundles between histone dimer pairs within the octameric core with inter-histone contacts being stabilized or destabilized depending on the preference for rotational state. We tested this prediction using hydrogen/deuterium exchange-mass spectrometry (HXMS). Strong protection of CENP-A nucleosomes (Figs. 2A, S3D, and S4) is conferred by CENP-CCD binding on peptides spanning helices that are predicted (3) to contact it (i.e. the α3 helix and C-terminal residues of CENP-A, the α2 helices of both H4 and H2A, and regions of H2A encompassing its acidic patch residues). In addition to the surface changes induced by CENP-CCD, there are internal changes to the nucleosome that we measure by HX (Figs. 2A,B and movie S1) that are consistent with the change in nucleosome shape that we observed by FRET (Fig. 1). The separation of H2A/H2B dimers in CENP-A nucleosomes lacking CENP-CCD (Fig. 1) is predicted to weaken an internal, intermolecular β-sheet that serves as the physical connection between the H2A subunit on one face of the nucleosome and the H4 subunit on the opposite face. When CENP-CCD binds to the CENP-A nucleosome, peptides spanning the corresponding β-sheet residues of both H2A and H4 exhibit extra protection from HX by 1–2 deuterons, where the same level of HX takes 5–10 times as long to occur than in CENP-A nucleosomes lacking CENP-CCD (Figs. 2, S5, and S6).
Figure 2. CENP-CCD rigidifies CENP-A nucleosomes.
(A) HXMS of all histone subunits of the CENP-A nucleosome from a single time point (104 s; see all time points in fig. S3). Each horizontal bar represents an individual peptide, and peptides are placed beneath schematics of secondary structural elements. (B) Regions showing substantial protection from HX mapped onto the structure of the CENP-A nucleosome (PDB 3AN2). (C and D) Comparison of representative peptides spanning the β-sheet region in histone H4 and histone H2A over the time course. The maximum number of deuterons possible to measure by HXMS for each peptide is shown by the dotted line. (E) The internal H4/H2A interface mapped onto the canonical nucleosome crystal structure (PDB 1KX5).
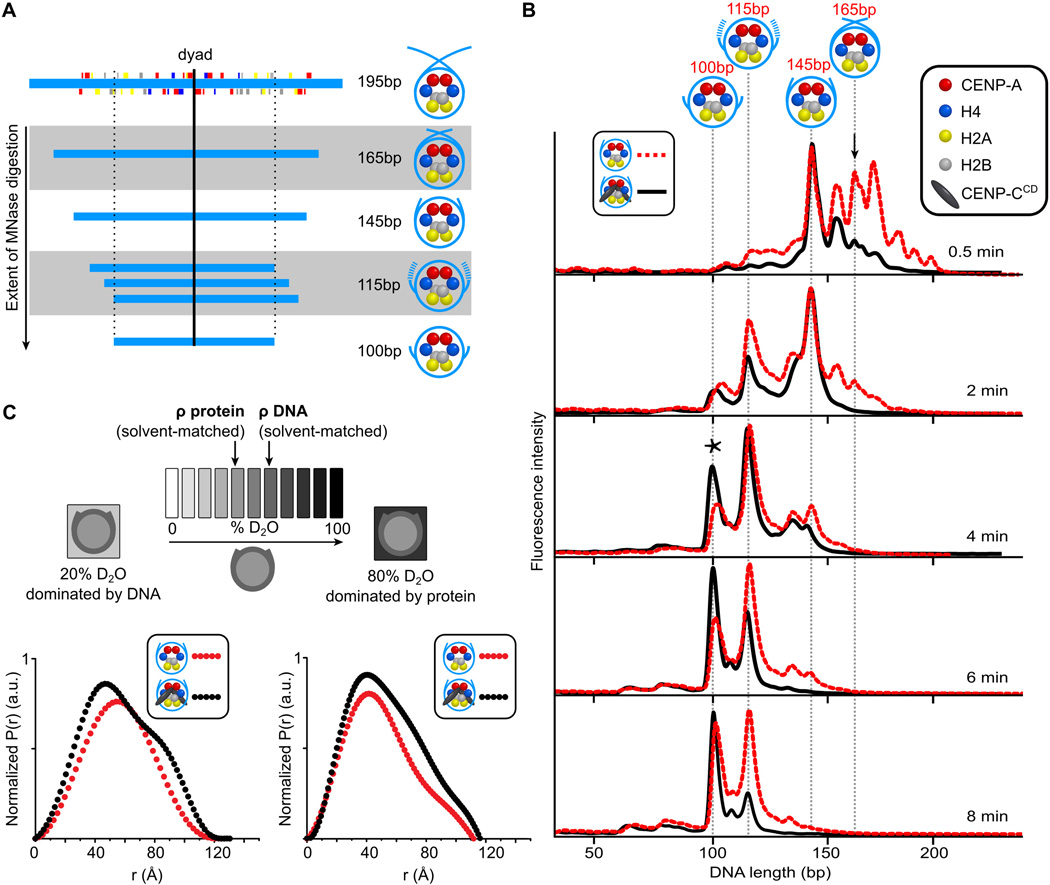
Since CENP-C might also affect the extent that DNA wraps the nucleosomes, we reconstituted CENP-A nucleosomes using an 195 bp DNA sequence from α-satellite DNA (9) that contains a contiguous sequence spanning the major binding site it occupies on human centromeres (7) (Fig. 3A). We first over-digested CENP-A nucleosomes and found very strong protection of 100 bp (fig. S9). Using a subsequent restriction digest of the 100 bp digestion product, we found that they were uniquely positioned with their dyad precisely where the same sized fragment previously mapped with native centromeric particles (7) (fig. S9). CENP-A-containing nucleosomes have many discrete intermediate digestion products before the strongly protected 100 bp fragment is generated (Figs. 3A,B and S10). When CENP-CCD is bound, digestion products larger than a nucleosome core particle (e.g. >145 bp where DNA strands could cross at ~165 bp for conventional nucleosomes (10)) are missing at early timepoints (Fig. 3B). This suggests that when CENP-CCD binds to the nucleosome the DNA above the dyad rarely crosses, as it would normally cross for conventional nucleosomes. Second, digestion to the 100 bp final fragment proceeds more quickly (Fig. 3B). Thus, transient unwrapping of two helical turns (i.e. ~20 bp) from each terminus of the nucleosome is enhanced when CENP-CCD is bound.
Figure 3. Alterations in the nucleosome terminal DNA upon CENP-CCD binding.
(A) Major MNase-digested DNA fragments observed for CENP-A nucleosomes assembled on its native centromere sequence. (B) MNase digestion profiles of CENP-A nucleosomes in the absence (red) and presence (black) of CENP-CCD. The black arrow (0.5 min) points to the 165 bp peak (DNA crossed at the dyad). The asterisk (4 min) denotes the final 100 bp peak. (C) Scheme of SANS contrast variation experiment together with paired distance distribution curves for CENP-A nucleosomes alone (red) and bound by CENP-CCD (black) in the indicated SANS contrast variation conditions.
To determine if CENP-CCD binding leads to a steady-state structural change of nucleosomal DNA, we used small-angle neutron scattering (SANS) with contrast variation. When CENP-CCD binds to CENP-A nucleosome core particles, the distance distribution profiles reflecting the shape in solution substantially redistribute for both the protein- and DNA-dominated measurements (Figs. 3C, S11, and Table S3). The increase in larger interatomic vectors for the protein component is expected to accompany an additional component (CENP-CCD). The pronounced redistribution of vectors to both smaller and larger distances in DNA dominated scattering when CENP-CCD is bound is attributed to compaction of the nucleosome core (smaller vectors), and opening of the nucleosome terminal DNA when CENP-CCD is bound (larger vectors).
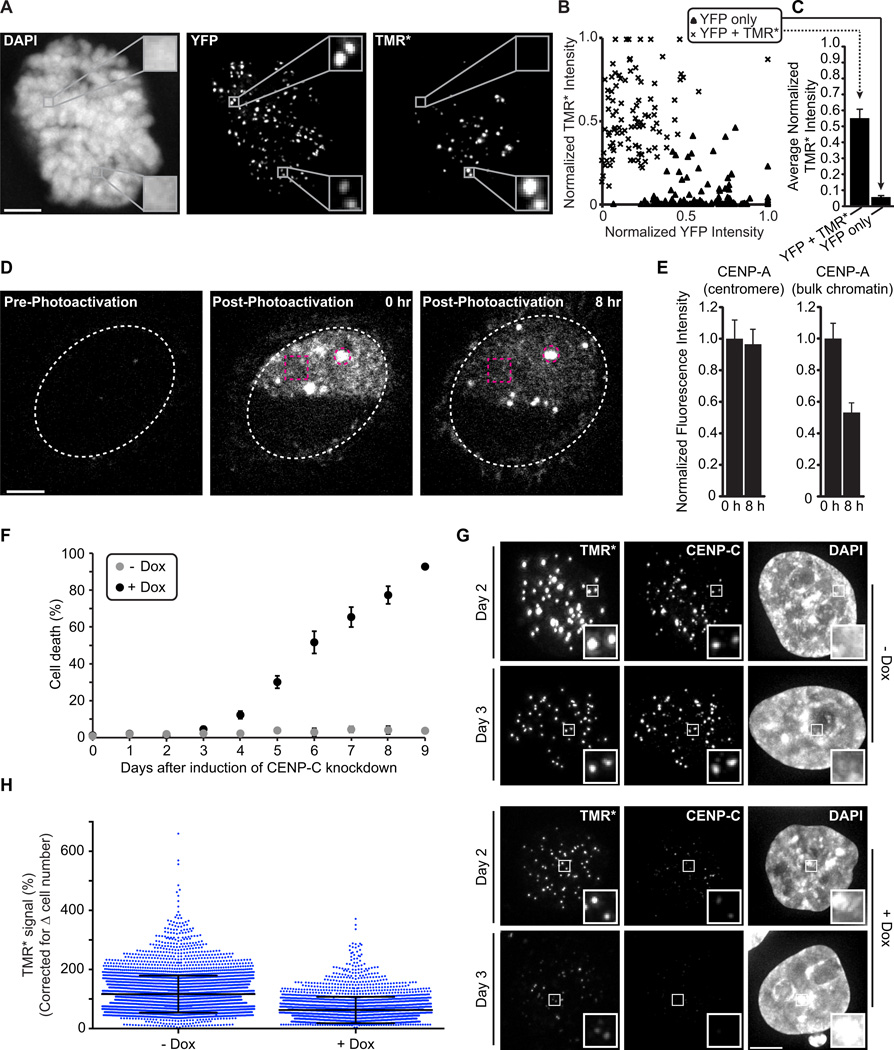
We took two complementary approaches in cells to determine whether CENP-A is stably retained at the centromere upon which it is initially deposited (see the legend for fig. S12 that describes the motivation for these experiments). First, we used cell cycle-synchronized fluorescence pulse labeling of CENP-A in ‘donor’ cells and subsequent cell fusion with an ‘acceptor’ cell line. The donor cells express SNAP-tagged CENP-A that has been pulse labeled with TMR-Star (TMR*) to irreversibly label CENP-A (11) prior to cell fusion. The acceptor cells express YFP-tagged CENP-A that is loaded at all centromeres, continuing even after fusion. At time points through the subsequent cell cycle (fig. S12) until the second mitosis (Fig. 4A), we observed no detectable exchange of the TMR* labeled donor CENP-A to the acceptor centromeres in a shared nucleoplasm. Quantitation of the fluorescence at each centromere in these heterokaryons yields a bimodal distribution. The donor centromere group with high TMR* and low YFP (Fig. 4B, ‘x’ symbols) has an average TMR* signal of 0.538 +/− 0.005 (normalized arbitrary units where the maximal measured TMR* signal in each heterokaryon equals 1; Fig. 4C), whereas the acceptor centromere group with high YFP and low TMR* (Fig. 4B, triangle symbols) has an average TMR* signal of 0.055 +/− 0.005 (Fig. 4C). These data indicate that once assembled at a centromere, an individual CENP-A molecule is stably maintained at that particular centromere.
Figure 4. Depletion of CENP-C reduces the high stability of CENP-A at centromeres.
(A–C) Cells expressing SNAP-tagged CENP-A were pulse labeled with TMR-Star (TMR*), then fused with cells expressing YFP-tagged CENP-A. Representative images (A) show a cell in the second mitosis after fusion; insets 3× magnification. X-means clustering was used to classify YFP only (triangles) or YFP and TMR* (‘x’ marks) centromeres (B), and mean (± s.e.m) TMR* intensity was calculated for each group (C). (D, E) Cells expressing high levels of CENP-A-PAGFP were photoactivated in bulk (box) and centromeric (circle) chromatin. Representative images (D) show a subset of centromeres in a single z-section. Fluorescence intensity was quantified at 0 and 8 hr after photoactivation (E, mean ± s.e.m). (F) CENP-C knockdown begins causing cell death 4 days post-induction (mean ± s.d.) (G, H) Cells with (+ Dox) and without (- Dox) CENP-C depletion were pulse labeled with TMR* (Day 2), and the relative CENP-A-SNAP signals were analyzed (Day 3). Quantification shows CENP-A-SNAP signal retained at day 3 (>2500 centromeres plotted with mean ± s.d.). Scale bars, 5 µm.
As a complementary approach to test CENP-A stability at individual centromeres, we used a photoactivatable version of CENP-A (CENP-A-PAGFP). We induced expression of CENP-A to the extent that it is present at locations throughout the nucleus, but with clear enrichment at centromeres, and then activated a defined region of each cell nucleus (Fig. 4D [0 hr post-photoactivation]). CENP-A-PAGFP signal is quantitatively retained at the activated centromeres and does not accumulate at unactivated centromeres (Fig. 4D,E), indicating that there is negligible exchange between centromeres, consistent with our cell fusion results. In contrast, CENP-A-PAGFP signal in bulk chromatin decays with ~half of the protein removed by 8 hr following photoactivation.
To test if CENP-C stabilizes CENP-A at centromeres, we combined SNAP labeling of CENP-A with CENP-C depletion (Fig. 4) for which we generated a cell line with a chromosomally integrated, doxycycline-inducible CENP-C shRNA cassette. In our SNAP system, CENP-C depletion leads to a dramatic decrease in the retention over 24 hr of the existing pool of CENP-A at centromeres (Fig. 4G,H). Without CENP-C depletion, the average retention of CENP-A is slightly >100% (112% +/− 63% s.d.), an increase that is explained by having a small pool of pre-nucleosomal CENP-A in the cell population that is labeled by the TMR* pulse and subsequently incorporated into centromeres. Nascent CENP-A deposition is also decreased when CENP-C is depleted (fig. S16C)—consistent with its proposed role in the CENP-A assembly reaction (12, 13) — but this would only impact incorporation of the small pre-nucleosomal pool in the CENP-A retention measurements (Fig. 4G,H). Thus, our findings implicate CENP-C in stabilizing CENP-A nucleosomes at centromeres. We cannot rule out the possibility that removal of CENP-C in turn removes another centromere component that stabilizes CENP-A nucleosomes, but we favor the idea that CENP-C is the key molecule for stabilizing CENP-A nucleosomes based on its direct binding to it.
CENP-A nucleosomes are highly stable at the centromeres upon which they are initially assembled. This stability is possible through collaboration with CENP-C. Along with the intranucleosomal rigidity of CENP-A and histone H4, where the key interfacial amino acids are important for accumulation at centromeres (6, 14, 15), the physical changes imposed by CENP-C combine to make CENP-A nucleosomes at centromeres very long-lived (fig. S21). Our data support a model of a steady-state octameric histone core where H2A/H2B dimers can exchange from either terminus of the CENP-A nucleosome. At the center, there is an essentially immobile (CENP-A/H4)2 heterotetramer (16) (Figs. 4 and S12–14). Thus, the physical properties related to CENP-A nucleosome stability at centromeres are tied to the intrinsic properties of the (CENP-A/H4)2 heterotetramer (6, 14, 15) and the extrinsic properties imposed by CENP-C (Figs. 1–3 and S21).
Supplementary Material
Acknowledgements
We thank our UPenn colleagues [J. Bell, C. Bialas, J. DeNizio, Y. Goldman, W. Hu, L. Lippert, L. Mayne, T. Panchenko, B. Türegün, and Z. Zhao] and S. Krueger and K. Sarachan (NIST) for assistance and advice with experiments, A. Straight (Stanford) for reagents and advice, and D. Cleveland (UCSD), J. Lippincott-Schwartz (NIH), K. Luger (Colorado State), and E. Makeyev (Nanyang Technical Univ.) for reagents. This work was supported by NIH grant GM082989 (B.E.B.), ERC grant ERC-2013-CoG-615638 (L.E.T.J.), predoctoral fellowships from AHA (E.M.S.) and NIH (CA186430; L.Y.G.), and a postdoctoral fellowship from ACS (N.S.). We acknowledge support from NIH grants GM008275 (UPenn Structural Biology Training Grant; L.Y.G. and E.M.S), GM008216 (UPenn Genetics Training Grant; S.J.F.), and GM007229 (UPenn Cell and Molecular Biology Training Grant; G.A.L.). This work utilized facilities supported in part by the NSF (Agreement #DMR-0944772). We acknowledge the support of NIST in providing the neutron research facilities used in this work.
Footnotes
Supplementary Materials:
Materials and Methods
Figures S1–S21
Tables S1–S3
Movie S1
References (17–56)
References
- 1.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato H, et al. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasson D, et al. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 9.Harp JM, et al. X-ray diffraction analysis of crystals containing twofold symmetric nucleosome core particles. Acta Crystallogr. D Biol. Crystallogr. 1996;52:283–288. doi: 10.1107/S0907444995009139. [DOI] [PubMed] [Google Scholar]
- 10.Kornberg RD. Structure of chromatin. Annu. Rev. Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 11.Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erhardt S, et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassett EA, et al. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 16.Bodor DL, Valente LP, Mata JF, Black BE, Jansen LET. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.