ABSTRACT
The capsule from Bacteroides, a common gut symbiont, has long been a model system for studying the molecular mechanisms of host-symbiont interactions. The Bacteroides capsule is thought to consist of an array of phase-variable polysaccharides that give rise to subpopulations with distinct cell surface structures. Here, we report the serendipitous discovery of a previously unknown surface structure in Bacteroides thetaiotaomicron: a surface layer composed of a protein of unknown function, BT1927. BT1927, which is expressed in a phase-variable manner by ~1:1,000 cells in a wild-type culture, forms a hexagonally tessellated surface layer. The BT1927-expressing subpopulation is profoundly resistant to complement-mediated killing, due in part to the BT1927-mediated blockade of C3b deposition. Our results show that the Bacteroides surface structure is capable of a far greater degree of structural variation than previously known, and they suggest that structural variation within a Bacteroides species is important for productive gut colonization.
IMPORTANCE
Many bacterial species elaborate a capsule, a structure that resides outside the cell wall and mediates microbe-microbe and microbe-host interactions. Species of Bacteroides, the most abundant genus in the human gut, produce a capsule that consists of an array of polysaccharides, some of which are known to mediate interactions with the host immune system. Here, we report the discovery of a previously unknown surface structure in Bacteroides thetaiotaomicron. We show that this protein-based structure is expressed by a subset of cells in a population and protects Bacteroides from killing by complement, a component of the innate immune system. This novel surface layer protein is conserved across many species of the genus Bacteroides, suggesting an important role in colonization and host immune modulation.
INTRODUCTION
Bacteria elaborate a broad array of capsules, structures that reside outside the cell wall and play roles in adhesion and immune modulation (1, 2). The capsular polysaccharides (CPSs) produced by species of Bacteroides, the dominant bacterial genus in the human gut (3–5), have served as a model system for understanding the role of the capsule in host-symbiont interactions (6). Bacteroides fragilis produces eight distinct capsular polysaccharides, PSA to PSH; with the exception of PSC, each is expressed in a phase-variable manner by a reversible inversion of repeat segments flanking the promoter of each polysaccharide locus (7). The configuration of the invertible CPS promoters is governed by a global Mpi (multiple-promoter invertase) from the Ssr family (8) as well as by enzymes in the Tsr family that are specific to the promoter in their immediate downstream region (9); UpxY family transcriptional antitermination factors and UpxZ family inhibitors block the simultaneous expression of two or more polysaccharides (10, 11). The capacity to produce multiple phase-variable CPSs with limited concurrent polysaccharide synthesis processes creates subpopulations of diverse surface structures and antigenicities that are thought to play an important role in the ability of Bacteroides to colonize the gastrointestinal (GI) tract (12–14). In light of the discoveries of immunomodulatory properties of one of the CPSs, polysaccharide A in Bacteroides fragilis (2, 6), we sought to determine whether there exist previously unknown surface structures in Bacteroides that may have other immunomodulatory properties.
RESULTS AND DISCUSSION
Serendipitous discovery of a cryptic surface layer protein in Bacteroides.
We began by systematically searching Bacteroides genome sequences for gene clusters predicted to encode factors that mediate microbe-host interactions (15). Three such gene clusters in Bacteroides thetaiotaomicron VPI-5482 (B. thetaiotaomicron 1949 [BT1949] to BT1954, BT2093 to BT2095, and BT4420 to BT4430) were predicted to contain genes that encode surface layer proteins, which vary widely in composition among microbial taxa but are known to play a role in processes central to microbe-host interaction, including biofilm formation, adhesion, and immune evasion (16). To determine whether these loci specify the production of a surface layer, we constructed strains in which BT1954, BT2095, or the entire BT1949-to-BT1957 locus was deleted. We then compared cell surface protein preparations of B. thetaiotaomicron and the mutants by SDS-PAGE, expecting to find a cell surface protein(s) that was present in the B. thetaiotaomicron preparation but missing from one or more of the mutants. No such proteins were found, but, to our surprise, we found reproducibly that one of the mutants, the ΔBT1954 strain, expressed very high levels of a new cell surface protein (Fig. 1A).
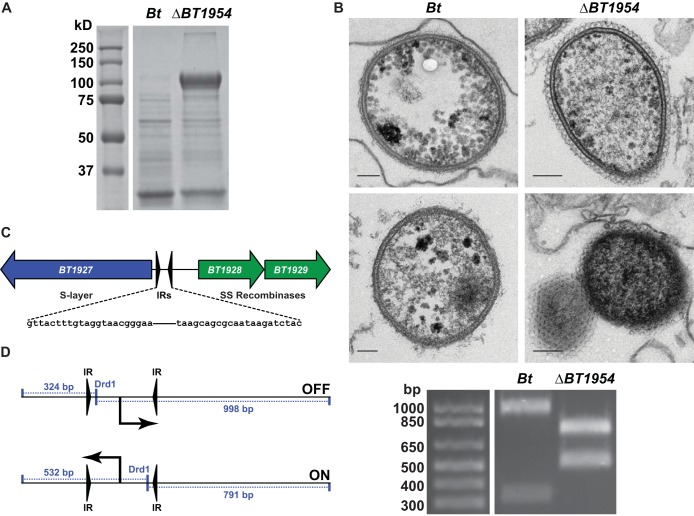
FIG 1 .
A phase-variable surface layer protein in B. thetaiotaomicron. (A) Proteins were precipitated from filtered culture supernatants of B. thetaiotaomicron (Bt) with trichloroacetic acid and subjected to SDS-PAGE. The ΔBT1954 strain culture supernatant harbors an abundant, high-molecular-mass band that is absent in B. thetaiotaomicron (see also Fig. S1 in the supplemental material). (B) B. thetaiotaomicron cells were fixed, cut into ultrathin sections, negative stained, and imaged through the use of a transmission electron microscope. The ΔBT1954 mutant (upper right and lower right) harbors a tessellated surface layer, adjacent to the outer membrane, which is absent in the parental strain (upper left and lower left). Each scale bar represents 100 nm. (C) Schematic of the BT1927-to-BT1929 locus, showing inverted repeat elements flanking the predicted BT1927 promoter. SS, site specific. (D) Schematic of the promoter orientation assay. DrdI-digested PCR products were separated by agarose gel electrophoresis. The inverted repeat element harboring the predicted BT1927 promoter is in opposite orientations in the parental strain and the ΔBT1954 mutant (see also Fig. S1).
Transmission electron micrographs (EM) of the ΔBT1954 mutant revealed a novel surface structure that resembles a crystalline surface layer. The hexagonal lattice extends ~500 Å beyond the outer membrane, and it appears to cover in a monolayer the entire length of the rod as well as both of its ends (Fig. 1B). We could not conclude from the EM images whether these mutants retain the electron-dense layer of capsular polysaccharides (17) characteristic of B. thetaiotaomicron.
As a proteinaceous surface layer has not yet been described in Bacteroides, we set out to determine the identity of its constituent protein(s). Cell-free culture fluid from log-phase ΔBT1954 mutant cells was acidified to precipitate proteins that were secreted or had dissociated from the cell material. We separated the proteins using denaturing polyacrylamide gel electrophoresis, and the most prominent band (apparent molecular mass = ~100 kDa) was excised and subjected to tandem mass spectrometry. Multiple peptide matches confirmed that it was BT1927, a 928-amino-acid (aa) protein of unknown function. (We suspect that the N terminus is annotated incorrectly in the database, as an earlier start codon would include an additional 12 amino acids that are highly conserved in the orthologs of BT1927. Residue numbers throughout the manuscript refer to the corrected open reading frame [ORF].) Although a related species of a different family of Bacteroidales (also a member of the human microbiota), Parabacteroides distasonis 8503, has been reported to possess a phase-variable S-layer glycoprotein (18), the unambiguous identification of a crystalline surface layer in a Bacteroides species is unprecedented.
Expression of BT1927 is controlled by an invertible promoter.
BT1927 resides in a locus adjacent to two site-specific recombinases, BT1928 and BT1929, and the 887 bp of intergenic sequence between BT1927 and BT1928 contains a 256-bp predicted promoter-containing element flanked by 22-bp inverted repeats (IRs) (Fig. 1C) (19). Similar invertible promoter elements are responsible for the phase-variable expression of other surface structures, including capsular polysaccharides (7–9, 18), indicating that the expression of this new type of surface structure might be controlled by a mechanism similar to that of the well-characterized capsular polysaccharides. Consistent with this view, a previously reported analysis of DNA sequencing reads from Bacteroides fragilis NCTC 9343 has shown that a promoter of similar architecture that resides adjacent to BF4087, a homolog of BT1927, exists in both orientations under conditions of laboratory culture and that the promoter’s orientation is controlled by the recombinase Tsr26, a homolog of BT1928 and BT1929 (9). In order to determine the orientation of the BT1927 promoter in the ΔBT1954 mutant, we amplified the IR element with primers that anneal beyond its 5′ and 3′ borders and digested the amplicon with the restriction enzyme DrdI, which has a recognition site inside the element that generates asymmetric fragments that are diagnostic of orientation (Fig. 1D). We found that in B. thetaiotaomicron, the promoter exists in one orientation (OFF, as described below), but in the ΔBT1954 mutant, the opposite orientation of the promoter is observed (ON, as described below) (Fig. 1D).
As the BT1927 locus operon is >36 kb away from BT1954, we next sought to determine whether BT1927 is related to BT1954. Two lines of evidence suggest that there is no direct relationship between the two proteins. (i) We rederived the ΔBT1954 mutant from the merodiploid (single-crossover) intermediate; none of the rederived mutants harbored the BT1927 promoter in the ON configuration. (ii) We independently constructed mutants in which the expression of BT1927 was “locked” on or off by mutating one of the half-sites of its invertible promoter (Fig. 2A; here, BT1927-ON and BT1927-OFF), and neither mutant harbored a polymorphism at the BT1954 locus. We conclude that (i) our initial ΔBT1954 mutant underwent a rare bottleneck event in which a BT1927-expressing variant was inadvertently isolated; (ii) BT1927 is unrelated to BT1954, although we cannot rule out the possibility of a high-level regulatory connection between the two loci; and (iii) our discovery of BT1927 was serendipitous. We therefore performed the rest of our experiments with the BT1927-ON and BT1927-OFF mutants, and BT1954 was not investigated further.
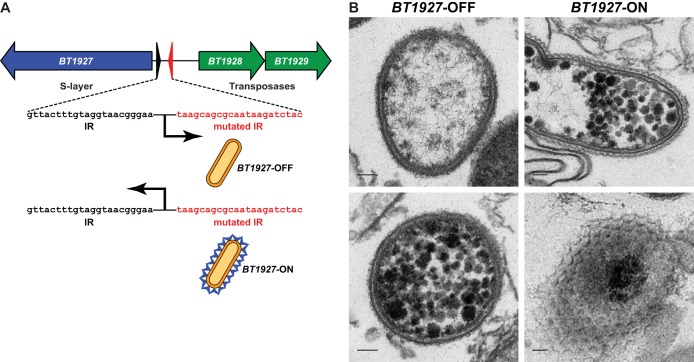
FIG 2 .
Construction and characterization of BT1927-ON and BT1927-OFF mutants. (A) One half-site of the inverted repeat surrounding the predicted BT1927 promoter was mutated to create strains in which this element could no longer flip, “locking” the expression of BT1927 on (BT1927-ON) or off (BT1927-OFF). (B) Cells of the BT1927-OFF (upper left and lower left) and BT1927-ON (upper right and lower right) mutants were prepared as described in the Fig. 1 legend and visualized by transmission electron microscopy, confirming the presence of the same surface structure in BT1927-ON that was observed in the ΔBT1954 mutant. Each scale bar represents 100 nm (see also Fig. S2 in the supplemental material).
BT1927 is secreted and localized to the cell surface.
We next sought to determine unambiguously that the BT1927 protein product is secreted and localized to the cell surface. The N terminus of BT1927 harbors a predicted signal peptide, and peptides from trypsin- and ArgC-digested BT1927 start with T26, suggesting that the protein is cleaved after D25 during the process of secretion. Three bioinformatic findings provided further evidence for the secretion of BT1927. (i) The N-terminal 48 aa of BT1927 are highly conserved across its homologs in Bacteroides spp. (75% to 100%), but the remaining 880 aa are less so (31% to 43%), indicating a conserved function for its putative N-terminal signal peptide (see Fig. S1A, Fig. S3, and Text S3 in the supplemental material). (ii) The conserved N-terminal peptide extends longer than a typical signal peptide, suggesting a function beyond that of a typical signal peptide (see Fig. S1B). (iii) BT1927 and its homologs are found in a location adjacent to a gene that encodes a predicted outer membrane beta-barrel protein, consistent with the combination of BT1927 and BT1926 (BT1927-BT1926) (and its orthologs) functioning as a two-partner secretion system (see Text S1 and Text S2 in the supplemental material).
Three lines of experimental evidence suggest that BT1927 is present on the cell surface. First, SDS-PAGE analysis of trichloroacetic acid (TCA)-precipitated proteins from cell-free culture fluid revealed a high-molecular-mass band in the BT1927-ON cells but not the BT1927-OFF cells; this band corresponds to BT1927 and is the same band that appeared in the ΔBT1954 mutant (see Fig. S2 in the supplemental material). Second, transmission electron micrographs of the BT1927-ON mutant show the same tessellated structure adjacent to the outer membrane as in the ΔBT1954 mutant, but this structure is absent in the BT1927-OFF mutant (Fig. 2B). Finally, we constructed mutants in the BT1927-ON and BT1927-OFF backgrounds in which a FLAG tag was appended in frame to the C terminus of BT1927. Immunocytochemical staining of BT1927-ON-FLAG and BT1927-OFF-FLAG cells confirmed that BT1927 is present on the surface of the former but not the latter mutant (Fig. 3A). Together, these results suggest that BT1927 is secreted through the inner and outer membranes, forming a novel surface structure adjacent to the outer membrane.
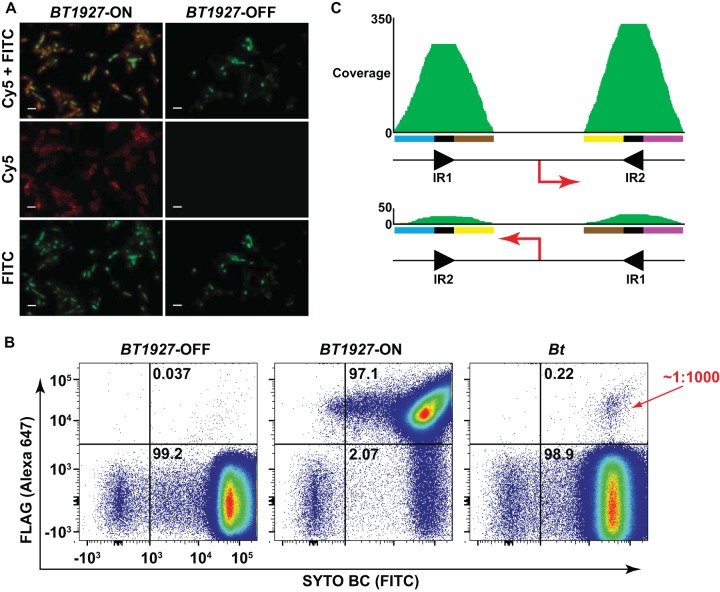
FIG 3 .
BT1927 is expressed in a subpopulation of B. thetaiotaomicron. (A) BT1927-ON-FLAG and BT1927-OFF-FLAG cells were washed, stained with an anti-FLAG antibody conjugated to Alexa 647 and the DNA stain SYTO BC, and visualized by fluorescence microscopy. Cell surface staining in the BT1927-ON-FLAG mutant but not the BT1927-OFF-FLAG mutant is consistent with localization of BT1927 to the cell surface. Each scale bar represents 2 µm. (B) BT1927-ON-FLAG, BT1927-OFF-FLAG, and BT1927-FLAG cells were washed, stained as described above, and analyzed by flow cytometry to measure the frequency of BT1927-expressing cells in each culture. As expected, the BT1927-expressing population (upper right quadrant) dominated the BT1927-ON culture and was nearly absent in the BT1927-OFF culture. The BT1927-FLAG culture reproducibly showed a BT1927-expressing subpopulation that represented ~1:1,000 cells in the population. (C) The frequency of the two promoter orientations of a BT1927 ortholog in a whole-genome shotgun metagenomic sequence sample from the NIH Human Microbiome Project was measured by recruiting raw sequence reads to each promoter orientation. A total of 92% of the reads were recruited to one (presumably OFF) orientation, and the remaining 8% of the reads were recruited to the opposite (presumably ON) orientation, suggesting that the BT1927-expressing cells might exist at a higher frequency in vivo than in vitro.
A subpopulation of a wild-type B. thetaiotaomicron culture expresses BT1927.
Having validated that BT1927-ON cells harbor a novel surface layer, we next asked what percentage of cells in a laboratory culture of B. thetaiotaomicron express BT1927. Because we were able to detect cells with the BT1927 promoter in the ON orientation using PCR amplification but not the PCR-based restriction digestion assay described above, we hypothesized that BT1927-expressing cells occur at a low frequency, requiring us to develop a more sensitive method of detection. Having previously constructed B. thetaiotaomicron mutants in which BT1927 is C-terminally FLAG tagged in the BT1927-ON and BT1927-OFF backgrounds, we next constructed a mutant in which BT1927 is C-terminally FLAG tagged in the parental B. thetaiotaomicron background (here, BT1927-FLAG). Cells from all three of the BT1927-FLAG strains were labeled with a monoclonal anti-FLAG antibody, and FLAG-positive (FLAG+) cells were quantified by flow cytometry. As expected, 93.2% to 97.1% of the cells in the BT1927-ON-FLAG population had detectable BT1927 expression, while expression in BT1927-OFF-FLAG cells was 0.04% to 0.08%. In a culture of BT1927-FLAG, we found reproducibly that 0.15% to 0.25% of the cells expressed BT1927, suggesting that cells expressing the surface layer form a minority of ~1:1,000 cells in a laboratory population of B. thetaiotaomicron (Fig. 3B).
The frequency of phase variants under conditions of laboratory culture is known to differ from that seen under conditions of growth in an animal host (20, 21). To determine the frequency of this phase variant in vivo, we next analyzed whole-genome-shotgun (WGS) metagenomic sequence data from Human Microbiome Project (HMP) stool samples in an effort to find a subject colonized predominantly by B. thetaiotaomicron. Although we found BT1927 in 52 (60%) of 86 stool samples, its coverage in those samples was not deep enough to detect both orientations. Instead, we found a subject colonized at a high level by a strain of Bacteroides fragilis that harbors a homolog of BT1927. We computationally recruited raw reads from this WGS sample to four loci corresponding to the two orientations of the invertible promoter; an average of 22 reads (~8% of total reads) were recruited to one orientation (presumably ON), while an average of 254 reads (~92% of total reads) were in the opposite orientation (presumably OFF). In order to compare this result to the frequency of each orientation in B. fragilis cultivated in vitro, we performed targeted amplicon sequencing on a region that encompasses the predicted invertible sites, including the B. fragilis ortholog of the BT1927 promoter. We used B. fragilis strain 638r, which harbors a BT1927 ortholog that is 100% identical at the nucleotide level to the B. fragilis locus found in the metagenomic data described above. From our data, we were able to recruit 74 reads to one orientation of the BT1927 ortholog promoter region (presumably ON; 0.008%) and 910,786 reads to the other orientation (presumably OFF; 99.992%) (see Table S2 and Text S3 in the supplemental material). This finding indicates that the frequency of BT1927 ortholog expression may be higher in vivo than in vitro (Fig. 3C).
BT1927-ON cells are resistant to complement-mediated killing.
Finally, we turned to the issue of whether the B. thetaiotaomicron subpopulation expressing a surface layer is functionally distinct from the majority of cells expressing a conventional polysaccharide capsule. To begin addressing this issue, we subjected the BT1927-ON and BT1927-OFF mutants to a set of assays relevant to colonization and growth in the mammalian gut: recognition by IgA, tropism in the small and large intestines, sensitivity to antimicrobial peptides, activation of TLR2 and TLR4, and sensitivity to lysis in serum.
Only one of these assays revealed a functional difference between BT1927-ON and BT1927-OFF: BT1927-ON cells were shown to be highly resistant to complement-mediated killing. When incubated with 20% human serum for 60 min, 48.18% ± 12% of the BT1927-ON cells survived compared to just 0.97% ± 0.18% of the BT1927-OFF cells (Fig. 4A); similar results were obtained by adjusting serum sources (human and rabbit) and concentrations. We note, however, that 20% human serum might be a superphysiological concentration of complement in the context of gut colonization.
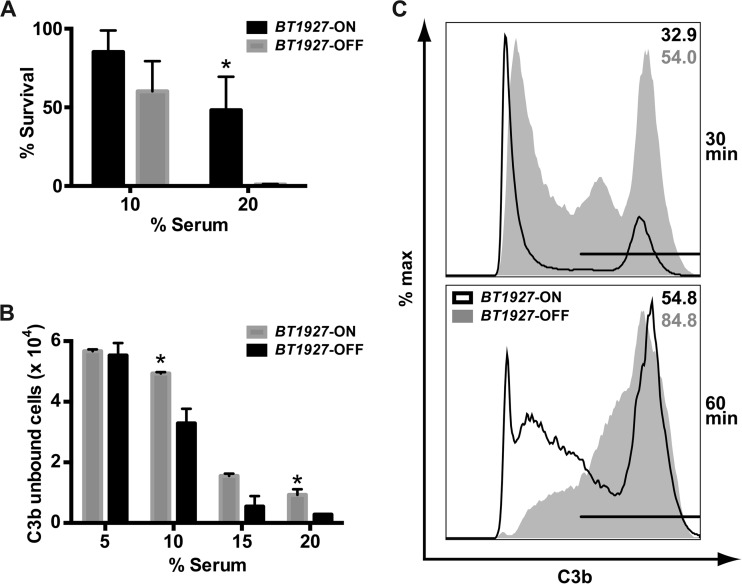
FIG 4 .
BT1927 confers resistance to complement-mediated killing. (A) BT1927-ON and BT1927-OFF cells were incubated in 10% or 20% human serum or 20% heat-inactivated serum at 37°C for 1 h, serially diluted, and plated. For each strain, the number of colonies that survived in untreated serum was counted and divided by the number of colonies that survived in BHI serum to determine the percentage of survival. In 20% serum, BT1927-ON cells were more resistant to serum-mediated lysis (48.18% ± 12% survival) than BT1927-OFF cells (0.97% ± 0.18% survival). *, P < 0.05 (Student’s t test). (B) Approximately the same numbers of BT1927-ON and BT1927-OFF cells were incubated in 5%, 10%, 15%, or 20% serum at 37°C for 30 min, washed, stained with FITC-conjugated anti-C3 F(ab′)2, and analyzed by flow cytometry. The bar graph shows the absolute numbers of events that were APC+ and FITC− (intact cells not associated with C3 or a C3 fragment). C3 or its fragments associated at lower efficiency with BT1927-ON cells than with BT1927-OFF cells, consistent with a blockade of C3b deposition by BT1927. *, P < 0.05 (Student’s t test). (C) The same experiment as that described for panel B was used to measure the distribution of FITC+ (C3 fragment-associated) and FITC− (not C3 fragment-associated) populations in the BT1927-ON and BT1927-OFF mutants. Samples were treated with 10% serum for 30 min (top panel) or 60 min (bottom panel). The results, which are representative of three independent experiments, show that BT1927-ON cells appear to activate complement with delayed kinetics compared with the BT1927-OFF cells, as measured by the presence of the C3 fragment. max, maximum.
To explore the molecular underpinnings of complement resistance in the BT1927-expressing subpopulation, we turned to the issue of what step in the complement cascade is blocked by BT1927-expressing cells. We tested the integrity of C3b deposition, a key step in the complement pathway that is known to be subverted by bacterial pathogens. We incubated the BT1927-ON and BT1927-OFF strains with human serum, labeled cells with fluorescein isothiocyanate (FITC)-conjugated anti-human C3 F(ab′)2, and performed flow cytometry to assess the level of C3 fragments associated with bacterial cells. As shown in Fig. 4B and C, C3 or its fragments associated with BT1927-ON cells at a lower efficiency than and with delayed kinetics relative to BT1927-OFF cells. These results suggest that BT1927-mediated blockade of C3b deposition is at least partly responsible for the resistance of the BT1927-expressing subpopulation to complement-mediated killing.
Previous work has shown that surface layer proteins from pathogenic bacteria such as Campylobacter fetus confer resistance to complement-mediated lysis (22, 23), one of several strategies used by pathogens to evade the complement system (24). However, B. thetaiotaomicron is a symbiont, not a pathogen or pathobiont, and is therefore unlikely to encounter the complement system in serum. Recent studies suggested that complement regulates the composition of the skin and oral communities (25–27) and that complement components are deposited into the lumen and epithelial surface of the murine and human GI tract (28–34). However, more work is needed to determine whether there exists a physiologically relevant interaction between complement and the microbiota under conditions of colonization and, if so, what role, if any, the B. thetaiotaomicron surface layer plays in an interaction with the complement system.
Our data show that Bacteroides can remodel its surface to express a previously unknown surface layer, and they suggest that Bacteroides populations harbor a greater diversity of subpopulations with distinct surface structures than was previously known. The BT1927-expressing subpopulation might be able to colonize a different niche in the mammalian gut, although further studies are needed to explore this idea and determine the biological significance of the surface layer. Further study of other phase-variable loci might also reveal new subpopulations of cells with distinct functions that are important for colonization and microbe-host interactions.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All bacterial strains used in this study are listed in Table S1 in the supplemental material. All Bacteroides thetaiotaomicron strains were grown at 37°C in brain heart infusion (BHI) agar supplemented with 10% horse blood, tryptone-yeast extract-glucose (TYG) medium, or Bacteroides minimal medium (MM) (35, 36) in an anaerobic chamber (Coy Laboratory Products) with a 5% H2, 20% CO2, and N2 (balance) gas mix. Escherichia coli strains were grown aerobically at 37°C in LB medium supplemented with ampicillin to select for the pExchange-tdk plasmid.
Construction of Bacteroides thetaiotaomicron mutants.
All plasmids and primers are listed in Tables S1 and S2 in the supplemental material. All mutants were created in the B. thetaiotaomicron VPI-5482 Δtdk background, which is referred to in the text as “B. thetaiotaomicron.” The ΔBT1954 mutant was constructed using the counterselectable allele exchange method described by Koropatkin and coworkers (37). Briefly, ~1-kb fragments upstream and downstream of the BT1954 gene were cloned and fused using primer pairs ΔBT1954 Sal-UF/UR and DF/XbaI-DR, respectively, and ligated into the suicide vector pExchange-tdk (obtained from Justin Sonnenburg, Stanford University). The resulting vector was electroporated into Escherichia coli S17-1 λ pir and then conjugated into B. thetaiotaomicron. Single recombinants were selected on BHI-blood agar plates containing 200 µg/ml gentamicin and 25 µg/liter erythromycin. Single recombinants were cultured in TYG medium overnight and then plated onto BHI-blood agar plates containing 200 µg/ml 5-fluoro-2-deoxyuridine (FUdR). Candidate BT1954 deletions were screened by PCR using the diagnostic primers listed in Table S2 and confirmed by DNA sequencing to identify isolates that had lost the gene.
BT1927-ON and BT1927-OFF mutants were constructed using a strategy adapted from Gunther et al. (38). We used the counterselectable allele exchange method described above, except that the insertion ligated into pExchange-tdk was made by fusing three fragments: (i) a 1-kb upstream fragment, (ii) the promoter region (in ON or OFF orientation) flanked by one native IR sequence and one mutated IR sequence consisting of random DNA sequence, and (iii) a 1-kb downstream fragment. The random sequence was generated using a random DNA sequence generator (http://www.faculty.ucr.edu/~mmaduro/random.htm) with the addition of a BglII site to facilitate screening by restriction digestion.
The FLAG epitope tag was inserted in frame at the 3′ end of the BT1927 coding sequence in the BT1927-ON and BT1927-OFF backgrounds using the method described above, except that the construct was made by fusing a 1-kb upstream fragment and a 1-kb downstream fragment harboring the FLAG sequence.
Diagnostic digestion to determine the orientation of the BT1927 promoter region.
The BT1927 promoter region was amplified by PCR using primers BT1927 IR1 and IR2 (see Table S2 in the supplemental material). The amplicon was cleaned using a Qiagen PCR purification kit and digested overnight with DrdI. The digested amplicon fragments were separated on a 1% agarose Tris-acetate-EDTA (TAE) gel and imaged using an Alpha Imager (Protein Simple).
Protein extraction and SDS-PAGE.
The B. thetaiotaomicron wild-type, ΔBT1954, BT1927-ON, and BT1927-OFF strains were cultured overnight in TYG medium, diluted 1:100 in fresh TYG medium, and grown to log phase (~6.5 h) the next day. The culture supernatant was filtered using a 0.22-µm-pore-size syringe filter (Millipore) and concentrated using a 10K-molecular-weight-cutoff Amicon Ultra-15 centrifugal filter (Millipore), and proteins were precipitated by addition of 1 vol of a trichloroacetic acid stock solution (500 g trichloroacetic acid dissolved in 350 ml H2O) to 4 vol of supernatant, followed by two cold acetone washes. The precipitate was resuspended in sample buffer and loaded onto a 10% acrylamide gel. Gels were stained with Coomassie blue for visualization and protein band excision.
Transmission electron microscopy sample preparation and imaging. (i) B. thetaiotaomicron culture and initial fixation.
B. thetaiotaomicron and mutant strains were cultured overnight in TYG medium in the anaerobic chamber at 37°C. The following day, saturated cultures were diluted 1:50 into 10 ml MM for 15 h at 37°C in the anaerobic chamber. After 15 h, 7.5 ml 4% glutaraldehyde–MM was mixed with 7.5 ml of the culture and fixed in the anaerobic chamber for 10 min with rocking. The mixture was centrifuged at 3,270 × g for 10 min, and the pellet was resuspended in 2 ml 2.5% glutaraldehyde–0.1 M sodium cacodylate buffer at pH 7.2 (abbreviated as “CB”). The resulting suspension was divided into aliquots, added to microcentrifuge tubes, and stored at 4°C until further processing.
(ii) Washing and postfixation.
For the remainder of the sample preparation procedure, all centrifugation steps were performed at ~21,000 × g for 1 min. The cell suspension in CB was centrifuged, and cells were resuspended in fresh CB and incubated for 10 min; this wash step was repeated for a total of 3 times. After the last wash step, the cells were resuspended in CB supplemented with 1% OsO4 and 1.6% potassium ferricyanide. Using a Pelco Biowave Pro microwave tissue processor (Ted Pella; abbreviated as “MW”), the cells were fixed with 2 cycles of the following procedure: 2 min of heating in a vacuum and 2 min of vacuum without heating. To remove the OsO4, the cells were resuspended in CB, heated for 40 s in the MW, and centrifuged, and the supernatant was removed. This wash step was repeated a total of 3 times.
(iii) Uranyl acetate en bloc staining.
To remove salts, cells were washed three times by centrifugation, resuspended in deionized (DI) water, and incubated for 10 min at room temperature. Cells were resuspended in 1% uranyl acetate prepared in DI water and stained in the MW using 2 cycles of the following program: 2 min heating in a vacuum and 2 min of vacuum without heating. Uranyl acetate was removed by resuspension in DI water and heating at 40 s in the MW a total of 3 times.
(iv) Dehydration and infiltration.
Dehydration consisted of the following procedure: centrifugation, resuspension of the cell pellet in 35% ethanol, and heating in the MW for 40 s. This process was repeated a total of 2 times each with 35%, 50%, 70%, 80%, 95%, and 100% ethanol, followed by one additional 40-s heating step with 100% ethanol and 40 s of heating with 100% acetone. The cells were pelleted, the acetone was decanted, and then the resin was infiltrated (recipe for 50 mg resin: 23.5 mg of Eponate 12 resin, 12.5 mg of dodecenyl succinic anhydride [DDSA], 14 mg of nadic methyl anhydride [NMA], and 0.75 ml of benzyldimethylamine [BDMA]) at an acetone/resin ratio of 3:1. This mixture was subjected to 2 cycles of 3 min of heating in a vacuum in the MW. The same procedure was repeated with a 1:1 mixture of acetone and resin and then a 1:3 mixture. Finally, cells were resuspended in pure resin, heated in a vacuum in the MW for 3 min, and then centrifuged; this final procedure was repeated a total of 3 times. Cells were placed in a beam capsule and heated in a 55°C oven for 2 days.
(v) Ultrathin sectioning and imaging.
The resin-embedded cells were cut into ultrathin sections using a diamond knife. Sectioned samples were inserted into a Pelco grid staining system. Samples were stained in 2% methanolic uranyl acetate (2% uranyl acetate dissolved in 70% methanol–H2O) for 5 min at room temperature, washed 5 times in DI water, and then stained in Reynolds lead citrate (recipe for 50 ml: 1.33 g lead nitrate and 1.76 g of sodium citrate dihydrate were dissolved in 30 ml DI H2O; 8 ml of 1 N NaOH was added, followed by 10 ml of DI H2O) for 5 min at room temperature. A total of 10 more washes were performed with DI water, and the sample was dried with a Kimwipe and imaged on an FEI Tecnai 12 transmission electron microscope. The images were cropped, and the size of each image was compressed using Adobe Photoshop. ImageJ was used to add the scale bar.
Immunofluorescent labeling and microscopy.
Immunofluorescent labeling was performed according to a protocol described by Moyes (39). Briefly, bacteria were cultured in TYG medium overnight in the anaerobic chamber at 37°C. Saturated cultures were diluted 1:50 in 10 ml MM and incubated for 15 h at 37°C in the anaerobic chamber. Following incubation, cultures were centrifuged at 3,720 × g for 10 min, washed with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), and resuspended to approximately 107 to 108 CFU/ml. A smear was prepared by spreading 20 µl of the resuspended cells using a sterile loop on a clean microscope slide, and then the smear was air dried in the fume hood, submerged in 95% ethyl alcohol (EtOH), and subjected to heat fixation by incubation in the 55°C oven for 10 min. Slides were incubated with an anti-FLAG antibody conjugated to Alexa Fluor 647 (Cell Signaling) (diluted 1:100 in PBS) for 30 min at 37°C, rinsed, and incubated in PBS for 30 min at room temperature. Labeled cells were counterstained with SYTO BC Bacteria Stain (Molecular Probes). Images were acquired using a 100× objective on a 6D high-throughput microscope at the Nikon Imaging Center of the University of California, San Francisco (UCSF). The images were cropped, and the size of each image was compressed using Adobe Photoshop. ImageJ was used to add the scale bar.
Flow cytometry to quantify BT1927-expressing cells.
B. thetaiotaomicron and mutant strains were grown in TYG medium overnight, diluted 1:100 in fresh TYG medium, and grown to the late log phase (~6.5 h). One milliliter of the culture was centrifuged for 2 min in a microcentrifuge at 6,000 × g, washed twice with PBS, and resuspended to approximately 50 × 106 cells/ml in Bacteria Staining buffer (BSB; 1% bovine serum albumin–0.025% sodium azide–PBS). In a V-bottom 96-well cell culture plate (Corning), 25 µl of the resuspended bacteria was mixed with 25 µl anti-FLAG Alexa Fluor 647-conjugated antibody (Cell Signaling) (diluted 1:25 in BSB) and incubated for 1 h at 37°C. The 96-well plates were centrifuged at 3,273 × g on a Beckman tabletop centrifuge for 4 min to pellet the cells and washed twice with 150 µl BSB. Cells were counterstained with SYTO BC Bacteria Stain (Molecular Probes) and analyzed on an LSRII flow cytometer. In order to minimize carryover of cells, 3 blank samples of PBS were included between samples. Forward scatter (FSC) and side scatter (SSC) data were set to logarithmic scale. Two populations (top and bottom) were visible on the FSC and SSC plots, where the bottom population was present even in sterile H2O, bleach, and PBS without any bacteria and the top population was present only in samples with bacteria. Therefore, a gate was set to exclude the bottom population and 500,000 events were collected for the top population. After data acquisition, the top B. thetaiotaomicron population was visualized on an allophycocyanin (APC)-versus-FITC plot and further separated by quadrants, which were set using the unstained sample. Spectral overlap was calculated from single-stained controls. All postacquisition analyses and compensations were performed using FlowJo (Tree Star).
Computational analysis of BT1927 and its orthologs in stool samples sequenced by the HMP.
The protein sequence of BT1927 was used as a query sequence for BLASTp searches of a translated database of assembled contigs from each of the first-visit stool samples sequenced by the Human Microbiome Project (HMP) (n = 86; available at the HMP Data Analysis and Coordinating Center [DACC]). BT1927 was deemed “present” in a sample when a protein sequence was detected as a hit, with an expectation value of <1.0e-50. The relatively low coverage of B. thetaiotaomicron in these samples (~20× to ~30×, as determined by coverage calculation for the BT1927 inverted repeat region in a subset of these samples) prevented us from determining the orientation of the BT1927 promoter region. We then performed a similar BLAST search to identify BT1927 orthologs from other Bacteroides spp. with high depth of coverage in the HMP samples. One sample, SRS016267, harbored a B. fragilis BT1927 ortholog at 477× coverage. Quality-filtered and trimmed reads of SRS016267 were obtained from the HMP DACC and used to determine the frequencies of both orientations of this gene’s promoter. Briefly, we divided the inverted repeat/promoter region of the BT1927 ortholog into four sequence blocks of 100 bp, where each block consisted of 18 bp of each of the inverted repeats flanked by 41 bp on each side, to determine its orientation. Two blocks corresponded to the upstream and downstream inverted repeat sequences of orientation 1, and the other two blocks corresponded to the upstream and downstream inverted repeat sequences of orientation 2. SRS016267 reads matching these blocks were identified using BLASTn with a cutoff expectation value of 1.0e-31, which we determined to be necessary to unequivocally assign the orientation. To further confirm their orientation, these reads were mapped to the corresponding sequence blocks using Geneious (with the following parameters: minimum percent identity at overlap, 90%; maximum percentage of mismatch per read, 20%), and the average coverage of each block was calculated only from the mapped reads.
Human complement killing assay.
B. thetaiotaomicron and mutant strains were cultured overnight in TYG medium. The next day, cells were harvested by centrifugation, washed once with PBS, and resuspended to 50 × 106 CFU/ml in PBS++ (PBS supplemented with 0.5 mM MgCl2 and 1 mM CaCl2). Cells were incubated in human serum and heat-inactivated serum at various concentrations in PBS++ at 37°C for 1 h. Following incubation, cells were diluted and plated on BHI–10% horse blood agar plates and incubated anaerobically until colonies appeared. Percent survival was calculated by dividing the average number of colonies that appeared on each human serum plate by the average number of colonies that appeared on the heat-inactivated serum plate and multiplying by 100. Error bars represent the standard deviation. Student’s t test was performed to calculate P values (*, P < 0.05).
Flow cytometry to determine C3 fragment presence in BT1927-ON and BT1927-OFF cells.
BT1927-ON and BT1927-OFF cells were cultured overnight in TYG medium. Cells were pelleted and washed once with PBS, and approximately the same numbers of cells were incubated in 20% human serum–PBS++ for 30 to 60 min at 37°C. Following incubation, cells were pelleted and washed 3 times with BSB. The cells were then stained with FITC-conjugated anti-human C3 F(ab′)2 (Protos Immunoresearch; catalog no. 365) at a 1:50 final concentration, washed 3 times with BSB, and stained with a 1:1,000 dilution of SYTO 59 (Molecular Probes) for 1 h at 37°C. Each sample was analyzed using an LSRII flow cytometer, with gating on the top population that appeared on the FSC/SSC plot determined as described earlier. Spectral overlap was calculated from single-stained controls, and a single-cell gate and an APC+ gate were further set using unstained controls. The absolute numbers of events that were APC+ and FITC− (intact cells that lacked C3 fragments) were plotted on the bar graph for each serum concentration. Student’s t test was performed to calculate the P value. The histogram was generated by plotting the fluorescence intensity of FITC versus the maximum percentage. Spectral overlap was calculated from single-stationed controls. All postacquisition analyses and compensations were performed using FlowJo (Tree Star).
SUPPLEMENTAL MATERIAL
BT1927 amino acid sequence analysis and its conservation among Bacteroides genomes. (A) As annotated in the NCBI protein database, BT1927 likely has an incorrect start site; there are likely 25 more aa at the N terminus. The corrected amino acid sequence of BT1927 was aligned with BT1927 orthologs in Bacteroides species using ClustalX. Note that the first ~50 amino acids are highly conserved. (B) The BT1927 amino acid sequence was aligned with a BT1927 homolog in B. thetaiotaomicron CAG:40, which showed almost a complete match for the first ~200 amino acids. (C) The amino acid composition of BT1927 was compared to the average percent amino acid composition of B. thetaiotaomicron proteins. Download
SDS-PAGE gel of BT1927-ON and BT1927-OFF. Surface proteins were isolated from culture supernatants of the BT1927-ON and BT1927-OFF mutants by TCA precipitation and were separated and visualized by SDS-PAGE. Download
Phylogenetic tree of Bacteroides with and without orthologs of BT1927. A phylogenetic tree of Bacteroides was generated using the JGI IMG website distance tree function based on 16S alignment. Genomes with an S-layer orthologs and homologs (E value cutoff of 1e-5) are colored pink. Strains with names with asterisks at the end have a conserved inverted repeat sequence (GTTAC[N7]GTAAC) 5′ upstream of the respective BT1927 orthologs. Strains with names with a plus sign at the end have a predicted site-specific recombinase nearby. The flipping of the inverted repeat elements has been verified experimentally in the following strains (highlighted in bold): B. thetaiotaomicron VPI-5482, B. fragilis 638R (current study), and B. fragilis YCH46 (19). Note that the tree is unrooted. Download
Bacterial strains, plasmids, and primers used in this study. (A) List of bacterial strains. (B) List of primers and their sequences. Restriction sites are underlined.
Frequency of orientations in B. fragilis 638R predicted invertible promoters. Targeted amplicon sequencing was performed on various predicted invertible promoter regions in the B. fragilis 638R genome, and the frequency of each orientation was determined by counting the number of reads that mapped specifically to one orientation. The frequency of each orientation (A), as well as the primers used (B), is shown for each site.
Supplemental text. Download
Supplemental References. Download
Supplemental Methods. Download
ACKNOWLEDGMENTS
We are indebted to Justin Sonnenburg (Stanford) and members of the Fischbach Group for helpful advice, and we are grateful to Jeff Johnson (UCSF) for help with mass spectrometry experiments. We thank Reena Zalpuri (UC Berkeley) for technical assistance with transmission electron microscopy, Chris Baker (UCSF) for help with flow cytometry analyses, Eric Chow for help with sequencing and library construction, and Arel Cordero for help with phylogenetic tree construction.
This work was supported by a Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Disease award (M.A.F.), a Medical Research Program grant from the W.M. Keck Foundation (M.A.F.), a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.), and NIH grants OD007290 (M.A.F.), GM081879 (M.A.F.), AI068730 (J.D.L.), and AI30040 (J.D.L.).
M.A.F. is on the scientific advisory board of NGM Biopharmaceuticals.
Footnotes
Citation Taketani M, Donia MS, Jacobson AN, Lambris JD, Fischbach MA. 2015. A phase-variable surface layer from the gut symbiont Bacteroides thetaiotaomicron. mBio 6(5):e01339-15. doi:10.1128/mBio.01339-15.
REFERENCES
- 1.Corbett D, Roberts IS. 2009. The role of microbial polysaccharides in host-pathogen interaction. F1000 Biol Rep 1:30. doi: 10.3410/B1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Gordon JI. 2009. The core gut microbiome, energy balance, and obesity. J Physiol 587:4153–4158 doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 7.Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 8.Coyne MJ, Weinacht KG, Krinos CM, Comstock LE. 2003. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc Natl Acad Sci U S A 100:10446–10451. doi: 10.1073/pnas.1832655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinacht KG, Roche H, Krinos CM, Coyne MJ, Parkhill J, Comstock LE. 2004. Tyrosine site-specific recombinases mediate DNA inversions affecting the expression of outer surface proteins of Bacteroides fragilis. Mol Microbiol 53:1319–1330. doi: 10.1111/j.1365-2958.2004.04219.x. [DOI] [PubMed] [Google Scholar]
- 10.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. 2009. A family of transcriptional antitermination factors necessary for synthesis of the capsular polysaccharides of Bacteroides fragilis. J Bacteriol 191:7288–7295. doi: 10.1128/JB.00500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatzidaki-Livanis M, Weinacht KG, Comstock LE. 2010. Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc Natl Acad Sci U S A 107:11976–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. 2008. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc Natl Acad Sci U S A 105:13099–13104. doi: 10.1073/pnas.0804220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coyne MJ, Comstock LE. 2008. Niche-specific features of the intestinal Bacteroidales. J Bacteriol 190:736–742. doi: 10.1128/JB.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu CH, Lee SM, VanLare JM, Kasper DL, Mazmanian SK. 2008. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci U S A 105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. 2014. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagan RP, Fairweather NF. 2014. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol 12:211–222. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- 17.Pumbwe L, Skilbeck CA, Wexler HM. 2006. The Bacteroides fragilis cell envelope: quarterback, linebacker, coach---or all three? Anaerobe 12:211–220. doi: 10.1016/j.anaerobe.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. 2007. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc Natl Acad Sci U S A 104:2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A 101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder JA, Lloyd AL, Lockatell CV, Johnson DE, Mobley HL. 2006. Role of phase variation of type 1 fimbriae in a uropathogenic Escherichia coli cystitis isolate during urinary tract infection. Infect Immun 74:1387–1393. doi: 10.1128/IAI.74.2.1387-1393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troy EB, Carey VJ, Kasper DL, Comstock LE. 2010. Orientations of the Bacteroides fragilis capsular polysaccharide biosynthesis locus promoters during symbiosis and infection. J Bacteriol 192:5832–5836. doi: 10.1128/JB.00555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaser MJ, Smith PF, Kohler PF. 1985. Susceptibility of Campylobacter isolates to the bactericidal activity of human serum. J Infect Dis 151:227–235. doi: 10.1093/infdis/151.2.227. [DOI] [PubMed] [Google Scholar]
- 23.Blaser MJ, Smith PF, Repine JE, Joiner KA. 1988. Pathogenesis of Campylobacter fetus infections. Failure of encapsulated Campylobacter fetus to bind C3b explains serum and phagocytosis resistance. J Clin Invest 81:1434–1444. doi: 10.1172/JCI113474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambris JD, Ricklin D, Geisbrecht BV. 2008. Complement evasion by human pathogens. Nat Rev Microbiol 6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chehoud C, Rafail S, Tyldsley AS, Seykora JT, Lambris JD, Grice EA. 2013. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc Natl Acad Sci U S A 110:15061–15066. doi: 10.1073/pnas.1307855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinicke AT, Herrmann A, Baer F, Sina C, Srinivas G, Kuenzel S, Baines J, Köhl J. 2012. C5aR regulates intestinal microbiota composition and controls the induction of gastrointestinal allergic hypersensitivity. Immunobiology 217:1147. doi: 10.1016/j.imbio.2012.08.052. [DOI] [Google Scholar]
- 27.Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K, Hashim A, Hoch S, Curtis MA, Nussbaum G, Lambris JD, Hajishengallis G. 2014. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe 15:768–778. doi: 10.1016/j.chom.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahrenstedt O, Knutson L, Nilsson B, Nilsson-Ekdahl K, Odlind B, Hällgren R. 1990. Enhanced local production of complement components in the small intestines of patients with Crohn’s disease. N Engl J Med 322:1345–1349. doi: 10.1056/NEJM199005103221903. [DOI] [PubMed] [Google Scholar]
- 29.Baklien K, Brandtzaeg P. 1974. Immunohistochemical localisation of complement in intestinal mucosa. Lancet 304:1087–1088. doi: 10.1016/S0140-6736(74)92198-9. [DOI] [PubMed] [Google Scholar]
- 30.Halstensen TS, Brandtzaeg P. 1991. Local complement activation in inflammatory bowel disease. Immunol Res 10:485–492. doi: 10.1007/BF02919746. [DOI] [PubMed] [Google Scholar]
- 31.Halstensen TS, Mollnes TE, Garred P, Fausa O, Brandtzaeg P. 1992. Surface epithelium related activation of complement differs in Crohn’s disease and ulcerative colitis. Gut 33:902–908. doi: 10.1136/gut.33.7.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belzer C, Liu Q, Carroll MC, Bry L. 2011. The role of specific IgG and complement in combating a primary mucosal infection of the gut epithelium. Eur J Microbiol Immunol (Bp) 1:311–318. doi: 10.1556/EuJMI.1.2011.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andoh A, Fujiyama Y, Sakumoto H, Uchihara H, Kimura T, Koyama S, Bamba T. 1998. Detection of complement C3 and factor B gene expression in normal colorectal mucosa, adenomas and carcinomas. Clin Exp Immunol 111:477–483. doi: 10.1046/j.1365-2249.1998.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laufer J, Oren R, Goldberg I, Horwitz A, Kopolovic J, Chowers Y, Passwell JH. 2000. Cellular localization of complement C3 and C4 transcripts in intestinal specimens from patients with Crohn’s disease. Clin Exp Immunol 120:30–37. doi: 10.1046/j.1365-2249.2000.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holdeman LV, Moore WEC. 1977. Anaerobe laboratory manual. Virginia Polytechnic Institute and State University, Blacksburg, VA. [Google Scholar]
- 37.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunther NW, Snyder JA, Lockatell V, Blomfield I, Johnson DE, Mobley HL. 2002. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect Immun 70:3344–3354. doi: 10.1128/IAI.70.7.3344-3354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moyes RB. 2009. Fluorescent staining of bacteria: viability and antibody labeling. Curr Protoc Microbiol Appendix 3:Appendix-3K. doi: 10.1002/9780471729259.mca03ks15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BT1927 amino acid sequence analysis and its conservation among Bacteroides genomes. (A) As annotated in the NCBI protein database, BT1927 likely has an incorrect start site; there are likely 25 more aa at the N terminus. The corrected amino acid sequence of BT1927 was aligned with BT1927 orthologs in Bacteroides species using ClustalX. Note that the first ~50 amino acids are highly conserved. (B) The BT1927 amino acid sequence was aligned with a BT1927 homolog in B. thetaiotaomicron CAG:40, which showed almost a complete match for the first ~200 amino acids. (C) The amino acid composition of BT1927 was compared to the average percent amino acid composition of B. thetaiotaomicron proteins. Download
SDS-PAGE gel of BT1927-ON and BT1927-OFF. Surface proteins were isolated from culture supernatants of the BT1927-ON and BT1927-OFF mutants by TCA precipitation and were separated and visualized by SDS-PAGE. Download
Phylogenetic tree of Bacteroides with and without orthologs of BT1927. A phylogenetic tree of Bacteroides was generated using the JGI IMG website distance tree function based on 16S alignment. Genomes with an S-layer orthologs and homologs (E value cutoff of 1e-5) are colored pink. Strains with names with asterisks at the end have a conserved inverted repeat sequence (GTTAC[N7]GTAAC) 5′ upstream of the respective BT1927 orthologs. Strains with names with a plus sign at the end have a predicted site-specific recombinase nearby. The flipping of the inverted repeat elements has been verified experimentally in the following strains (highlighted in bold): B. thetaiotaomicron VPI-5482, B. fragilis 638R (current study), and B. fragilis YCH46 (19). Note that the tree is unrooted. Download
Bacterial strains, plasmids, and primers used in this study. (A) List of bacterial strains. (B) List of primers and their sequences. Restriction sites are underlined.
Frequency of orientations in B. fragilis 638R predicted invertible promoters. Targeted amplicon sequencing was performed on various predicted invertible promoter regions in the B. fragilis 638R genome, and the frequency of each orientation was determined by counting the number of reads that mapped specifically to one orientation. The frequency of each orientation (A), as well as the primers used (B), is shown for each site.
Supplemental text. Download
Supplemental References. Download
Supplemental Methods. Download