ABSTRACT
The herpes simplex virus 1 (HSV-1) capsid is a massive particle (~200 MDa; 1,250-Å diameter) with T=16 icosahedral symmetry. It initially assembles as a procapsid with ~4,000 protein subunits of 11 different kinds. The procapsid undergoes major changes in structure and composition as it matures, a process driven by proteolysis and expulsion of the internal scaffolding protein. Assembly also relies on an external scaffolding protein, the triplex, an α2β heterotrimer that coordinates neighboring capsomers in the procapsid and becomes a stabilizing clamp in the mature capsid. To investigate the mechanisms that regulate its assembly, we developed a novel isolation procedure for the metastable procapsid and collected a large set of cryo-electron microscopy data. In addition to procapsids, these preparations contain maturation intermediates, which were distinguished by classifying the images and calculating a three-dimensional reconstruction for each class. Appraisal of the procapsid structure led to a new model for assembly; in it, the protomer (assembly unit) consists of one triplex, surrounded by three major capsid protein (MCP) subunits. The model exploits the triplexes’ departure from 3-fold symmetry to explain the highly skewed MCP hexamers, the triplex orientations at each 3-fold site, and the T=16 architecture. These observations also yielded new insights into maturation.
IMPORTANCE
This paper addresses the molecular mechanisms that govern the self-assembly of large, structurally complex, macromolecular particles, such as the capsids of double-stranded DNA viruses. Although they may consist of thousands of protein subunits of many different kinds, their assembly is precise, ranking them among the largest entities in the biosphere whose structures are uniquely defined to the atomic level. Assembly proceeds in two stages: formation of a precursor particle (procapsid) and maturation, during which major changes in structure and composition take place. Our analysis of the HSV procapsid by cryo-electron microscopy suggests a hierarchical pathway in which multisubunit “protomers” are the building blocks of the procapsid but their subunits are redistributed into different subcomplexes upon being incorporated into a nascent procapsid and are redistributed again in maturation. Assembly is a highly virus-specific process, making it a potential target for antiviral intervention.
INTRODUCTION
As with other herpesviruses, the virion of herpes simplex virus 1 (HSV-1) is composed of a nucleocapsid that is surrounded by an amorphous layer of proteins called the tegument and enclosed in an envelope studded with glycoproteins (1, 2). The capsid is first assembled as a precursor particle or procapsid that undergoes irreversible changes in structure and composition as it matures. In this, as in other distinctive features, herpesvirus capsid assembly resembles that of tailed double-stranded DNA (dsDNA) bacteriophages, suggesting common evolutionary origins (3–7).
The HSV-1 procapsid has a spherical shell that consists of 150 hexamers and 11 pentamers of the major capsid protein (MCP), 320 triplexes, and a single dodecamer of the portal protein. It overlies a thick-walled (~250-Å) inner shell (8, 9) made up of ~1,900 copies of the scaffolding protein (10) (see Fig. S1 [inset] in the supplemental material). About 10% of the scaffolding subunits have the viral protease and a linker fused to their N terminus. In maturation, the protease is activated and processes its polyprotein and the scaffolding protein. As DNA is packaged, the processed scaffolding protein is expelled and the capsid morphology converts from spherical to polyhedral (i.e., its facets flatten). Although the protease is required if a procapsid is to mature, and the portal and terminase complex (the DNA packaging motor) are essential if DNA is to be packaged, four proteins suffice to produce a correctly formed T=16 procapsid shell. These proteins are the MCP, the two triplex subunits, and the scaffolding protein. Their morphogenetic mechanism is the main focus of the present study.
Triplexes consist of two UL18 subunits (34 kDa) and one UL38 subunit (50 kDa), and they occupy all 3-fold positions (local and global) on the icosahedral surface lattice, where they coordinate interactions between hexamers and pentamers of the MCP. These capsomers are separated by 20-Å gaps (9, 11), but in maturation, direct contacts are established between them as the contiguous capsid “floor” is established. In the mature capsid, the triplexes remain associated with the 3-fold lattice sites, where they now serve as stabilizing clamps, similarly to gpD of phage lambda (12, 13) or gp.soc of phage T4 (14–16).
Recently, several herpesvirus capsid reconstructions have been reported at resolutions below 10 Å (17–19). These results were achieved by imaging mature, DNA-filled capsids inside intact virions. However, mature capsid structures provide little direct information on assembly, on account of the structural changes that take place during maturation. Earlier reconstructions of the procapsid and maturation intermediates have been limited to resolutions of 20 to 30 Å (11, 20). In large part, this barrier has been attributable to difficulty in isolating procapsids, which are extremely labile; moreover, they are metastable and tend to embark on maturation, even in the absence of proteolytic activity (21, 22). This gives rise to structural heterogeneity that limits the resolution of cryo-electron microscopy (cryo-EM) reconstructions.
In the present study, we sought to achieve a more detailed structural account of the procapsid and maturation intermediates. To this end, we developed a new isolation procedure and exploited recent technical developments in cryo-EM to collect a large data set of 12,000 micrographs (~100,000 particles) on a microscope equipped with a “direct detector” camera and automated data collection software (see Materials and Methods). As analysis proceeded, it became apparent that many of the particles exhibited visible departures from icosahedral symmetry. The remaining particles were classified. In addition to the naive procapsid, four other states were sufficiently populated to yield reconstructions at resolutions of 11 Å to 16 Å. In addition, cryo-electron tomography was used to investigate the structure of the internal scaffolding shell. These data have led to a new model of procapsid assembly based on a protomer (assembly unit) consisting of one triplex plus three MCP subunits. As assembly proceeds, the three MCP subunits from a given protomer are incorporated into three neighboring capsomers (each a hexamer or pentamer of the MCP). The scaffolding shell guides the curvature of the growing surface shell. This model rationalizes certain departures from symmetry in the procapsid structure—viz., the intrinsic asymmetry of the triplexes and the distortions of the MCP hexamers—and assigns them roles in specifying the T=16 geometry of the procapsid.
RESULTS
A gentle protocol for procapsid purification.
Procapsids were produced using the m100 virus mutant, which lacks the viral protease (23). Alternatively, the temperature-sensitive protease mutant ts.Prot.A can be used (24). With circumspect handling, procapsids can be extracted from infected cells, but they are extremely labile and do not withstand any gradient purification. Previously, in order to obtain adequate yields of procapsids, a monoclonal antibody against the MCP was used to concentrate the particles (24). However, this procedure results in aggregation in three dimensions and a low yield of particles suitably distributed on cryo-EM grids. To tackle this problem, we developed a new purification protocol (see Materials and Methods); in brief, we optimized a differential centrifugation procedure using a combination of gentle pelleting and filtering to obtain a concentrated suspension of procapsids from nuclear lysates (see Materials and Methods). The sample was harvested at 12 to 14 h postinfection and frozen immediately after purification, at about 18 to 20 h postinfection.
Five relatively long lived maturation intermediates.
When prepared for cryo-EM, these isolates gave monolayer distributions of procapsids suitable for automated data collection. However, many (40 to 50%) of the particles were visibly distorted (see Fig. S1 in the supplemental material). Discarding them left a total of ~50,000 particles, and these were subjected to iterative classification. In this procedure, not only must different views be identified but also different conformers, each represented by a current density map. In a given cycle, each particle is assigned to the reference map with which it has the highest correlation. A new set of reference maps is then calculated, and the procedure is repeated until convergence. We started with the 17 previously reported intermediates (11, 25) as reference maps. However, after three cycles of classification and reconstruction, most classes were sparsely populated, limiting these reconstructions to low resolution. Accordingly, the number of classes was reduced to 10 and then further reduced to five. In each class, the top 50% of particles, as ranked by correlation coefficients, were used to calculate the reconstruction. This strategy led to a distribution with 3,000 to 6,000 particles per class and reconstructions with resolutions of 11 Å to 16 Å (Table 1). It is likely that the number of distinct intermediates is greater than five, but the density maps described here (Fig. 1) represent relatively stable (i.e., long-lived) staging posts on the maturation pathway. The sequential ordering of the five reconstructions is based on the time course experiment previously reported (11) in which the waxing and waning of the various classes of procapsid were monitored. The improved resolution made it possible to segment the maps into their molecular constituents and to place the crystal structure of the HK97 capsid protein in the MCP floor domains. This analysis yielded new information about the movements of the capsid proteins during maturation (see below).
TABLE 1 .
Statistics for the five density maps
| Map no. | Resolution (Å) | No. of particles |
|---|---|---|
| 1 (procapsid) | 14 | 3,881 |
| 2 | 16 | 6,286 |
| 3 | 15 | 4,175 |
| 4 | 12 | 5,096 |
| 5 (capsid) | 11 | 3,530 |
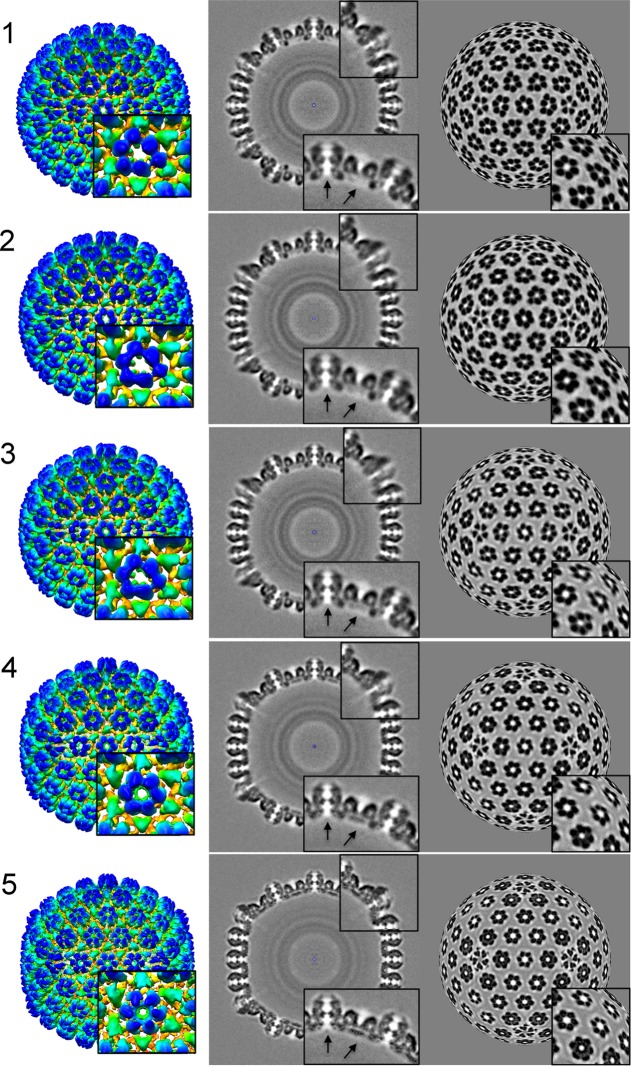
FIG 1 .
Structures of five intermediates (1 to 5) in HSV-1 procapsid maturation. Map 1 represents the earliest procapsid captured, and map 5 shows an almost-mature capsid. The capsids are viewed along a 2-fold axis. (Left) Rendering of the outer surface, color-coded radially from yellow to blue. The procapsid is 1,250 Å in diameter. The blow-up at bottom right is centered on the P-hexamer, i.e., the hexamer closest to the vertex. (Middle) Central sections. The blow-ups are centered on the pentamer (top) and the E-hexamer (edge hexamer) and P-hexamer (bottom). The E-hexamer is centered on a 2-fold axis at the middle of an edge. The two arrows point to a region where substantial changes take place in the transcapsomer pore (left) and the initially empty region which is filled in by “floor” density in the mature capsid. (Right) Concentric spherical layers of density inside the capsid correspond to regions occupied by different segments of the scaffolding protein. (Right) Spherical sections at a radius of 600 Å, illustrating how the major departures from 6-fold symmetry in the earliest hexamers are resolved (i.e., become symmetric) in the mature capsid.
Shifts in the positions and orientations of MCP subunits during procapsid maturation.
Maturation of the HSV-1 capsid is very similar to that of dsDNA bacteriophages, including HK97, T4, and P22, among others (reviewed in reference 26). All of these viruses have capsid proteins based on the same distinctive fold, termed the HK97 fold after the system in which it was discovered (27). This fold is embellished with an N-terminal scaffolding domain in HK97, with insertion domains in T4 (28) and P22 (29) and with a large (120-kDa) C-terminal appendage in HSV-1 (30) (Fig. 1 to 3). In the HSV-1 MCP, the “floor” domain has the HK97 fold while the “tower” appendage, with its middle and tip domains, protrudes outward (Fig. 2). A crystal structure has been determined for the tip domain (604 amino acids [aa] out of 1,374 total) (31).
FIG 3 .
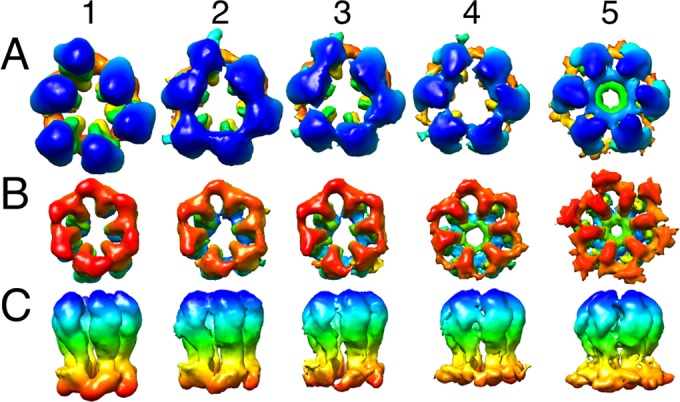
Rearrangement of the P-hexamer during maturation. The panels show segmented hexamers from maps 1 to 5. (A) View from the outside. (B) View from inside the capsid. (C) Side view. The radius-dependent color coding is as in Fig. 2. The initial major departure from 6-fold symmetry in the external protrusions (blue [A]) is less evident in the floor domains (red [B]).
FIG 2 .
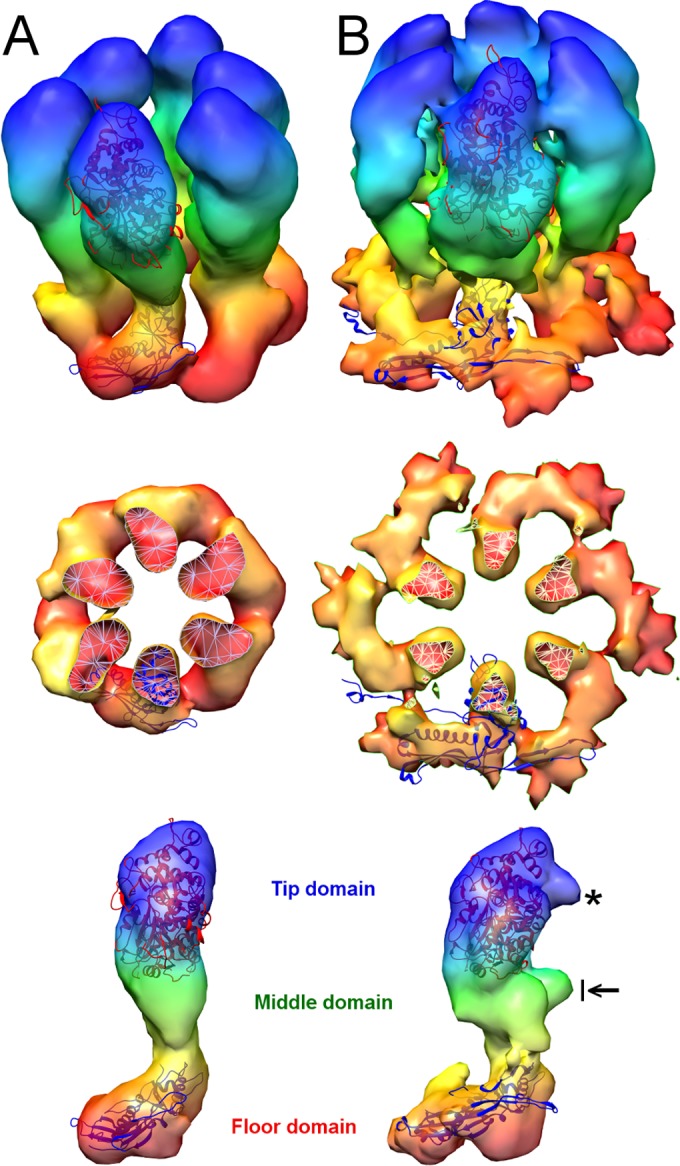
Conformational changes in the P-hexamer during maturation. (A) Image from map 1. (B) Image from map 5. Radial color coding goes from red (inner surface) to blue (outer surface). The crystal structure of the tip domain (PDB 1N07) fits well into both density maps, indicating that the tip and middle domains rotate as a single rigid body during this transition. HK97 capsid protein structures have been fitted into the floor domains. Prohead I (the HK97 procapsid; PDB 3QPR) was used for map 1, and head II (mature capsid; PDB 2FT1) was used for map 5. The middle panels show cutaway views down the 6-fold axis and highlight the rotation and outward movement of the floor domains. At bottom are side views of the MCP subunit, giving its domain organization. The black arrow points to the “drawbridge” domain, an outcrop of density from the middle domain. The asterisk points out a protrusion in the tip domain, an MCP-MCP interaction region in a mature capsomer.
In the procapsid, the six MCP subunits per hexamer are distributed asymmetrically around its central axis. There are three different kinds of hexamers, distinguished by their positions on the icosahedral surface lattice—P (peripentonal), E (edge), and C (central)—and by their structures (Fig. 4). P-hexamers have a particularly distorted appearance (Fig. 1 to 3). With the current density maps, it was possible to segment capsomers into individual subunits and show that, despite the asymmetry of a given hexamer, its six MCP subunits are consistent in shape, and it is their positioning, i.e., rigid body-like shifts and tilts relative to a 6-fold-symmetric ring, that is responsible for the observed departures from 6-fold symmetry (Fig. 2 and 3).
FIG 4 .
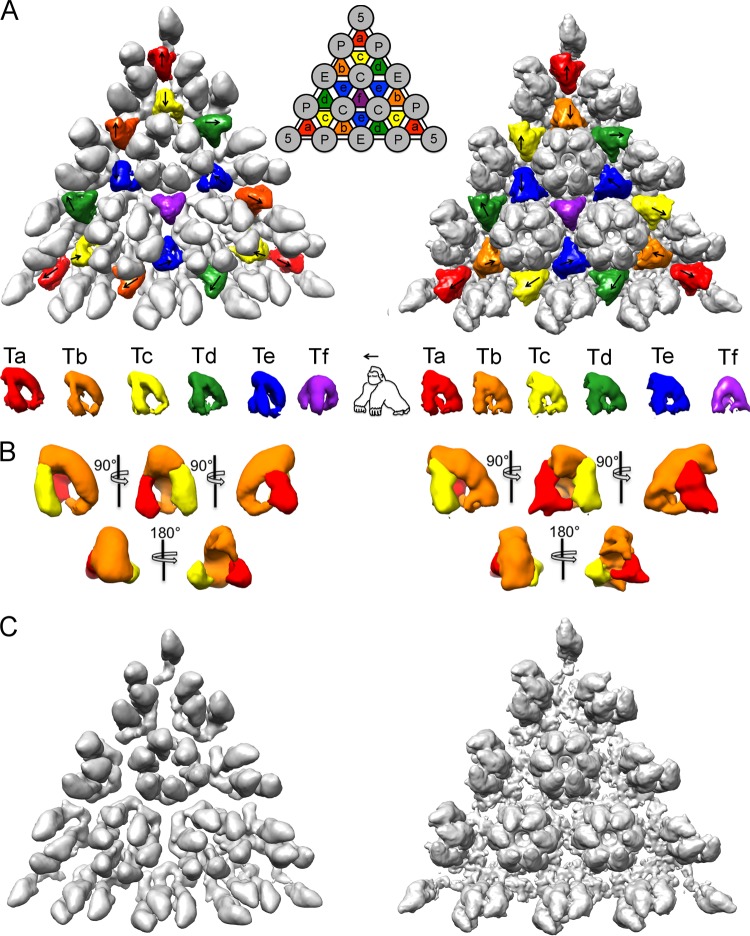
Structure and orientations of the triplexes in the procapsid and the mature capsid. (A) An icosahedral facet from map 1 is shown at left, and one from map 5 is shown at right. The six quasiequivalent triplexes, Ta through Tf (nomenclature according to Heymann et al. [11]), segmented out from the respective maps, are aligned in a row underneath. Morphologically, the triplex resembles a gorilla. In the upper panels, an arrow indicates the directionality of each triplex. (B) Several views of the averaged procapsid triplex (left) and the mature capsid triplex (right). At the current resolutions, the various triplexes in each capsid are close to isomorphous, so they could be averaged together without loss of information. Similarly, they change little during maturation. The averaged triplexes were segmented into three regions that were assigned to two UL18 subunits (yellow and red) and one UL38 subunit (orange) (see text). (C) Monochrome view of the same triangular facets as in panel A but with the triplexes excised, highlighting the gaps between adjacent capsomers in the procapsid and the extent of floor reorganization during maturation.
In state 1 (of 5), putatively the earliest procapsid, the majority of intersubunit contacts within a capsomer are between adjacent floor domains. During maturation, MCP subunits swivel so that some contacts between neighboring floor domains within a hexamer are broken and some new interactions are engaged (Fig. 2 and 3). These rearrangements also result in formation of the contiguous floor (Fig. 4C), in which adjacent capsomers are connected. In contrast to the hexamers, the pentamers do not change during maturation and the disposition of their floor domains remains essentially the same throughout the transition. However, pushed by reorganization of the surrounding hexamers, the pentamers move radially, as the particle transforms into the angular (polyhedral) form of the mature capsid (see Movie S1 in the supplemental material).
In summary, several striking changes take place in the maturing surface shell. In addition to formation of the floor, the hexamers become 6-fold symmetric, the axial pores through the capsomers narrow down, and the capsid angularizes. Movements of the “drawbridge” domains constrict the pore (the drawbridge domain is an outcrop of the middle domain [Fig. 2B, arrow]), and the outermost “tip” domains move closer together, narrowing this part of the pore (Fig. 1 and 2). Both of the latter changes take place relatively late in maturation and are most apparent in maps 4 and 5.
As the procapsid matures, the triplexes change their interactions but not their shape.
Despite the many maturation-related behaviors that it shares with phages, there is no overall expansion of the HSV-1 procapsid. In effect, this procapsid is preexpanded, with the triplexes acting as spacers between capsomers (Fig. 4C). Just as the increase in size of maturing phage capsids is primarily due to rotations of the MCP subunits (32), those of the maturing HSV-1 procapsid also undergo substantial rotations as the floor domains move into the spaces underlying the triplexes (Fig. 4C).
There are six quasiequivalent triplexes per asymmetric unit of the T=16 surface lattice, labeled Ta to Tf in the schematic insert in Fig. 4A. Due to the imposition of icosahedral symmetry in the reconstructions, the Tf triplex centered on the 3-fold axis is (artifactually) 3-fold symmetric. The other five triplexes exhibit a pronounced directionality, i.e., departure from 3-fold symmetry, and have defined orientations relative to the frame of reference given by an icosahedral facet, marked with arrows in Fig. 4A (left panel). An identical pattern of triplex directionality is observed in the mature capsid (Fig. 4A, right panel). (Although, to our knowledge, this pattern has not been previously described, it has also been observed independently by J. F. Conway [personal communication].) It follows that triplex directionality is specified during procapsid assembly and does not change upon maturation. We propose below (Discussion) an assembly model in which the directionality of the triplexes plays a key role.
At the current resolution, triplexes Ta to Te are closely similar in structure (they can be superimposed pairwise with correlation coefficients of >0.97). Moreover, they do not change perceptibly as the procapsid matures (Fig. 4A [bottom row] and 4B). In overall morphology, the triplex resembles a “gorilla,” with two similar “arms” that we assign to the two UL18 subunits and a “back” and “head” that we assign to UL38 (Fig. 4B). Otherwise described, the triplex has the form of an asymmetric tripod. We are not yet able to delineate the connection between the two UL18 subunits, which, on their own, have been shown to dimerize in vitro (33). However, the UL18 subunits probably extend from the “arms” into the “head” region and make contact with each other. This assignment is consistent with previous triplex segmentations in mature-state reconstructions of several different herpesviruses (17–19). Moreover, the UL38 homologs are smaller in cytomegalovirus (CMV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) than in HSV-1 (~35 kDa versus ~50 kDa), and the triplex density in those capsids lacks the “head” of the HSV-1 “gorilla,” consistent with the assignment given here.
Although there is little variation in triplex structure, there are substantial changes in their interactions with the surrounding MCPs as the procapsid matures. Appraisal of the points of contact between triplexes and MCPs reveals interaction areas in addition to the four previously described (11) (Fig. 5). In the procapsid, these are c1 to c3 (named consistently with Fig. 6 of reference 11) between the triplex and the middle domain of neighboring MCPs. During maturation, the c2 connection is severed due to the increased separation of the MCPs, and a new connection, c4, is formed next to c3 on the same MCP. In addition, as maturation proceeds, the three subunits of a given triplex contact the floor domains of the three surrounding MCPs, establishing connections c5 to c7 (Fig. 5).
FIG 5 .
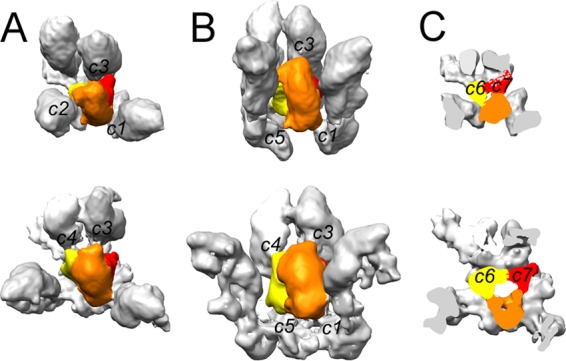
Contacts between the Tc triplex and surrounding MCPs. MCPs are in gray, and the triplexes are segmented into the two UL18 subunits (yellow and red) and one UL38 subunit (orange). The top views (A), side views (B), and cutaway views (C) of the triplex with surrounding MCPs are shown for the procapsid (map 1, top) and the nearly mature capsid (map 5, bottom). c1 through c7 denote intermolecular contacts (see text).
A periodicity in the scaffolding shell.
Formation of the procapsid is also guided by coassembly of the outer shell with an inner shell of scaffolding proteins. (Parenthetically, we note that bacteriophage ϕX174, a much smaller [T=1] particle, also has inner and outer scaffolding proteins [34], but there is little reason to suppose that they operate as in the HSV-1 system.) The protease, located at the N-terminal end of the polyprotein, protrudes into the cavity inside the inner shell (10). The inner shell is thick-walled (~250 Å; we equate this dimension with the length of the scaffolding protein), and it appears in the reconstructions as a set of concentric spherical shells (Fig. 1, middle column). The densest shell peaks at a radius of about 280 Å, and this is likely to be the site of predominant nearest-neighbor interactions. However, the shells disclose no information about the in-plane packing of scaffolding protein protomers.
To explore the possibility that offsets in register between the outer and inner shells may be responsible for lateral (in-plane) smearing of inner shell density, we addressed this problem by cryo-electron tomography, which affords density maps of individual particles (1). Some 700 procapsids were extracted from 34 tomograms, their icosahedral orientations were determined, and the maps were averaged. However, no substructure was observed in the inner shell, other than the observed spherical stratification (Fig. 6, top). Since these alignments were dominated by the MCP/triplex shell (as in the cryo-EM reconstructions), we masked it out and aligned the remaining inner shells relative to each other, without applying symmetry. The top correlation-ranked 50% of these reconstructions were combined to give a final rendering of the inner shell (Fig. 6, bottom). As in the cryo-EM reconstructions, the densest feature is a layer at a radius of ~250 Å, but again, there is no structural differentiation within this layer. However, the region outside this layer is resolved into two strata: in the outer one, at a radius of ~450 Å, an 80-Å in-plane repeat is resolved (arrows in Fig. 6, bottom). The possible significance of this repeat is considered further in the Discussion.
FIG 6 .
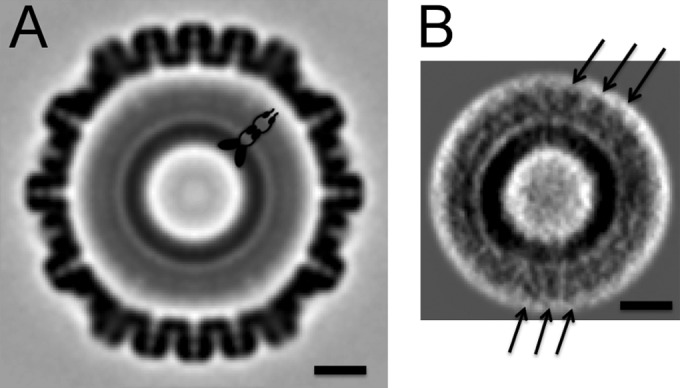
Tomographic reconstruction of the HSV-1 procapsid with and without symmetrization. Left, central section of procapsid map obtained by averaging and symmetrizing multiple tomograms. A schematic model of an elongated scaffolding protein dimer is drawn. Right, central section through the scaffolding core reconstruction obtained by averaging without applying symmetry. The arrows indicate 8-nm repeats in the outer part of the scaffolding shell. Bars, 20 nm.
DISCUSSION
Particle stability and conformational diversity.
In this study, we produced preparations of HSV-1 procapsids that made suitable specimens for high-throughput cryo-EM. However, a substantial fraction turned out to be distorted and could not be used for reconstruction. These distortions may reflect the response of fragile particles to physical stresses (e.g., surface tension), and/or they may represent particles in which icosahedral symmetry is not maintained throughout maturation but the transformation propagates out from an initiation site or sites. (Despite the absence of protease, structural transformation initiates spontaneously over a period of hours to days in a stochastic process. It is likely that DNA packaging in vivo causes capsid maturation to proceed more rapidly.) In our earlier work in which an antibody was used to concentrate the procapsids, there was a lower incidence of distorted particles and late-stage intermediates (9, 24). In retrospect, it may be that the antibody had a beneficial stabilizing effect on the procapsids and may also have restrained them from embarking on maturation.
In an earlier analysis, 5,000 procapsid images were divided into 17 classes (11). However, most of these classes were sparsely populated and could yield only low-resolution reconstructions. In the present analysis, we started with ~100,000 procapsids and the 17 previously reported models. As before, the exact number of distinct classes could not be rigorously determined. As a pragmatic measure intended to maximize resolution, we reduced the number of classes and increased the numbers of particles in them, recognizing that this may concede some heterogeneity on a scale too small to be evident from visual inspection. We finished up with five classes and resolutions of 11 to 16 Å, approximately twice as high as in previous work. In the following discussion, we assume that state 1 represents the naive procapsid, state 5 represents the nearly mature state, and states 2 to 4 represent relatively long-lived intermediates, in exploring their implications for the assembly mechanism and maturation dynamics.
A model for HSV-1 procapsid assembly.
Assembly of an HSV-1 procapsid entails the ordered aggregation of some 4,000 protein subunits (reviewed in reference 35). For a fully fledged procapsid, these are of 11 different kinds, but just four gene products—the MCP, the two triplex subunits, and a scaffolding protein—suffice to produce a basic procapsid with a geometrically correct T=16 shell. With complex assembly systems, an efficient strategy is first to produce subassemblies (protomers), which then assemble into higher-order structures. This modus operandi has been demonstrated in the relatively simple case of the HK97 procapsid (a T=7 particle), where the protomers are hexamers and pentamers of the same protein, an MCP-scaffolding protein fusion (36, 37). The proposition that a scenario of this kind applies to the HSV-1 procapsid raises the following questions. (i) How many different kinds of protomers are involved? (ii) How many conformational variants of each protomer are there in the preassembly pool? (iii) In what order are the protomers incorporated into nascent particles? (In principle, the fewer the protomers, the less the need for regulatory supervision.) While hexamers and pentamers of the MCP are natural candidates, this model is undermined by two considerations: the fact that these capsomers do not make nearest-neighbor contacts in the procapsid (Fig. 4C) and the extreme variations in structure exhibited by assembled hexamers. On the other hand, several considerations suggest that the triplexes play an important role: they are essential for correct assembly (21, 22), they occupy strategic positions in the procapsid surface lattice, and the quasiequivalent versions are all closely similar in structure (Fig. 4A).
Drawing primarily on observed features of the procapsid, we propose that the assembly unit (protomer) consists of a triplex surrounded by three MCP subunits and that the T=16 procapsid shell is built from 320 copies of this protomer (Fig. 7). To date, such a protomer has not been isolated outside the context of a capsid. Each icosahedral facet accommodates 16 protomers of six quasiequivalent kinds: three each for Ta to Te and one for Tf. (We name the protomers according to the triplexes that they contain.) Despite little sequence similarity, we infer that there is enough structural similarity between UL38 and UL18 that both proteins can bind an MCP subunit (hence, three per triplex) but also sufficient difference between them for the protomers to depart significantly from 3-fold symmetry. Strikingly, four protomers (Ta, Tb, Tc, and Td) are essentially superimposable (they align with pairwise correlations of >0.95 [see Fig. S2 in the supplemental material]). As the Te protomer includes an MCP that is part of the 2-fold-symmetrized E-hexamer, it is also compromised by the application of icosahedral symmetry.
FIG 7 .
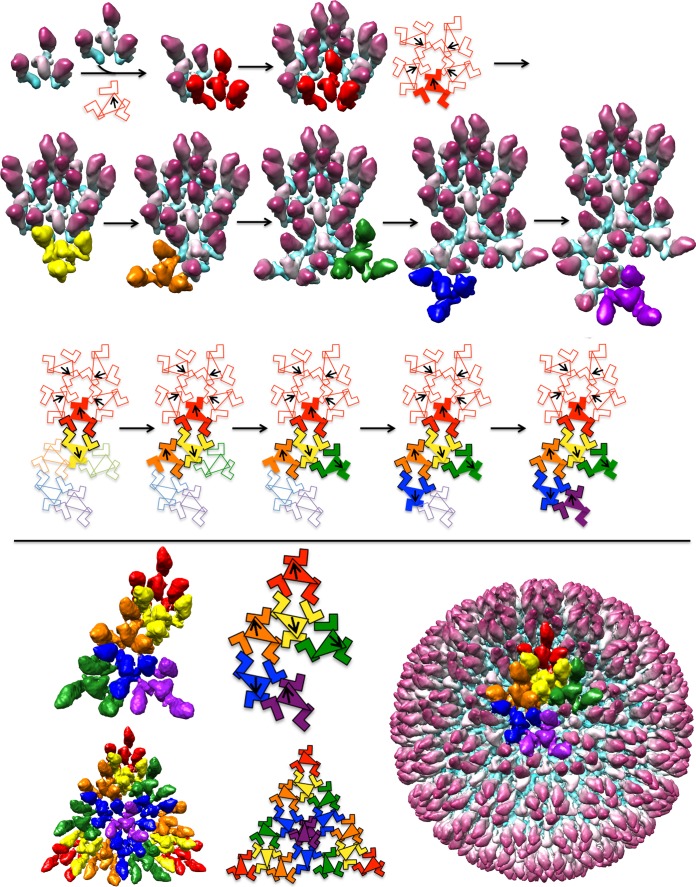
An assembly pathway for the HSV-1 procapsid. The top panel shows assembly of the procapsid asymmetric unit from protomers, each consisting of 3 MCPs surrounding a triplex. The final asymmetric unit consists of six protomers, each colored according to the respective triplex, as in Fig. 4. Ta, red; Tb, orange; Tc, yellow; Td, green; Te, blue; Tf, magenta. The pathway shown starts with five protomers associating around a 5-fold axis. The orientations of the triplexes are shown on the schematic (bottom row) with arrows, as in Fig. 4. The schematic illustrates the sequential addition of protomers, as shown in the surface renderings (above). Upon addition of each protomer, two new MCP-MCP contacts are formed and highlighted as black outlines on the schematic. The colors in the schematic correspond to the density colors shown as surface rendering. The lower panel shows the asymmetric unit, the icosahedral triangle, and the outer surface of the complete procapsid. The sequence illustrated here is not exclusive; some other starting assemblies and sequences of protomer addition could also lead into productive assembly pathways.
We envisage that procapsid assembly starts with five protomers clustering with Ta directionality around a 5-fold axis (Fig. 7, first row). The next step involves adding a ring of protomers oriented so as to have Tc triplexes, as shown in Fig. 7 (second row, leftmost panel). Due to its pseudo-3-fold nature, there are three different settings in which a Tc protomer could be added. On examining the procapsid structure, we noticed that one of these possibilities supports more MCP-MCP interactions between the two interacting protomers (Fig. 8). We suppose that it may be selected on that basis. Continuing, the rest of the asymmetric unit can be built up in similar fashion (Fig. 7, second row, second to fifth panels). This outgrowth to complete an asymmetric unit, starting from an initial pentamer of protomers, is illustrated schematically in Fig. 7, third row. Further addition of appropriately oriented protomers leads to a complete procapsid (Fig. 7, fourth row). The essential features of this model are that it explains both the observed directionality pattern of the triplexes and the skewed/distorted nature of MCP hexamers. Also noteworthy is that each of the three MCP subunits in a given protomer becomes incorporated into a different capsomer (hexamer or pentamer) in the nascent procapsid.
FIG 8 .
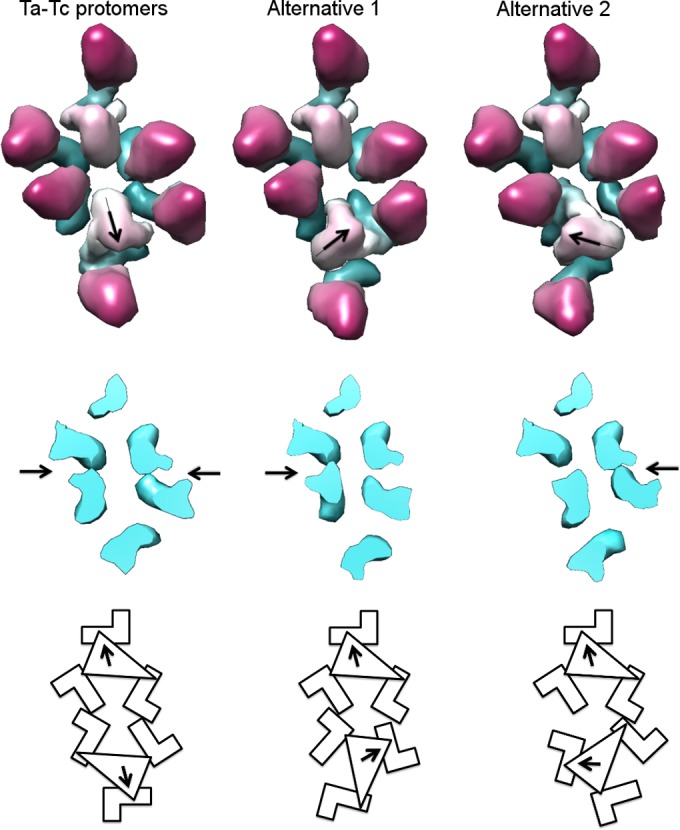
Directionality of triplex association. Two triplex-MCP protomers—Ta and Tc—are shown, associating in three different ways. The Ta-Tc association, observed in the density maps, is shown on the left; two alternative 3-fold-related positions are shown in the middle and on the right. Middle row, cutaway views, showing MCP floor domain associations. The arrows indicate interactions made between the neighboring MCPs. The arrangement (at left) best supports the formation of new MCP-MCP contacts. Bottom row, schematic diagram of the three potential modes of interaction.
In the mature capsid, the four protomer-equivalents (a triplex plus three MCPs) that can be distinguished without compromise from imposed icosahedral symmetry are markedly different from each other (see Fig. S2 in the supplemental material). These differences come about when the protruding MCP towers (middle plus tip domains) of a given protomer move away from their triplex, and six or five of these protrusions (from different protomers) cluster into hexameric or pentameric rings, respectively.
Generalization of the model to accommodate a scaffold and a portal.
Thus far, we have not considered the internal scaffold. It has been observed that the inner (scaffold) and outer (MCP/triplex) shells coassemble approximately in unison, i.e., incomplete particles appear to have about the same amount of inner and outer shell (24). This suggests that the scaffolding shell guides the curvature of the growing procapsid via a network of interactions between the MCP floor domains and the underlying scaffolding subunits. That these interactions are weak is suggested by the nonregistration of the inner and outer shells (Fig. 6 and related Discussion). Transient oligomers have been shown to play a role in the assembly of other capsids (38, 39). The HSV-1 procapsid has 960 MCP subunits. The most recent value reported for the copy number of the scaffolding proteins is 1,918 ± 170 (10). Given evidence that the scaffolding protein forms dimers (40), this would be consistent with one dimer per MCP subunit. Thus, one could imagine expanding the protomer by adding three scaffolding protein dimers, one to each MCP. (There are also indications that the scaffolding protein forms a complex with MCP at an earlier stage of assembly in the cytoplasm and escorts it into the nucleus [41].) We suspect that the ~80-Å repeat detected in the scaffolding may reflect the average spacing between protomers at that radial level in the inner shell of the procapsid.
In herpesviruses and dsDNA phages alike, the procapsid has a dodecameric ring at the portal vertex. Although the HSV-1 portal protein UL6 is not essential for assembly (hence, a procapsid lacking a portal is possible), it is likely to be involved in initiating procapsid assembly (42). In the absence of a specific nucleus, such as the portal, several starting points of assembly are possible, but some of these might lead to geometric barriers, resulting in a failure to incorporate the next protomer. The presence of portal might serve a nucleus that provides a direction for growth, limiting other nonproductive assemblies. Despite the absence of direct experimental evidence, it is nevertheless noteworthy that simply omitting one MCP subunit from each of five Ta protomers and organizing these five reduced protomers around the portal ring would afford a nucleation complex from which outgrowth of the procapsid surface lattice could then proceed as outlined above. In this complex, the portal ring would essentially replace an MCP pentamer (see Fig. S3 in the supplemental material).
MATERIALS AND METHODS
Propagation of mutant virus.
Stocks of M100 virus (a gift of M. Gao, Bristol-Myers Squibb) were generated by infecting monolayers of the complementary cell line F3 (a gift of P. Desai, Johns Hopkins University). F3 monolayers were grown to 75% confluency and infected at a multiplicity of infection (MOI) of 0.1 PFU/cell. At 36 h postinfection, infected-cell medium was clarified and mixed with polyethylene glycol 8000 (PEG 8000) and NaCl to give final concentrations of 7% and 2.3%, respectively. After gentle overnight stirring at 4°C, the PEG precipitate was pelleted and resuspended in small volumes of PBS, aliquoted, and stored frozen at −80°C. The titers of these stocks were determined on both complementary F3 cells and noncomplementary Vero cells. Stocks having a titer ratio of 104-fold or greater between the two cell types were used to produce procapsids.
Procapsid production.
One 75-cm2 flask of Vero cells was infected with M100 stock at an MOI of 5 PFU/cell. Attachment was allowed to take place for 45 min at room temperature. Unattached virus was then removed by washing the monolayer with 10 ml of PBS and replacing it with 15 ml of overlay medium (minimum essential medium [MEM] supplemented with 1% fetal bovine serum [FBS] and penicillin-streptomycin [Pen-Strep]). Infection was allowed to proceed for 12 to 14 hours at 37°C, at which time the infected monolayer was scraped off and pelleted at ~200 × g in a clinical centrifuge for 5 min at room temperature. Beyond this point, the entire procedure was conducted at room temperature, using phosphate-buffered saline (PBS) supplemented with complete protease inhibitor plus EDTA (Roche Diagnostics). The pellet was resuspended in 10 ml of PBS, repelleted, and then gently resuspended in 1.0 ml of PBS and transferred to a 1.5-ml microcentrifuge tube. One hundred milliliters of 10% Triton X-100 was added to the cell suspension, mixed by inversion, and incubated for 20 min, promoting the release of procapsids from infected cell nuclei. After nuclei were removed by centrifugation at 850 × g for 4 min, the supernatant was transferred to a 1.5-ml microcentrifuge tube. Procapsids were purified in four pelleting steps in an Eppendorf microcentrifuge (model 5424), followed by use of a spin filter. The first pelleting step was at 5,000 rpm for 4 min. After the supernatant was transferred to a new tube, the tube containing the pellet was inverted, allowed to drain, and then gently resuspended with 100 μl of PBS. The supernatant from this step was recentrifuged at 6,000 rpm for 5 min: this supernatant was transferred to a fresh tube, and the pellet was drained and resuspended as in the previous step. This process was repeated two more times for speeds of 8,000 rpm and 10,000 rpm. Pellets that appeared turquoise were combined, sonicated in a bath sonicator (Branson; model HD-50) for 1 s, and centrifuged at 845 × g for 3 min to remove aggregates. This supernatant was centrifuged in a 300,000-molecular-weight (MW)-cutoff Nanosep centrifugal filter (Pall) at 5,000 rpm until the volume was reduced to 50 μl. Three hundred microliters of PBS were added to the spin filter, and the sample was recentrifuged until the volume was reduced to 50 μl. This, the final procapsid preparation, was then processed for cryo-EM within 18 to 20 hours postinfection.
Cryo-electron microscopy.
Typically, a 3-μl drop of sample was applied to a glow-discharged Quantifoil holey grid which had been overlaid with a thin layer of carbon, incubated for 40 s, and then blotted and flash-frozen using a Leica EM GP PlungeFreezer. About 12,000 micrographs were collected on a Titan Krios microscope operated at 300 kV at the FEI Nanoport (Acht, Netherlands), using EPU automation software to operate a Falcon II camera. Images were recorded at 2.3 Å/pixel. The final set of particles was picked manually from 9,945 micrographs.
Image processing and reconstruction.
Boxing, defocus determination, and computational processing were carried out using the Bsoft package (43). The computational selection and classification procedures utilized were based on correlation coefficients calculated in reciprocal space between fast Fourier transforms (FFTs) of the images and reference map projections. Seventeen previously reported intermediates (11) were used as initial reference maps. Thereafter, newly calculated reconstructions were used as references for the next cycle. The particles were allowed to undergo redistribution in successive cycles. After the first three iterations, it was observed that only 10 out of 17 classes contained more than 300 particles. Accordingly, the other seven reference maps were omitted and particles from these classes were allowed to undergo redistribution among the remaining 10 reference maps (classes). After 10 cycles of reconstruction and refinement, the resolutions of 5 out of 10 reconstructions had significantly improved, whereas the resolutions of the other five classes were still limited to 20 Å. Thus, the number of references was further reduced to five, and the selection criteria allowed only 50% of the top-scoring particles, as ranked by correlation coefficients, to be used in the reconstructions. The resolutions of the five reconstructions (Table 1) were calculated using the Fourier shell correlation criterion with a threshold of 0.5.
Structure fitting and figure preparation.
HK97 capsid protein structures as found in prohead I (PDB 3QPR) and head II (PDB 2FT1) were fitted into maps 1 and 5, respectively, using UCSF Chimera (44). SEGGER, implemented in UCSF Chimera, was used for map segmentation. Figures were prepared using UCSF Chimera.
Cryo-electron tomography.
Grids prepared as described above were imaged in an FEI Titan Krios microscope operated at 300 kV. Tilt series were collected using FEI Tomography 4 operated in batch mode at ×29,000 magnification, giving a final pixel size of 0.96 nm after 2-fold binning. In each tilt series, images were recorded at 1.5° increments over an angular range of approximately −55° to +55°. The electron dose was ~1 e−/Å2 per image, for a cumulative dose of ~70 e−/Å2 per tilt series. Tilt series target defocus ranged from −4 μm to −8 μm, corresponding to first contrast transfer function zeros from (28 Å)−1 to (40 Å)−1. Tomogram reconstruction was done using IMOD (45). Subtomogram alignment and averaging used Bsoft routines (43), modified as needed and wrapped into Python scripts. The top 75% of subvolumes, ranked according to cross-correlation coefficient, were used to calculate the final density map of the scaffolding core. Although classification of the particles was attempted, limited resolution and particle numbers made it difficult to achieve a stable result, and the averaged structure shown represents a superposition of all classes.
SUPPLEMENTAL MATERIAL
Cryo-electron micrograph of purified HSV-1 procapsids. Red arrows indicate procapsids deemed to be significantly distorted. Black arrows indicate procapsids deemed not to be distorted, according to visual assessment. The particles are 1,250 Å in diameter. The inset at top right is a schematic map of the procapsid proteins. UL19 is the major capsid protein; UL38 and UL18 are the triplex proteins; UL6 is the connector; UL26.5 and UL26 are the scaffolding proteins. VP24 (small yellow oval) is the protease. Download
Superposition of the four triplex-MCP units (each colored red, blue, yellow, and orange) in the procapsid map (left) and capsid map (right). Download
Alternative nucleation complexes. (A) Fivefold ring of Ta protomers. (B) Fivefold ring of Ta protomers, each lacking an MCP subunit, surrounding a 12-fold ring of portal protein. Download
This movie was made by combining, in order, central sections through the reconstructions of the five conformational states of the maturing HSV-1 capsid that were captured in this study. The capsids are viewed along a 2-fold axis of symmetry. Download
ACKNOWLEDGMENTS
We thank Kasim Sader of the FEI Nanoport facility in Acht, Netherlands, for data collection and James Conway for helpful discussions.
This work was supported by the Intramural Research Program of NIAMS, with additional support from an NIGMS PRAT fellowship (to A.A.A.).
Footnotes
Citation Aksyuk AA, Newcomb WW, Cheng N, Winkler DC, Fontana J, Heymann JB, Steven AC. 2015. Subassemblies and asymmetry in assembly of herpes simplex virus procapsid. mBio 6(5):e01525-15. doi:10.1128/mBio.01525-15.
REFERENCES
- 1.Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 2.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Fokine A, Rossmann MG. 2014. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 4:e28281. doi: 10.4161/bact.28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrix RW, Johnson JE. 2012. Bacteriophage HK97 capsid assembly and maturation. Adv Exp Med Biol 726:351–363. doi: 10.1007/978-1-4614-0980-9_15. [DOI] [PubMed] [Google Scholar]
- 5.Rixon FJ, Schmid MF. 2014. Structural similarities in DNA packaging and delivery apparatuses in herpesvirus and dsDNA bacteriophages. Curr Opin Virol 5:105–110. doi: 10.1016/j.coviro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol 15:227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steven AC, Spear PG. 1997. Herpesvirus capsid assembly and envelopment, p 312–351. In Chiu W, Burnett RM, Garcea RL (ed), Structural biology of viruses. Oxford University Press, New York, NY. [Google Scholar]
- 8.Newcomb WW, Cockrell SK, Homa FL, Brown JC. 2009. Polarized DNA ejection from the herpesvirus capsid. J Mol Biol 392:885–894. doi: 10.1016/j.jmb.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol 263:447–462. doi: 10.1016/S0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 10.Newcomb WW, Trus BL, Cheng N, Steven AC, Sheaffer AK, Tenney DJ, Weller SK, Brown JC. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol 74:1663–1673. doi: 10.1128/JVI.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol 10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- 12.Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. 2008. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure 16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Plückthün A, Wlodawer A. 2000. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat Struct Biol 7:230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- 14.Ishii T, Yamaguchi Y, Yanagida M. 1978. Binding of the structural protein soc to the head shell of bacteriophage T4. J Mol Biol 120:533–544. doi: 10.1016/0022-2836(78)90352-2. [DOI] [PubMed] [Google Scholar]
- 15.Qin L, Fokine A, O’Donnell E, Rao VB, Rossmann MG. 2010. Structure of the small outer capsid protein, Soc: a clamp for stabilizing capsids of T4-like phages. J Mol Biol 395:728–741. doi: 10.1016/j.jmb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steven AC, Greenstone HL, Booy FP, Black LW, Ross PD. 1992. Conformational changes of a viral capsid protein. Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, co-operativity, and super-stabilization by soc binding. J Mol Biol 228:870–884. doi: 10.1016/0022-2836(92)90871-G. [DOI] [PubMed] [Google Scholar]
- 17.Dai X, Gong D, Wu TT, Sun R, Zhou ZH. 2014. Organization of capsid-associated tegument components in Kaposi’s sarcoma-associated herpesvirus. J Virol 88:12694–12702. doi: 10.1128/JVI.01509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. 2013. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol 425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou ZH, Hui WH, Shah S, Jih J, O’Connor CM, Sherman MB, Kedes DH, Schein S. 2014. Four levels of hierarchical organization, including noncovalent chainmail, brace the mature tumor herpesvirus capsid against pressurization. Structure 22:1385–1398. doi: 10.1016/j.str.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng N, Trus BL, Belnap DM, Newcomb WW, Brown JC, Steven AC. 2002. Handedness of the herpes simplex virus capsid and procapsid. J Virol 76:7855–7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatman JD, Preston VG, Nicholson P, Elliott RM, Rixon FJ. 1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol 75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 22.Thomsen DR, Roof LL, Homa FL. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol 68:2442–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao M, Matusick-Kumar L, Hurlburt W, DiTusa SF, Newcomb WW, Brown JC, McCann PJ, Deckman I, Colonno RJ. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J Virol 68:3702–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb WW, Homa FL, Thomsen DR, Booy FP, Trus BL, Steven AC, Spencer JV, Brown JC. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol 263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 25.Heymann JB, Conway JF, Steven AC. 2004. Molecular dynamics of protein complexes from four-dimensional cryo-electron microscopy. J Struct Biol 147:291–301. doi: 10.1016/j.jsb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Häuser R, Blasche S, Dokland T, Haggård-Ljungquist E, von Brunn A, Salas M, Casjens S, Molineux I, Uetz P. 2012. Bacteriophage protein-protein interactions. Adv Virus Res 83:219–298. doi: 10.1016/B978-0-12-394438-2.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 28.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA 102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W. 2011. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci USA 108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardone G, Heymann JB, Cheng N, Trus BL, Steven AC. 2012. Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions, p 423–439. In Rossmann MG, Rao V (ed), Viral molecular machines. Springer, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. 2003. Structure of the herpesvirus major capsid protein. EMBO J 22:757–765. doi: 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. 2001. Virus maturation involving large subunit rotations and local refolding. Science 292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- 33.Spencer JV, Newcomb WW, Thomsen DR, Homa FL, Brown JC. 1998. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol 72:3944–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fane BA, Prevelige PE Jr. 2003. Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem 64:259–299. doi: 10.1016/S0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 35.Baines JD. 2011. Herpes simplex virus capsid assembly and DNA packaging: a present and future antiviral drug target. Trends Microbiol 19:606–613. doi: 10.1016/j.tim.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Oh B, Moyer CL, Hendrix RW, Duda RL. 2014. The delta domain of the HK97 major capsid protein is essential for assembly. Virology 456-457:171–178. doi: 10.1016/j.virol.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Z, Hendrix RW. 1995. Assembly in vitro of bacteriophage HK97 proheads. J Mol Biol 253:74–85. doi: 10.1006/jmbi.1995.0537. [DOI] [PubMed] [Google Scholar]
- 38.Johnson JM, Tang J, Nyame Y, Willits D, Young MJ, Zlotnick A. 2005. Regulating self-assembly of spherical oligomers. Nano Lett 5:765–770. doi: 10.1021/nl050274q. [DOI] [PubMed] [Google Scholar]
- 39.Tresset G, Decouche V, Bryche JF, Charpilienne A, Le Coeur C, Barbier C, Squires G, Zeghal M, Poncet D, Bressanelli S. 2013. Unusual self-assembly properties of norovirus Newbury2 virus-like particles. Arch Biochem Biophys 537:144–152. doi: 10.1016/j.abb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Pelletier A, Dô F, Brisebois JJ, Lagacé L, Cordingley MG. 1997. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J Virol 71:5197–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson P, Addison C, Cross AM, Kennard J, Preston VG, Rixon FJ. 1994. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J Gen Virol 75:1091–1099. doi: 10.1099/0022-1317-75-5-1091. [DOI] [PubMed] [Google Scholar]
- 42.Newcomb WW, Thomsen DR, Homa FL, Brown JC. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J Virol 77:9862–9871. doi: 10.1128/JVI.77.18.9862-9871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heymann JB, Belnap DM. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol 157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cryo-electron micrograph of purified HSV-1 procapsids. Red arrows indicate procapsids deemed to be significantly distorted. Black arrows indicate procapsids deemed not to be distorted, according to visual assessment. The particles are 1,250 Å in diameter. The inset at top right is a schematic map of the procapsid proteins. UL19 is the major capsid protein; UL38 and UL18 are the triplex proteins; UL6 is the connector; UL26.5 and UL26 are the scaffolding proteins. VP24 (small yellow oval) is the protease. Download
Superposition of the four triplex-MCP units (each colored red, blue, yellow, and orange) in the procapsid map (left) and capsid map (right). Download
Alternative nucleation complexes. (A) Fivefold ring of Ta protomers. (B) Fivefold ring of Ta protomers, each lacking an MCP subunit, surrounding a 12-fold ring of portal protein. Download
This movie was made by combining, in order, central sections through the reconstructions of the five conformational states of the maturing HSV-1 capsid that were captured in this study. The capsids are viewed along a 2-fold axis of symmetry. Download