Abstract
Mammalian hibernation is a unique strategy for winter survival in response to limited food supply and harsh climate, which includes resistance to cardiac arrhythmias. We previously found that hibernating woodchucks (Marmota monax) exhibit natural resistance to Ca2+ overload-related cardiac dysfunction and nitric oxide (NO)-dependent vasodilation, which maintains myocardial blood flow during deep torpor (DT). Since, the cellular/molecular mechanisms mediating the protection are less clear, the goal of this study was to investigate dynamic seasonal changes in the heart proteome and reveal related signaling networks which are involved in establishing cardioprotection in woodchucks during hibernation. This was accomplished using isobaric tags for a relative and absolute quantification (iTRAQ) approach. The most significant seasonal changes observed were upregulation of the anti-oxidant, catalase, and inhibition of endoplasmic reticulum (ER) stress response by downregulation of GRP78, mechanisms which could be responsible for the adaptation and protection in hibernating animals. Furthermore, protein networks pertaining to NO signaling, acute phase response, CREB and NFAT transcriptional regulations, protein kinase A and α-adrenergic signaling were dramatically upregulated during hibernation. These adaptive mechanisms in hibernators may provide new directions to protect myocardium of non-hibernating animals, especially humans, from cardiac dysfunction induced by hypothermic stress and myocardial ischemia.
Keywords: iTRAQ, true hibernation, cardioprotection
INTRODUCTION
Mammalian hibernation is an energy-conservation strategy that allows various small animals to survive winter under conditions of low temperatures and food scarcity. Hibernators such as woodchucks (Marmota monax) undergo a remarkable phenotypic switch that involves physiological, morphological, and biochemical changes in response to environmental stresses 1. During hibernation, hibernators sink into DT where metabolic rate is below 5% of the normal rate, body temperature typically falls to 32–50 °F, and physiological functions, as well as protein turnover, are profoundly depressed. During torpor, to match decreased metabolic demand, heart rate, mean arterial pressure, and maximum and minimum left ventricle (LV) pressure change rates (dP/dt) are significantly decreased in woodchucks 2. However, myocardial blood flow is actually maintained, which is in apparent contradiction to the concept of decreased myocardial blood flow matching reduced metabolic demand2. Major cardiac stress adaptation during hibernation includes resistance to fibrillation/arrhythmias 3, dynamic maintenance of conduction and repolarization patterns through improved gap junction functions 4, maintenance of Na+/K+ ion homeostasis 5 and improved Ca2+ handling 6. Despite the dramatic physiological adaptation of harsh environment during hibernation, the molecular basis for the adaptive mechanisms in hibernating animals is not well known. We have previously observed that woodchucks demonstrate epigenetic changes from summer to winter in response to ischemic injury7, 8. Although there is now a considerable amount of information related to various aspects of the morphological, physiological, and biochemical changes that are associated with the dramatic adaptation of hibernating myocardium 2–6, the cellular and molecular basis of mammalian hibernation, especially in cardioprotection, are still poorly understood. It is likely that novel mechanisms are involved in cardioprotection, but are not yet identified.
Proteomics approaches are effective at identifying new protein signaling networks, which cannot be readily identified with targeted biochemical approaches. Using a label-free approach, Shao et al. discovered that proteins involved in translation, protein turnover, mRNA processing and oxidative phosphorylation significantly changed in the livers of ground squirrels (Urocitellus parryii) throughout the torpor-arousal cycle during hibernation 9. In contrast, Martin et al. used the two-dimensional differential gel electrophoresis (DIGE) approach to identify protein differences in intestines of ground squirrels (Ictidomys tridecemlineatus) between sham-treated animals with those exposed to ischemia-reperfusion (I/R) 10; these proteomics profiles of intestines were able to distinguish among the sham-treated summer and hibernating samples, as well as I/R-induced proteomic changes between summer and hibernating animals. More recently Grabek et al. discovered the importance of proteins involved in ATP-conserving mechanisms for winter cardioprotection in the hearts of 13-lined ground squirrels 11. In the present study, we conducted two independent sets of iTRAQ quantitative proteomics experiments in order to discover hibernation and DT-specific cardiac proteomic changes in Marmota monax; summer animals were served as the controls in each set of experiments; the first set involved non-hibernating winter animals fed and housed at room temperature and the summer control animals, and the second set involved winter animals in DT and the summer control animals. The proteomics results from these two studies were compared to distinguish DT-specific protein network changes from simply seasonal protein changes. Because the woodchuck is not a widely studied animal model, a complete set of gene and protein sequences are not yet available. Although the availability of high resolution tandem mass spectrometers has enabled the acquisition of high resolution MS/MS spectra, software packages for high throughput “deep” de novo sequencing of shotgun proteomics data and subsequent assembly of peptides into protein sequences are not yet robust enough for routine shotgun proteomics studies of rare species. Consequently, we designed the iTRAQ proteomics approach to search the MS/MS spectra obtained from woodchuck peptides against both rodent and human protein sequences and quantified over 4,000 unique proteins. Based on these quantitative proteomics studies, a surprisingly small number of protein networks are altered specifically during hibernation and DT, suggesting that a select scope of mechanisms may be important for determining cardiac protection in hibernating animals. Development of therapeutics that can recapitulate protein signaling networks similar to the ones found in hibernating animals may lead to better treatments of vascular human diseases.
MATERIALS AND METHODS
Reagents
All chemical reagents were purchased from Sigma (St. Louis, MO, USA) unless otherwise indicated. Sequencing grade trypsin, triethylammonium bicarbonate (TEAB), NP-40, Triton X-100, protease inhibitor cocktail, phosphatase inhibitor cocktail, phosphate buffer saline (PBS), KH2PO4, KCl, formic acid (FA), BCA protein assay kit were purchased from Thermo Scientific (Rockford, IL). iTRAQ 8-plex kit containing tris(2-carboxyethyl) phosphine (TCEP), methyl methanethiosulfonate (MMTS) and 8-plex iTRAQ reagents were obtained from AB SCIEX, (Foster City, CA). Acetonitrile (ACN) and HPLC-grade water were purchased from J. T. Baker, (Phillipsburg, NJ, USA).
Animal models
Animals used in this study were maintained in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 8th Edition) and the Institutional Animal Care and Use Committee at UMDNJ-New Jersey Medical School. Woodchucks used in this study were obtained from Northeastern Wildlife, Idaho. The following groups of animals were analyzed in this study:
DT Group
During winter in Dec – Jan, a subgroup of the male and female woodchucks (n=3–4 were used for different experiments due to limited availability of this animal model) was instrumented (see Supplemental Materials and Methods for details) for telemetry (Model: d70-pctp, Data Science International (DSI), St. Paul, MN) and LV pressure measurements using a micromanometer (Konigsberg Instruments, Inc., Pasadena, CA). After 2 weeks recovery, the woodchucks were placed in the hibernaculum (40–42 °F) (see Supplemental Materials and Methods) without food to allow the animals to go into DT. The animals were monitored for electrocardiography (ECG or EKG) via a telemetry system with d70-pctp as a transmitter and rmc-1 as a DSI receiver. After the animals demonstrated a prolonged DT (heart rate <25 bpm for 2 days and no response to mild external stimuli), the animals were performed for either physiological analysis or euthanized with sodium pentobarbital (100 mg/kg, i.v. to effect) for heart tissue collection.
WRT Group
Heart samples were also collected from woodchucks in winter which were fed and housed at room temperature (WRT, Dec – Jan, n=3–4), thus not experiencing DT.
SM Group
As the base comparison group for the two iTRAQ studies, heart samples were collected from woodchucks in summer (SM, June – July, n=3–5) for both iTRAQ proteomics and downstream biochemical studies described in this study.
NDT Group
For Western blotting analysis, an additional set of heart tissues was also collected from woodchucks in winter (Dec – Jan) which were placed in the hibernaculum at cold temperature for 10 days without food, but failed to enter DT (NDT, n=3). This group was analyzed to determine whether prolonged DT has an effect on cardiac proteins in winter woodchucks exposed to winter conditions.
After the hearts were excised, the blood was quickly washed out from the heart with saline, and the tissue samples were frozen at −80 °C prior to analysis.
Protein extraction and iTRAQ labeling
A total of two sets of 8-plex iTRAQ reagents were used for labeling 11 samples in the two separate experiments (see experimental design in Supplemental Table 1). The hearts were first rinsed with PBS containing a protease inhibitor cocktail on ice to remove any residual blood prior to protein extraction. Proteins were extracted from the 30 mg of heart tissue from each animal using 0.5 ml of lysis buffer containing 100 mM TEAB, 1% NP40, 1% Triton X-100, a protease inhibitor cocktail and a phosphatase inhibitor cocktail. BCA protein assay was used for protein concentration measurements. Following the manufacturer’s instructions (AB SCIEX), 100 μg of protein from each sample was first reduced by 5 mM TCEP at 60 ºC for 1 h, then alkylated with 10 mM MMTS at RT for 10 min and followed by trypsin digestion (1:10 by weight, enzyme to protein ratio) overnight. For both sets of quantitative experiments, peptides derived from the 4 summer woodchucks (SM1 to SM4) were labeled with iTRAQ reagents 113, 114, 115 and 116, whereas peptides derived from 4 winter room temperature woodchucks (WRT1 to WRT4) were labeled with iTRAQ reagents 117, 118, 119 and 121 in the first experiment, and 3 deep torpor woodchucks (DT1 to DT3) were labeled with iTRAQ reagents 117, 118, and 121 in the second experiment, due to limited availability of these hibernating animals at the time of the experiment.
Liquid Chromatography and Tandem Mass Spectrometry (LC/MS/MS)
For each of the two quantitative proteomics experiments, the iTRAQ-labeled peptides were first combined according to the experimental design outlined in Supplemental Table 1 and were then fractionated by strong cation exchange chromatography (SCX) on a BioCAD™ Perfusion Chromatography System (AB SCIEX). A polysulfoethyl A column (4.6 mm × 200 mm, 5 μm, 300 Å, Poly LC Inc., Columbia, MD, USA) was used with a 1-hr binary gradient consisting of Mobile phase A (10 mM KH2PO4 and 25% acetonitrile (ACN), pH 2.7) and Mobile phase B (600 mM KCl, 10 mM KH2PO4 and 25% ACN, pH 2.7), as described previously 12. For each SCX separation, 10 fractions with similar peptide complexities were collected. The peptides in the SCX fractions were desalted using PepClean™ C18 spin columns (Pierce, Rockford, IL, USA) prior to the LC/MS/MS analysis by reversed phase liquid chromatography (RPLC) on an Ultimate 3000 LC system (Dionex, Sunnyvale, CA, USA) coupled with an LTQ Orbitrap Velos tandem mass spectrometer (Thermo Scientific). In brief, the peptides were separated by a C18 RPLC column (75 μm × 150 mm, 3 μm, 100 Å, C18, Dionex, Sunnyvale, CA, USA) at 250 nl/min using an 180-min gradient consisted of solvent A (2% ACN and 0.1% FA) and solvent B (85% ACN and 0.1% FA): 0–140 min, from 3% to 25% B; 140–160 min, from 25% to 45% B and 160 to 170 min, from 45% to 95% B. The eluted peptides were introduced into the Orbitrap via a Proxeon nano electrospray ionization source with a spray voltage of 2 kV and a capillary temperature of 275 ºC. MS spectra were acquired in the positive ion mode with a scanning mass range of m/z 350–2,000 and a resolution of 60,000 full-width at half maximum (FWHM). The Higher Energy Collision Dissociation (HCD) MS/MS spectra were acquired in a data-dependent manner. For WRT vs. SM iTRAQ Experiment 1, 10 most abundant ions were selected for HCD fragmentation per MS scan in the Orbitrap at a resolution of 7,500 FWHM. The normalized collision energy was set to 40. For the DT vs. SM iTRAQ experiment 2, 10 most abundant ions were selected for collision-induced dissociation (CID) fragmentation in the ion trap followed by HCD fragmentation in the Orbitrap. The normalized collision energies were set to 35 for CID and 45 for HCD. The precursor isolation width was set at 2 amu, and a minimum ion threshold count was set as 3,000. The lock mass feature was engaged for accurate mass measurements. A m/z 445.120030 corresponding to polysiloxane ion was used as the lock mass for internal calibration.
Bioinformatics Analysis
The MS/MS spectra from all the analyses were searched against either a UniRef100 human (120,982 entries), mouse (82,522 entries) or rat (51,862 entries) database (downloaded on January 20th, 2012), using both Mascot (V.2.3) and SEQUEST search engines via the Proteome Discoverer platform (V. 1.3, Thermo Scientific). The precursor mass error window was set as 10 ppm and MS/MS error tolerance was set as 0.1 Da for HCD spectra and 0.5 Da for CID spectra with up to one missed cut. Methionine oxidation and tyrosine 8-plex iTRAQ labeling were set as variable modifications, whereas N-terminus and lysine side chain 8-plex iTRAQ labeling and cysteine MMTS conjugation were set as fixed modifications. The resulting .dat files from Mascot and .msf files from Proteome Discoverer were filtered with Scaffold (V3.3.2, Proteome Software, Inc., Portland, OR) for protein identification and quantification analyses. All peptides were identified with at least 95% confidence interval value (C.I. value) as specified by the Peptide Prophet algorithm and less than 1% false discovery rate (FDR) based on forward/reverse database searches 13. Proteins were considered confidently identified with at least one unique peptide, and an experiment-wide FDR of no more than 1.5%. Proteins that share the same peptides and could not be differentiated based on MS/MS analysis alone were grouped together to reduce the redundancy, using Scaffold. Relative quantification of proteins was determined with Scaffold Q+ module in a normalized log2-based relative iTRAQ ratio format, with iTRAQ 113 tag as the reference denominator. The average protein expression ratios between SM and WRT groups, or between SM and DT groups were calculated as the following: (average of the four WRT or the three DT ratios) / (average of the four SM ratios specific to each experiment). Significance of protein expression changes in p-values were calculated using a two-tailed Student’s T-test for each protein, with the four WRT or three DT ratios comparing to their corresponding four SM ratios. The proteins with a greater than 20% changes and p-values 0.05 are considered as significantly changed based on our previously determined analytical variations14. A heatmap for clustering DT/SM and WRT/SM iTRAQ protein expression ratios was created using TM4 microarray software 15. For functional analysis of the affected protein networks, the significantly changed proteins were submitted to Ingenuity (http://www.ingenuity.com/) for pathway analysis.
Western blotting
Twenty μg of proteins/lane from each sample were separated using 10% SDS-PAGE gels and transferred onto nitrocellulose membranes (Biorad, Hercules, CA, USA), which were probed with the primary antibodies at 4 ºC overnight: anti-catalase (1:5000, Abcam, Cambridge, MA, USA); anti-GRP78 (1:1000, Stressgen Bioreagents, Brussels, Belgium); anti-Troponin I (1:1000, Fitzgerald, Acton, MA, USA); anti-ATM (1:2000, Sigma-Aldrich, St Louis, MO, USA); anti-PI3 Kinase p110α (1:1000, Cell Signaling, Danvers, MA, USA); anti-PI3 Kinase p85 (1:1000, Cell Signaling); anti-PKA 2β (1:1000, Abcam); anti-Protein Kinase A regulatory Iα (1:1000, Abcam); anti-AKAP2 (1:500, Assay Biotechnology, Sunnyvale, CA, USA); anti-CaMKII (1:500, Abcam); anti-beta-catenin E5 (1:500, Santa Cruz, Santa Cruz, CA, USA) and anti-actin (Santa Cruz). Secondary antibodies, including a goat anti-rabbit IgG (1:5000, Biorad) or a goat anti-mouse IgG (1:5000, Biorad) were used for the visualization of the membrane with enhanced chemiluminescent substrate (PerkinElmer, Waltham, MA, USA).
RESULTS AND DISCUSSION
Hibernating animals have unique cardiac physiology
Physiological and hemodynamic data indicate that hibernating woodchucks entering DT have a distinct cardiac functional profile compared to the animals analyzed in summer (SM) or housed at room temperature in winter (WRT) (Fig. 1). As expected, the body weights (BW) were significantly lower in woodchucks in winter, even when they were housed at RT, when compared to summer (Fig. 1A); DT resulted in further significant reduction in their BW (Fig. 1A). By comparison, both body temperatures and heart rates were not altered between SM and WRT, while the body temperatures and heart rates were dramatically reduced in DT (Fig. 1, B and C). Physiologically, DT resulted in a trend, albeit insignificant, decrease in left ventricle (LV) systolic pressure (LVSP, Fig. 1D); on the other hand, DT caused a significant decrease in maximum LV pressure change rate, dP/dt (Fig. 1E). These data correlated well with the notion that the woodchucks exhibit a suppression of LV systolic function to match decreased metabolic demand during winter particularly when they enter the phase of torpor.
Figure 1. Comparison of cardiac function among woodchucks living in the summer, winter at room temperature (RT) and winter with DT.
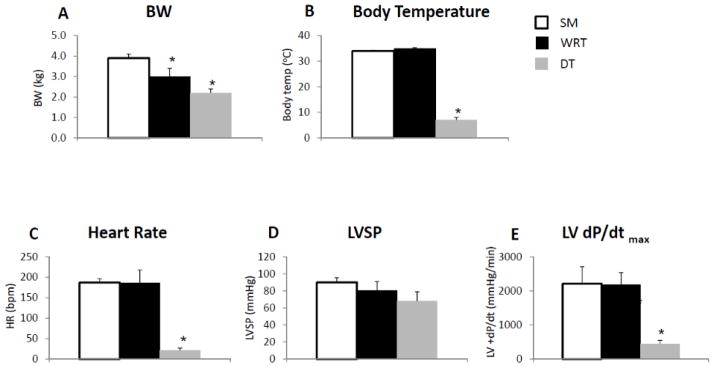
(A) Average woodchuck body weight (BW) was lower during winter at RT. A further BW reduction during DT was also observed. (B) The body temperature was dramatically lower during DT only. (C) The heart rate was dramatically reduced during DT only. (D) DT resulted in a trend (but not significant) towards a decrease in LV systolic pressure (LVSP). (E) DT resulted in a significant decrease in maximum LV dP/dt. All measurements were performed in woodchucks in summer (n=5), winter at RT with food (n=4), and winter during DT (n=4). *p<0.05 vs. summer. Data are expressed as mean±SEM.
Cross-species database searches can effectively identify woodchuck proteins
In order to discover proteomic changes in hibernating woodchucks, we conducted two sets of iTRAQ analyses: in Experiment 1, to identify seasonal proteomic changes by comparing proteomics profiles between WRT vs. SM and in Experiment 2, to compare the differentially expressed proteins between DT vs. SM (Supplemental Table 1). Since a comprehensive protein database for woodchucks is not available, we used a cross-species database search strategy for the identification of woodchuck proteins. For each set of iTRAQ experiments, the MS/MS spectra were searched independently against either the UniRef100 human (120,982 sequences), mouse (82,522 sequences) or rat (51,862 sequences) database. Positive protein identifications were based on the forward/reverse sequence database search routine, filtered at the protein FDR ≤1.5% via Scaffold. Each identified protein contains at least one unique peptide with a C. I. value ≥95%. Despite the vast differences in the sequence database sizes, similar numbers of proteins were identified across the three species, ranging from low to mid 4,000’s (Table 1, Supplemental Tables 2A and 3A; also provided to the readers are proteins identified with at least two peptides, Supplemental Tables 2B and 3B), while slightly more unique peptides were identified using the human relative to rodent databases. Regardless of the database searched, 5–10% more proteins were discovered in Experiment 1 over Experiment 2, with 1,161 proteins found in both experiments, a relatively small number, likely due to both the different animals and LC/MS/MS analytical methods involved. Overall, it appeared that these protein sequences are conserved between woodchucks and either human, mouse or rat proteins to render this cross-species database search strategy effective.
Table 1.
Cross-species database search for the identification of woodchuck cardiac proteins.
| Experimental Design a | Human | Mouse | Rat | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein | Peptide | FDR | Protein | Peptide | FDR | Protein | Peptide | FDR | |
| Experiment 1 | 4,014 | 9,455 | 1.2% | 4,048 | 9,372 | 1.7% | 4,079 | 9,120 | 1.5% |
| Experiment 2 | 4,293 | 8,742 | 0.5% | 4,226 | 8,608 | 1.4% | 4,381 | 8,530 | 1.3% |
The raw data from Experiments 1 and 2 were independently searched against human, mouse and rat sequences in the UniRef 100 database, using the search results combined from Mascot (v2.3) and Sequest search engines through the Proteome Discoverer software (v1.3). The resulting files were imported into Scaffold for both qualitative and quantitative analyses. The number of proteins and peptides identified from each database search are listed in this table, following stringent data filtering described in Materials and Methods.
Identification of torpor-specific proteomic changes
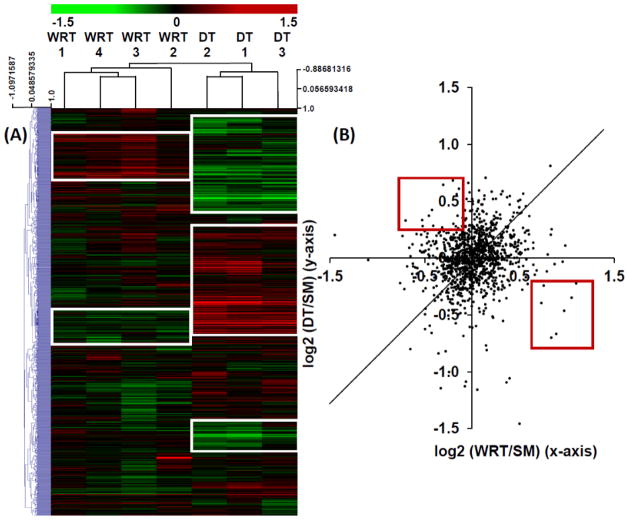
In order to identify hibernation-specific changes of protein networks, we performed a hierarchical cluster analysis of the 1,161 proteins common to both iTRAQ experiments, using MeV v.4.7.1 software suite 16. The resulting heatmap clearly indicates that numerous protein clusters were uniquely increased (red) or decreased (green) in DT vs. SM experiment; while the same proteins in the WRT vs. SM experiment were either unchanged or altered in the opposite direction, compared to the DT vs. SM experiment (Fig. 2A, boxed regions). In fact, when iTRAQ ratios from the common proteins in both experiments were analyzed with a scatter plot, we discovered that when compared with summer animals, most protein expressions were not dramatically altered in the winter, regardless of whether they underwent DT; and among the ones that were substantially altered, ~ 50% DT-specific proteomic changes were distinctive from the WRT-specific changes (Fig. 2B, red boxed regions).
Figure 2. Heatmap clustering of WRT and DT cardiac iTRAQ proteomic ratios relative to SM.
A) A two-colored heatmap representation of the log2-transformed iTRAQ ratios among the common proteins identified from both Experiments 1 (WRT/SM, Supplemental Table 4A) and 2 (DT/SM, Supplemental Table 5A). Proteins upregulated over SM are displayed in red, and the downregulated proteins are shown in green. Hierarchical clustering of the iTRAQ ratios was performed by Pearson correlation with average linkage. This analysis demonstrates good concordance among the biological replicates. Distinctive DT and WRT-modulated protein networks are shown in white squares. (B) A scatter plot of the iTRAQ expression ratios among the common proteins quantified in both DT/SM and WRT/SM experiments. Although iTRAQ ratios of many proteins shared similar expression trends between DT/SM and WRT/SM samples, only a few proteins had opposite expression patterns between DT/SM and WRT/SM experiments (red box).
Based on our previous analysis of iTRAQ analytical variation 14 on our instruments, the proteins with iTRAQ ratios beyond 20% of the normalized population means and p-values ≤0.05 are considered as significantly changed. Consequently, we found 455 proteins in WRT vs. SM (Experiment 1, Supplemental Table 4A), and 174 proteins in DT vs. SM (Experiment 2, Supplemental Table 5A) were significantly changed (also provided to the readers are more confidently identified proteins, with at least two matched peptides, Supplemental Tables 4B and 5B). Given the dramatic cardiac functional changes which occur during torpor, it is surprising that less than 5% of the identified proteins were significantly altered in DT animals, suggesting the importance of key protein networks in determining cardiac protective mechanisms during torpor.
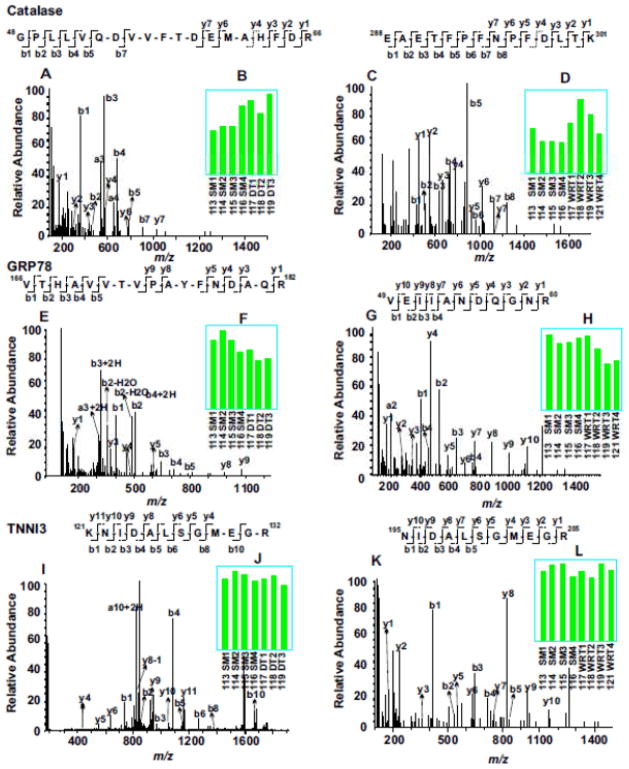
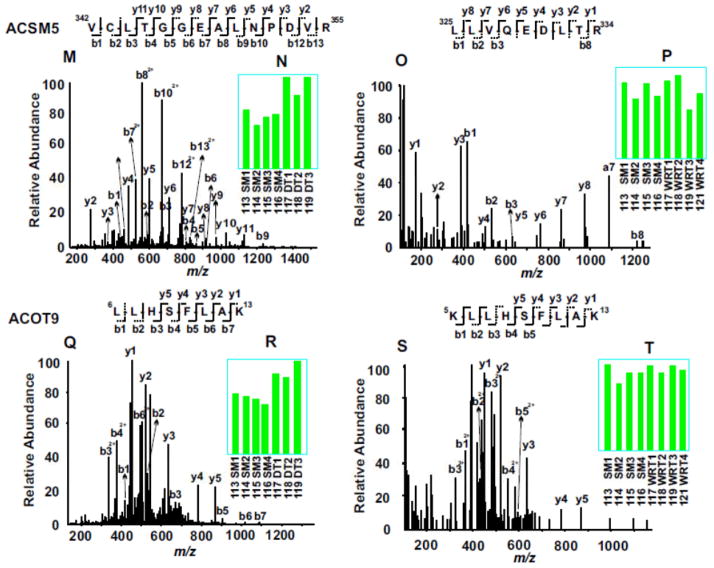
Evaluation of representative MS/MS spectra confirmed the detailed distinction of winter-related seasonal proteomic changes from DT-specific alternation of proteins (Fig. 3). For example, both catalase and 78 kDa glucose-regulated protein (GRP78) were seasonally altered in winter compared to summer, with the expression levels of catalase increased (Fig. 3, A to D) and GRP78 decreased (Fig. 3, E to H). In contrast, like most identified proteins in both experiments, troponin I type 3 (TNNI3) was not altered by either seasonal changes or DT (Fig. 3, I to L). On the other hand, lipid metabolic enzymes, acyl-coenzyme A synthetase medium-chain family member 5 (ACSM5) and mitochondrial acyl-coenzyme A thioesterase 9 (ACOT9) were only elevated during DT, but not in winter animals fed and held at room temperature (Fig. 3, M to T). It is clear from these two experiments that the availability of the 8-plex iTRAQ reagents for incorporating relatively large numbers of animal samples in each multi-plexing expression profiling experiment is crucial for discovering significant proteomics changes, since the random variations among different animals are unavoidable. Furthermore, validation of key protein expression changes by iTRAQ with alternative, largely antibody-based approaches is also important for understanding the biological significance of the protein networks affected.
Figure 3. Example of iTRAQ analyses of woodchuck cardiac proteomic changes among SM, WRT and DT animal groups.
MS/MS spectra (A, C, E, G, I, K, M, O, Q and S) enabled confident identifications of peptide sequences based on the determination of streches of continuous series of y ions and b ions. The bar graph inserts (B, D, F, H, J, L, N, P, R and T) indicate the normalized intensities of the iTRAQ tags observed in each MS/MS spectrum. Compared to SM, catalase was up regulated in both DT and NDT groups, whereas 78 kDa glucose-regulated protein (GRP78) was down regulated in both groups. Troponin I type 3(TNNI3) did not change among all groups. Compared to SM, both acyl-coenzyme A synthetase (ACSM5) and acyl-coenzyme A thioesterase 9 (ACOT9) were up regulated only in DT groups, but not in WRT groups.
Validation of iTRAQ-derived protein changes by Western blotting
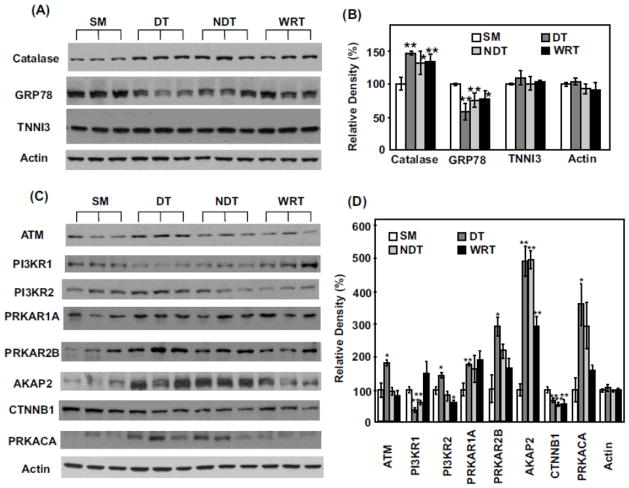
Following iTRAQ analyses, we performed additional Western blotting in order to verify key proteomic differences discovered by iTRAQ analyses. In addition to analyzing the SM, DT and WRT groups, we also included a fourth group of heart proteins obtained from woodchucks in winter, housed at cold temperature with food, but failing to entering deep torpor status (NDT). This last group of animals may allow us to further discover hibernation-specific proteomic changes, beyond the impact of season and environmental factors, such as temperature. Similar to the discovery made with iTRAQ analysis when compared to the summer group, the upregulation of anti-oxidant catalase and the downregulation of ER stress regulator GRP78 were most likely a seasonal phenomenon occurring in the winter (Fig. 4, A to B), regardless of the actual temperatures of the habitats or torpor status. Overexpression of catalase and inhibition of ER stress response by downregulation of GRP78 have been reported to be involved in cardioprotection17–23. Thus, the seasonal change in protein expression of catalase and GRP78 in woodchucks could be intrinsic adaptive mechanisms responsible for the changed phenotype from summer to winter in hibernating animals.
Figure 4. Western blot validation of iTRAQ proteomics changes among select proteins.
(A) Each lane represents the proteins extracted from the heart of a single distinct woodchuck. Three samples each from SM, DT, NDT and WRT groups are blotted with the antibodies against catalase, GRP78, tropoinin I (TNNI3) and actin (as the loading controls), respectively. (B) The blot densities of Catalase, GRP78, TNNI3 and actin were determined using Quantity One software (Bio-Rad). (C) Western blots and (D) blot densities determined by Quantity One for ATM, PI3KR1, PI3KR2, PRKAR1, PRKAR2B, AKAP2, CTNNB1, PRKACA and actin are shown. (D) All densities were normalized to SM1 intensities for each protein for the statistical analysis. Significant expression difference from SM was determined by Students’ t-tests. *p < 0.05 and ** p < 0.02. Data are expressed as mean (SD).
By comparison, significant increase of ataxia telangiectasia mutated (ATM), phosphoinositide-3-kinase, regulatory subunit 2 (PI3KR2), cAMP-dependent protein kinase type IIβ regulatory chain (PRKAR2B) and cAMP-dependent protein kinase catalytic α (PRKACA) appeared to be most dramatically affected by DT (Fig. 4, C to D). In addition, the decrease of phosphoinositide-3-kinase, regulatory subunit 1 (PI3KR1) may be related largely to low environmental temperature, since its levels were significantly decreased in both DT and NDT groups, but were not reduced in WRT (Fig. 4, C to D). On the other hand, A-kinase anchor protein 2 (AKAP2) appeared to be significantly induced both seasonally and by cold temperature (Fig. 4, C to D). PI3K is a well known kinase which regulates cell survival, cell growth, cell cycle entry, and cell migration through its downstream targets24. The downstream targets including Akt, PKC, p70S6K, and ERK, have been demonstrated to play a role in mediating cardioprotection and limiting myocardial damage induced by ischemia24–26. In addition, since hibernating animals induce adaptive cardiac hypertrophy due to increased heart contractility during DT27, 28, the proteins which regulate hypertrophy could be upregulated. PI3K/AKT, p70S6K and ERK are well known pathways for mediating physiological hypertrophy29–31, therefore, the upregulation of PI3K-related protein networks likely contribute to enhanced cardiac hypertrophy.
Identification of hibernation- regulated protein networks
In order to discover novel protein networks that are uniquely regulated during hibernation, we performed a comparative bioinformatics analysis of the significantly changed proteins from the two iTRAQ analyses. Similar to the elegant work published by Russeth et al32, it is essential to use multiple protein database search engines to achieve a broad proteome coverage for the analysis of non-model organisms. However, despite the recent advancements in mass spectrometry sensitivities, proteomics approaches, including the current study, still cannot match the depths of genomics studies for revealing expression changes of many low abundance molecules during hibernation33, 34. It is likely that, with more high quality nucleic acid sequence data obtained from deep sequencing studies and the improvement in bioinformatics technologies, proteomics methods can achieve much higher proteome coverages in the near future, by directly searching the MS/MS spectra against proteins predicted from deep sequencing approaches.
Despite these limitations, our study has both confirmed previous findings and revealed some novel insights regarding the effect of hibernation on protein networks. Not surprisingly, some of the protein networks affected by hibernation discovered from this study (see examples in Supplemental Table 6) have been reported previously. For example, down regulation of fatty acid synthesis and upregulation of fatty acid catabolism during hibernation have been reported from previous proteomics and genomics studies of the tissues of hibernating arctic ground squirrels9, 35 and another proteomics study of the hearts of ground squirrels11.
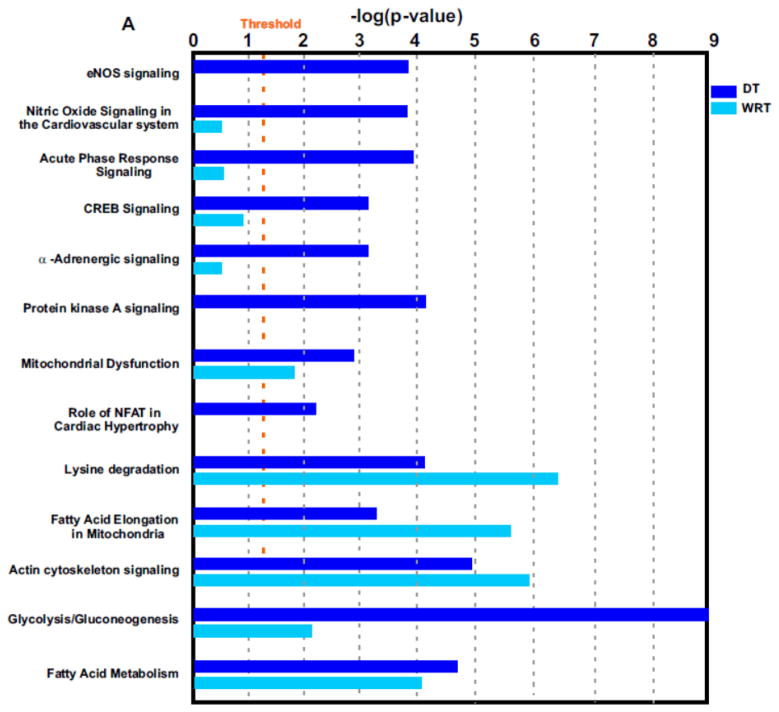
Interestingly, it appeared that protein networks pertaining to NO signaling, acute phase response, CREB and NFAT transcriptional regulations, protein kinase A (PKA) signaling and α-adrenergic signaling were more dramatically altered during DT but not altered seasonally in the winter (Fig. 5A and Supplemental Table 6). A beneficial role for NO during ischemia has been clearly demonstrated2, 36–38. We previously found that during DT, the myocardial blood flow was surprisingly maintained although the blood flow to the visceral organs, e.g. to the kidneys, was decreased2. The maintenance of myocardial blood flow was apparently involved in NO-dependent vasodilation2. However, we do not know which NO synthase isoform mediates these effects in previous observations. Interestingly, in the current study, we found that endothelial nitric oxide synthase (eNOS) signaling was upregulated during DT (Fig. 5A to B). According to the analysis of NO protein signaling networks during DT (Fig. 5B), we hypothesize that NO is produced in the cardiovascular system by eNOS, following agonist induction of intracellular [Ca2+], and downstream caveolin-eNOS protein complex formation.
Figure 5. Identification of distinctive protein signaling networks modulated during DT by Ingenuity Pathways Analysis.
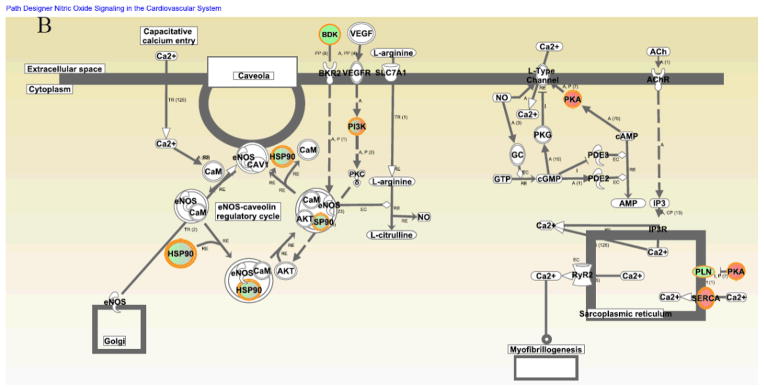
(A) Significantly altered protein networks from DT/SM (dark blue) and WRT/SM (light blue) experiments. The significantly changed proteins (p-values less than 0.05 with the DT/SM or WRT/SM protein iTRAQ ratios smaller than 0.8 or greater than 1.2) from Experiments 1 and 2 were analyzed by Ingenuity Pathways Analysis software (IPA; Ingenuity Systems, Mountain View, CA; www.ingenuity.com). Canonical pathways which contains well-characterized metabolic and cell signaling pathways were extracted from the Ingenuity Pathway Knowledge base (Supplemental Table 6). The resulting protein networks were compared between DT/SM (dark blue) and WRT/SM (light blue) experiments. -Log10 (p-values) are plotted in the X-axis. Select protein networks are shown on the Y-axis. The threshold (orange line) indicates the p-value of 0.05, above which indicates significant enrichment of protein networks. (B). Modulation of nitric oxide (NO) protein signaling networks in DT. NO is produced in the cardiovascular system by endothelial nitric oxide synthase (eNOS), following agonist induction of intracellular [Ca2+], and downstream caveolin-eNOS protein complex formation. Other proteins in this network include HSP90, Akt, and CaM. Cardiac NO regulates targets such as the L-type Ca2+ channels via cGMP-dependent protein kinase (PKG), the cGMP-stimulated phosphodiesterase (PDE2) and the cGMP-inhibited PDE (PDE3). There is also evidence that NO may modulate the function of the ryanodine receptor Ca2+ release channel (RyR2). Proteins identified in this study are shown in red (increased over summer) or green (downregulated from summer), or green filled circle with red border (different isoforms have opposite regulation).
In addition, from the network analysis, selective downstream protein network changes in DT animals suggested that CREB protein network activation may be important for cardiac protection during torpor. Several in vitro studies have also shown the roles of CREB in the regulation of cardiac function. In one study, the hypertrophic agonist, phenylephrine, promotes phosphorylation of CREB in adult rat cardiac myocytes through α- and β-adrenergic receptors, which plays an important role in the hypertrophic response39. Another study indicates that preconditioning stimulated the induction of thioredoxin. Subsequently, thioredoxin can be translocated into the nucleus and activates CREB via phosphorylation for a delayed induction of mitochondrial anti-apoptotic Bcl-2 and anti-oxidative MnSOD40. A more mechanistic study demonstrates that the cardioprotection induced by pharmacological preconditioning with resveratrol is mediated by the activation of CREB through the adenosine A3 receptor by Akt-dependent (PI3K-Akt-CREB-Bcl2) and Akt-independent (ERK/p38MAPK-MSK1-CREB-Bcl2) pathways41–43. Moreover, a recent study demonstrated that CREB activation mediates ischemic preconditioning44. Thus, CREB protein network activation could be involved in cardiac hypertrophy and ischemic preconditioning in hibernating animals in winter.
CONCLUSIONS
Hibernating mammals are models with natural myocardial protection, which can provide mechanisms involved in cardioprotection. Proteomics analysis demonstrated that hibernating woodchucks may “prepare” for winter by evoking their intrinsic cardioprotective proteomic mechanisms, which include upregulation of the anti-oxidant, catalase, and inhibition of ER stress response by downregulation of GRP78, as well as selective activation of NO signaling, acute phase response CREB and NFAT transcriptional regulation, protein kinase A and α-adrenergic signaling networks for cardiac cell survival during torpor. These cellular and molecular mechanisms involved in natural resistance to cardiac stress in hibernating mammals potentially provide new strategies to protect myocardium of non-hibernating animals, especially humans, from cardiac dysfunction induced by hypothermic stress and myocardial ischemia.
Supplementary Material
Supplemental Table 1. iTRAQ experimental design
Supplemental Table 2A. All proteins identified in SM vs. WRT experiment.
Supplemental Table 2B. All identified proteins in SM vs WRT with at least 2 unique peptides identified from each protein.
Supplemental Table 3A. All proteins identified in SM vs. DT experiment.
Supplemental Table 3B. All identified proteins in SM vs DT with at least 2 unique peptides identified from each protein.
Supplemental Table 4A. Significant protein expression difference between SM and WRT groups.
Supplemental Table 4B. Significant protein expression difference between SM and WRT groups with at least 2 unique peptides identified from each protein.
Supplemental Table 5A. Significant protein expression difference between SM and DT groups.
Supplemental Table 5B. Significant protein expression difference between SM and DT groups with at least 2 unique peptides identified from each protein.
Supplemental Table 6: Comparison of select enriched protein networks by Ingenuity Pathway Analysis between SM/DT and SM/WRT groups.
Acknowledgments
The Orbitrap mass spectrometer used in this study is supported in part by a NIH grant NS046593 to HL for the support of a UMDNJ Neuroproteomics Core Facility and LY is supported by NIH grants 1R01HL091781-01.
ABBREVIATIONS
- BW
body weight
- C.I. value
confidence interval value
- dP/dt
rate of left ventricular pressure change
- DT
woodchucks lived in winter at low temperature with deep torpor
- ECG
electrocardiography
- FA
formic acid
- FDR
false discovery rate
- FWHM
full-width at half maximum
- HCD
higher energy collision dissociation
- I/R
ischemic reperfusion
- iTRAQ
isobaric tags for relative and absolute quantification
- LV
left ventricle
- MMTS
methyl methanethiosulfonate
- MS/MS
tandem mass spectrometry
- NDT
winter woodchucks at low temperature and no food without deep torpor
- NO
nitric oxide
- RPLC
reversed phase liquid chromatography
- SCX
strong cation exchange
- SM
woodchucks in summer
- TCEP
tris(2-carboxyethyl) phosphine
- WRT
woodchucks lived in winter at room temperature with food without deep torpor
Footnotes
Author Contributions
L.Y. and H.L. designed most experiments and wrote the paper. S.F.V advised on the design of woodchuck grant proposal and study. D.E.V revised the manuscript. H.L., T.L., W.C., M.J., R.K.K., and L.Y. performed the experiments and analyzed the data.
References
- 1.Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83 (4):1153–81. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- 2.Kudej RK, Vatner SF. Nitric oxide-dependent vasodilation maintains blood flow in true hibernating myocardium. J Mol Cell Cardiol. 2003;35 (8):931–5. doi: 10.1016/s0022-2828(03)00174-3. [DOI] [PubMed] [Google Scholar]
- 3.Johansson BW. The hibernator heart--nature’s model of resistance to ventricular fibrillation. Cardiovasc Res. 1996;31 (5):826–32. doi: 10.1016/0008-6363(95)00192-1. [DOI] [PubMed] [Google Scholar]
- 4.Fedorov VV, Glukhov AV, Sudharshan S, Egorov Y, Rosenshtraukh LV, Efimov IR. Electrophysiological mechanisms of antiarrhythmic protection during hypothermia in winter hibernating versus nonhibernating mammals. Heart Rhythm. 2008;5 (11):1587–96. doi: 10.1016/j.hrthm.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamm KE, Zatzman ML, Jones AW, South FE. Maintenance of ion concentration gradients in the cold in aorta from rat and ground squirrel. Am J Physiol. 1979;237 (1):C17–22. doi: 10.1152/ajpcell.1979.237.1.C17. [DOI] [PubMed] [Google Scholar]
- 6.Yatani A, Kim SJ, Kudej RK, Wang Q, Depre C, Irie K, Kranias EG, Vatner SF, Vatner DE. Insights into cardioprotection obtained from study of cellular Ca2+ handling in myocardium of true hibernating mammals. Am J Physiol Heart Circ Physiol. 2004;286 (6):H2219–28. doi: 10.1152/ajpheart.01096.2003. [DOI] [PubMed] [Google Scholar]
- 7.Peppas AP, Yan L, TSY, Vatner DE, Vatner SF, Kudej RK. Seasonal variation in ischemia tolerance in a true mammalian hibernator. Circulation. 2009;120 (S):846. [Google Scholar]
- 8.Xie LH, Yan L, KKR, Peppas AP, Zhao Z, Ge H, Feflova N, You B, Shen YT, Vatner DE, FVS Arrhythmia protection insights from a hibernating animal. Circulation. 2009;120 (S):674. [Google Scholar]
- 9.Shao C, Liu Y, Ruan H, Li Y, Wang H, Kohl F, Goropashnaya AV, Fedorov VB, Zeng R, Barnes BM, Yan J. Shotgun proteomics analysis of hibernating arctic ground squirrels. Mol Cell Proteomics. 9(2):313–26. doi: 10.1074/mcp.M900260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin SL, Epperson LE, Rose JC, Kurtz CC, Ane C, Carey HV. Proteomic analysis of the winter-protected phenotype of hibernating ground squirrel intestine. Am J Physiol Regul Integr Comp Physiol. 2008;295 (1):R316–28. doi: 10.1152/ajpregu.00418.2007. [DOI] [PubMed] [Google Scholar]
- 11.Grabek KR, Karimpour-Fard A, Epperson LE, Hindle A, Hunter LE, Martin SL. Multistate proteomics analysis reveals novel strategies used by a hibernator to precondition the heart and conserve ATP for winter heterothermy. Physiol Genomics. 43(22):1263–75. doi: 10.1152/physiolgenomics.00125.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T, D’Mello V, Deng L, Hu J, Ricardo M, Pan S, Lu X, Wadsworth S, Siekierka J, Birge R, Li H. A multiplexed proteomics approach to differentiate neurite outgrowth patterns. J Neurosci Methods. 2006;158 (1):22–9. doi: 10.1016/j.jneumeth.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ai N, Krasowski MD, Welsh WJ, Ekins S. Understanding nuclear receptors using computational methods. Drug Discov Today. 2009;14 (9–10):486–94. doi: 10.1016/j.drudis.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J, Qian J, Borisov O, Pan S, Li Y, Liu T, Deng L, Wannemacher K, Kurnellas M, Patterson C, Elkabes S, Li H. Optimized proteomic analysis of a mouse model of cerebellar dysfunction using amine-specific isobaric tags. Proteomics. 2006;6 (15):4321–34. doi: 10.1002/pmic.200600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34 (2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 16.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 17.Turdi S, Han X, Huff AF, Roe ND, Hu N, Gao F, Ren J. Cardiac-specific overexpression of catalase attenuates lipopolysaccharide-induced myocardial contractile dysfunction: role of autophagy. Free Radic Biol Med. 53(6):1327–38. doi: 10.1016/j.freeradbiomed.2012.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Ge W, Zhang Y, Han X, Ren J. Cardiac-specific overexpression of catalase attenuates paraquat-induced myocardial geometric and contractile alteration: role of ER stress. Free Radic Biol Med. 49(12):2068–77. doi: 10.1016/j.freeradbiomed.2010.10.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin F, Lennon-Edwards S, Lancel S, Biolo A, Siwik DA, Pimentel DR, Dorn GW, Kang YJ, Colucci WS. Cardiac-specific overexpression of catalase identifies hydrogen peroxide-dependent and -independent phases of myocardial remodeling and prevents the progression to overt heart failure in G(alpha)q-overexpressing transgenic mice. Circ Heart Fail. 3(2):306–13. doi: 10.1161/CIRCHEARTFAILURE.109.864785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Yu A, Saari JT, Kang YJ. Repression of hypoxia-reoxygenation injury in the catalase-overexpressing heart of transgenic mice. Proc Soc Exp Biol Med. 1997;216 (1):112–6. doi: 10.3181/00379727-216-44162b. [DOI] [PubMed] [Google Scholar]
- 21.Tao J, Zhu W, Li Y, Xin P, Li J, Liu M, Redington AN, Wei M. Apelin-13 protects the heart against ischemia-reperfusion injury through inhibition of ER-dependent apoptotic pathways in a time-dependent fashion. Am J Physiol Heart Circ Physiol. 301(4):H1471–86. doi: 10.1152/ajpheart.00097.2011. [DOI] [PubMed] [Google Scholar]
- 22.Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, Kawase I, Hirota H. AMP-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress. Mol Cell Biol. 2005;25 (21):9554–75. doi: 10.1128/MCB.25.21.9554-9575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XY, Yang CT, Zheng DD, Mo LQ, Lan AP, Yang ZL, Hu F, Chen PX, Liao XX, Feng JQ. Hydrogen sulfide protects H9c2 cells against doxorubicin-induced cardiotoxicity through inhibition of endoplasmic reticulum stress. Mol Cell Biochem. 363(1–2):419–26. doi: 10.1007/s11010-011-1194-6. [DOI] [PubMed] [Google Scholar]
- 24.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296 (5573):1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 25.Ban K, Cooper AJ, Samuel S, Bhatti A, Patel M, Izumo S, Penninger JM, Backx PH, Oudit GY, Tsushima RG. Phosphatidylinositol 3-kinase gamma is a critical mediator of myocardial ischemic and adenosine-mediated preconditioning. Circ Res. 2008;103 (6):643–53. doi: 10.1161/CIRCRESAHA.108.175018. [DOI] [PubMed] [Google Scholar]
- 26.Ravingerova T, Matejikova J, Neckar J, Andelova E, Kolar F. Differential role of PI3K/Akt pathway in the infarct size limitation and antiarrhythmic protection in the rat heart. Mol Cell Biochem. 2007;297 (1–2):111–20. doi: 10.1007/s11010-006-9335-z. [DOI] [PubMed] [Google Scholar]
- 27.Tessier SN, Storey KB. Myocyte enhancer factor-2 and cardiac muscle gene expression during hibernation in thirteen-lined ground squirrels. Gene. 501(1):8–16. doi: 10.1016/j.gene.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Wickler SJ, Hoyt DF, van Breukelen F. Disuse atrophy in the hibernating golden-mantled ground squirrel, Spermophilus lateralis. Am J Physiol. 1991;261 (5 Pt 2):R1214–7. doi: 10.1152/ajpregu.1991.261.5.R1214. [DOI] [PubMed] [Google Scholar]
- 29.McMullen JR, Shioi T, Huang WY, Zhang L, Tarnavski O, Bisping E, Schinke M, Kong S, Sherwood MC, Brown J, Riggi L, Kang PM, Izumo S. The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110alpha) pathway. J Biol Chem. 2004;279 (6):4782–93. doi: 10.1074/jbc.M310405200. [DOI] [PubMed] [Google Scholar]
- 30.Luo J, McMullen JR, Sobkiw CL, Zhang L, Dorfman AL, Sherwood MC, Logsdon MN, Horner JW, DePinho RA, Izumo S, Cantley LC. Class IA phosphoinositide 3-kinase regulates heart size and physiological cardiac hypertrophy. Mol Cell Biol. 2005;25 (21):9491–502. doi: 10.1128/MCB.25.21.9491-9502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molkentin JD. Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc Res. 2004;63 (3):467–75. doi: 10.1016/j.cardiores.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 32.Russeth KP, Higgins L, Andrews MT. Identification of proteins from non-model organisms using mass spectrometry: application to a hibernating mammal. J Proteome Res. 2006;5 (4):829–39. doi: 10.1021/pr050306a. [DOI] [PubMed] [Google Scholar]
- 33.Brauch KM, Dhruv ND, Hanse EA, Andrews MT. Digital transcriptome analysis indicates adaptive mechanisms in the heart of a hibernating mammal. Physiol Genomics. 2005;23 (2):227–34. doi: 10.1152/physiolgenomics.00076.2005. [DOI] [PubMed] [Google Scholar]
- 34.Hampton M, Melvin RG, Kendall AH, Kirkpatrick BR, Peterson N, Andrews MT. Deep sequencing the transcriptome reveals seasonal adaptive mechanisms in a hibernating mammal. PLoS One. 6(10):e27021. doi: 10.1371/journal.pone.0027021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan J, Barnes BM, Kohl F, Marr TG. Modulation of gene expression in hibernating arctic ground squirrels. Physiol Genomics. 2008;32 (2):170–81. doi: 10.1152/physiolgenomics.00075.2007. [DOI] [PubMed] [Google Scholar]
- 36.Huang CH, Vatner SF, Peppas AP, Yang G, Kudej RK. Cardiac nerves affect myocardial stunning through reactive oxygen and nitric oxide mechanisms. Circ Res. 2003;93 (9):866–73. doi: 10.1161/01.RES.0000097762.64561.D2. [DOI] [PubMed] [Google Scholar]
- 37.Kim SJ, Ghaleh B, Kudej RK, Huang CH, Hintze TH, Vatner SF. Delayed enhanced nitric oxide-mediated coronary vasodilation following brief ischemia and prolonged reperfusion in conscious dogs. Circ Res. 1997;81 (1):53–9. doi: 10.1161/01.res.81.1.53. [DOI] [PubMed] [Google Scholar]
- 38.Kudej RK, Kim SJ, Shen YT, Jackson JB, Kudej AB, Yang GP, Bishop SP, Vatner SF. Nitric oxide, an important regulator of perfusion-contraction matching in conscious pigs. Am J Physiol Heart Circ Physiol. 2000;279 (1):H451–6. doi: 10.1152/ajpheart.2000.279.1.H451. [DOI] [PubMed] [Google Scholar]
- 39.Markou T, Hadzopoulou-Cladaras M, Lazou A. Phenylephrine induces activation of CREB in adult rat cardiac myocytes through MSK1 and PKA signaling pathways. J Mol Cell Cardiol. 2004;37 (5):1001–11. doi: 10.1016/j.yjmcc.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Chiueh CC, Andoh T, Chock PB. Induction of thioredoxin and mitochondrial survival proteins mediates preconditioning-induced cardioprotection and neuroprotection. Ann N Y Acad Sci. 2005;1042:403–18. doi: 10.1196/annals.1338.034. [DOI] [PubMed] [Google Scholar]
- 41.Das S, Tosaki A, Bagchi D, Maulik N, Das DK. Potentiation of a survival signal in the ischemic heart by resveratrol through p38 mitogen-activated protein kinase/mitogen- and stress-activated protein kinase 1/cAMP response element-binding protein signaling. J Pharmacol Exp Ther. 2006;317 (3):980–8. doi: 10.1124/jpet.105.095133. [DOI] [PubMed] [Google Scholar]
- 42.Das S, Tosaki A, Bagchi D, Maulik N, Das DK. Resveratrol-mediated activation of cAMP response element-binding protein through adenosine A3 receptor by Akt-dependent and -independent pathways. J Pharmacol Exp Ther. 2005;314 (2):762–9. doi: 10.1124/jpet.105.084285. [DOI] [PubMed] [Google Scholar]
- 43.Das S, Cordis GA, Maulik N, Das DK. Pharmacological preconditioning with resveratrol: role of CREB-dependent Bcl-2 signaling via adenosine A3 receptor activation. Am J Physiol Heart Circ Physiol. 2005;288 (1):H328–35. doi: 10.1152/ajpheart.00453.2004. [DOI] [PubMed] [Google Scholar]
- 44.Marais E, Genade S, Lochner A. CREB activation and ischaemic preconditioning. Cardiovasc Drugs Ther. 2008;22 (1):3–17. doi: 10.1007/s10557-007-6078-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. iTRAQ experimental design
Supplemental Table 2A. All proteins identified in SM vs. WRT experiment.
Supplemental Table 2B. All identified proteins in SM vs WRT with at least 2 unique peptides identified from each protein.
Supplemental Table 3A. All proteins identified in SM vs. DT experiment.
Supplemental Table 3B. All identified proteins in SM vs DT with at least 2 unique peptides identified from each protein.
Supplemental Table 4A. Significant protein expression difference between SM and WRT groups.
Supplemental Table 4B. Significant protein expression difference between SM and WRT groups with at least 2 unique peptides identified from each protein.
Supplemental Table 5A. Significant protein expression difference between SM and DT groups.
Supplemental Table 5B. Significant protein expression difference between SM and DT groups with at least 2 unique peptides identified from each protein.
Supplemental Table 6: Comparison of select enriched protein networks by Ingenuity Pathway Analysis between SM/DT and SM/WRT groups.