ABSTRACT
Morphological switch is tightly coupled with the pathogenesis of many dimorphic fungal pathogens. Cryptococcus neoformans, the major causative agent of cryptococcal meningitis, mostly presents as the yeast form but is capable of switching to the hyphal form. The filamentous form has long been associated with attenuated virulence, yet the underlying mechanism remains elusive. We previously identified the master regulator Znf2 that controls the yeast-to-hypha transition in Cryptococcus. Activation of Znf2 promotes hyphal formation and abolishes fungal virulence in vivo. Here we demonstrated that the cryptococcal strain overexpressing ZNF2 elicited strong and yet temporally confined proinflammatory responses in the early stage of infection. In contrast, exacerbated inflammation in mice infected with the wild-type (WT) strain showed that they were unable to control the infection. Animals inoculated with this filamentous Cryptococcus strain had fewer pulmonary eosinophils and CD11c+ CD11b+ cells than animals inoculated with WT yeast. Moreover, mice infected with this strain developed protective Th1- or Th17-type T cell responses. These findings suggest that the virulence attenuation of the filamentous form is likely due to its elicitation of protective host responses. The antivirulence effect of Znf2 was independent of two previously identified factors downstream of Znf2. Interestingly, mucosal immunizations with high doses of ZNF2-overexpressing cells, either in the live or heat-killed form, offered 100% protection to the host from a subsequent challenge with the otherwise lethal clinical strain H99. Our results demonstrate that heat-resistant cellular components presented in cryptococcal cells with activated ZNF2 elicit protective host immune responses. These findings could facilitate future research on novel immunological therapies.
IMPORTANCE
Cryptococcal meningitis is one of the leading causes of death among AIDS patients. This disease presents a severe threat to public health. The current antifungal regimens are unsatisfactory in controlling or clearing the pathogen Cryptococcus neoformans. Immunotherapies and/or vaccines could be a promising approach to prevent or manage this deadly disease. However, the lack of understanding of host-pathogen interactions during cryptococcal infection greatly hampers the development of effective immunotherapies. In this study, we discovered that inoculation of cryptococcal cells with activated Znf2, a morphogenesis regulator and an antivirulence factor, could shift the host pathological Th2 responses to the protective Th1 or Th17 responses. Importantly, we discovered that vaccination with either the viable or heat-killed form of ZNF2-overexpressing cells protected animals from the otherwise lethal infection by the highly virulent clinical strain. Our study suggests that the fungal cellular component(s) of the ZNF2-overexpressing strain may provide potential vaccine candidate(s) for controlling the fatal disease.
INTRODUCTION
Morphotype transition is critical in many aspects for fungal pathogens to cause disease. It could increase the fitness of microbes in the physiological conditions of the host or their resistance to antimicrobial host responses. Alterations in cell surface molecules during morphogenesis, for example, could shape the immune recognition of fungal pathogens (1–13). Cryptococcus neoformans, although not considered a classic dimorphic fungus, is able to switch from the yeast morphotype to the hyphal morphotype. Earlier studies indicated an inverse relationship between filamentation and virulence (14–19). However, the mechanisms controlling the phase transition and virulence in Cryptococcus remain to be defined. Research on the dimorphism-associated host-pathogen interactions in the classic dimorphic fungal pathogens have provided a more comprehensive understanding of the antifungal immune response and facilitated the development of novel immunotherapy for various mycoses. For instance, the morphotype-specific adhesin Bad1 of Blastomyces dermatitidis blocks the activation of T cells (20), and the corresponding deletion mutant strain evokes a protective immune response and serves as a live-attenuated fungal cell vaccine to protect the host from lethal infection by the wild-type strain (21–26). The Candida albicans hypha-specific surface protein Als3 mediates in vivo attachment and invasion (27), and an anti-Candida vaccine designed based on Als3 is now in clinical trials (28). Vaccination is an effective strategy to prevent infections from a plethora of microbial pathogens, particularly those that frequently interact with us.
Humans are potentially exposed to Cryptococcus neoformans from the environment via respiration starting in early childhood (29). Once established in the lungs, the fungal cells may remain latent for months or even decades (29–33). Upon activation of cryptococcal infection due to impaired or suppressed host immunity, this fungus often disseminates to the central nervous system (30, 34), causing fatal cryptococcal meningitis (34–36). Cryptococcal meningitis is one of the leading causes of death among immunocompromised individuals (37), and this pathogen also infects people with no known immune defects (38–40).
Efficient clearance of Cryptococcus is dependent on Th1-polarized cell-mediated immunity. The hallmark cytokine of Th1 responses, gamma interferon (IFN-γ), is associated with classical activation of macrophages and is indispensable in protection against Cryptococcus (41). In contrast, Th2 responses exacerbate the disease and promote the secretion of interleukin 4 (IL-4), IL-5, and IL-13, which are associated with alternative activation of macrophages, pulmonary eosinophilia, and IgE expression during Cryptococcus infection (41–47). These nonprotective Th2 cell responses are mediated by interferon regulatory factor 4 (IRF4)-dependent CD11b+ CD11c+ conventional dendritic cells in a murine inhalation model of cryptococcosis (48). Given the association between the filamentous morphotype and virulence attenuation in Cryptococcus, we decided to focus on the impact of Cryptococcus morphotype on the host responses.
We recently discovered that the morphogenesis regulator Znf2 determines hyphal development in Cryptococcus (49, 50). Activation of ZNF2 by a constitutive promoter of the GPD1 gene (PGPD1-ZNF2) (the strain with PGPD1-ZNF2 [PGPD1-ZNF2 strain] or the ZNF2-overexpressing strain) promotes hyphal formation both in vitro and in vivo and abolishes the ability of the highly virulent clinical and reference serotype A strain H99 to cause fatal disease in animals (50). Given that drastically lowering the H99 inoculum (10-fold or even 100-fold reduction) still causes fatal disease with only slightly prolonged median survival time (51) and that this PGPD1-ZNF2 strain grows normally in vitro under conditions that are relevant to those in the host (host-relevant conditions), the attenuation of cryptococcal virulence by the activation of Znf2 is not likely due to a simple growth defect of the PGPD1-ZNF2 strain in vivo. We hypothesize that the cryptococcal cells with activated ZNF2 shape the host immune response, particularly cell-mediated immunity, toward a host-protective type. In this case, ZNF2-overexpressing strains may serve as vaccine agents to protect the host from subsequent lethal infection. Our study examined these hypotheses and further explored the contributions of two known downstream factors of Znf2 to its powerful antivirulence effect. Our results demonstrate that heat-resistant cellular components presented in the hyphal mutant cells are likely responsible for eliciting protective host immune responses.
RESULTS
The PGPD1-ZNF2 strain induced temporally strong inflammatory responses during early stages of infection.
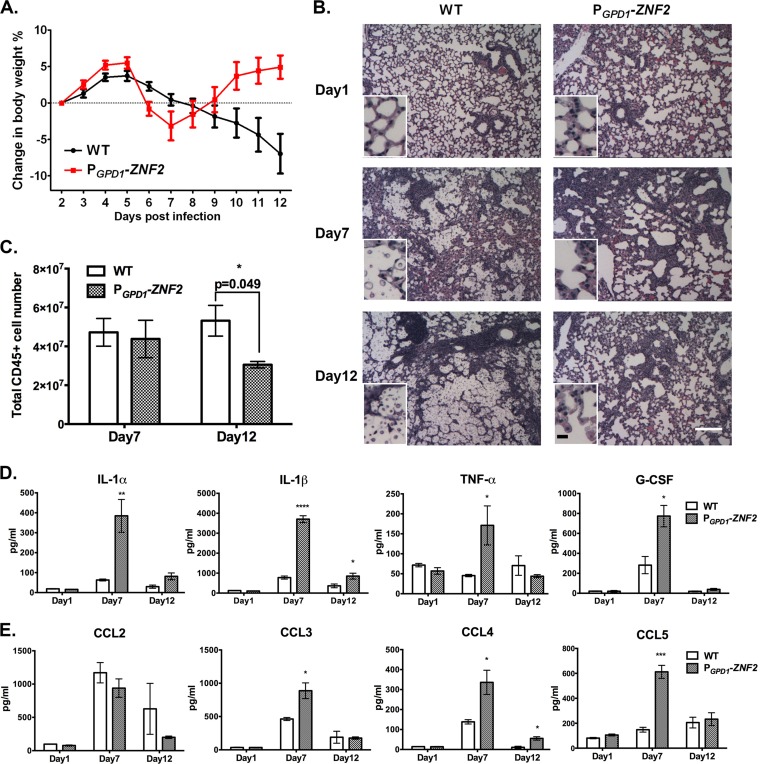
We previously reported that the PGPD1-ZNF2 strain did not cause any fatality in the infected mice up to 60 days postinfection when we ended the study (50). All the mice were healthy and active by the termination point. Interestingly, we noticed that these mice experienced a significant loss of body weight around 1 week postinfection and then regained it after an additional 5 days (Fig. 1A). In contrast, animals infected with the wild-type (WT) H99 strain experienced a gradual decline in body weight (Fig. 1A) and eventually succumbed to the infection. According to studies of pulmonary bacterial infections (52), we speculated that the temporary body weight loss might reflect strong inflammatory responses occurring in the host.
FIG 1 .
The PGPD1-ZNF2 strain elicits strong but temporary inflammatory response. (A) Curves of body weight changes in animals infected with the wild-type (WT) H99 strain or the PGPD1-ZNF2 strain (data shown are from one experiment with eight mice in each group). (B) H&E staining of the lungs of mice infected with the wild-type H99 strain or the PGPD1-ZNF2 strain (magnification of ×4). The small insets display images at a higher magnification (×40). Many fungal cells lodge at alveoli of lungs in wild-type strain-infected animals, whereas few cells of the mutant strain can be observed. White bar, 200 µm; black bar, 8 µm. (C) Total pulmonary CD45+ leukocyte count on day 7 and day 12 postinfection. (D and E) Proinflammatory cytokines (D) and chemokines (E) recovered from the supernatant of the homogenized lungs (n = 4). Values that are significantly different from the value for mice infected with the WT strain are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Therefore, we decided to examine the levels of pulmonary inflammation in these two groups of animals. We picked the examination time points of day 1, day 7 (when the animals had the lowest body weight), and day 12 (when the animals had gained back the body weight) postinfection. At day 7, we observed similar levels of immune cell infiltration in the lungs of mice infected with either the PGPD1-ZNF2 strain or the WT strain (Fig. 1B and C). This was surprising, given that the fungal burden in the group infected with the PGPD1-ZNF2 strain was about 100-fold lower than that of the group infected with the WT strain (see Fig. S1 in the supplemental material). Therefore, animals infected with the hyphal cells were more efficient in recruiting immune cells than the animals infected with the wild-type strain (WT control group). Interestingly, the PGPD1-ZNF2 strain-infected group showed significantly lower levels of pulmonary CD45+ leukocytes than those of the WT control group on day 12 despite their similar levels on day 7 (Fig. 1C), suggesting a divergence of inflammatory progression. To test this hypothesis, we examined the dynamics of total lung cytokines. On day 1, all cytokines were at very low concentrations. On day 7, animals infected with the PGPD1-ZNF2 strain presented much higher levels of the proinflammatory cytokines tested (IL-1α, IL-1β, tumor necrosis factor alpha [TNF-α], and granulocyte colony-stimulating factor [G-CSF]) and the majority of the chemokines tested (CC chemokine ligand 3 [CCL3], CCL4, and CCL5) than those infected with the WT strain (Fig. 1D and E). Notably, the levels of proinflammatory cytokines and chemokines in the PGPD1-ZNF2 strain-infected group decreased drastically on day 12 compared to the PGPD1-ZNF2 strain-infected mice on day 7. These observations indicate that the PGPD1-ZNF2 strain elicited strong, yet temporally confined, inflammatory responses.
Given that the PGPD1-ZNF2 strain did not show any apparent defect in growth or stress tolerance in host-relevant conditions (50), our observations suggest that mice infected with the PGPD1-ZNF2 strain are able to restrict rapid fungal proliferation, probably by eliciting effective immune responses during the early stages of infection.
Infection by the PGPD1-ZNF2 strain helps recruit distinct cell subsets to the lungs.
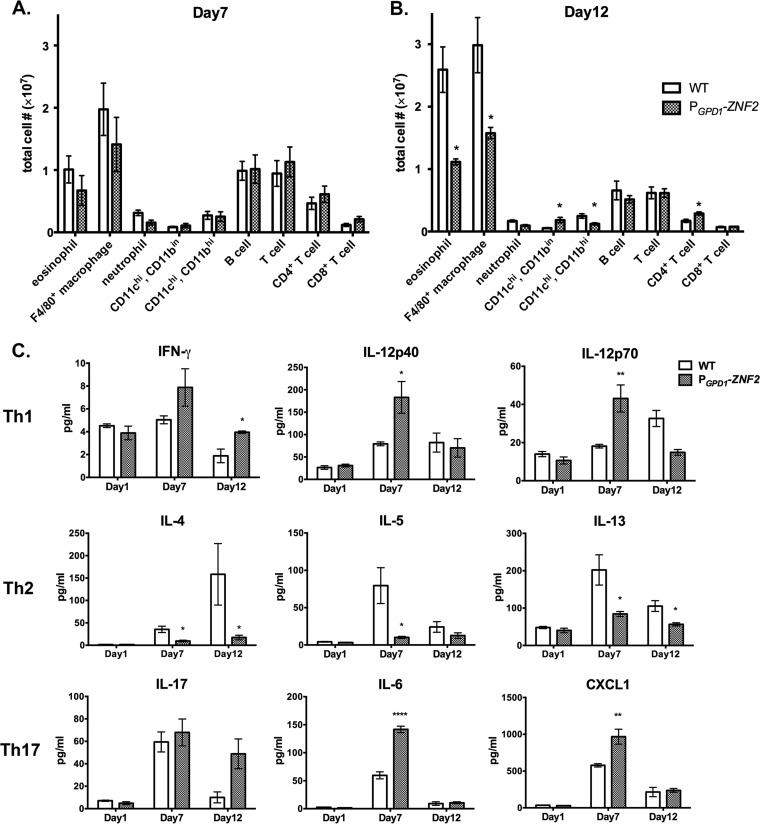
In order to define the pulmonary cellular infiltrates in response to the PGPD1-ZNF2 strain infection in mice, we performed flow cytometry with total lung cells on day 7 and day 12 postinfection. We found no significant difference in the total CD45+ leukocytes or any other cell subset between the groups infected with the wild-type strain and the PGPD1-ZNF2 strain on day 7 (Fig. 2A and 1C). On day 12, however, we observed significant differences in several cell infiltrate subsets. First, mice infected with the PGPD1-ZNF2 strain showed significantly fewer eosinophils and F4/80+ macrophages than the mice infected with the WT strain (Fig. 2B), which might contribute to the lower levels of total pulmonary CD45+ leukocytes observed in this group at this time point (Fig. 1C). Second, the PGPD1-ZNF2 strain-infected group had a significant increase of CD11c positive, CD11b intermediate (CD11c+ CD11bin) cells, while the WT strain-infected group had an increase of CD11c+ CD11b+ cells (Fig. 2B). Pulmonary eosinophilia, alternative activation of macrophages, and the development of CD11c+ CD11b+ conventional dendritic cells are linked to the pathogenic Th2 immune responses during cryptococcal infections (41, 44, 48). Although we did not examine the quality of the infiltrated macrophages, the decrease of eosinophilia and CD11b+ CD11c+ cells might be beneficial to the animals in the PGPD1-ZNF2 strain-infected group. In addition to the differential recruitment of macrophages and myeloid cells, infection with the PGPD1-ZNF2 strain induced higher recruitment of CD4+ T cells on day 12 (Fig. 2B). These observations lead us to hypothesize that the quality of cell-mediated immunity upon infection with the PGPD1-ZNF2 strain is skewed toward a protective response. This is in contrast to the infection with the parental H99 strain that appears to induce a response incapable of protecting the host (Fig. 1).
FIG 2 .
The PGPD1-ZNF2 strain elicits a distinct pattern of pulmonary cellular infiltration and T cell response. (A and B) Subsets of pulmonary cells on day 7 (A) and day 12 (B) from the lungs of mice infected with 105 WT H99 strain or the PGPD1-ZNF2 strain (data shown are from three experiments using four mice in each group for each time point). (C) Th1, Th2, and Th17 response-related cytokines recovered from the supernatants of the homogenized lungs (four mice in each group). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
We then examined the levels of cytokines involved in the cell-mediated immune response. We observed that animals infected with the PGPD1-ZNF2 strain showed a significant increase in the levels of IL-12p40 and IL-12p70 on day 7, as well as IFN-γ on day 12 (Fig. 2C), suggesting the development of Th1-type cell-mediated immunity. In contrast, the levels of major Th2 cytokines IL-4, IL-5, and IL-13 were significantly increased in animals infected with the WT parental H99 strain than in the animals infected with the PGPD1-ZNF2 strain. There was also a trend of increased IL-17A production on day 12 (P = 0.063) and significantly higher levels of Th17-related cytokines (IL-6 and CXC chemokine ligand 1 [CXCL1]) in the PGPD1-ZNF2 strain-infected group. The Th17 response was previously found to be not critical but supportive for the defense against cryptococcal infections (53, 54). Collectively, the results of analyses of the infected lungs indicate that the PGPD1-ZNF2 strain likely altered the polarization of cell-mediated immune response during infection toward the Th1 or Th17 (Th1/Th17) type while suppressing the Th2 type.
The PGPD1-ZNF2 strain induces protective cell-mediated immunity.
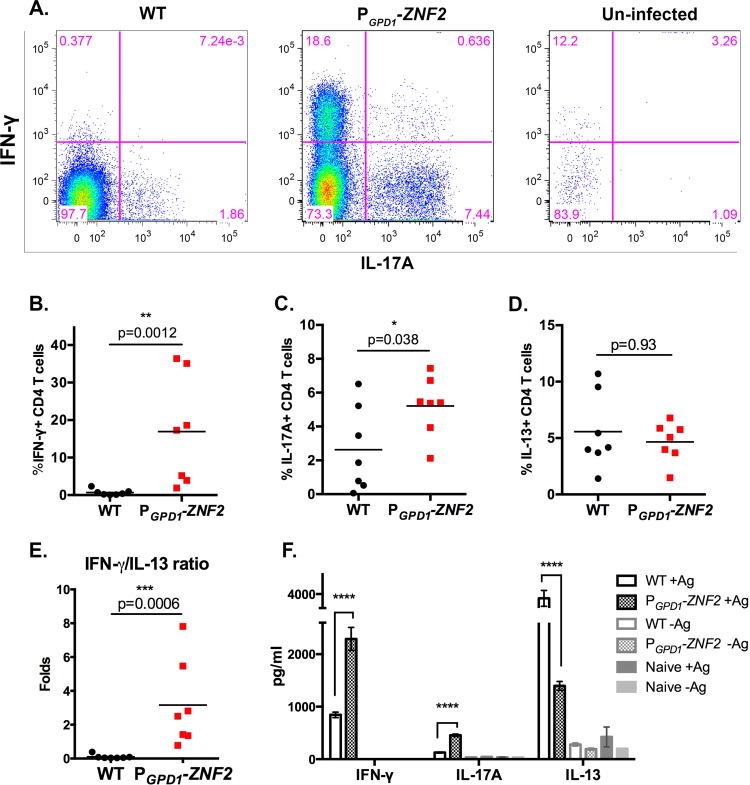
To examine whether the encounter with the PGPD1-ZNF2 Cryptococcus cells steered the host to differentiate protective T helper cells, we collected cells from bronchoalveolar lavage fluid (BALF) samples of animals infected with the wild-type H99 strain or the PGPD1-ZNF2 strain on day 7 postinfection. The polarization of CD4+ T cells toward Th1, Th17, and Th2 responses was examined by the intracellular staining for the signature IFN-γ, IL-17A, and IL-13 cytokines, respectively. Strikingly, CD4+ T cells recovered from the airways of the mice infected with the PGPD1-ZNF2 strain produced significantly higher levels of IFN-γ and IL-17A (Fig. 3A to C) than CD4+ T cells recovered from mice infected with the WT strain. The phenotype was consistent with the analysis of global cytokine levels in lung tissue (Fig. 2C). These data suggest that infection with the PGPD1-ZNF2 strain enhanced the differentiation of beneficial Th1 and Th17 cells. Although the frequencies of IL-13-producing CD4+ T cells were similar in both groups of infected mice (Fig. 3D), the IFN-γ/IL-13 ratio was significantly higher in the PGPD1-ZNF2 strain-infected group (Fig. 3E). Therefore, these findings indicate that the activation of ZNF2 in Cryptococcus promotes a host response dominated by protective Th1 cells.
FIG 3 .
The PGPD1-ZNF2 strain alters the polarization of cell-mediated immunity. (A) CD4+ T cell gate, intracellular staining of IFN-γ and IL-17A of uninfected mice and mice infected with the WT or PGPD1-ZNF2 strain. (B to D) Frequencies of cells positive for IFN-γ (B), IL-17A (C), and IL-13 (D) out of total CD4+ T cells (n = 7). (E) IFN-γ/IL-13 ratio of CD4+ T cells. In panels B to E, each symbol represents the value for an individual mouse, and the short horizontal line represents the mean value for the group of mice. (F) ELISA of IFN-γ, IL-17A, and IL-13 levels from purified mediastinal lymph node CD4+ T cells restimulated with Cryptococcus antigens (+Ag) (data shown are from one experiment using seven mice in each group). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To further examine the possible differential activation of Cryptococcus-specific CD4+ T cells in the two groups of infected mice, we examined CD4+ T cell responses in the lung-draining mediastinal lymph node (MLN) on day 7 postinfection. Purified MLN CD4+ T cells were restimulated by co-culture with naive T cell-depleted splenocytes in the presence or absence of cryptococcal antigens. The response of Cryptococcus-specific CD4+ T cells was examined by measuring cytokine secretion in the culture supernatants in an enzyme-linked immunosorbent assay (ELISA). We included cocultures of purified CD4+ T cells with splenocytes without Cryptococcus antigen as controls for the specificity of the response. As expected, cytokines were minimally produced without the addition of Cryptococcus antigens (Fig. 3F). In the presence of Cryptococcus antigens, however, the CD4+ T cells isolated from the PGPD1-ZNF2 strain-infected mice produced significantly higher levels of IFN-γ and IL-17A and significantly lower levels of IL-13 than the CD4+ T cells isolated from WT strain-infected mice (Fig. 3F). Taken together, these results strongly support the hypothesis that the PGPD1-ZNF2 cryptococcal strain directs the host cell-mediated immune response toward the protective Th1/Th17 type.
Vaccination with live PGPD1-ZNF2 cells protects the host from a subsequent challenge by the otherwise lethal wild-type strain.
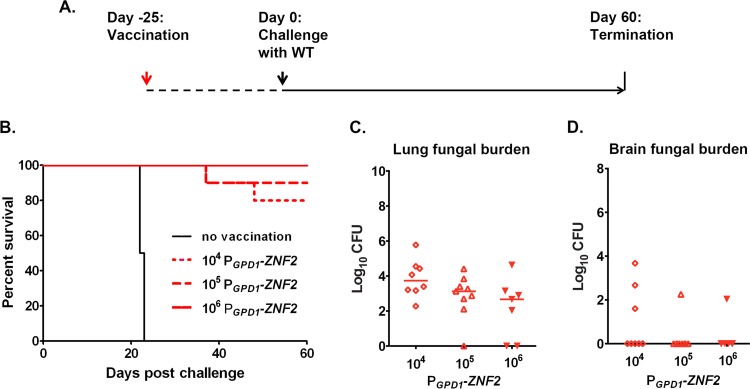
The aforementioned evidence indicates that the immune response that developed in mice infected with the PGPD1-ZNF2 strain could potentially be protective against a subsequent challenge by WT cryptococcal cells. This prompted us to examine whether this strain could serve as a live-attenuated cryptococcal cell vaccine to protect the host against a subsequent lethal infection. First, we performed a pilot study and tested two different inoculation dosages of the live PGPD1-ZNF2 cells (1 × 103 and 1 × 104 CFU per animal). We found that the PGPD1-ZNF2 strain was partially protective and that the protection was dose dependent (see Fig. S2 in the supplemental material). The animals challenged on day 25 postimmunization exhibited a better level of protection than those challenged on day 48 postimmunization (Fig. S2). We also performed a coinfection experiment where mice were inoculated with the PGPD1-ZNF2 and WT cells mixed at a 9:1 ratio. We did not observe any protection compared with the infection with WT cells alone. This suggests that the protective effect could be achieved only sequentially, as observed for most vaccines currently in clinical use. On the basis of these results, we tested higher doses of the PGPD1-ZNF2 strain (1 × 104, 1 × 105, and 1 × 106 CFU per mouse) for the vaccination and challenged these mice together with the nonvaccinated control mice with wild-type strain H99 (1 × 104 cells per animal) on day 25 postvaccination (Fig. 4A). As expected, all the mice in the unimmunized control group died within 4 weeks after the challenge (Fig. 4B). Strikingly, 80%, 90%, and 100% of the mice immunized with the PGPD1-ZNF2 strain at a dose of 1 × 104, 1 × 105, and 1 × 106 CFU, respectively, survived without any moribund symptoms by 60 days postinfection with WT cells when we terminated the experiment (Fig. 4B). Thus, the PGPD1-ZNF2 strain provided dose-dependent protection to the host against otherwise lethal challenges with WT cells.
FIG 4 .
The PGPD1-ZNF2 strain serves as a live-attenuated fungal cell vaccine. (A) Strategy of vaccination. (B) Survival curves plotted against time upon challenge with WT H99 (data shown are from two experiments with 10 mice in each group for the 104 and 105 groups and 7 mice in the 106 group). (C and D) Fungal burden in the lungs (C) and brains (D) from the animals that survived 60 days after challenge in each vaccination group.
We further examined the fungal burden in lungs and brains of the protected immunized mice at the endpoint of the experiment. We did not detect any cryptococcal cells in the brains of the majority of mice that survived the WT cryptococcal challenge for 60 days (Fig. 4D). The lungs of animals immunized with the higher dose of the PGPD1-ZNF2 cells contained lower fungal burdens (Fig. 4C), consistent with the dose-dependent protection for survival. We were able to recover some PGPD1-ZNF2 cells, though at very low numbers, from several of the surviving animals at the end point of the experiment. This finding indicated that inducing a protective immune response did not require total clearance of the live-attenuated cells. Taken together, the data demonstrate that immunization with the PGPD1-ZNF2 strain protects animals from the subsequent lethal infection and restrains proliferation of WT H99 cells in the lung. Such an effective immunization even led to the clearance of the highly virulent H99 cells from several mice in the experiment.
The antivirulence effect of Znf2 remains in the absence of two Znf2 downstream factors.
As the master regulator of morphogenesis, Znf2 controls the expression of multiple factors (49, 50, 55). It is possible that some of its downstream factors account for its antivirulence property. Two factors, Pum1 and Cfl1, are the major downstream factors contributing to the ZNF2-controlled filamentation in vitro in the serotype D XL280 background (50, 55, 56). Deletion of the CFL1 or PUM1 gene alone leads to less robust hyphal growth in strain XL280. Disruption of both genes severely reduces the hyphal formation in vitro, even when Znf2 is activated (55). Given that serotype A isolates represent the vast majority of Cryptococcus clinical isolates, we decided to investigate the contributions of Pum1 and Cfl1 to the antivirulence effect of Znf2 in a serotype A background.
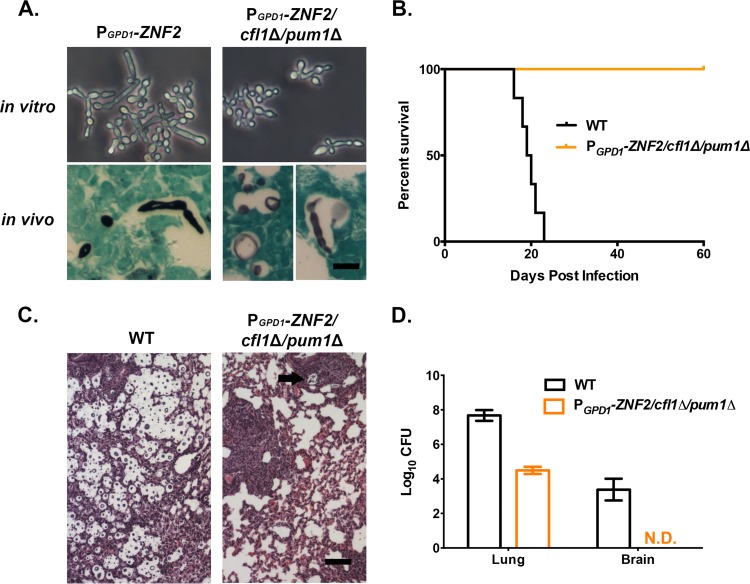
We deleted both CFL1 and PUM1 genes in the PGPD1-ZNF2 strain in the H99 background. As expected, the robustness of hyphal formation was significantly reduced in the absence of CFL1 and PUM1 in vitro (Fig. 5A, top panels), consistent with our previous observations of the serotype D background (55). However, we noticed that the degree of reduction in filamentation in vitro of this PGPD1-ZNF2 cfl1Δ pum1Δ strain in the H99 background was not as dramatic as we previously observed for the serotype D background (55). We then tested the virulence and looked at the morphology of this mutant strain in vivo in a murine model of cryptococcosis. Grocott’s methenamine silver (GMS) staining of the infected lung tissues indicated that the PGPD1-ZNF2 cfl1Δ pum1Δ strain displayed a mixed morphology in vivo (Fig. 5A, bottom panels). All mice infected with this PGPD1-ZNF2 cfl1Δ pum1Δ mutant strain survived and appeared healthy when we terminated the experiment 60 days postinfection (Fig. 5B). This was similar to what we observed for the PGPD1-ZNF2 strain. In contrast, animals infected with the same inoculum of wild-type H99 strain died within 23 days postinfection (Fig. 5B). Consistent with the survival data, our histological examination of the lung tissues suggested that the PGPD1-ZNF2 cfl1Δ pum1Δ strain elicited strong inflammation in the lungs (Fig. 5C), which again resembled the phenotype of lung infection by the PGPD1-ZNF2 strain.
FIG 5 .
Two downstream targets of Znf2, CFL1 and PUM1, do not contribute to Znf2’s antivirulence effect. (A) Morphology of the PGPD1-ZNF2 strain and the PGPD1-ZNF2 cfl1Δ pum1Δ strain in vitro (24 h of incubation in DMEM plus 10% FBS at 37°C under 5% CO2) and in vivo (GMS staining of lung tissues from the infected mice). Bar, 10 µm. (B) Survival curves of the PGPD1-ZNF2 cfl1Δ pum1Δ strain and wild-type H99 strain (the results for one experiment are shown, with eight mice in each group). (C) H&E staining of the lung tissues. Pseudohyphal cells were enveloped by the well-developed bronchus-associated lymphoid tissue (black arrow). Bar, 100 µm. (D) Lung and brain fungal burden of animals infected with the PGPD1-ZNF2 cfl1Δ pum1Δ strain and wild-type H99 strain (one experiment, with four mice in each group). N.D., not detected.
We further examined the fungal burden in lungs and brains at 10 days postinfection. The mice infected with the PGPD1-ZNF2 cfl1Δ pum1Δ strain had lung fungal burdens about 3 orders of magnitude lower than those in the H99-infected animals (Fig. 5D). Furthermore, no fungal cells were recovered from the brains of animals infected with the mutant strain (Fig. 5D). All the in vivo phenotypes observed for the PGPD1-ZNF2 cfl1Δ pum1Δ strain resembled what we previously observed for the parental PGPD1-ZNF2 strain (50). The results indicate that Cfl1 and Pum1 do not contribute to the antivirulence effect of Znf2.
Cellular components of ZNF2-overexpressing strains provide vaccination effect to the host.
Two hypotheses could explain the antivirulence effect of Znf2. First, the active metabolism of cryptococcal cells transitioning from the yeast form to the hyphal form may be directly associated with the virulence attenuation and vaccination effect observed earlier (Fig. 4). Alternatively, cell components that are prominently presented in ZNF2-overexpressing strains may serve as potent antigens to elicit a strong protective host response. The first hypothesis would require the presence of viable ZNF2-overexpressing cells for protection. Given that we were able to recover low levels of fungal cells from seven out of eight lungs from mice infected with the PGPD1-ZNF2 strain on day 60 postinoculation (see Fig. S3 in the supplemental material), it was reasonable to speculate that most of the immunized mice still had viable PGPD1-ZNF2 cells at the time of challenge by the WT strain on day 25 or day 48 after inoculation with the PGPD1-ZNF2 strain.
The second hypothesis, however, would predict that it was the alteration in cellular components enacted by Znf2 (e.g., cell surface remodeling) that provided potent antigens to elicit a strong protective host response. In such a scenario, a component(s) from the ZNF2-overexpressing cells could exert a similar effect on the host regardless of whether the strain remained viable or whether it retained the ability to proliferate.
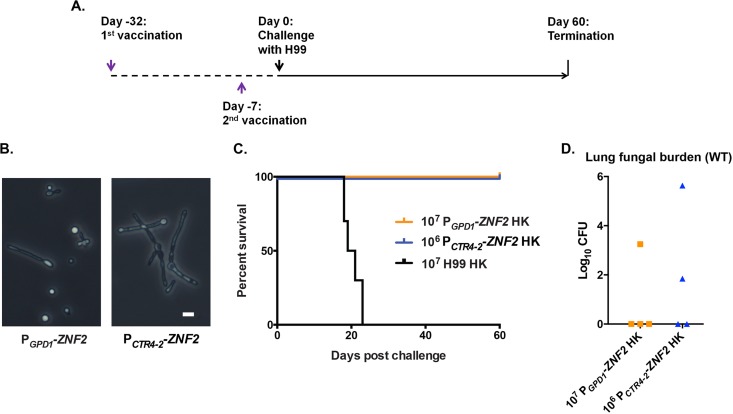
Previous studies have demonstrated that the heat-killed wild-type strain could not provide protection (57, 58). Our pilot experiment showed that one-time inoculation with the heat-killed PGPD1-ZNF2 strain at a dose of 1 × 104 or 1 × 105 CFU/animal offered no protection (see Fig. S4 in the supplemental material). Given that the live PGPD1-ZNF2 strain provided dose-dependent protection (Fig. 4), we decided to examine the protective effect of heat-killed ZNF2-overexpressing strains with higher doses (106 or 107). For this purpose, we chose to employ two strains with different promoters driving the expression of ZNF2. One was the PGPD1-ZNF2 strain used in this study and the previous study (50). The PGPD1-ZNF2 strain had constitutively active expression of ZNF2, and the level of its expression was modestly high (50). Consequently, this strain displayed mixed morphotypes (yeast, pseudohyphae, and hyphae) under both in vitro growth conditions and in animal lungs during infection (50). The second strain was the PCTR4-2-ZNF2 strain, where the inducible promoter of the copper transporter CTR4 drove the expression of ZNF2. The expression of ZNF2 could be induced to such a high level by copper limitation that the whole population could turn filamentous (55) (Fig. 6B). The live PCTR4-2-ZNF2 cells were not used earlier to study the effect of Znf2 on virulence because of the necessity of manipulating copper levels in the host. However, for the purpose of using heat-inactivated cells in this experiment, the manipulation of copper levels in the host was not required. We included heat-killed wild-type H99 cells at a vaccination dose of 1 × 107 as a control.
FIG 6 .
Heat-killed cells of two different ZNF2-overexpressing strains offer protection to the host. (A) Strategy of vaccination. (B) Cellular morphology of the PGPD1-ZNF2 strain and the PCTR4-2-ZNF2 strain. Bar, 5 µm. (C) Survival curves of mice vaccinated by heat-killed (HK) cells of strain H99 (n = 10), PGPD1-ZNF2 (n = 4), or PCTR4-2-ZNF2 (n = 4). (E) Fungal burden in the lungs of immunized animals at the end of the experiment (60 days postchallenge).
To achieve a better protection effect using inviable cells, we decided to modify the vaccination strategy. Here we vaccinated the animals with two doses of heat-killed cells, one at day −32 and one at day −7 (day 0 was the day the mice were challenged with lethal clinical strain H99). Then, we challenged the immunized mice with live WT H99 (1 × 104 cells/animal) 7 days after the booster dose (Fig. 6C). Excitingly, both heat-killed ZNF2-overexpressing strains protected the mice from the otherwise lethal challenge, while the heat-killed WT cells did not offer any protection (Fig. 6C). This finding suggests that changes in cell components caused by the activation of ZNF2, rather than the persistence of the mutant cells in vivo, were responsible for the protective effect. We terminated the experiment at day 60 postchallenge and evaluated the organ fungal burden. The wild-type H99 cells were cleared from more than half of the immunized animals during the period of this experiment, suggesting that the immunization could help animals clear or restrict cryptococcal proliferation in the lungs (Fig. 6D). More importantly, none of the animals had any brain infection at the termination point of this experiment. Taken together, our studies suggest that heat-resistant fungal components from two hyphal mutant strains could offer promising protection from the otherwise lethal cryptococcal infections.
DISCUSSION
Morphological switch is tightly linked to pathogenesis in many pathogenic fungi. In Cryptococcus, Znf2 provides the underlying molecular link between morphology and pathogenesis. However, how Znf2 impacts cryptococcal pathogenesis and whether its antivirulence effect could be useful for translational studies remain unknown.
The activation of Znf2 abolishes cryptococcal virulence, but it does not reduce cryptococcal fitness or stress tolerance. This differs from many other mutations that attenuate fungal virulence. Interestingly, during the survival study, we observed one episode of relapse in one mouse infected with the live PGPD1-ZNF2 strain. This animal lost about one-third of its body weight within a week at around week 4 postinfection, but it gradually recovered its original body weight after another 2 weeks. Except for the weight loss, this animal did not show any moribund symptoms during the time course of the experiment. Nonetheless, it showed a relatively high lung fungal burden (~106 CFU) and brain dissemination when we terminated the experiment (see Fig. S3 [asterisks] in the supplemental material). The observation of relapse reinforced the notion that the PGPD1-ZNF2 strain can proliferate in vivo and that it possesses the ability to disseminate to the central nervous system (CNS). Therefore, we believe that the protective host response elicited by the PGPD1-ZNF2 cells, rather than any growth defect of the PGPD1-ZNF2 strains in vivo, is mainly responsible for the containment of the infection.
It is important to note that the host response elicited by the PGPD1-ZNF2 cells is stage dependent, which is critical for protection. For instance, at day 7, the infection led to an influx of CD45+ leukocytes to the primary infectious site and dramatically enhanced the secretion of major proinflammatory cytokines. During the second week of infection, however, the level of inflammation was reduced. The mitigation of host inflammatory responses is essential to the protective effect, as strong and prolonged inflammation is detrimental or even fatal: mutants (e.g., rim101Δ mutants) that elicit exacerbated and persistent pulmonary inflammation could cause even earlier death in animals than the highly virulent wild-type H99 strain does (59, 60).
We speculate that this rapid decrease of inflammation resulted from effective antigen presentation. The flow cytometry data showed a different pattern of CD11c+ cell subsets in mice infected with the PGPD1-ZNF2 strain compared to the WT strain on day 12, but not on day 7 (Fig. 2B). Mice infected with the PGPD1-ZNF2 strain presented significantly higher CD11c+ CD11bin cells and lower levels of the pathological CD11c+ CD11b+ conventional dendritic cells (DCs). Therefore, we propose that the increased CD11c+ CD11bin cells are associated with a protective response. Accordingly, the antigen-presenting cells (APCs) that interacted with the PGPD1-ZNF2 cells directed a type 1 T helper cell differentiation in the host (Fig. 3A). A thorough investigation into the different APC subsets generated during the PGPD1-ZNF2 strain infection will help us better understand the protective immune response in the future.
Another exciting finding is that the ZNF2-overexpressing cells, in the viable or heat-killed form, can protect the host from challenge with the lethal wild-type strain. To our knowledge, no virulence-attenuated Cryptococcus strain reported so far provides such a level of protection. The only exception is the viable form of the H99 strain expressing the protective cytokine IFN-γ (H99γ): the viable H99γ can provide 100% protection to animals, yet the heat-killed H99γ fails (61–63). This current discovery suggests that a fungal factor(s) controlled by Znf2 is capable of initiating potent protective responses. Surprisingly, two factors contributing to the Znf2-mediated filamentation in vitro, Cfl1 and Pum1, are not required for the antivirulence effect of Znf2. Animals infected with the PGPD1-ZNF2 cfl1Δ pum1Δ strain survived through the study, as we observed for the PGPD1-ZNF2 strain. One plausible explanation is that Cfl1 and Pum1 are present at levels too low in the yeast form of Cryptococcus to serve as effective antigens that elicit protective host responses against yeast infections. Therefore, the major factors contributing to the antivirulence effect of Znf2 are yet to be identified. The successful protection offered by the two different types of heat-killed ZNF2-overexpressing cells suggests that characterization of the cellular component(s) of the ZNF2-overexpressing cells is the key to pinpoint the cryptococcal factors that elicit host protective immune responses. Additionally, it would be interesting to compare the type of T cell response induced by the live mutant cells with that induced by heat-killed cells and examine whether a similar Th1/Th17-type cell-mediated immunity is present in both.
Altogether, our results unveiled the effect of Znf2 on host-pathogen interactions during Cryptococcus infection. More importantly, the protective effect of ZNF2-overexpressing cells provides a promising lead for future development of vaccination agents against this fatal disease.
MATERIALS AND METHODS
Ethics statement.
This study was performed according to the guidelines of NIH and Institutional Animal Care and Use Committee (IACUC). The animal models and procedures used have been approved by the IACUCs at Texas A&M University (protocols 2011-22 and 2014-0049), Rutgers University (12020D0615), and the University of Texas at San Antonio (MU021).
Mutant generation and strain growth conditions.
The Cryptococcus strains used in the study are the wild-type H99 strain (64), the PGPD1-ZNF2 strain (LW10) (50), the PCTR4-2-ZNF2 strain (LW30) (50), and the PGPD1-ZNF2 cfl1Δ pum1Δ strain (generated for this study). All Cryptococcus strains were maintained on yeast extract-peptone-dextrose (YPD) medium at 30°C unless indicated otherwise. The ZNF2oe (oe stands for overexpressing) cfl1Δ pum1Δ triple mutant strain is constructed by replacing the CFL1 gene and the PUM1 gene with the hygromycin (HYG) and the nourseothricin (NAT) drug cassette, respectively, in the PGPD1-ZNF2 strain (LW10) background. The construct generation and cryptococcal biolistic transformation were performed essentially as we described previously (55). Primers used for making the knockout constructs are listed in Table S1 in the supplemental material.
To culture the fungal strains for infection, fungal strains were streaked out from −80°C freezer stock and inoculated on YPD agar plates at 30°C for 2 days. Then, the fungal cells were inoculated in 2 to 3 ml YPD liquid medium at 30°C with shaking at 250 rpm overnight. When grown under these conditions, PGPD1-ZNF2 and PGPD1-ZNF2 cfl1Δ pum1Δ strains would form clumps. In order to obtain single-cell suspensions, overnight liquid cultures were allowed to sit on a bench for 15 to 20 min to let the large clumpy cells precipitate. The upper layer of the liquid culture was collected and centrifuged at 3,000 rpm for 2 min, and the cell pellets were used for the following washing and counting steps. In this case, more than 95% of the collected cells would be yeast or small butterfly-shaped cells. The actual inoculation should be verified by plating the cell suspension on YPD agar plates and counting the number of CFU after incubation.
Murine model of cryptococcosis.
Female A/J mice of 8 to 10 weeks old were purchased from the Jackson Laboratory (Bar Harbor, ME). In survival assays, 8 to 10 mice were assigned to each group. To prepare fungal cells for infection, strains were cultured in RPMI 1640 medium overnight at 37°C with shaking at 250 rpm to reduce cell aggregation. The fungal cells were washed with sterile 0.9% saline three times, and the final concentration of the cell suspension was adjusted with saline to 2 × 106 cell/ml. Mice were sedated with ketamine and xylazine via intraperitoneal injection and then inoculated intranasally with 50 µl fungal cell suspension as previously described (51, 65). After infection, animals were weighed daily and monitored twice a day for progression of disease, including weight loss, gait changes, labored breathing, or fur ruffling. Moribund mice were sacrificed. The change of body weight of each animal was calculated as follows: [(weight on day X − weight on day 2)/weight on day 2] × 100%. The resulting data on body weight change were then plotted against time.
Cytokine analysis.
The lungs of infected animals were dissected on the indicated days (indicated on the figures) and homogenized in 1 ml ice-cold antiprotease buffer solution containing phosphate-buffered saline (PBS), protease inhibitors (inhibiting cysteine, serine, and other metalloproteinases), and 0.05% Triton X-100. The mixtures were centrifuged at 800 × g for 5 min at 4°C. The supernatant of each sample was collected for cytokine analysis using the Bio-Plex protein array system (Luminex-based technology; Bio-Rad Laboratories, Hercules, CA) according to the instructions provided by the manufacturer (66).
Histology assays.
The animals infected with different Cryptococcus strains were sacrificed on the indicated days (indicated on the figures), and their lungs and brains were harvested. The organs were fixed in 10% formalin, embedded in paraffin, sliced into 5-µm-thick sections, and processed either with hematoxylin-and-eosin (H&E) staining for bronchus-associated lymphoid tissue, or with Grocott’s methenamine silver (GMS) staining for the fungal morphology observation in vivo.
Organ fungal burden analysis.
The infection procedure is the same as in the survival study. At the time of sacrifice, the lungs and brains were dissected. The dissected organs were homogenized in 2 ml cold PBS buffer with the same setting for each type of organ. The tissue suspensions were serially diluted (10 times), plated onto YNB agar medium, and incubated at 30°C for 2 days such that the colonies became visible so that the number of CFUs could be counted.
Pulmonary leukocyte isolation.
Lungs were excised at the indicated time points (indicated in the figures) and digested enzymatically at 37°C for 30 min in 10 ml of digestion buffer (RPMI 1640 medium and 1 mg/ml of collagenase type IV [Sigma-Aldrich, St. Louis, MO]) with intermittent (every 10 min) stomacher homogenizations. The enzymatically digested tissues were then successively filtered through sterile nylon filters of various pore sizes (70 and 40 µm) (BD Biosciences) and washed with sterile Hanks balanced salt solution (HBSS) to enrich for leukocytes. Erythrocytes were lysed by incubation in NH4Cl buffer (0.859% NH4Cl, 0.1% KHCO3, 0.0372% Na2EDTA [pH 7.4]; Sigma-Aldrich) for 3 min on ice followed by the addition of a 10-fold excess of PBS. The resulting leukocyte population was then collected by centrifugation (800 × g) for 5 min, washed twice with sterile PBS, resuspended in sterile PBS plus 2% heat-inactivated fetal bovine serum (FBS) (fluorescence-activated cell sorting [FACS] buffer) and enumerated in a hemacytometer using trypan blue dye exclusion. Flow cytometric analysis was used to determine the percentage of each leukocyte population as well as the absolute number of total leukocytes (CD45+) within the lung cell suspension for standardization of hemacytometer counts.
Total lung cell surface staining and flow cytometry.
Standard methodology was employed for the direct immunofluorescence of pulmonary leukocytes. Briefly, in 96-well U-bottom plates, 100 µl containing 1 × 106 cells in PBS plus 2% FBS (FACS buffer) were incubated with 50 µl of Fc Block (BD Biosciences) diluted in FACS buffer for 5 min to block nonspecific binding of antibodies to cellular Fc receptors. Subsequently, an optimal concentration of fluorochrome-conjugated antibodies (eBioscience [San Diego, CA] and Molecular Probes [Eugene, OR]) (between 0.06 and 0.5 µg/1 × 106 cells in 50 µl of FACS buffer) were added in various combinations to allow for dual or triple staining experiments, and plates were incubated at 4°C for 30 min. Following incubation, the cells were washed three times with FACS buffer, and cells were fixed in 200 µl of 2% ultrapure formaldehyde (Polysciences, Inc., Warrington, PA) diluted in FACS buffer (fixation buffer). Cells incubated with either FACS buffer alone or single-fluorochrome-conjugated antibodies were used to determine positive staining and spillover/compensation calculations, and the flow cytometer determined the background fluorescence. The samples were analyzed using a BD FACSArray flow cytometer (BD Biosciences), and FlowJo was used to determine cell populations. Dead cells were excluded on the basis of forward angle and 90° light scatter. For data analyses, 30,000 events (cells) were evaluated from a predominantly leukocytic population identified by back-gating from stained CD45+ cells. The absolute number of total leukocytes was quantified by multiplying the total number of cells observed by hemacytometer counting by the average percentage of CD45+ cells determined by flow cytometry. The absolute number of each leukocyte subset (Ly6G+, F4/80+ MHCII+ [MHCII stands for major histocompatibility complex class II], CD11c+ CD11bin, CD11c+ CD11b+, CD19+, Siglec-F+ CD11bin, CD4+ CD3+, and CD8+ CD3+) was determined by multiplying the percentage of each gated population by the total number of CD45+ cells.
BALF cell stimulation and intracellular staining.
Bronchoalveolar lavage fluid (BALF) samples were harvested 7 days after inoculation, and cells were plated in a round-bottom 96-well plate where they were reactivated with a leukocyte activation cocktail with GolgiPlug (BD Pharmingen) in RPMI 1640 medium containing 10% fetal calf serum (FCS) and penicillin-streptomycin (PenStrep). The cells were then washed and stained for cell surface markers and fixed in 1% paraformaldehyde (PFA) overnight. Then, cells were permeabilized for intracellular staining and analyzed in a BD LSR II flow cytometer.
CD4+ T cell restimulation from mediastinal lymph nodes.
Mediastinal lymph nodes were harvested 7 days after inoculation, and cells from the same group were pooled and used for CD4+ T cell enrichment by a negative-sorting CD4+ isolation kit (Miltenyi Biotec). Then, 2 × 105 isolated CD4+ T cells were cocultured with 3 × 105 T cell-depleted splenocytes, with the addition of voriconazole (final concentration of 1.25 µg/ml) or live H99 cells as a source of Cryptococcus antigen in RPMI 1640 medium containing 10% FCS and PenStrep. The culture was maintained for 3 days, and the supernatant was harvested to measure cytokine production according to the manufacturer’s instructions for the Ready-SET-Go! (eBioscience) enzyme-linked immunosorbent assay (ELISA) kits.
T cell-depleted splenocytes were generated by obtaining a single-cell suspension of a naive spleen and treated with red blood cell (RBC) lysis buffer to remove red blood cells. After the cells were washed with PBS, splenocytes were incubated at 37°C in 5 ml of RPMI 1640 medium (10% FCS and PenStrep) containing anti-Thy1.2 antibody and Low-ToxM rabbit complement (Cedarlane) to remove T cells. Finally, the cells were washed and counted.
Vaccination.
For the live-cell vaccination, mice were vaccinated intranasally with different doses (104, 105, and 106) of the live PGPD1-ZNF2 cells. The concentration of fungal cell in suspension was adjusted according to the required inocula to meet the volume requirement of 50 µl per inoculation. The vaccinated groups and the naive group (negative control) were challenged with 1 × 104 wild-type H99 cells 25 or 48 days postvaccination. For the heat-killed cell vaccination, H99 and PGPD1-ZNF2 cells were cultured in YPD liquid medium at 30°C with shaking (250 rpm) overnight. The PCTR4-2-ZNF2 cells were cultured in YPD liquid medium plus 200 µM copper chelator bathocuproine disulfonate (BCS) at 30°C overnight without shaking. Cells of the three fungal strains were precipitated at 3,000 rpm for 2 min and washed twice with sterile PBS. The concentration of fungal cell suspension was adjusted according to the required inocula to meet the volume requirement of 50 µl per inoculation. The cell suspension with the correct concentration was then aliquoted into 1.7-ml tubes and heated in a boiling water bath for 20 min. The viability of the cells after such treatment was examined by plating the processed cell suspension on YPD agar plate; no colonies were recovered after incubating at 30°C for 3 days. Mice were vaccinated intranasally with the heat-killed fungal strains at day −32. Mice were vaccinated again with the same dose of heat-killed fungal strains at day −7. All the animals were challenged with 104 live H99 cells via intranasal inoculation 32 days after the first vaccination. Animals after the live H99 cell challenge were weighed and monitored daily for disease progression, and moribund mice were sacrificed. All the animals were terminated on day 60 after challenge with live H99.
Statistical analysis.
Statistical significance of the survival data for the different groups was assessed by the Gehan-Breslow Wilcoxon test. Multiple t tests were applied in the grouped cytokine profiling/cell infiltration data. The intracellular staining data were analyzed by nonparametric Mann-Whitney test. All statistical analyses were performed using the GraphPad Prism 6 program, with P values lower than 0.05 considered statistically significant.
SUPPLEMENTAL MATERIAL
Fungal burden of lungs infected with 105 WT H99 or the PGPD1-ZNF2 strain on day 1, day 7, and day 12 postinoculation (four mice in each group). Download
The protective effect of vaccination by the PGPD1-ZNF2 strain is dose dependent in preliminary tests (four mice in each group). Download
Lung and brain fungal burdens of the animals infected with the PGPD1-ZNF2 strain on day 60 postinfection. The red line shows the initial inoculation dose of cryptococcal cells in lungs. The two asterisks indicate two individual mice that relapsed. Download
No protection effect of heat-killed PGPD1-ZNF2 at a dose of 1 × 104 or 1 × 105 CFU/animal (data shown are from one experiment with eight mice per group). The two experimental groups displayed the exact same dynamics in survival, so the curves overlapped. Download
Primers and cryptococcal strains used in this study.
ACKNOWLEDGMENTS
We thank Linqi Wang for helpful suggestions and Dylan Foyle for his assistance in the animal studies.
We gratefully acknowledge the financial support from the National Institute of Allergy and Infectious Diseases (grants R01AI097599 and R21AI107138 to X.L., grant R01AI071752 to F.L.W., and grant R01AI114647 to A.R.). X.L. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Zhai B, Wozniak KL, Masso J, Upadhyay S, Hole C, Rivera A, Wormley FL, Jr, Lin X. 2015. Development of protective inflammation and cell-mediated immunity against Cryptococcus neoformans after exposure to hyphal mutants. mBio 6(5):e01433-15. doi:10.1128/mBio.01433-15.
Contributor Information
Andrew Alspaugh, Duke University Medical Center.
Françoise Dromer, Institut Pasteur.
REFERENCES
- 1.Bozza S, Gaziano R, Spreca A, Bacci A, Montagnoli C, di Francesco P, Romani L. 2002. Dendritic cells transport conidia and hyphae of Aspergillus fumigatus from the airways to the draining lymph nodes and initiate disparate Th responses to the fungus. J Immunol 168:1362–1371. doi: 10.4049/jimmunol.168.3.1362. [DOI] [PubMed] [Google Scholar]
- 2.d’Ostiani CF, Del Sero G, Bacci A, Montagnoli C, Spreca A, Mencacci A, Ricciardi-Castagnoli P, Romani L. 2000. Dendritic cells discriminate between yeasts and hyphae of the fungus Candida albicans. Implications for initiation of T helper cell immunity in vitro and in vivo. J Exp Med 191:1661–1674. doi: 10.1084/jem.191.10.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moyes DL, Runglall M, Murciano C, Shen C, Nayar D, Thavaraj S, Kohli A, Islam A, Mora-Montes H, Challacombe SJ, Naglik JR. 2010. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe 8:225–235. doi: 10.1016/j.chom.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seider K, Heyken A, Lüttich A, Miramón P, Hube B. 2010. Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr Opin Microbiol 13:392–400. doi: 10.1016/j.mib.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Kanetsuna F, Carbonell LM. 1971. Cell wall composition of the yeastlike and mycelial forms of Blastomyces dermatitidis. J Bacteriol 106:946–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rappleye CA, Eissenberg LG, Goldman WE. 2007. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc Natl Acad Sci U S A 104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holbrook ED, Rappleye CA. 2008. Histoplasma capsulatum pathogenesis: making a lifestyle switch. Curr Opin Microbiol 11:318–324. doi: 10.1016/j.mib.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Klein BS, Tebbets B. 2007. Dimorphism and virulence in fungi. Curr Opin Microbiol 10:314–319. doi: 10.1016/j.mib.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemecek JC, Wüthrich M, Klein BS. 2006. Global control of dimorphism and virulence in fungi. Science 312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Köhler J, Fink GR. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 11.Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen VQ, Sil A. 2008. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci U S A 105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webster RH, Sil A. 2008. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc Natl Acad Sci U S A 105:14573–14578. doi: 10.1073/pnas.0806221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin X. 2009. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect Genet Evol 9:401–416. doi: 10.1016/j.meegid.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Zimmer BL, Hempel HO, Goodman NL. 1983. Pathogenicity of the hyphae of Filobasidiella neoformans. Mycopathologia 81:107–110. doi: 10.1007/BF00436987. [DOI] [PubMed] [Google Scholar]
- 16.Shadomy HJ, Utz JP. 1966. Preliminary studies on a hypha-forming mutant of Cryptococcus neoformans. Mycologia 58:383–390. doi: 10.2307/3756912. [DOI] [PubMed] [Google Scholar]
- 17.Shadomy HJ, Lurie HI. 1971. Histopathological observations in experimental cryptococcosis caused by a hypha-producing strain of Cryptococcus neoformans (Coward strain) in mice. Sabouraudia 9:6–9. doi: 10.1080/00362177185190031. [DOI] [PubMed] [Google Scholar]
- 18.Lurie HI, Shadomy HJ. 1971. Morphological variations of a hypha-forming strain of Cryptococcus neoformans (Coward strain) in tissues of mice. Sabouraudia 9:10–14. [DOI] [PubMed] [Google Scholar]
- 19.Chung S, Mondon P, Chang YC, Kwon-Chung KJ. 2003. Cryptococcus neoformans with a mutation in the tetratricopeptide repeat-containing gene, CCN1, causes subcutaneous lesions but fails to cause systemic infection. Infect Immun 71:1988–1994. doi: 10.1128/IAI.71.4.1988-1994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandhorst TT, Roy R, Wüthrich M, Nanjappa S, Filutowicz H, Galles K, Tonelli M, McCaslin DR, Satyshur K, Klein B. 2013. Structure and function of a fungal adhesin that binds heparin and mimics thrombospondin-1 by blocking T cell activation and effector function. PLoS Pathog 9:e1003464. doi: 10.1371/journal.ppat.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wüthrich M, Filutowicz HI, Klein BS. 2000. Mutation of the WI-1 gene yields an attenuated Blastomyces dermatitidis strain that induces host resistance. J Clin Invest 106:1381–1389. doi: 10.1172/JCI11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wüthrich M, Warner T, Klein BS. 2005. IL-12 is required for induction but not maintenance of protective, memory responses to Blastomyces dermatitidis: implications for vaccine development in immune-deficient hosts. J Immunol 175:5288–5297. doi: 10.4049/jimmunol.175.8.5288. [DOI] [PubMed] [Google Scholar]
- 23.Wüthrich M, Gern B, Hung CY, Ersland K, Rocco N, Pick-Jacobs J, Galles K, Filutowicz H, Warner T, Evans M, Cole G, Klein B. 2011. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J Clin Invest 121:554–568. doi: 10.1172/JCI43984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wüthrich M, Krajaejun T, Shearn-Bochsler V, Bass C, Filutowicz HI, Legendre AM, Klein BS. 2011. Safety, tolerability, and immunogenicity of a recombinant, genetically engineered, live-attenuated vaccine against canine blastomycosis. Clin Vaccine Immunol 18:783–789. doi: 10.1128/CVI.00560-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wüthrich M, Ersland K, Sullivan T, Galles K, Klein BS. 2012. Fungi subvert vaccine T cell priming at the respiratory mucosa by preventing chemokine-induced influx of inflammatory monocytes. Immunity 36:680–692. doi: 10.1016/j.immuni.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wüthrich M, Brandhorst TT, Sullivan TD, Filutowicz H, Sterkel A, Stewart D, Li M, Lerksuthirat T, LeBert V, Shen ZT, Ostroff G, Deepe GS Jr, Hung CY, Cole G, Walter JA, Jenkins MK, Klein B. 2015. Calnexin induces expansion of antigen-specific CD4+ T cells that confer immunity to fungal ascomycetes via conserved epitopes. Cell Host Microbe 17:452–465. doi: 10.1016/j.chom.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Filler SG. 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards JE., Jr 2012. Fungal cell wall vaccines: an update. J Med Microbiol 61:895–903. doi: 10.1099/jmm.0.041665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L, Niang R, Casadevall A. 2001. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107:E66. doi: 10.1542/peds.107.5.e66. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Hermoso D, Janbon G, Dromer F. 1999. Epidemiological evidence for dormant Cryptococcus neoformans infection. J Clin Microbiol 37:3204–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dromer F, Ronin O, Dupont B. 1992. Isolation of Cryptococcus neoformans var. gattii from an Asian patient in France: evidence for dormant infection in healthy subjects. J Med Vet Mycol 30:395–397. doi: 10.1080/02681219280000511. [DOI] [PubMed] [Google Scholar]
- 32.Lin X, Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol 60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 33.Goldman DL, Lee SC, Mednick AJ, Montella L, Casadevall A. 2000. Persistent Cryptococcus neoformans pulmonary infection in the rat is associated with intracellular parasitism, decreased inducible nitric oxide synthase expression, and altered antibody responsiveness to cryptococcal polysaccharide. Infect Immun 68:832–838. doi: 10.1128/IAI.68.2.832-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casadevall A, Perfect JR. 1998. Cryptococcus neoformans. ASM Press, Washington, DC. [Google Scholar]
- 35.Bicanic T, Harrison TS. 2004. Cryptococcal meningitis. Br Med Bull 72:99–118. doi: 10.1093/bmb/ldh043. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell TG, Perfect JR. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev 8:515–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, Kwon-Chung KJ. 2008. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis 14:755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain N, Wickes BL, Keller SM, Fu J, Casadevall A, Jain P, Ragan MA, Banerjee U, Fries BC. 2005. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J Clin Microbiol 43:5733–5742. doi: 10.1128/JCM.43.11.5733-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang W, Fa Z, Liao W. 2015. Epidemiology of Cryptococcus and cryptococcosis in China. Fungal Genet Biol 78:7–15. doi: 10.1016/j.fgb.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Arora S, Hernandez Y, Erb-Downward JR, McDonald RA, Toews GB, Huffnagle GB. 2005. Role of IFN-gamma in regulating T2 immunity and the development of alternatively activated macrophages during allergic bronchopulmonary mycosis. J Immunol 174:6346–6356. doi: 10.4049/jimmunol.174.10.6346. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Wang F, Tompkins KC, McNamara A, Jain AV, Moore BB, Toews GB, Huffnagle GB, Olszewski MA. 2009. Robust Th1 and Th17 immunity supports pulmonary clearance but cannot prevent systemic dissemination of highly virulent Cryptococcus neoformans H99. Am J Pathol 175:2489–2500. doi: 10.2353/ajpath.2009.090530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voelz K, Lammas DA, May RC. 2009. Cytokine signaling regulates the outcome of intracellular macrophage parasitism by Cryptococcus neoformans. Infect Immun 77:3450–3457. doi: 10.1128/IAI.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piehler D, Stenzel W, Grahnert A, Held J, Richter L, Köhler G, Richter T, Eschke M, Alber G, Müller U. 2011. Eosinophils contribute to IL-4 production and shape the T-helper cytokine profile and inflammatory response in pulmonary cryptococcosis. Am J Pathol 179:733–744. doi: 10.1016/j.ajpath.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandez Y, Arora S, Erb-Downward JR, McDonald RA, Toews GB, Huffnagle GB. 2005. Distinct roles for IL-4 and IL-10 in regulating T2 immunity during allergic bronchopulmonary mycosis. J Immunol 174:1027–1036. doi: 10.4049/jimmunol.174.2.1027. [DOI] [PubMed] [Google Scholar]
- 46.Olszewski MA, Huffnagle GB, McDonald RA, Lindell DM, Moore BB, Cook DN, Toews GB. 2000. The role of macrophage inflammatory protein-1 alpha/CCL3 in regulation of T cell-mediated immunity to Cryptococcus neoformans infection. J Immunol 165:6429–6436. doi: 10.4049/jimmunol.165.11.6429. [DOI] [PubMed] [Google Scholar]
- 47.Olszewski MA, Huffnagle GB, Traynor TR, McDonald RA, Cook DN, Toews GB. 2001. Regulatory effects of macrophage inflammatory protein 1alpha/CCL3 on the development of immunity to Cryptococcus neoformans depend on expression of early inflammatory cytokines. Infect Immun 69:6256–6263. doi: 10.1128/IAI.69.10.6256-6263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiesner DL, Specht CA, Lee CK, Smith KD, Mukaremera L, Lee ST, Lee CG, Elias JA, Nielsen JN, Boulware DR, Bohjanen PR, Jenkins MK, Levitz SM, Nielsen K. 2015. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS Pathog 11:e1004701. doi: 10.1371/journal.ppat.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet 6:e1000953. doi: 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Zhai B, Lin X. 2012. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog 8:e1002765. doi: 10.1371/journal.ppat.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhai B, Wu C, Wang L, Sachs MS, Lin X. 2012. The antidepressant sertraline provides a promising therapeutic option for neurotropic cryptococcal infections. Antimicrob Agents Chemother 56:3758–3766. doi: 10.1128/AAC.00212-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Heeckeren AM, Tscheikuna J, Walenga RW, Konstan MW, Davis PB, Erokwu B, Haxhiu MA, Ferkol TW. 2000. Effect of Pseudomonas infection on weight loss, lung mechanics, and cytokines in mice. Am J Respir Crit Care Med 161:271–279. doi: 10.1164/ajrccm.161.1.9903019. [DOI] [PubMed] [Google Scholar]
- 53.Wozniak KL, Hardison SE, Kolls JK, Wormley FL. 2011. Role of IL-17A on resolution of pulmonary C. neoformans infection. PLoS One 6:e17204. doi: 10.1371/journal.pone.0017204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardison SE, Wozniak KL, Kolls JK, Wormley FL Jr. 2010. Interleukin-17 is not required for classical macrophage activation in a pulmonary mouse model of Cryptococcus neoformans infection. Infect Immun 78:5341–5351. doi: 10.1128/IAI.00845-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Tian X, Gyawali R, Upadhyay S, Foyle D, Wang G, Cai JJ, Lin X. 2014. Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Pathog 10:e1004185. doi: 10.1371/journal.ppat.1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Tian X, Gyawali R, Lin X. 2013. Fungal adhesion protein guides community behaviors and autoinduction in a paracrine manner. Proc Natl Acad Sci U S A 110:11571–11576. doi: 10.1073/pnas.1308173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy JW, Schafer F, Casadevall A, Adesina A. 1998. Antigen-induced protective and nonprotective cell-mediated immune components against Cryptococcus neoformans. Infect Immun 66:2632–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy KM, Reiner SL. 2002. The lineage decisions of helper T cells. Nat Rev Immunol 2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 59.O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. 2013. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. mBio 4:e00522-12. doi: 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wormley FL Jr, Cox GM, Perfect JR. 2005. Evaluation of host immune responses to pulmonary cryptococcosis using a temperature-sensitive C. neoformans calcineurin A mutant strain. Microb Pathog 38:113–123. doi: 10.1016/j.micpath.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Wormley FL Jr, Perfect JR, Steele C, Cox GM. 2007. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect Immun 75:1453–1462. doi: 10.1128/IAI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hardison SE, Ravi S, Wozniak KL, Young ML, Olszewski MA, Wormley FL Jr. 2010. Pulmonary infection with an interferon-gamma-producing Cryptococcus neoformans strain results in classical macrophage activation and protection. Am J Pathol 176:774–785. doi: 10.2353/ajpath.2010.090634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perfect JR, Lang SD, Durack DT. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol 101:177–194. [PMC free article] [PubMed] [Google Scholar]
- 65.Zhai B, Zhu P, Foyle D, Upadhyay S, Idnurm A, Lin X. 2013. Congenic strains of the filamentous form of Cryptococcus neoformans for studies of fungal morphogenesis and virulence. Infect Immun 81:2626–2637. doi: 10.1128/IAI.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hardison SE, Herrera G, Young ML, Hole CR, Wozniak KL, Wormley FL Jr. 2012. Protective immunity against pulmonary cryptococcosis is associated with STAT1-mediated classical macrophage activation. J Immunol 189:4060–4068. doi: 10.4049/jimmunol.1103455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fungal burden of lungs infected with 105 WT H99 or the PGPD1-ZNF2 strain on day 1, day 7, and day 12 postinoculation (four mice in each group). Download
The protective effect of vaccination by the PGPD1-ZNF2 strain is dose dependent in preliminary tests (four mice in each group). Download
Lung and brain fungal burdens of the animals infected with the PGPD1-ZNF2 strain on day 60 postinfection. The red line shows the initial inoculation dose of cryptococcal cells in lungs. The two asterisks indicate two individual mice that relapsed. Download
No protection effect of heat-killed PGPD1-ZNF2 at a dose of 1 × 104 or 1 × 105 CFU/animal (data shown are from one experiment with eight mice per group). The two experimental groups displayed the exact same dynamics in survival, so the curves overlapped. Download
Primers and cryptococcal strains used in this study.