FIG 1 .
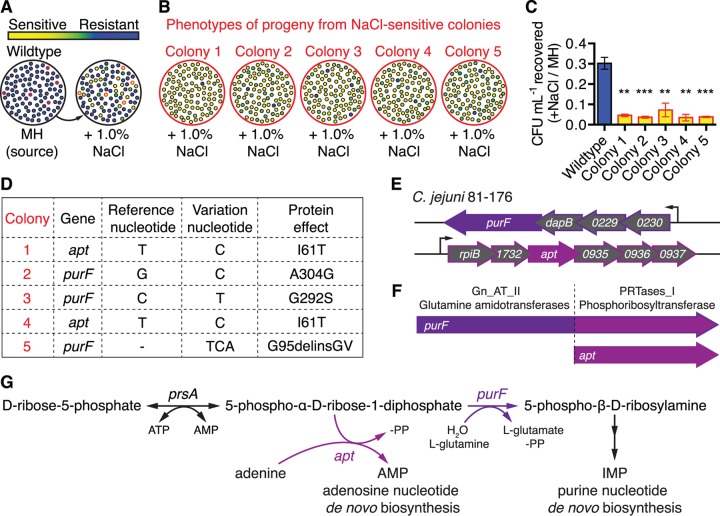
Heritable colony stress phenotype variation linked with purF and apt mutations. (A) Schematic illustration depicting 100 single colonies. Blue indicates growth/stress resistance and yellow indicates defective growth/stress sensitivity of replicated colonies grown on Mueller-Hinton (MH) agar with 1.0% NaCl (hyperosmotic stress). Colonies with red outlines on both plates indicate NaCl-sensitive isolates designated for heritability testing. (B) Heritability assessment. Colonies outlined in red in panel A were selected from the MH agar-only plate and correspond to the five NaCl-sensitive isolates also outlined in red on MH agar with 1.0% NaCl. These clones were propagated on MH agar only and then tested for heritability of the NaCl-sensitivity defect. One hundred progeny were tested on MH agar with 1.0% NaCl; shown is a schematic illustration of progeny phenotypes, color coded as shown by the key in panel A. (C) Relative levels of NaCl stress sensitivity of colony isolates compared to that of the parental wild type. Mean results with standard errors of the means (SEM) from three independent experiments are shown, presented as the ratio of CFU recovered on MH agar with 1.0% NaCl versus CFU recovered on MH agar only: **, P ≤ 0.01; ***, P ≤ 0.001. (D) Whole-genome sequencing identification of a single mutation with 100% variant frequency in either purF or apt in each sensitive strain, with anticipated protein effect. (E) Genomic loci and operons of purF and apt. (F) Conserved domains of purF and apt. (G) Purine biosynthesis pathway schematic with purine substrates involved in purF and apt reactions.