ABSTRACT
The human fungal pathogen Cryptococcus neoformans is capable of infecting a broad range of hosts, from invertebrates like amoebas and nematodes to standard vertebrate models such as mice and rabbits. Here we have taken advantage of a zebrafish model to investigate host-pathogen interactions of Cryptococcus with the zebrafish innate immune system, which shares a highly conserved framework with that of mammals. Through live-imaging observations and genetic knockdown, we establish that macrophages are the primary immune cells responsible for responding to and containing acute cryptococcal infections. By interrogating survival and cryptococcal burden following infection with a panel of Cryptococcus mutants, we find that virulence factors initially identified as important in causing disease in mice are also necessary for pathogenesis in zebrafish larvae. Live imaging of the cranial blood vessels of infected larvae reveals that C. neoformans is able to penetrate the zebrafish brain following intravenous infection. By studying a C. neoformans FNX1 gene mutant, we find that blood-brain barrier invasion is dependent on a known cryptococcal invasion-promoting pathway previously identified in a murine model of central nervous system invasion. The zebrafish-C. neoformans platform provides a visually and genetically accessible vertebrate model system for cryptococcal pathogenesis with many of the advantages of small invertebrates. This model is well suited for higher-throughput screening of mutants, mechanistic dissection of cryptococcal pathogenesis in live animals, and use in the evaluation of therapeutic agents.
IMPORTANCE
Cryptococcus neoformans is an important opportunistic pathogen that is estimated to be responsible for more than 600,000 deaths worldwide annually. Existing mammalian models of cryptococcal pathogenesis are costly, and the analysis of important pathogenic processes such as meningitis is laborious and remains a challenge to visualize. Conversely, although invertebrate models of cryptococcal infection allow high-throughput assays, they fail to replicate the anatomical complexity found in vertebrates and, specifically, cryptococcal stages of disease. Here we have utilized larval zebrafish as a platform that overcomes many of these limitations. We demonstrate that the pathogenesis of C. neoformans infection in zebrafish involves factors identical to those in mammalian and invertebrate infections. We then utilize the live-imaging capacity of zebrafish larvae to follow the progression of cryptococcal infection in real time and establish a relevant model of the critical central nervous system infection phase of disease in a nonmammalian model.
INTRODUCTION
As an AIDS-associated pathogen and with an enlarging immunosuppressed population, Cryptococcus neoformans is presently the leading invasive fungal pathogen worldwide by incidence and mortality rates (1). While the disease is initially established in the lung, it is not until it acts on its unique propensity to invade the central nervous system (CNS) that the disease becomes a major life-threatening fungal disease. Clinical presentation most commonly coincides with an established CNS infection, and subsequent mortality rates remain high in both resource-available and resource-limited countries.
Robust but expensive vertebrate model systems such as mice and rabbits have been well established for studying cryptococcal pathogenesis. These models have been effectively used in pathogenesis and treatment studies. For instance, the rabbit model of cryptococcal meningitis, where cryptococci are introduced intracisternally into the subarachnoid space, facilitates dynamic studies of genetic, transcriptional, and biochemical analyses of wild-type and mutant cryptococcal strains during CNS infection (2). The mouse inhalation model allows for detailed evaluation of the host immune response to cryptococcal infection as well as assessment of cryptococcal virulence determinants of wild-type or mutant strains. However, neither mammalian model allows real-time visualization of the dynamic interaction between innate immune cells and yeasts at distinct sites within the host, and neither model allows easy interrogation of large numbers of wild-type strains and mutants for screening and comparative purposes (3, 4).
Several nonmammalian models of C. neoformans pathogenesis have been reported to allow the analysis of cryptococcal pathogenesis through survival and cryptococcal growth assays in invertebrates, namely, Caenorhabditis elegans (worm), Galleria mellonella (wax moth), and Drosophila melanogaster (fruit fly) (5–8). Although these invertebrate models allow rapid analysis of cryptococcal virulence determinants and host genetics to various degrees, there are important limitations. First, the invertebrate nature of these hosts precludes investigations into complex multicellular interactions and specific body sites of infection necessary for investigating processes such as CNS entry and growth within granulomas. Second, anatomical differences between these invertebrate models and mammals limit the information gathered through live imaging of infectious disease progression.
The zebrafish (Danio rerio) is a powerful vertebrate model platform that has been applied to investigate host-pathogen interactions. Zebrafish larvae are genetically tractable, optically transparent, and possess several functional organ systems early in development, such as a circulatory system and innate immune system. Infections of zebrafish larvae essentially bridge the technical simplicity of invertebrate models with the tissue and cell type complexity of mammalian models (9). A larval zebrafish model of mucosal Candida albicans infection has recently been established. It critically allows the visualization of Candida-host interactions at this important but previously unimaged stage of yeast infection. With this basic platform work, the power of zebrafish live-imaging tools for exploring fungal-host interactions was elegantly demonstrated (10).
We propose that live imaging of transparent zebrafish larval will help address key gaps in our knowledge of cryptococcal virulence. Important stages of pathogenesis that are not well understood include the formation of cryptococcomas, the formation in vivo of Titan cells, and the mechanism of blood-brain barrier invasion responsible for cryptococcal meningitis—the most dangerous presentation of cryptococcal infection.
Here we have utilized infection of zebrafish larvae to interrogate the host and pathogen genetics that contribute to cryptococcal disease. We find in vivo that there is early recruitment of macrophages that phagocytose infecting yeast and provide initial protection. Furthermore, we report the use of hematogenous infection of zebrafish larvae as a simple visual platform for assessing C. neoformans invasion across the vertebrate blood-brain barrier and emphasize the relevance of host processes conserved with mammalian immune responses to cryptococcal disease, including macrophages and granulomas.
RESULTS
Macrophages are essential for protecting zebrafish from cryptococcal disease progression.
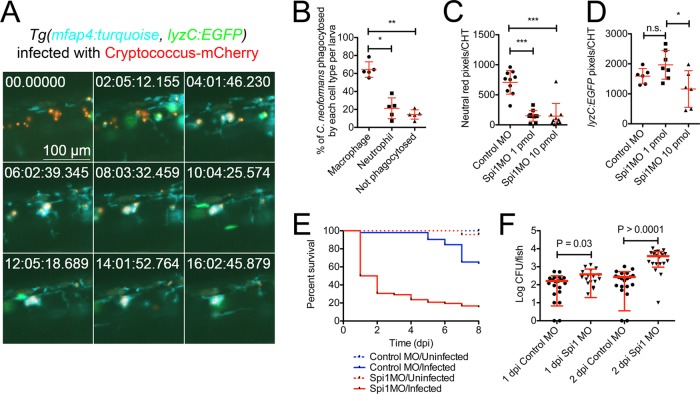
As C. neoformans enters a human host, it typically first encounters resident macrophages. Macrophages have been recognized as the primary cell type responsible for containment of C. neoformans in experimentally infected mouse lungs (4). To determine whether macrophages play a similar role in a zebrafish model of cryptococcal infection, we assessed the dynamics of macrophage and neutrophil recruitment after injection of fluorescent C. neoformans cells into the caudal vein of zebrafish larvae 2 days postfertilization (dpf). To simultaneously image cryptococci, macrophages, and neutrophils, we made use of double transgenic Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) animals, in which macrophages are marked by expression of turquoise from the mfap4 promoter and neutrophils are marked by the expression of enhanced green fluorescent protein (EGFP) from the lyzC promoter (11, 12). Macrophages rapidly phagocytosed the majority of C. neoformans cells following injection with the majority of injected C. neoformans cells observed within mfap4-expressing macrophages by 13 h postinfection and only very rare neutrophil phagocytosis of yeast observed during these experiments (Fig. 1A and B). Detailed observation of leukocyte movement around engulfed cryptococcus cells revealed a persistent clustering of macrophages around infected macrophages, consistent with the appearance of a classic cryptococcoma in mammals as primarily composed of macrophages (see Movie S1 in the supplemental material).
FIG 1 .
Macrophages are responsible for containing acute C. neoformans infection. (A) Still images spaced 2 h apart from time-lapse imaging from a Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans starting at 1 h postinfection (neutrophils in green, macrophages in blue, and Cryptococcus in red) (full video available as Movie S1 in the supplemental material). The field of view follows a single cluster of yeast cells that are phagocytosed by macrophages by the 4-h mark of the recording. (B) Quantification of the phagocytic fate of C. neoformans yeast cells at 13 h postinfection in Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larvae assessed by manual counting. Repeated-measures analysis of variance (ANOVA). The data represent counts from five individual larvae infected with 15 to 30 C. neoformans cells each. (C) Quantification of macrophage numbers by fluorescent pixel count in the caudal hematopoietic tissue (CHT) of uninfected 3-dpf larvae injected with a standard dose of 1 pmol and a high dose of 10 pmol Spi1 morpholino (MO) and soaked in neutral red. ANOVA with Tukey posttest. ***, P < 0.001. (D) Quantification of neutrophil numbers by fluorescent pixel count in the caudal hematopoietic tissue (CHT) of uninfected 3-dpf Tg(lyzC:EGFP) larvae injected with a standard dose of 1 pmol and high dose of 10 pmol Spi1 morpholino. ANOVA with Tukey posttest. *, P < 0.05. (E) Survival of Spi1 knockdown larvae following infection with 50 C. neoformans cells. Log-rank test between infected conditions. P < 0.0001. Data are representative of two experiments each with 15 to 20 larvae per experimental condition. (F) C. neoformans burden in Spi1 knockdown larvae at 1 and 2 days postinfection. t test. Data are pooled from two experiments each with 10 to 20 larvae per experimental condition.
Conversely, we observed an initial burst of neutrophils to the site of injection consistent with a response to injection trauma but did not observe clustering or retention of neutrophils around infected macrophages. The defensive role of neutrophils in mammalian models against cryptococcus is unclear. The zebrafish neutrophils appear to be able to recognize C. neoformans as foreign but are largely not retained at the site of infection, nor do they appear to be the primary cell type responsible for containment of the yeast. This is likely due to the production of known cryptococcal virulence factors such as capsule (13). In multiple time-lapse experiments, we largely observed macrophage interactions with the yeast. Infrequently, lyzC-expressing neutrophils were observed to phagocytose C. neoformans. In one example, an lyzC-expressing neutrophil phagocytosed C. neoformans yeast, migrated away from a nascent granuloma, and was then in turn phagocytosed by an mfap4-expressing macrophage nearby a second nascent granuloma (see Movie S2 in the supplemental material). In a second example, we observed a lyzC-expressing neutrophil with internalized C. neoformans traverse the trunk tissue over 20 h before being phagocytosed by an mfap4-expressing macrophage nearby a nascent granuloma (see Movie S3 in the supplemental material). These rare interactions with neutrophils eventually resulted in macrophage engulfment, consistent with a possible role for efferocytosis during the host response to C. neoformans.
Serendipitous observation of a Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) animal that had been inoculated with C. neoformans directly into the trunk vacuole, a fluid-filled tube that is continuous with the brain ventricles, revealed uninhibited C. neoformans division in the vacuoles. Our observation suggests that the vacuoles represent an immunologically privileged site as lyzC-expressing neutrophils and mfap4-expressing macrophages were only recruited to yeast cells residing outside the vacuoles (see Movie S4 in the supplemental material). We hypothesized that the lack of macrophage ingress at these sites allowed the rapid outgrowth of C. neoformans not seen in other anatomical sites where macrophages are able to reach and surround the yeasts.
To experimentally test this hypothesis, we depleted zebrafish macrophages by morpholino knockdown of the Spi1/Pu.1 transcription factor (14). As high levels of Pu.1 expression favor macrophage differentiation and intermediate levels favor neutrophil production, a titrated knockdown of Spi1 with a 1-pmol dose of morpholino arrests myelopoiesis and delays development of macrophages, while neutrophil populations remain largely intact (Fig. 1C and D). Higher doses of morpholino (10 pmol) ablate both macrophage and neutrophil populations (Fig. 1C and D). To test the role of macrophages, we used the lower dose, in which only macrophage development was compromised. These larvae were extremely susceptible to mortality following infection and additionally had a higher fungal burden by 2 days postinfection (dpi), demonstrating a lack of host control of yeast growth (Fig. 1E and F) and, consistent with our in vivo imaging studies, implicating macrophages as critical to initial control of infection.
Cryptococcus neoformans replicates and establishes persistent infection in zebrafish larvae.
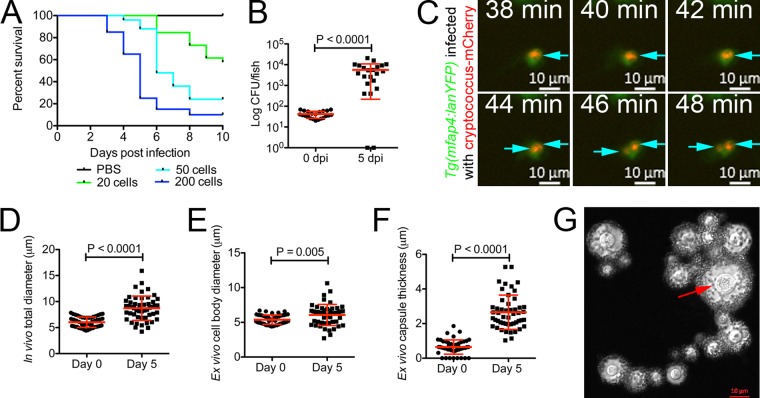
We inoculated 2-dpf zebrafish larvae with a range of C. neoformans doses from 20 to 200 yeast cells and observed dose-dependent mortality (Fig. 2A). C. neoformans can replicate in zebrafish larvae. The cryptococcal burden was ~2 log CFU higher at 5 dpi than immediately following infection with 50 ± 10 yeast cells (Fig. 2B). The times to death of 50% of larvae (TD50s) for infection with 20 ± 5, 50 ± 10, and ~200 yeast cells were 10, 6, and 5 days postinfection, respectively. Thus, inoculum size has an impact on disease, but robust disease and host response can be established with a small burden of yeast cells.
FIG 2 .
Cryptococcus replicates and causes disease in zebrafish larvae. (A) Survival of zebrafish larvae infected with a range of C. neoformans doses. Data are representative of two experiments each with 15 to 20 larvae per experimental condition. (B) C. neoformans burden at 0 and 5 days postinfection in larvae infected with a dose of 50 CFU. Data are representative of two experiments, each with at least 20 larvae per experimental condition. t test with Welch’s correction. (C) Still images from time-lapse imaging of a Tg(mfap4:lanYFP) larva infected with mCherry-expressing C. neoformans starting at 1 day postinfection (macrophages in green and Cryptococcus in red) (full video available as Movie S5 in the supplemental material). Arrows indicate individual yeast cells throughout the time series, with replication appearing to occur between 42 and 44 min. (D) Cryptococcal diameter measured by transmitted light microscopy from the starting inoculum and yeast cells isolated from 5-dpi larvae. t test with Welch’s correction. (E) Cell body diameter measured from India-ink-stained C. neoformans from the starting inoculum and yeast cells isolated from 5-dpi larvae. t test with Welch’s correction. (F) Capsule thickness measured from India-ink-stained C. neoformans from the starting inoculum and yeast cells isolated from 5-dpi larvae. t test with Welch’s correction. (G) Image of C. neoformans isolated from homogenized 5-dpi larvae contrasted by standard India ink preparation. The red arrow indicates a large yeast cell measured at 10 µm representative of observed giant cells recovered from zebrafish infections.
Since macrophages appeared to be the primary intracellular residence of C. neoformans in zebrafish larvae, we next performed high-resolution microscopy focusing on yeast cells that were intracellular within macrophages. In spite of the macrophage-dependent restriction of growth, yeast persisted and indeed replicated within macrophages in vivo (Fig. 2C; see Movie S5 in the supplemental material). This intracellular division recapitulated the characteristic asymmetrical mother-daughter cell division that can be seen in in vitro growth. In addition to expanding capsule diameter, regulation of cell size has been a documented response to conditions experienced during infection by C. neoformans (15, 16). Bright-field microscopy of infected larvae suggested that C. neoformans increased in size as infection progressed (Fig. 2D). We used an India ink stain to facilitate measurement of capsule diameter and yeast cell sizes. These observations demonstrated an in vivo increase in yeast cell body size to the threshold of Titan cell diameter and further the production of capsule enlargement during infection of zebrafish larvae (Fig. 2E, F, and G). This production of capsule is a key characteristic of infection in other animal models and provides a mechanism to explain the observation that lyzC-expressing zebrafish neutrophils rarely interact with C. neoformans in vivo.
Cryptococcal mutant phenotypes are recapitulated in the zebrafish infection model.
The small size and versatility of live-imaging zebrafish larvae make them an ideal host for performing high-throughput host-pathogen interaction screens such as libraries of yeast mutants or multiple mutant or wild-type strains in a mutagenized host platform (17–19). Therefore, to test the robustness of the zebrafish model for mammalian virulence determinants, we infected zebrafish larvae with several C. neoformans mutants chosen for their description as virulence or fitness factors in mice to verify that the zebrafish infection model recapitulates findings from standard vertebrate models. (i) Capsule formation is a characterizing feature of cryptococcus that becomes enlarged within specific sites within the host and is essential for human disease production. The capsule surrounds the cell wall and is a critical virulence factor with a multitude of effects in the host, including preventing phagocytosis by macrophages (16). cap64 is a key capsule formation gene: deletion of this gene, cap64Δ, has been reported to compromise capsule formation and cryptococcal virulence in mammals (20). (ii) PLB1 encodes a secreted phospholipase B that may function in multiple processes that contribute to disease within the host, including survival in macrophages and/or possible nutrient acquisition through breakdown of host phospholipids of the phagosome, and the plblΔ deletion mutant is avirulent in the murine system (21, 22). (iii) The ability to adapt to growth at 37°C is a common attribute of all human pathogens. The trehalose pathway is critical for the ability of this yeast to grow at 37°C (23, 24). The examined tps1Δ strain lacks the ability to produce trehalose and is unable to survive at high temperatures in vitro and in vivo (23, 25). In vivo, the tps1Δ strain is rapidly cleared from infected mice. However, the tps1Δ mutant is also avirulent in the C. elegans model system (25), suggesting roles beyond adaptation to elevated temperatures since C. elegans is maintained at room temperature. (iv) URE1 encodes a urease enzyme that has been shown to promote cryptococcal survival by altering the host immune response to pulmonary infections and to increase the rate of hematogenous dissemination in mice (24, 26–28). However, the urelΔ mutant survives normally when directly inoculated into the CNS of rabbits and mice (26, 28). Additionally, there are several documented cases of atypical urease-negative isolates causing serious disease in AIDS patients, suggesting that urease could be a site-specific virulence factor (29, 30).
Compared to larvae infected with the H99 (wild-type) strain, larvae infected with each of the four mutant strains survived better in an 8-day survival study, suggesting that these virulence factors are conserved between mammalian and teleost cryptococcal infection (Fig. 3A). We next examined fungal burden in 5-dpi larvae, prior to the divergence of mutant survival rates from wild-type survival rates, to determine if the increased survival of these mutants was caused by reduced cryptococcal proliferation (Fig. 3B). We observed significantly reduced cryptococcal burden in larvae infected with either the plb1Δ or tps1Δ mutant strain, confirming a range of roles for these virulence factors in cryptococcal pathogenesis across a broad range of host species. Interestingly, like other invertebrate models, the tps1Δ mutation showed that its impact on virulence or fitness was due to more than its importance in high-temperature (37°C) growth since zebrafish larvae are incubated at 32°C.
FIG 3 .
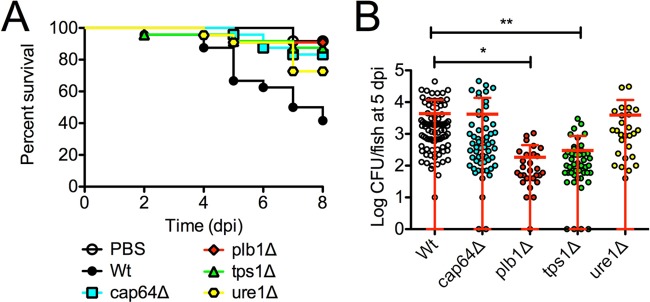
Cryptococcal mutant phenotypes are recapitulated in zebrafish larvae. (A) Survival of zebrafish larvae infected with mutant strains of C. neoformans. Data are representative of two experiments, each with 22 larvae per C. neoformans genotype. Log-rank test between mutant and wild-type genotypes, cap64Δ, P = 0.003; plb1Δ, P = 0.0007; tps1Δ, P = 0.001; ure1Δ, P = 0.03. (B) C. neoformans burden at 5 days postinfection in larvae infected with mutant strains of C. neoformans H99. Data are pooled from 3 experiments, each with at least 15 larvae per C. neoformans genotype. One-way ANOVA with Tukey’s posttest. *, P < 0.05; **, P < 0.01.
On the other hand, despite survival attenuation of the host, burdens in larvae infected with either cap64Δ or ure1Δ mutants were comparable to those of larvae infected with the H99 strain. These findings suggest larval zebrafish are either more tolerant of these mutants, the site of critical infection may be different, or the larvae are able to curtail the growth of these mutants just below a lethal threshold of yeast burden, resulting in increased survival at the later time points. This model system can recapitulate the mammalian virulence composite in either fungal burden or survival times with well-characterized cryptococcal mutants.
A zebrafish assay for brain invasion.
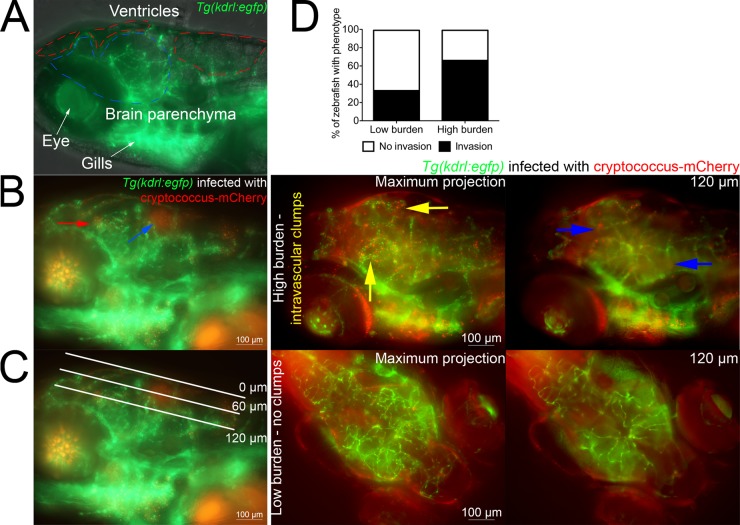
Invasion of the CNS is a major component of microbial pathogenesis; however, direct determination of rates of microbial brain invasion in mammalian infection models is intrinsically laborious, and comparable structures are not present in the invertebrate model organisms (27). Zebrafish possess a functional blood-brain barrier that is first established at 3 dpf and then matures over the next week of development (31, 32). We took advantage of the optical transparency of the zebrafish head to perform a noninvasive live-imaging assay for cryptococcal invasion of the brain parenchyma from the bloodstream of Tg(kdrl:EGFP)s843 animals, where the blood vessels are marked by EGFP expression (Fig. 4A). In a procedural equivalent to mouse tail vein injections, transgenic 2-dpf zebrafish were infected with C. neoformans by caudal vein microinjection, resulting in colonization of the cranial vasculature, the brain ventricles, and brain tissue by 4 dpi (Fig. 4B).
FIG 4 .
Assessment of cryptococcal brain tissue invasion in zebrafish larvae. (A) Lateral z axis projection of the head of an uninfected 6-dpf Tg(kdrl:EGFP)s843 larva (blood vessels in green). The boundaries of the ventricles and brain parenchyma are marked by red and blue dashed lines, respectively. (B) Lateral z-axis projection of the head of a 4-dpi Tg(kdrl:EGFP)s843 larva (blood vessels in green and Cryptococcus in red). The blue arrow indicates C. neoformans colonization of hindbrain ventricle, and the red arrow indicates C. neoformans in the brain parenchyma. (C) Lateral z-axis projection of the head of a 4-dpi Tg(kdrl:EGFP)s843 larva (blood vessels in green and Cryptococcus in red). White lines indicate the approximate depth of optical sections used throughout Fig. 4 and 6. The 0-µm edge is stereotypically set at the dorsal-most blood vessel, and a 60-µm depth stereotypically traverses ventricle spaces dorsal to the brain tissue, which is reached at around 100 to 120 µm in depth. (D) Comparison of C. neoformans brain tissue invasion rates between larvae with sporadic colonization of cranial vasculature (low burden) and larvae with clumps of yeast cells within cranial vasculature (high burden). In example images, blood vessels are in green and Cryptococcus is in red. Yellow arrows in example images indicate clumps of intravascular yeast, and blue arrows indicate extravasated yeast in the brain tissue.
We next switched from a conventional lateral imaging orientation to a dorsal orientation to better define the bloodstream, brain ventricle, and brain parenchyma compartments where C. neoformans resides (Fig. 4C). By observing a number of animals with different levels of cryptococcal burden in cerebral blood vessels, we found a positive correlation between local infection burden levels of cranial vessels versus invasion into the brain parenchyma (Fig. 4D).
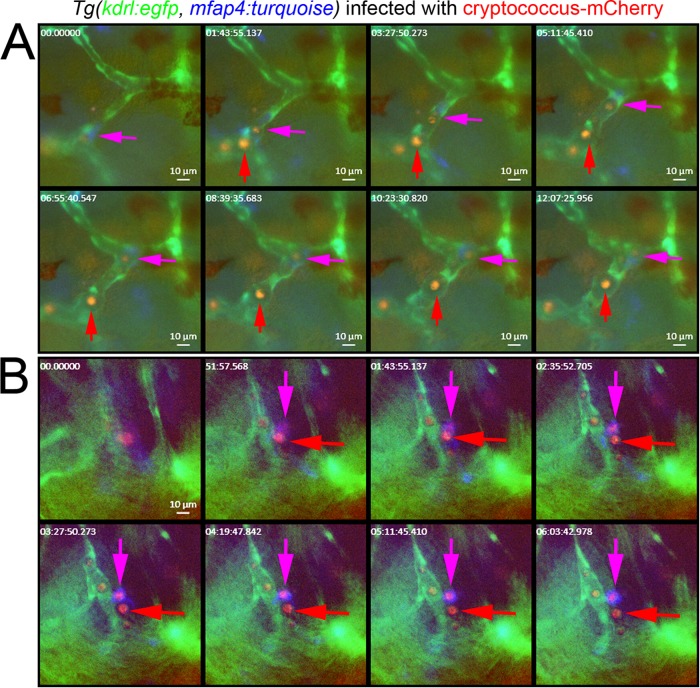
We performed time-lapse imaging of the cranial vasculature, macrophages, and yeast cells in double transgenic Tg(kdrl:EGFPs843, mfap4:turquoisext27) larvae hematogenously infected with C. neoformans. By 4 dpi, we were able to observe intramacrophage and extracellular C. neoformans cells free in circulation and lodged in the vasculature (Fig. 5A; see Movie S6 in the supplemental material). Close examination of these apparently extracellular circulating yeast cells revealed a circular zone of vascular distention around many yeast cells consistent with the production of capsule and avoidance of phagocytosis.
FIG 5 .
(A) Still images from the brain of a 5-dpi Tg(kdrl:EGFPs843, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans by caudal vein injection (blood vessels in green, macrophages in blue, and Cryptococcus in red). The full time-lapse video is available as Movie S7 in the supplemental material. A purple arrow indicates a macrophage with phagocytosed yeast cells moving inside cranial blood vessels. A red arrow indicates presumptively extracellular yeast cells moving inside cranial blood vessels. Note the nonfluorescent area of vessel distension that follows presumptively extracellular yeast cells. (B) Still images from the brain of a 5-dpi Tg(kdrl:EGFPs843, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans by caudal vein injection (blood vessels in green, macrophages in blue, and Cryptococcus in red). Full time-lapse imaging is available as Movie S7 in the supplemental material. A purple arrow indicates a macrophage with a phagocytosed yeast cell located in brain tissue adjacent to cranial blood vessel. A red arrow indicates a presumptively extracellular yeast cell located in brain tissue.
Further imaging also identified invasive C. neoformans cells lodged in the brain tissue in double transgenic Tg(kdrl:EGFPs843, mfap4:turquoisext27) larvae (Fig. 5B). Brain-tissue-resident yeast cells that were at this point outside macrophages did not elicit a strong macrophage migration or phagocytosis (see Movie S7 in the supplemental material). Furthermore, brain-tissue-resident yeast cells that were contained in mfap4-expressing macrophages generally remained static throughout imaging. This was in striking contrast to the tissue-resident yeast cells in earlier experiments that were carried by migrating macrophages or formed the focus of nascent granulomas, suggesting that extracellular yeast cells in zebrafish brain tissue may be masked from zebrafish macrophages through previously documented mechanisms, such as capsule production early in infection.
Recapitulation of a known blood-brain barrier invasion-deficient mutant phenotype in mice.
Since the blood-brain barrier has not reached full maturation by the end of our experimental window, we wished to functionally assess the blood-brain barrier in the context of a mutant known to be deficient in invasion of the mouse blood-brain barrier. The C. neoformans fnx1Δ mutant is deficient for a multidrug resistance-like protein that may facilitate yeast-endothelial cross talk. This mutant was previously characterized as having a deficiency in a competitive murine blood-brain barrier transmigration assay and in vitro reduced microvascular entrapment and transcytosis across immortalized human brain capillary endothelial cells (33).
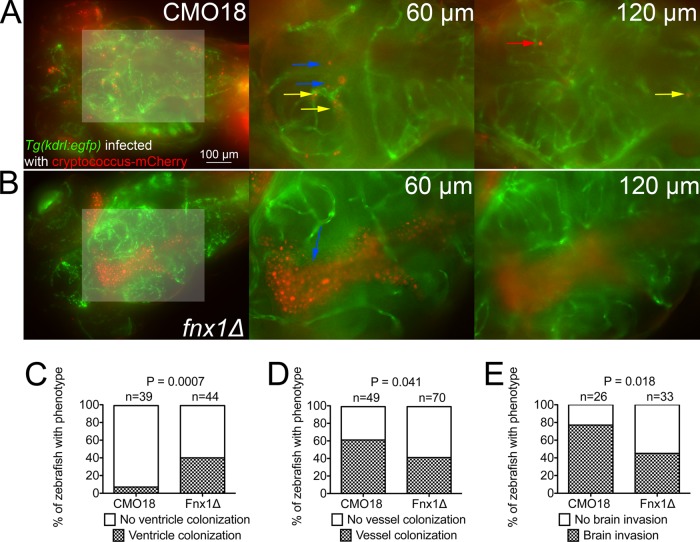
Transgenic Tg(kdrl:EGFP)s843 zebrafish larvae were infected with either the fnx1Δ mutant strain or the parental CMO18 strain. We did not observe any obvious differences in infection burden until around 4 to 5 dpi, when we observed frequent brain ventricle fungal overgrowth only in animals infected with the fnx1Δ mutant (Fig. 6A and B; see Movies S8 and S9 in the supplemental material). We quantified the appearance of the ventricle overgrowth phenotype in 5-dpi larvae and found that infection with the fnx1Δ mutant resulted in ventricle colonization at a rate over 4× that of the parental strain (Fig. 6C).
FIG 6 .
The C. neoformans fnx1Δ mutant strain is deficient in invading the brain parenchyma. (A) Dorsal-viewpoint images taken from a z-axis stack of a 5-dpi Tg(kdrl:EGFPs843) larva infected with an mCherry-expressing C. neoformans CMO18 strain by caudal vein injection (left). Shown is a z-axis projection with a lightened box indicating a magnified region of interest (shown to the right) of 2× digital magnification slices at ventricle (60-µm) and brain tissue (120-µm) depths of focus. Blue arrows indicate C. neoformans cells located in the brain ventricle, red arrows indicate C. neoformans cells located in the brain parenchyma, and yellow arrows indicate C. neoformans cells located in the cranial vessels. Distances indicate the depth of image plane from the dorsal edge of the head. The full stack is available as Movie S8 in the supplemental material. (B) Dorsal-viewpoint images taken from a z-axis stack of a 5-dpi Tg(kdrl:EGFPs843) larva infected with an mCherry-expressing C. neoformans fnx1Δ mutant by caudal vein injection (left). Shown is a z axis projection with a lightened box indicating the magnified region of interest (shown to the right) of 2× digital magnification slices at ventricle (60-µm) and brain tissue (120-µm) depths of focus. Blue arrows indicate C. neoformans cells located in the brain ventricle. Distances indicate depths of the image planes from the dorsal edge of the head. The full stack is available as Movie S9 in the supplemental material. (C) Quantification of head ventricle C. neoformans colonization in 4-dpi larvae infected with the C. neoformans fnx1Δ mutant by caudal vein injection. Fisher’s exact test. Data are pooled from 3 replicates each with 10 to 20 larvae per genotype. (D) Quantification of cranial blood vessel C. neoformans colonization in 5-dpi larvae infected with the C. neoformans fnx1Δ mutant by caudal vein injection. Fisher’s exact test. Data are pooled from 4 replicates each with 10 to 20 larvae per genotype. (E) Quantification of brain tissue invasion in 5-dpi larva infected with the C. neoformans fnx1Δ mutant by caudal vein injection. Fisher’s exact test. Data are pooled from 3 replicates each with 10 to 20 larvae per genotype.
Strikingly, despite clearly having a much higher burden of C. neoformans around the brain in larvae infected with the fnx1Δ mutant, we observed a markedly reduced rate of cranial blood vessel colonization by the fnx1Δ mutant consistent with in vitro assays of microvascular entrapment (Fig. 6D).
Correlating with the reduced rate of cranial vascular localization, we observed a lower rate of brain tissue invasion by the fnx1Δ mutant than in the CMO18 parental strain (Fig. 6E). Taken together, these observations confirm a role for FNX1 in aiding the colonization of the cerebral vasculature and promoting the subsequent invasion of the central nervous system across model systems.
DISCUSSION
We have reported multiple assays for applying the highly tractable zebrafish platform as a model of C. neoformans pathogenesis. We show that the innate immune system of zebrafish larvae responds to C. neoformans primarily with a phagocytic macrophage response that is essential for containing an acute infection. Virulence and disease progression in the zebrafish model are dependent on conserved cryptococcal processes, including pathways related to the evasion of the host immune response. The live-imaging assay systems we have described will provide a novel platform for investigating the intersection of cryptococcal and host genetics and cell behaviors in intact vertebrates.
Our imaging experiments tracking fluorescently labeled innate immune cells and C. neoformans cells confirmed that, as predicted from murine experiments, zebrafish macrophages are the dominant phagocytic cell type that interacts with and contains C. neoformans during acute infection (13). Our observations also demonstrate a conserved host response driven by macrophages resulting in nascent cryptococcomas in zebrafish larvae infected with C. neoformans even during our relatively short window of infection. Although this macrophage predominance is a fairly common response to microbial infection in zebrafish, differential roles have been observed for macrophages and neutrophils in containing fungal infections in zebrafish (34, 35). The primary response also includes macrophage aggregates consistent with the granulomas observed in mammals. It will be intriguing to determine if the postulated immunomodulatory role of neutrophils in acute murine C. neoformans infections is conserved across species by further live imaging of the dynamic immune response of zebrafish larvae manipulated to have high or low numbers of neutrophils.
The visual accessibility of the zebrafish larvae also allowed us the opportunity to visualize intramacrophage yeast cell division in vivo, demonstrating that C. neoformans is able to replicate inside zebrafish macrophages. Since effective chemotherapy for invasive C. neoformans can lead to dangerous sequelae due to the massive release of fungal immunostimulants and development of immune reconstitution inflammatory syndrome (IRIS), there is a need to better characterize and modulate the host response to fungal pathogens. As demonstrated by our Spi1 knockdown study and by others, including Brothers et al., the zebrafish host is amenable to genetic manipulation to identify the host factors necessary for combating and controlling fungal pathogenesis (34). The zebrafish may thus present a tractable tool for investigating the role of specific host factors that recognize and respond to general and virulence-specific cryptococcal factors.
C. neoformans has been observed to physically respond to the infection milieu by enlarging both capsule and overall yeast cell size. We also observed the presence of large cells, also termed Titan or giant cells during zebrafish infection, with yeast cells above 10 µm in size. It is posited that these large cells are important in the early stages of infection as a means to avoid engulfment by immune cells and demonstrate the stress that these yeast cells are undergoing in the host (15). To clearly define the large cells as Titan/giant cells, additional characterization is needed to evaluate the genomic content, cell wall thickness, and overall size. However, the high fecundity of zebrafish should provide an excellent in vivo platform for harvesting and defining whether these large cells that we have observed are truly Titan/giant cells. Furthermore, the live-imaging capacity of the zebrafish platform provides a unique system to image the interaction between Titan/giant cells and specific elements of the host innate immune system.
The reproduction of increased host survival in zebrafish larvae infected with several C. neoformans mutants from independent virulence pathways demonstrates that known virulence pathways are conserved during infection of this nonmammalian vertebrate host. These experiments set an important proof of principle and suggest that investigation of mutant interactions with the immune system in vivo by live imaging will be feasible to aid our understanding of how cryptococcal virulence pathways promote pathogenesis. The magnitude of the burden differences in the mutants may be limited by the short time frame over which we performed the zebrafish larval infections. Modifying the inoculum and extending the duration of infection for burden analysis might reveal larger absolute burden differences and additional congruence between the model systems used.
The discrepancy between the increased survival of the cap64Δ and ure1Δ mutants compared to the wild-type strain and the similar fungal burdens of all three genotypes at 5 dpi was surprising as mortality had previously appeared to be tightly associated with fungal overgrowth in our dose-response infections. We speculate that this discrepancy could be caused by reduced immunopathology following infection with these mutants resulting in an uncoupling of the burden-mortality relationship. Another hypothesis is that the growth of these mutants is controlled at the 5-dpi time point by a host immune response that is either independent of or a slower version of the response that rapidly controls the plb1Δ and tps1Δ mutants. Alternatively, the mutants may have body site differences in growth and entry—exemplified by the normal growth of the urelΔ mutant when directly inoculated into the mammalian CNS (26, 28). Thus, we postulate that studying a range of mutants in the zebrafish model will be useful to better understand the contribution of certain virulence pathways to cryptococcal pathogenesis, and we can watch these yeasts in real time as they produce disease.
CNS involvement is an important stage of C. neoformans pathogenesis that has severe consequences for the host. The zebrafish is an emerging model for the study of blood-brain barrier development and function (36, 37). Here, we have demonstrated that C. neoformans migrates to the CNS of larval zebrafish and that a cryptococcal mutant known to be defective in mammalian blood-brain barrier invasion is also defective in invading the zebrafish brain. Since many of the fnx1Δ mutant animals displaying the vesicle overgrowth phenotype did not have colonization of their cranial blood vessels, we hypothesize that ventricle colonization may utilize a different fungal pathogenicity program than the blood-brain barrier.
Future investigation will be designed to image invasion events in vivo to answer the important question of if and why infected blood-borne macrophages would migrate across the blood-brain barrier—is it an inbuilt macrophage program triggered by C. neoformans (i.e., the Trojan Horse hypothesis)? Alternatively, live imaging may reveal the blood-brain barrier invasion in zebrafish to be a consequence of having a large accumulation of yeast cells in the blood compartment resulting in transcytosis across the barrier or simply bursting of the vessel into the brain. The platform described here can be readily applied to studying a range of laboratory-generated mutants and clinical/environmental isolates to rapidly identify cryptococcal determinants of CNS pathology as well as the cellular mechanisms of CNS invasion.
MATERIALS AND METHODS
Zebrafish (Danio rerio) husbandry.
All zebrafish husbandry and experimental procedures were performed in accordance and compliance with policies approved by the Duke University Institutional Animal Care and Use Committee (protocol A180-11-07). Clutches of eggs were collected from natural spawning and raised in filtered fish system water at 28°C. Pigment development was halted in 1-dpf embryos by the addition of 1-phenyl-2-thiourea (PTU [final concentration, 45 µg/ml]) (Sigma-Aldrich, St. Louis, MO). Other strains used in this study include those with the genotpes Tg(lyzC:EGFP)nz117, Tg(kdrl:EGFP)s843, Tg(mfap4:turquoise)xt27, and Tg(mpeg1:lanYFP)xt2.
Labeling of cryptococcus strains.
Labeling of the Cryptococcus strains was performed by biolistic transformation of plasmid pCN52 or pCN53 containing a mCherry-RAS1 fusion (38). Random integration of this reporter was performed by selection on yeast extract-peptone-dextrose (YPD) containing 100 µg/ml nourseothricin (NAT; Werner BioAgents, Jena-Cospeda, Germany) or 200 µg/ml Geneticin (NEO; Thermo, Fisher Scientific, Grand Island, NY). The Cryptococcus neoformans var. grubii H99 GFP-RAS1 and H99 mCherry-RAS1 strains were kindly provided by Connie Nichols.
C. neoformans var. grubii strains and growth conditions.
Strains used in this study include wild-type H99 and CM108, and the tps1Δ (25), cap64Δ (39), ure1Δ (28), plb1Δ (21), and fnx1Δ (33) mutants (33, 40). A single colony of each strain was inoculated into 5 ml YPD and grown overnight at 30°C with shaking at 225 rpm. Cells were harvested by centrifugation for 5 min at 3000 rpm, washed twice with 25 ml PBS, and suspended in 1 ml phosphate-buffered saline (PBS). A dose of 7 × 107 CFU/ml was prepared to achieve a cell count of ~50 cells per injection. Injected larvae were screened postinjection to confirm the injection dose. Doses ranging from 3 × 107 to 8 × 107 CFU/ml were prepared for the dose curve experiment to achieve the appropriate number of yeast cells for inoculation.
Infection by microinjection.
Embryos were anesthetized at 2 dpf with tricaine (MS-222; Sigma-Aldrich, St. Louis, MO) (final concentration, 160 µg/ml) and injected with C. neoformans in an injection bolus of 10 to 20 nl into the caudal vein. Infected embryos were then recovered back to filtered fish system water supplemented with PTU and raised at 32°C as an intermediate temperature between standard zebrafish (28°C) and cryptococcal (37°C) growth temperatures that is above ambient temperature and well tolerated by larval zebrafish. Embryos that were physically damaged by injection handling were discarded and excluded from further analysis.
Live imaging.
Conventional microscopy and time-lapse fluorescence microscopy were carried out on a Zeiss observer Z1 inverted microscope. Embryos were anesthetized with 160 µg/ml tricaine and mounted in 3% (wt/vol) methylcellulose or 1% low-melting-point agarose for static microscopy. Embryos for time-lapse microscopy were anesthetized with 120 µg/ml tricaine, mounted in 0.75% low-melting-point agarose containing between 100 and 140 µg/ml tricaine in 96-well plates, and immersed in filtered fish system water supplemented with PTU. Images were processed with ImageJ (NIH) and Photoshop CS4 (Adobe).
Morpholino knockdown.
Modified antisense oligonucleotides designed to knock down expression of Spi1 were injected into 1- to 4-cell-stage embryos. The sequences of the morpholinos used in this study are as follows: Spi1 morpholino sequence, 5′ GATATACTGATACTCCATTGGTGGT 3′, and control morpholino, 5′ CCTCTTACCTCAGTTACAATTTATA 3′. Morpholinos were injected at either 1 or 10 pmol per embryo.
Recovery of C. neoformans for capsule size measurement.
Zebrafish were euthanized in fish water containing 300 µg/ml tricaine, washed 2 times with PBS, and transferred individually to a 2-ml screw-cap tube with an O-ring. To each tube, a 6.35-mm steel bead and 1 ml of PBS were added. Fish were homogenized by a 15-s pulse using a Mini-Beadbeater-16 (BioSpec Products). To isolate Cryptococcus cells for imaging of capsule and cell size, a monoclonal antibody (kindly provided by Arturo Casadevall) was labeled with goat anti-mouse IgG magnetic beads (New England Biolabs catalog no. S1431S) according to the manufacturer’s instructions. The magnetic beads coated with the 18B7 antibody were diluted such that few beads were observed per yeast cell. The magnetic beads were incubated with the fish lysate at room temperature for 5 min, and beads were separated to the side of the tube for 5 min. The lysate was removed, and beads were washed 3 times with 1 ml of PBS. The beads were resuspended in 20 µl of PBS. Samples were prepared with India ink staining and imaged on a Zeiss Axio Imager A1 microscope with an AxioCam MRM digital camera. Image analysis was performed using ImageJ software.
Quantification of morpholino hematopoietic effects.
Morpholino-injected Tg(lyzC:EGFP)nz101 embryos were raised to 3 dpf, soaked in 2.5 µg/ml neutral red (Sigma-Aldrich, St. Louis, MO) for 6 h, and imaged for red and green fluorescence.
Enumeration of CFU.
Zebrafish were euthanized in fish water containing 300 µg/ml tricaine, washed 2 times with 25 ml PBS, and transferred individually to a 2-ml screw-cap tube with an O-ring. To each tube, a 6.35-mm steel bead (BioSpec Products, Bartlesville, OK) and 1 ml of PBS were added. Fish were homogenized by a 15-s pulse using a Mini-Beadbeater-16 (BioSpec Products, Bartlesville, OK). Serial dilutions were performed, and 100 µl of the neat solution and 1/10 dilutions were plated onto YPD containing 100 µg/ml chloramphenicol. Plates were incubated at 30°C for 3 days. Colonies were enumerated after 3 days of growth.
Statistical analysis.
Comparison of fungal burdens was performed using unpaired two-tailed t test. Values resulting in P values of <0.05 relative to the control were considered significantly different. Zebrafish survival was plotted using Kaplan-Meier survival curves and analyzed by log rank test using GraphPad Prism. Survival curves resulting in P values of <0.05 relative to the control were considered significantly different. Error bars represent 1 standard deviation.
SUPPLEMENTAL MATERIAL
Time-lapse imaging of z-axis projection of nascent granuloma formation in an Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans. The movie covers a 16-h period from 1 h postinfection. Stills from the movie are available in Fig. 1A. Download
Time-lapse imaging of z-axis projection of leukocyte-yeast interactions in a Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans. The movie covers a 16-h period from 1 day postinfection. An arrow at the beginning indicates a neutrophil with internalized C. neoformans that will migrate, apoptose, and be phagocytosed by a macrophage. The specimen is oriented anterior to posterior from bottom right to top left and dorsal-ventral from bottom left to top right. Download
Time-lapse imaging of z-axis projection of leukocyte-yeast interactions in an Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans. The movie covers a 23-h period from 1 day postinfection. The white arrow at the start of the movie indicates a neutrophil that is phagocytosed by a macrophage at the red arrow around the 20-h mark. Download
Time-lapse imaging of a misinjected Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva injected with mCherry-expressing C. neoformans into the cerebrospinal fluid (CSF) instead of the tissue. The movie covers a 24-h period from 1 h postinfection. Download
Time-lapse imaging of z-axis projection of an mCherry-expressing C. neoformans yeast cell dividing inside a macrophage within a Tg(mfap4:lanYFP) larva. The movie covers a 2-h period at 1 day postinfection. Stills from the movie are available in Fig. 2C. Download
Time-lapse imaging of z-axis projection from the head region of a 5-dpi Tg(kdrl:EGFPs843, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans by caudal vein injection with yeast cells located only within blood vessels from Fig. 5A (blood vessels in green, macrophages in blue, and Cryptococcus in red). Download
Time-lapse imaging of z-axis projection from the head region of a 5-dpi Tg(kdrl:EGFPs843, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans by caudal vein injection with yeast cells located in the brain parenchyma and within blood vessels from Fig. 5B (blood vessels in green, macrophages in blue, and Cryptococcus in red). Download
Full stack of dorsal-viewpoint images taken from a z-axis stack of a 5-dpi Tg(kdrl:EGFPs843) larva infected with mCherry-expressing C. neoformans strain CMO18 by caudal vein injection from Fig. 6A. Download
Full stack of dorsal-viewpoint images taken from a z-axis stack of a 5-dpi Tg(kdrl:EGFPs843) larva infected with mCherry-expressing C. neoformans fnx1Δ mutant by caudal vein injection from Fig. 6B. Download
ACKNOWLEDGMENTS
This work was supported by the Duke University Center for AIDS Research (CFAR), a National Institutes of Health (NIH)-funded program (5P30 AI064518), by an Australian National Health and Medical Research Council C. J. Martin Early Career Fellowship (SHO), by a Mallinckrodt Scholar Award, a Searle Scholar Award, a Vallee Foundation Young Investigator Award, and an NIH Director’s New Innovator Award (1DP2-OD008614) to D.M.T., by Public Health Service grants AI73896 and AI93257 to J.R.P., and by a Medicine Research Collaboration Award to J.R.P. and D.M.T.
We thank Andrew Alspaugh for helpful discussions and comments on the manuscript, Dana Sisk for zebrafish facility management, Mark Cronan and Eric Walton for providing transgenic zebrafish lines, and Vinicius Ponzio for technical assistance.
Footnotes
Citation Tenor JL, Oehlers SH, Yang JL, Tobin DM, Perfect JR. 2015. Live imaging of host-parasite interactions in a zebrafish infection model reveals cryptococcal determinants of virulence and central nervous system invasion. mBio 6(5):e01425-15. doi:10.1128/mBio.01425-15.
REFERENCES
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Lee A, Toffaletti DL, Tenor J, Soderblom EJ, Thompson JW, Moseley MA, Price M, Perfect JR. 2010. Survival defects of Cryptococcus neoformans mutants exposed to human cerebrospinal fluid result in attenuated virulence in an experimental model of meningitis. Infect Immun 78:4213–4225. doi: 10.1128/IAI.00551-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, Kronstad JW, Perfect JR. 2011. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2(3):e00103-11. doi: 10.1128/mBio.00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 68:4225–4237. doi: 10.1128/IAI.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apidianakis Y, Rahme LG, Heitman J, Ausubel FM, Calderwood SB, Mylonakis E. 2004. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot Cell 3:413–419. doi: 10.1128/EC.3.2.413-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desalermos A, Tan X, Rajamuthiah R, Arvanitis M, Wang Y, Li D, Kourkoumpetis TK, Fuchs BB, Mylonakis E. 2015. A multi-host approach for the systematic analysis of virulence factors in Cryptococcus neoformans. J Infect Dis 211:298–305. doi: 10.1093/infdis/jiu441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci U S A 99:15675–15680. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tobin DM, May RC, Wheeler RT. 2012. Zebrafish: a see-through host and a fluorescent toolbox to probe host-pathogen interaction. PLoS Pathog 8:e1002349. doi: 10.1371/journal.ppat.1002349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gratacap RL, Rawls JF, Wheeler RT. 2013. Mucosal candidiasis elicits activation of NF-kappaB, proinflammatory gene expression and localized neutrophilia in a transparent vertebrate host. Dis Model Mech 6:1260–1270. doi: 10.1242/dmm.012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall C, Flores MV, Storm T, Crosier K, Crosier P. 2007. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol 7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oehlers SH, Cronan MR, Scott NR, Thomas MI, Okuda KS, Walton EM, Beerman RW, Crosier PS, Tobin DM. 2015. Interception of host angiogenic signalling limits mycobacterial growth. Nature 517:612–615. doi: 10.1038/nature13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voelz K, May RC. 2010. Cryptococcal interactions with the host immune system. Eukaryot Cell 9:835–846. doi: 10.1128/EC.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. 2005. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell 8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog 6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 68:133–216. doi: 10.1016/S0065-2164(09)01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvalho R, de Sonneville J, Stockhammer O, Savage ND, Veneman WJ, Ottenhoff TH, Dirks RP, Meijer AH, Spaink HP. 2011. A high-throughput screen for tuberculosis progression. PLoS One 6:e16779. doi: 10.1371/journal.pone.0016779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tobin DM, Vary JC Jr, Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, Moens CB, Ramakrishnan L. 2010. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140:717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiles TJ, Norton JP, Russell CW, Dalley BK, Fischer KF, Mulvey MA. 2013. Combining quantitative genetic footprinting and trait enrichment analysis to identify fitness determinants of a bacterial pathogen. PLoS Genet 9:e1003716. doi: 10.1371/journal.pgen.1003716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang YC, Penoyer LA, Kwon-Chung KJ. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun 64:1977–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, Leidich SD, Casadevall A, Ghannoum MA, Perfect JR. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol 39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- 22.Chrisman CJ, Albuquerque P, Guimaraes AJ, Nieves E, Casadevall A. 2011. Phospholipids trigger Cryptococcus neoformans capsular enlargement during interactions with amoebae and macrophages. PLoS Pathog 7:e1002047. doi: 10.1371/journal.ppat.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngamskulrungroj P, Himmelreich U, Breger JA, Wilson C, Chayakulkeeree M, Krockenberger MB, Malik R, Daniel HM, Toffaletti D, Djordjevic JT, Mylonakis E, Meyer W, Perfect JR. 2009. The trehalose synthesis pathway is an integral part of the virulence composite for Cryptococcus gattii. Infect Immun 77:4584–4596. doi: 10.1128/IAI.00565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osterholzer JJ, Surana R, Milam JE, Montano GT, Chen GH, Sonstein J, Curtis JL, Huffnagle GB, Toews GB, Olszewski MA. 2009. Cryptococcal urease promotes the accumulation of immature dendritic cells and a non-protective T2 immune response within the lung. Am J Pathol 174:932–943. doi: 10.2353/ajpath.2009.080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petzold EW, Himmelreich U, Mylonakis E, Rude T, Toffaletti D, Cox GM, Miller JL, Perfect JR. 2006. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect Immun 74:5877–5887. doi: 10.1128/IAI.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olszewski MA, Noverr MC, Chen GH, Toews GB, Cox GM, Perfect JR, Huffnagle GB. 2004. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am J Pathol 164:1761–1771. doi: 10.1016/S0002-9440(10)63734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi M, Li SS, Zheng C, Jones GJ, Kim KS, Zhou H, Kubes P, Mody CH. 2010. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J Clin Invest 120:1683–1693. doi: 10.1172/JCI41963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect Immun 68:443–448. doi: 10.1128/IAI.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bava AJ, Negroni R, Bianchi M. 1993. Cryptococcosis produced by a urease negative strain of Cryptococcus neoformans. J Med Vet Mycol 31:87–89. doi: 10.1080/02681219380000091. [DOI] [PubMed] [Google Scholar]
- 30.Ruane PJ, Walker LJ, George WL. 1988. Disseminated infection caused by urease-negative Cryptococcus neoformans. J Clin Microbiol 26:2224–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie J, Farage E, Sugimoto M, Anand-Apte B. 2010. A novel transgenic zebrafish model for blood-brain and blood-retinal barrier development. BMC Dev Biol 10:76. doi: 10.1186/1471-213X-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming A, Diekmann H, Goldsmith P. 2013. Functional characterisation of the maturation of the blood-brain barrier in larval zebrafish. PLoS One 8:e77548. doi: 10.1371/journal.pone.0077548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tseng HK, Liu CP, Price MS, Jong AY, Chang JC, Toffaletti DL, Betancourt-Quiroz M, Frazzitta AE, Cho WL, Perfect JR. 2012. Identification of genes from the fungal pathogen Cryptococcus neoformans related to transmigration into the central nervous system. PLoS One 7:e45083. doi: 10.1371/journal.pone.0045083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brothers KM, Gratacap RL, Barker SE, Newman ZR, Norum A, Wheeler RT. 2013. NADPH oxidase-driven phagocyte recruitment controls Candida albicans filamentous growth and prevents mortality. PLoS Pathog 9:e1003634. doi: 10.1371/journal.ppat.1003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. 2011. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117:e49–e56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim BJ, Hancock BM, Bermudez A, Del Cid N, Reyes E, van Sorge NM, Lauth X, Smurthwaite CA, Hilton BJ, Stotland A, Banerjee A, Buchanan J, Wolkowicz R, Traver D, Doran KS. 2015. Bacterial induction of Snail1 contributes to blood-brain barrier disruption. J Clin Invest 125:2473–2483. doi: 10.1172/JCI74159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Leeuwen LM, van der Kuip M, Youssef SA, de Bruin A, Bitter W, van Furth AM, van der Sar AM. 2014. Modeling tuberculous meningitis in zebrafish using Mycobacterium marinum. Dis Model Mech 7:1111–1122. doi: 10.1242/dmm.015453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selvig K, Ballou ER, Nichols CB, Alspaugh JA. 2013. Restricted substrate specificity for the geranylgeranyltransferase-I enzyme in Cryptococcus neoformans: implications for virulence. Eukaryot Cell 12:1462–1471. doi: 10.1128/EC.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moyrand F, Janbon G. 2004. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37°C and for capsule biosynthesis. Eukaryot Cell 3:1601–1608. doi: 10.1128/EC.3.6.1601-1608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse imaging of z-axis projection of nascent granuloma formation in an Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans. The movie covers a 16-h period from 1 h postinfection. Stills from the movie are available in Fig. 1A. Download
Time-lapse imaging of z-axis projection of leukocyte-yeast interactions in a Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans. The movie covers a 16-h period from 1 day postinfection. An arrow at the beginning indicates a neutrophil with internalized C. neoformans that will migrate, apoptose, and be phagocytosed by a macrophage. The specimen is oriented anterior to posterior from bottom right to top left and dorsal-ventral from bottom left to top right. Download
Time-lapse imaging of z-axis projection of leukocyte-yeast interactions in an Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans. The movie covers a 23-h period from 1 day postinfection. The white arrow at the start of the movie indicates a neutrophil that is phagocytosed by a macrophage at the red arrow around the 20-h mark. Download
Time-lapse imaging of a misinjected Tg(lyzC:EGFPNZ117, mfap4:turquoisext27) larva injected with mCherry-expressing C. neoformans into the cerebrospinal fluid (CSF) instead of the tissue. The movie covers a 24-h period from 1 h postinfection. Download
Time-lapse imaging of z-axis projection of an mCherry-expressing C. neoformans yeast cell dividing inside a macrophage within a Tg(mfap4:lanYFP) larva. The movie covers a 2-h period at 1 day postinfection. Stills from the movie are available in Fig. 2C. Download
Time-lapse imaging of z-axis projection from the head region of a 5-dpi Tg(kdrl:EGFPs843, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans by caudal vein injection with yeast cells located only within blood vessels from Fig. 5A (blood vessels in green, macrophages in blue, and Cryptococcus in red). Download
Time-lapse imaging of z-axis projection from the head region of a 5-dpi Tg(kdrl:EGFPs843, mfap4:turquoisext27) larva infected with mCherry-expressing C. neoformans by caudal vein injection with yeast cells located in the brain parenchyma and within blood vessels from Fig. 5B (blood vessels in green, macrophages in blue, and Cryptococcus in red). Download
Full stack of dorsal-viewpoint images taken from a z-axis stack of a 5-dpi Tg(kdrl:EGFPs843) larva infected with mCherry-expressing C. neoformans strain CMO18 by caudal vein injection from Fig. 6A. Download
Full stack of dorsal-viewpoint images taken from a z-axis stack of a 5-dpi Tg(kdrl:EGFPs843) larva infected with mCherry-expressing C. neoformans fnx1Δ mutant by caudal vein injection from Fig. 6B. Download