Abstract
Secreted frizzled-related proteins (SFRPs), the first identified Wnt antagonists, have been well recognized as tumor suppressors in multiple human cancers through suppressing the Wnt/β-catenin pathway. To better elucidate the mechanisms of SFRPs involved in the carcinogenesis of oral submucous fibrosis (OSF), one of the precancerous lesions of oral squamous cell carcinoma (OSCC), we investigated expression and localization of SFRP1, SFRP5, and β-catenin in normal oral epithelium, OSF, and OSCC tissues. We found that SFRP1 and SFRP5 were readily expressed in normal oral mucous tissues but gradually decreased in OSF early, moderately advanced, and advanced tissues and rarely expressed in OSCC tissues. We found the changes of SFRP1 localization and SFRP5 localization from nucleus to cytoplasm in the carcinogenesis of OSF. There is a significant association among reduced SFRP1, SFRP5, and cytoplasmic/nuclear β-catenin expression, which is correlated with higher tumor grade and stage of OSCC. We further found that SFRP1 and SFRP5 were frequently methylated in OSCC cases with betel quid chewing habit but not in normal oral mucous and different stages of OSF tissues, suggesting that methylation of SFRP1 and SFRP5 is tumor specific in the carcinogenesis of OSF. Taken together, our data demonstrated that reduced SFRP1 and SFRP5 by promoter methylation could lead to cytoplasmic/nuclear accumulation of β-catenin and tumor progression. The changes of SFRPs and β-catenin localization, as well as SFRPs’ methylation, could be useful predictors or biomarkers of OSF malignant progression and prognosis.
Keywords: Wnt antagonist, methylation, tumor suppressor gene, Wnt signaling, OSF, OSCC
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common head and neck malignancies, characterized by high mortality rate and poor prognosis.1,2 OSCC development is a multistep process caused by accumulated genetic and epigenetic alterations; it further leads to deregulation of key cellular signaling pathways, thus contributing to tumor initiation and progression.3,4 Several risk factors have been identified to be involved in oral carcinogenesis, such as betel quid or areca nut chewing, tobacco smoking, alcohol consumption, and viral infections.
OSCC is preceded by some precancerous lesions, including oral leukoplakia, oral lichen planus, and oral submucous fibrosis (OSF).2,4 Seven to thirty percent of OSF cases develop OSCC based on epidemiology studies and are always coincident with OSCC clinically.5,6 Betel quid chewing is a critical risk factor for OSF.7,8 However, few studies have been performed to elucidate the key genes and cell signaling pathways deregulated at the early stage of OSCC.
Wnt/β-catenin signaling is one of the most crucial pathways involved in the development of multiple malignancies, including OSCC, through regulating cell proliferation, differentiation, adhesion, stemness, metastasis, and invasion.9–11 Multiple intracellular and extracellular components concordantly regulate Wnt/β-catenin signaling pathway in tumorigenesis.12 β-Catenin is the key mediator of canonical Wnt signaling, and its activation contributes to tumor development and progression. β-Catenin could be released from the membrane and translocated into the nucleus, which further binds to TCF/LEF-1 family of transcription factors and activates target genes, resulting in Wnt signaling pathway activation. In OSCC, aberrant expression of β-catenin was associated with poor survival and nodal metastases, as well as increased mortality risk.13–18
In addition, a group of secreted Wnt antagonists have been implicated in the regulation of Wnt/β-catenin-signaling pathway, including Wnt inhibitory factor 1, secreted frizzled-related protein (SFRP), and the Dickkopf families. The SFRP family contains five glycoproteins (SFRP1, -2, -3, -4, and -5) and inhibits the Wnt signaling pathway through binding WNT ligand and competing with frizzled (Fz) receptors.19 Reduced SFRPs (SFRP1, -2, -4, and -5) in messenger RNA (mRNA) levels has been identified in a variety of malignancies including OSCC.20,21 However, little is known about SFRPs and β-catenin expression and their clinical significance in the carcinogenesis of OSF.
In this study, we selected two (SFRP1 and SFRP5) of the five SFRP members as representatives to investigate expression of SFRPs and their roles in OSF and OSCC pathogenesis. We examined the expression of SFRPs and β-catenin in normal oral mucosa, OSF, and OSCC tissues, as well as the correlation with clinicopathological features. We also investigated related regulatory mechanism of SFRPs reduction in OSF progression.
Materials and methods
Tissue specimens
OSCC (n=55), OSF (n=45), and normal oral mucosa (n=15) tissue specimens were obtained at the time of surgical resection at Xiangya Second Hospital and Xiangya Hospital, Central South University (Changsha, People’s Republic of China) and Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, People’s Republic of China) from January 2013 to June 2014. The informed consents had been obtained from patients under a protocol reviewed and approved by the institutional review boards of the Xiangya School of Medicine or Shanghai Jiao Tong University School of Medicine. The clinical diagnosis and pathologic stage of OSF were determined in terms of the Pingborg criteria.22 OSCC was diagnosed according to World Health Organization criteria of 2005. Fifteen normal specimens were obtained from healthy oral mucosa. Forty-five cases of OSF were incident, newly diagnosed without OSCC or neoplastic disease. OSF was classified into the following three grades: early stage (E, n=15), moderately advanced stage (M, n=15), and advanced stage (A, n=15). All collected tissues were divided into two parts, one of which was frozen immediately at −80°C after careful removal of the tumor mass, OSF tissue in epithelium layer and grossly normal-looking epithelium and the other part was fixed in 4% buffered formalin solution for pathologic diagnosis and immunohistochemical staining. Clinicopathological staging of OSCC was determined by the tumor node metastasis (TNM) classification of the International Union Against Cancer in 2009. The results of immunostained formalin-fixed, paraffin-embedded sections were evaluated separately by two pathologists (Lizhen Wang and Zhigang Yao at the Department of Oral Pathology).
Immunohistochemistry
Immunohistochemical staining was performed on 4 µm serial sections from formalin-fixed, paraffin-embedded specimens. After deparaffinization and hydration, the slides were treated with endogeneous peroxidase in 3% H2O2 for 20 minutes. The sections were then blocked for 30 minutes at 37°C with 1.5% blocking serum in phosphate-buffered saline before reacting with SFRP1 (1:100 dilution, AP9037A; Abgent [Abgent Biotech, Co. Ltd., Suzhou, People’s Republic of China]), SFRP5 (1:100 dilution, AP6991C; Abgent [Abgent Biotech, Co. Ltd., Suzhou, People’s Republic of China]), and β-catenin antibodies (1:100 dilution, 51067-2-AP; Proteintech [Proteintech group, Inc., Chicago, IL, USA]) at 4°C in a moist chamber overnight. Negative control slides were duplicate sections in the absence of primary antibodies. For evaluating SFRP1, SFRP5, and β-catenin expression, a scoring method was used. A mean percentage of positive cells were determined by the examination of 500 cells in at least five areas at ×400 magnification. Cells were assigned to one of the five following categories according to the percentage of positive cells (PP): 0) <5%; 1) 5%–24%; 2) 25%–49%; 3) 50%–75%; and 4) >75%. The intensity of the SFRP1, SFRP5, and β-catenin staining (SI) was then scored as follows: negative, − (0); weak, + (1); moderate, ++ (2); and intense, +++ (3). The final immunoreactive score (IRS = SI + PP) was as follows: −, 0 and 1; +, 2 and 3; ++, 4 and 5; and +++, 6 and 7. The stained tissues were scored blindly, regarding clinical patient data. Statistical analyses were performed with SPSS 17.0 software. Statistical significance was evaluated by the chi-square (χ2) test. The significance level was set at 0.05.
Reverse-transcription polymerase chain reaction
Total RNAs were extracted using Trizol Legend (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instruction. Reverse-transcription polymerase chain reaction (RT-PCR) was performed with the use of a kit from Promega Corporation (Fitchburg, WI, USA). Real-time PCR was performed to detect SFRP1 and SFRP5 expression, according to the manufacturer’s protocol (HT7500 system; Thermo Fisher Scientific). Primers for amplifying SFRP1 and SFRP5 mRNA sequences were synthesized as described previously.23 The 497 bp mRNA of SFRP1 was amplified by PCR with forward primer 5′-CCAGCGAGTACGACTACGTGAGCTT and reverse primer 5′-CTCAGATTTCAACTCGTTGTCACAGG. The 546 bp mRNA of SFRP5 was amplified by PCR with forward primer 5′-TGCGCCCAGTGTGAGATGGAGCAC and reverse primer 5′-CCCATCCCTTAGGCCTTGTGCCAGT. GAPDH was used as an internal control, the forward primer: 5′-ATCTCTGCCCCCTCTGCTGA-3′ and the reverse primer 5′-GATGACCTTGCCCACAGCCT-3′. PCR amplification was performed with denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds in 32 cycles. The PCR products were visualized on 2% agarose gels under ultraviolet transillumination.
Methylation-specific PCR
Bisulfite modification of DNA and methylation-specific PCR were performed as described previously.24–27 The bisulfite-treated DNA was amplified with the methylation-specific primer sets:23 SFRP1-Nm3: 5′-TGTAGTTTTCGGAGTTAGTGTCGCGC, SFRP1-m2: 5′-CCTACGATCGAAAACGACGCGAACG or the unmethylation-specific primer set SFRP1-u1: 5′-GTTT TGTAGTTTTTGGAGTTAGTGTTGTGT, SFRP1-u2: 5′-CTCAACCTACAATCAAAAACAACACAAACA. SFRP5-Nm3: 5′-AGATTTGGCGTTGGGCGGGACGTTC, SFRP5-Nm4: 5′-ACTCCAACCCGAACCTCGCCGTACG or the unmethylation-specific primer set SFRP1-u1: 5′-ATTTGGTGTTGGGTGGGATGTTT, SFRP1-u2: 5′-CCAACCCAAACCTCACCATACA. Methylation-specific PCR was done using AmpliTaq Gold (Applied Bio-systems, Foster City, CA, USA) (methylation-specific primer: annealing temperature 60°C, 40 cycles and unmethylation-specific primer: annealing temperature 58°C, 40 cycles).
Results
Reduced expression of SFRP1 and SFRP5 in the carcinogenesis of OSF
We first examined protein expression levels of SFRP family members (SFRP1 and SFRP5) by immunohistochemistry staining using SFRP1- and SFRP5-specific antibodies in normal oral mucous samples, OSF samples, and OSCC cases with betel quid chewing habit. SFRP1 and SFRP5 expressions were detected in all samples at variable intensities (Figures 1 and 2). Twelve of 15 (80.0%) normal oral mucous tissues showed nuclear SFRP1 positivity, whereas 30 of the 45 (66.7%) OSF tissues showed cytoplasmic SFRP1 expression, including tissues from 12 of the 15 (80.0%) early stage, 10 of the 15 (66.7%) moderately advanced stage, and 8 of the 15 (53.3%) advanced stage, as well as 24 of the 55 (43.6%) OSCC tissues (Figure 1 and Table 1). SFRP5 nuclear expression was shown in 86.7% (13/15) of normal oral mucous tissues, whereas cytoplasmic SFRP5 was detected in 73.3% (33/45) of OSF tissues, including 86.7% (13/15) of early stage tissues, 73.3% (11/15) of moderately advanced stage tissues, and 60.0% (9/15) of advanced stage tissues, as well as 41.8% (23/55) of OSCC tissues (Figure 2 and Table 2).
Figure 1.
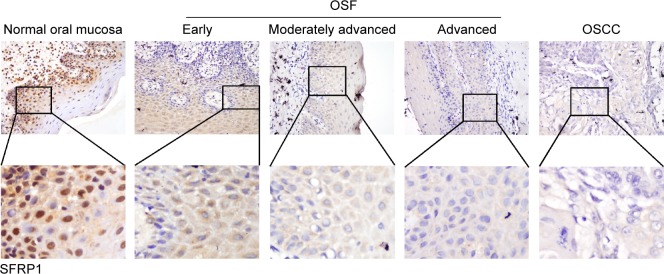
Immunohistochemical staining of SFRP1 in normal oral mucosa, OSF, and OSCC tissues.
Notes: Normal oral tissues exhibited strongly positive SFRP1 protein expression in the nucleus. OSF early stage and moderately advanced stage tissues showed SFRP1-positive expression in the cytoplasm and nucleus. OSF advanced stage tissues showed weak SFRP1 expression in cytoplasmic parts. Primary OSCC showed very weak cytoplasmic immunoreaction for SFRP1. Original magnification was ×400.
Abbreviations: SFRP, secreted frizzled-related protein; OSF, oral submucous fibrosis; OSCC, oral squamous cell carcinoma.
Figure 2.
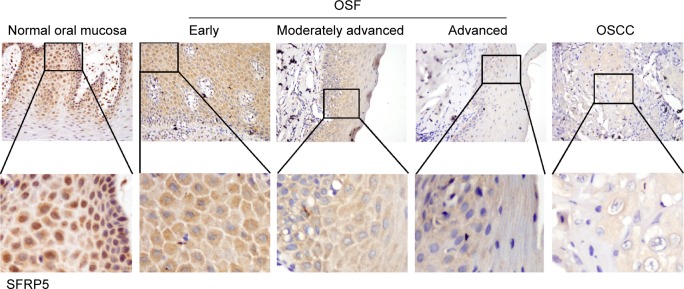
Examination of SFRP5 expression in normal oral mucosa, OSF, and OSCC tissues.
Notes: Normal oral mucous tissues exhibited strongly positive SFRP5 protein expression in the nucleus. OSF early stage and moderately advanced stage tissues showed SFRP5-positive expression in the cytoplasm and nucleus. OSF advanced stage tissues showed weak SFRP5 expression in cytoplasmic parts. Primary OSCC showed very weak cytoplasmic immunoreaction for SFRP5. Original magnification was ×400.
Abbreviations: SFRP, secreted frizzled-related protein; OSF, oral submucous fibrosis; OSCC, oral squamous cell carcinoma.
Table 1.
SFRP1 expression in the carcinogenesis of OSF
| Group | n | SFRP1
|
SFRP1 expression (%) | Mean SFRP1 score | |||
|---|---|---|---|---|---|---|---|
| − | + | ++ | +++ | ||||
| Normal | 15 | 3 | 1 | 5 | 6 | 80.0 | 4.47 |
| OSF | 45 | 15 | 13 | 8 | 9 | 66.7 | 3.09 |
| E | 15 | 3 | 3 | 4 | 5 | 80.0 | 4.07 |
| M | 15 | 5 | 5 | 3 | 2 | 66.7 | 2.87 |
| A | 15 | 7 | 5 | 1 | 2 | 53.3 | 2.33 |
| OSCC | 55 | 31 | 11 | 8 | 5 | 43.6 | 2.05 |
Abbreviations: SFRP, secreted frizzled-related protein; OSF, oral submucous fibrosis; E, early stage of OSF; M, moderately advanced stage of OSF; A, advanced stage of OSF; OSCC, oral squamous cell carcinoma.
Table 2.
SFRP5 expression in the carcinogenesis of OSF
| Group | n | SFRP5
|
SFRP5 expression (%) | Mean SFRP5 score | |||
|---|---|---|---|---|---|---|---|
| − | + | ++ | +++ | ||||
| Normal | 15 | 2 | 1 | 5 | 7 | 86.7 | 4.80 |
| OSF | 45 | 12 | 14 | 11 | 8 | 73.3 | 3.22 |
| E | 15 | 2 | 4 | 4 | 5 | 86.7 | 4.13 |
| M | 15 | 4 | 5 | 4 | 2 | 73.3 | 3.07 |
| A | 15 | 6 | 5 | 3 | 1 | 60.0 | 2.47 |
| OSCC | 55 | 32 | 10 | 8 | 5 | 41.8 | 2.00 |
Abbreviations: SFRP, secreted frizzled-related protein; OSF, oral submucous fibrosis; E, early stage of OSF; M, moderately advanced stage of OSF; A, advanced stage of OSF; OSCC, oral squamous cell carcinoma.
The average values of SFRP1 and SFRP5 expression varied in different tissue samples, with mean score of 4.47 and 4.80 in normal oral mucous tissues, respectively, 3.09 and 3.22 in OSF tissues, as well as 2.05 and 2.00 in OSCC tissues, respectively (Tables 1 and 2). SFRP1 and SFRP5 expressions were gradually reduced in normal oral mucous tissues, OSF tissues, and OSCC tissues (P=0.01125, P=0.00046). These data suggest that SFRP1 and SFRP5 are downregulated in the carcinogenesis of OSF.
Association of SFRP1, SFRP5, and β-catenin expressions and the correlation with clinicopathological features
To determine whether decreased SFRP1 and SFRP5 would cause Wnt/β-catenin signaling pathway activation, we further examined β-catenin expression in normal oral mucous samples, OSF samples, and OSCC cases with betel quid chewing habit. In normal oral mucous and OSF tissues, homogeneous membranous staining of β-catenin was observed, while increased cytoplasmic/nuclear (C/N) staining of β-catenin was shown in OSCC tissues, without membranous staining (Figure 3). However, we did not find significantly increased β-catenin expression in OSCC tissues, compared with normal oral mucous and OSF tissues (P>0.05).
Figure 3.
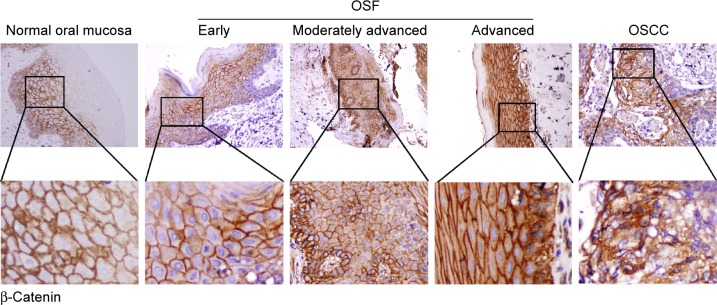
Detection of β-catenin expression in normal oral mucosa, OSF, and OSCC tissues.
Notes: Normal oral mucous and OSF tissues exhibited strongly positive β-catenin protein expression in the membrane. Primary OSCC showed positive cytoplasmic/nuclear localization of β-catenin. Original magnification was ×400.
Abbreviations: OSF, oral submucous fibrosis; OSCC, oral squamous cell carcinoma.
We further analyzed the association of SFRP1 and SFRP5 expression with the distribution of β-catenin in OSCC cases with betel quid chewing habit. In OSCC tissues with reduced SFRP1 expression, 25 of the 31 samples (80.6%) were classified as showing C/N β-catenin staining and 6 of the 31 cases (19.4%) as showing membranous β-catenin staining, whereas in OSCC tissues with decreased SFRP5 expression, 84.4% (27/32) of OSCC samples displayed C/N β-catenin staining and 15.6% (5/32) of samples showed membranous β-catenin staining. There was significant association between loss of SFRP1, SFRP5, and β-catenin C/N expressions (Table 3; P=0.00709; P=0.00950).
Table 3.
Association of SFRP1, SFRP5 expression, and immunostaining results of β-catenin in OSCC
| SFRPs expression | n | β-Catenin
|
P-value | |
|---|---|---|---|---|
| Mem | C/N stain | |||
| SFRP1 | ||||
| + | 24 | 13 | 11 | 0.00709 |
| − | 31 | 6 | 25 | |
| SFRP5 | ||||
| + | 23 | 11 | 12 | 0.00950 |
| − | 32 | 5 | 27 | |
Abbreviations: SFRP, secreted frizzled-related protein; OSCC, oral squamous cell carcinoma; Mem, membranous stain; C/N, cytoplasmic/nuclear.
The association among SFRP1, SFRP5, and β-catenin expressions, as well as clinicopathological characteristics was examined. No significant association was found between SFRP1 and SFRP5 expressions with any of the clinical parameters investigated (Table 4), whereas there was significant correlation between primary tumor and TNM stage with β-catenin expression (P=0.00798). β-Catenin-positive expression was observed in 100% (43/43) of OSCC patients with T3 + T4 and late TNM stage (III–IV) and 75.0% (9/12) of OSCC patients with T1 + T2 and early TNM stage (I–II).
Table 4.
SFRP1, SFRP5, and β-catenin expression and association with clinicopathological features in OSCC
| Clinicopathological features | Total (n) | SFRP1
|
SFRP5
|
β-Catenin
|
|||
|---|---|---|---|---|---|---|---|
| + | − | + | − | + | − | ||
| Age (years) | |||||||
| <50 | 43 | 16 | 27 | 15 | 28 | 40 | 3 |
| ≥50 | 12 | 8 | 4 | 8 | 4 | 12 | 0 |
| Sex | |||||||
| Male | 52 | 24 | 28 | 23 | 29 | 49 | 3 |
| Female | 3 | 0 | 3 | 0 | 3 | 3 | 0 |
| Tumor site | |||||||
| Tongue | 40 | 20 | 20 | 18 | 22 | 38 | 2 |
| Others | 15 | 4 | 11 | 5 | 10 | 14 | 1 |
| Primary tumor | |||||||
| T1 + T2 | 12 | 5 | 7 | 8 | 4 | 9 | 3 |
| T3 + T4 | 43 | 19 | 24 | 15 | 28 | 43* | 0 |
| TNM stage | |||||||
| I + II | 12 | 6 | 6 | 7 | 5 | 9 | 3 |
| III + IV | 43 | 18 | 25 | 16 | 27 | 43* | 0 |
| Differentiation grade | |||||||
| Well | 20 | 11 | 9 | 9 | 11 | 17 | 3 |
| Moderately poor | 35 | 13 | 22 | 14 | 21 | 35 | 0 |
Note:
P=0.00798.
Abbreviations: SFRP, secreted frizzled-related protein; OSCC, oral squamous cell carcinoma; TNM, tumor node metastasis.
SFRP1 and SFRP5 are downregulated at mRNA levels in the carcinogenesis of OSF
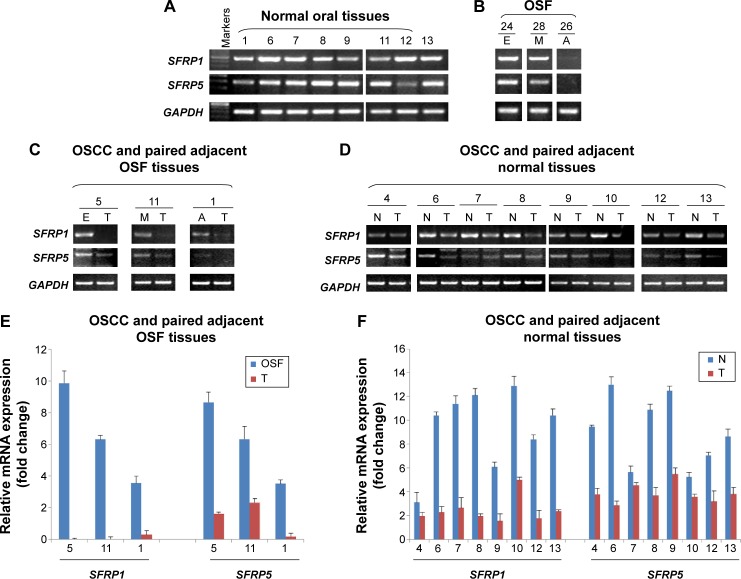
We also examined SFRP1 and SFRP5 expressions at mRNA levels in normal oral mucous tissues, OSF tissues, OSCC, and their paired adjacent tissues by semiquantitative RT-PCR. We found that SFRP1 and SFRP5 were readily expressed in normal oral mucous tissues (Figure 4A) and OSF early stage tissues, but decreased in OSF moderately advanced stage tissues, whereas rarely expressed in OSF advanced stage tissues (Figure 4B). We also detected SFRP1 and SFRP5 expression in OSCC and their adjacent OSF or normal oral mucous tissues. Results showed that SFRP1 and SFRP5 were downregulated in OSCC tissues, compared with their paired adjacent OSF or normal mucous tissues (Figure 4C and D). Real-time RT-PCR further confirmed reduced expression of SFRP1 and SFRP5 in OSCC tissues, compared with their adjacent normal or OSF tissues (Figure 4E). Therefore, SFRP1 and SFRP5 mRNA expression levels are decreased in the carcinogenesis of OSF.
Figure 4.
Detection of SFRP1 and SFRP5 mRNA expression in normal oral mucosa, OSF, and OSCC tissues.
Notes: Semiquantitative RT-PCR examined SFRP1 and SFRP5 expression in (A) normal oral mucosa tissues, (B) OSF tissues, (C) OSCC and paired adjacent OSF tissues, and (D) OSCC and paired adjacent normal oral mucous tissues. GAPDH was used as an internal control. (E, F) Quantitative real-time RT-PCR was used to confirm SFRP1 and SFRP5 expression in representative samples from OSCC and paired adjacent OSF or normal tissues. The expression level of each sample was normalized to internal control GAPDH. Results are represented as mean ± SD and are based on three independent experiments.
Abbreviations: SFRP, secreted frizzled-related protein; mRNA, messenger RNA; OSF, oral submucous fibrosis; OSCC, oral squamous cell carcinoma; RT-PCR, reverse-transcription polymerase chain reaction; SD, standard deviation; E, early stage of OSF; M, moderately advanced stage of OSF; A, advanced stage of OSF; N, normal oral tissues; T, OSCC.
Promoter methylation of SFRP1 and SFRP5 in the carcinogenesis of OSF
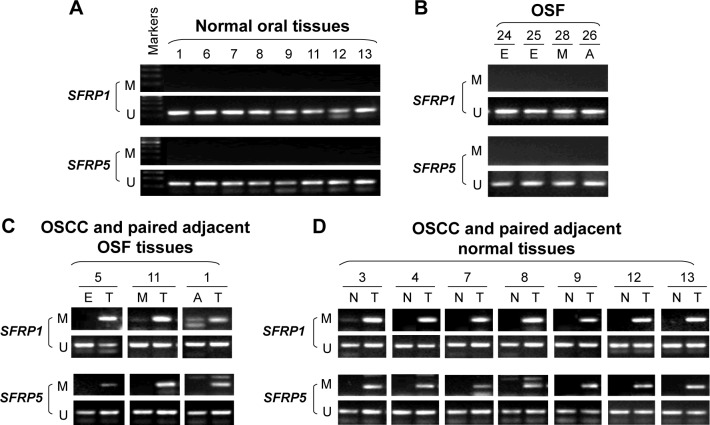
We further investigated possible regulatory mechanism of SFRP1 and SFRP5 reduction in the carcinogenesis of OSF. As promoter methylation mediates tumor suppressor genes transcriptional repression, we next detected promoter methylation of SFRP1 and SFRP5 in normal oral mucous and OSF tissues, OSCC, and their paired adjacent OSF or normal tissues. We found that SFRP1 and SFRP5 methylation was not detected in ten normal oral tissues and ten OSF tissues from early stage, moderately advanced stage, and advanced stage (Figure 5A and B). We also found that SFRP1 and SFRP5 were frequently methylated in OSCC tumor tissues but barely methylated in their paired adjacent OSF and normal oral mucous tissues (Figure 5C and D). These data suggest that promoter methylation of SFRP1 and SFRP5 is tumor-specific event in the carcinogenesis of OSF.
Figure 5.
Promoter methylation of SFRP1 and SFRP5 in normal oral mucosa, OSF, and OSCC tissues.
Notes: MSP was used to detect SFRP1 and SFRP5 methylation in (A) normal oral mucosa tissues, (B) OSF tissues, (C) OSCC and paired adjacent OSF tissues, and (D) OSCC and paired adjacent normal tissues.
Abbreviations: SFRP, secreted frizzled-related protein; OSF, oral submucous fibrosis; OSCC, oral squamous cell carcinoma; MSP, methylation-specific polymerase chain reaction; M, methylated; U, unmethylated; E, early stage of OSF; M, moderately advanced stage of OSF; A, advanced stage of OSF; N, normal oral tissues; T, OSCC.
Discussion
SFRPs are the largest family of Wnt-negative modulators and the first Wnt antagonists to be identified. SFRP proteins contain ~300 amino acids, including a cysteine-rich domain (CRD) in N-terminal domain and a hydrophilic heparin-binding region in C-terminal domain. Its CRD domain has ten conserved cysteine residues, with high homology to the extracellular CRD domain of the Fz receptors.19 Thus, SFRPs as secreted glycoproteins could bind directly to Wnt ligands or Fz receptors, leading to the suppression of Wnt/β-catenin signaling.
Epigenetic silencing of Wnt antagonists has been well-documented in human malignancies.28 Promoter methylation is the major regulatory mechanism. Deregulation of SFRPs by promoter CpG methylation has been reported in multiple human cancers, including oral,20 nasopharyngeal, esophageal, lung, gastric, colorectal, hepatocellular, breast, ovarian, endometrial, cervical, renal, and bladder carcinomas.19 Methylated SFRP1, SFRP2, SFRP4, and SFRP5 could mediate tumor cell proliferation, differentiation, apoptosis, and progression.19 Loss of SFRP1 expression in the early stage of breast cancer is associated with poor prognosis, thus is a novel prognostic early marker for breast cancer.29–31 Therefore, deregulation of Wnt antagonists SFRPs could be an early event in the multiple-step tumorigenesis of OSCC. However, there is no report about SFRPs expression in OSCC, especially in the precancerous lesion (OSF). SFRP2, SFRP4, and SFRP5 have been found to be reduced by promoter methylation in OSCC samples,20,21 whereas one group reported that SFRP1 demethylated in OSCC.21 Here, we selected SFRP1 and SFRP5, two representative members of SFRP family, to study the roles of SFRP family members in the carcinogenesis of OSCC. We first found that SFRP1 and SFRP5 were downregulated in the precancerous OSF and OSCC with varied protein levels.
Activation and delocalization of β-catenin in precancerous oral lesions and oral cancers have been found, suggesting the importance of Wnt/β-catenin signaling pathway in OSCC tumorigenesis. We thus evaluated whether reduced SFRP1 and SFRP5 would activate Wnt/β-catenin signaling pathway. We found significant association between decreased SFRP1, SFRP5, and β-catenin C/N expression in the carcinogenesis of OSF, which was correlated with poor prognosis of OSCC. Infrequent mutations of β-catenin, Adenomatous polyposis coli, and Axin1 were reported in oral cancer,32,33 which indicates that epigenetic silencing of Wnt signaling components plays an important role in OSCC pathogenesis.21 We further investigated the regulatory mechanism of SFRP1 and SFRP5 downregulation in the carcinogenesis of OSF. We found that promoter methylation mediated their silencing in OSCC patient tissues with betel quid chewing habit but not in normal oral mucous tissues and their paired adjacent normal or OSF tissues, which are consistent with other reports of SFRPs methylation in OSCC from the population of Western.
Conclusion
In summary, this is the first report to show downregulation of Wnt antagonists, SFRP1 and SFRP5, at protein and mRNA levels in the carcinogenesis of OSF. Deregulation of SFRP1 and SFRP5 by promoter methylation, coupled with activated β-catenin, may result in OSF canceration and OSCC progression. Tumor-specific methylation of SFRPs in OSCC will help to develop biomarker for early detection and personalized medicine of OSCC patients.
Acknowledgments
This article was supported by the National Natural Science Foundation of China (number 81202133), the Fifth Outstanding Youth Fund of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, and research grant (number 15411950301) from Science and Technology Commission of Shanghai Municipality.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Braakhuis BJ, Brakenhoff RH, Leemans CR. Head and neck cancer: molecular carcinogenesis. Ann Oncol. 2005;16(suppl 2):ii249–ii250. doi: 10.1093/annonc/mdi713. [DOI] [PubMed] [Google Scholar]
- 3.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008;87(1):14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- 5.Tilakaratne WM, Klinikowski MF, Saku T, Peters TJ, Warnakulasuriya S. Oral submucous fibrosis: review on aetiology and pathogenesis. Oral Oncol. 2006;42(6):561–568. doi: 10.1016/j.oraloncology.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Wollina U, Verma SB, Ali FM, Patil K. Oral submucous fibrosis: an update. Clin Cosmet Investig Dermatol. 2015;8:193–204. doi: 10.2147/CCID.S80576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Reichart PA. A review of betel quid chewing, oral cancer and precancer in Mainland China. Oral Oncol. 2007;43(5):424–430. doi: 10.1016/j.oraloncology.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Pillai R, Balaram P, Reddiar KS. Pathogenesis of oral submucous fibrosis. Relationship to risk factors associated with oral cancer. Cancer. 1992;69(8):2011–2020. doi: 10.1002/1097-0142(19920415)69:8<2011::aid-cncr2820690802>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 9.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 10.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 11.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17(1):45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Moles MA, Ruiz-Avila I, Gil-Montoya JA, Plaza-Campillo J, Scully C. β-catenin in oral cancer: an update on current knowledge. Oral Oncol. 2014;50(9):818–824. doi: 10.1016/j.oraloncology.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Alvarado CG, Maruyama S, Cheng J, et al. Nuclear translocation of beta-catenin synchronized with loss of E-cadherin in oral epithelial dysplasia with a characteristic two-phase appearance. Histopathology. 2011;59(2):283–291. doi: 10.1111/j.1365-2559.2011.03929.x. [DOI] [PubMed] [Google Scholar]
- 15.Kaur J, Sawhney M, DattaGupta S, et al. Clinical significance of altered expression of beta-catenin and E-cadherin in oral dysplasia and cancer: potential link with ALCAM expression. PLoS One. 2013;8(6):e67361. doi: 10.1371/journal.pone.0067361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laxmidevi LB, Angadi PV, Pillai RK, Chandreshekar C. Aberrant beta-catenin expression in the histologic differentiation of oral squamous cell carcinoma and verrucous carcinoma: an immunohistochemical study. J Oral Sci. 2010;52(4):633–640. doi: 10.2334/josnusd.52.633. [DOI] [PubMed] [Google Scholar]
- 17.Bezdekova M, Brychtova S, Sedlakova E, et al. Immunohistochemical assessment of E-cadherin and beta-catenin in trichofolliculomas and trichoepitheliomas. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2007;151(2):251–255. doi: 10.5507/bp.2007.042. [DOI] [PubMed] [Google Scholar]
- 18.Mahomed F, Altini M, Meer S. Altered E-cadherin/beta-catenin expression in oral squamous carcinoma with and without nodal metastasis. Oral Dis. 2007;13(4):386–392. doi: 10.1111/j.1601-0825.2006.01295.x. [DOI] [PubMed] [Google Scholar]
- 19.Surana R, Sikka S, Cai W, et al. Secreted frizzled related proteins: implications in cancers. Biochim Biophys Acta. 2014;1845(1):53–65. doi: 10.1016/j.bbcan.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Sogabe Y, Suzuki H, Toyota M, et al. Epigenetic inactivation of SFRP genes in oral squamous cell carcinoma. Int J Oncol. 2008;32(6):1253–1261. doi: 10.3892/ijo_32_6_1253. [DOI] [PubMed] [Google Scholar]
- 21.Pannone G, Bufo P, Santoro A, et al. WNT pathway in oral cancer: epigenetic inactivation of WNT-inhibitors. Oncol Rep. 2010;24(4):1035–1041. doi: 10.3892/or.2010.1035. [DOI] [PubMed] [Google Scholar]
- 22.Gupta PC, Sinor PN, Bhonsle RB, Pawar VS, Mehta HC. Oral submucous fibrosis in India: a new epidemic? Natl Med J India. 1998;11(3):113–116. [PubMed] [Google Scholar]
- 23.Li L, Zhang Y, Fan Y, et al. Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes. Epigenomics. 2015;7(2):155–173. doi: 10.2217/epi.14.79. [DOI] [PubMed] [Google Scholar]
- 24.Qiu GH, Tan LK, Loh KS, et al. The candidate tumor suppressor gene BLU, located at the commonly deleted region 3p21.3, is an E2F-regulated, stress-responsive gene and inactivated by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Oncogene. 2004;23(27):4793–4806. doi: 10.1038/sj.onc.1207632. [DOI] [PubMed] [Google Scholar]
- 25.Ying J, Li H, Seng TJ, et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25(7):1070–1080. doi: 10.1038/sj.onc.1209154. [DOI] [PubMed] [Google Scholar]
- 26.Ai L, Tao Q, Zhong S, et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27(7):1341–1348. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 27.Chan SL, Cui Y, van Hasselt A, et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab Invest. 2007;87(7):644–650. doi: 10.1038/labinvest.3700547. [DOI] [PubMed] [Google Scholar]
- 28.Ying Y, Tao Q. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics. 2009;4(5):307–312. doi: 10.4161/epi.4.5.9371. [DOI] [PubMed] [Google Scholar]
- 29.Yang ZQ, Liu G, Bollig-Fischer A, Haddad R, Tarca AL, Ethier SP. Methylation-associated silencing of SFRP1 with an 8p11-12 amplification inhibits canonical and non-canonical WNT pathways in breast cancers. Int J Cancer. 2009;125(7):1613–1621. doi: 10.1002/ijc.24518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veeck J, Niederacher D, An H, et al. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25(24):3479–3488. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 31.Klopocki E, Kristiansen G, Wild PJ, et al. Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol. 2004;25(3):641–649. [PubMed] [Google Scholar]
- 32.Iwai S, Katagiri W, Kong C, Amekawa S, Nakazawa M, Yura Y. Mutations of the APC, beta-catenin, and axin 1 genes and cytoplasmic accumulation of beta-catenin in oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2005;131(12):773–782. doi: 10.1007/s00432-005-0027-y. [DOI] [PubMed] [Google Scholar]
- 33.Yeh KT, Chang JG, Lin TH, et al. Correlation between protein expression and epigenetic and mutation changes of Wnt pathway-related genes in oral cancer. Int J Oncol. 2003;23(4):1001–1007. [PubMed] [Google Scholar]