Abstract
Over the past decade, inhibition of the kinase activities of oncogenic proteins using small molecules and antibodies has been a mainstay of our anticancer drug development effort, resulting in several Food and Drug Administration–approved cancer therapies. The clinical effectiveness of kinase-targeted agents has been inconsistent, mostly because of the development of resistance. The expression and function of oncoproteins and tumor suppressors are regulated by numerous posttranslational protein modifications including phosphorylation, ubiquitination, and acetylation; hence, targeting specific posttranslational protein modifications provides for an attractive strategy for anticancer drug development. The present review discusses the hypothesis that targeted degradation of an oncoprotein may overcome many of the shortcomings seen with kinase inhibitors and that the approach would enable targeted inhibition of oncogenic proteins previously thought to be undruggable.
Introduction
The molecular complexity of cancer is reflected by the ever-increasing list of genetic drivers of oncogenesis. In preclinical models, the targeted inhibition of oncogenic pathways has been an effective strategy in many types of cancer; however, the clinical success of these drugs has been limited to a handful of targets and diseases. Because aberrant kinase activity is linked to oncogenesis, adenosine triphosphate–competitive inhibitors such as erlotinib represent the mainstay of our current drug development pipeline. Unfortunately, many oncogenic targets are thought to be undruggable using these traditional drug design strategies, and the development of drug resistance to existing targeted agents is a significant problem. Novel strategies to target oncogenic drivers are greatly needed.
X-ray crystallography, nuclear magnetic resonance spectroscopy, and molecular modeling have elucidated the three-dimensional structures of many protein targets. Insight into the structural basis of kinase function as well as the structural requirements for protein-ligand and protein-protein interactions has opened the door for novel therapeutic strategies. The identification of specific protein domains involved in chaperone binding, ubiquitination, and dimer formation allows for the development of novel agents that target oncoprotein stability and induce degradation. These strategies have the potential to overcome resistance seen with traditional kinase inhibitors.
In this article, we will review previous attempts at targeting various protein posttranslational modifications including phosphorylation. We then present an argument that the targeted degradation of an oncoprotein has several advantages over the mere inhibition of kinase activity as this strategy has the potential to affect the cellular processes of a protein that are not related to kinase activity. Furthermore, we describe how undruggable proteins such as KRAS may be targeted with this approach.
Background: Targeting Posttranslational Modifications for Cancer Therapy
Although several posttranslational modifications affect the function of an oncoprotein, almost all drug development efforts have focused on the altering the attachment of phosphate groups by protein kinases. This protein modification controls many important aspects of protein activity, localization, and stability. Many very successful drugs, particularly tyrosine kinase inhibitors, have been developed using this strategy (Table 1). For example, imatinib (Gleevec, Novartis) revolutionized the treatment of chronic myeloid leukemia and gastrointestinal stromal tumors [1]. Also, epidermal growth factor receptor (EGFR) inhibitors including erlotinib (Tarceva, Genentech) have become standard of care for subsets of patients with non–small cell lung cancer and colorectal cancer [2]. Serine/threonine kinase inhibitors and multikinase inhibitors have also been developed. Vemurafenib (Zelboraf, Daiichi Sankyo) is a serine/threonine kinase inhibitor that is Food and Drug Administration (FDA) approved for patients with metastatic BRAFV600E mutant melanoma [3]. Sorafenib (Nexavar, Bayer) inhibits the serine/threonine kinase RAF and the tyrosine kinases platelet-derived growth factor receptor and vascular endothelial growth factor receptor. It has FDA approval for the treatment of advanced-stage renal cell carcinoma and hepatocellular carcinoma [4].
Table 1.
List of Posttranslational Protein Modifications and FDA-Approved Agents as Anticancer Agents
| Agent | Target | Disease Site | Clinical Results | Year FDA Approval |
|---|---|---|---|---|
| Tyrosine kinase inhibitors | ||||
| Trastuzumab | HER2 | Metastatic HER2 + gastric cancer | OS improved 11.7 to 13.1 mo | 2010 |
| Adjuvant therapy for HER2 +, LN + breast cancer | DFS HR 0.48 | 2006 | ||
| Metastatic HER2 + breast cancer | TTP improved 4.6 to 7.6 mo | 1998 | ||
| Imatinib | BCR-ABL c-Kit | Adjuvant therapy for GIST | Improved OS with 36 mo vs 12 mo HR 0.45 | 2008/2012 |
| Unresectable or metastatic GIST | ORR 38% | 2002 | ||
| CML | Hematologic response 88% (chronic phase) | 2001 | ||
| Gefitinib | EGFR | NSCLC | ORR 10.6% | 2003 |
| Two failed clinical trials | Revoked 2005 | |||
| Bevacizumab | VEGFR | Metastatic cervical cancer | OS improved 12.9 to 16.8 mo | 2014 |
| Platinum-resistant ovarian cancer | PFS improves 3.4 to 6.8 mo | 2014 | ||
| Renal cell carcinoma | PFS improved 5.4 to 10.2 mo | 2009 | ||
| Refractory high-grade glioma | No phase III data with non–bevacizumab-containing arm | 2009 | ||
| Metastatic renal cell carcinoma | PFS improved 5.4 to 10.2 mo | 2009 | ||
| Metastatic breast cancer | PFS improved 5.8 to 11.3 mo | 2008 | ||
| No survival benefit | Revoked 2011 | |||
| Nonsquamous NSCLC | OS improved 10.3 to 12.3 mo | 2006 | ||
| Second-line metastatic colorectal cancer | OS improved 10.8 to 13.0 mo | 2006 | ||
| First-line metastatic colorectal cancer | ORR improved 35% to 45% | 2004 | ||
| Cetuximab | EGFR | K-ras wild type, EGFR-expressing metastatic colorectal cancer | OS improved 19.5 to 23.5 mo in K-ras wild-type tumors | 2012 |
| Metastatic head and neck cancer | Improved OS 18.2 to 19.1 mo | 2012 | ||
| Head and neck cancer with radiation therapy | OS improved 29.3 to 49.0 mo | 2006 | ||
| EGFR-expressing metastatic colorectal cancer | ORR 23% combined with irinotecan | 2004 | ||
| Erlotinib | EGFR | Metastatic NSCLC with EGFR mutation | Improved PFS 5.2 to 10.4 mo | 2013 |
| Maintenance treatment of NSCLC | Improved PFS HR 0.71 | 2010 | ||
| Unresectable pancreatic cancer | OS improved 6.0 to 6.4 mo | 2005 | ||
| Refractory NSCLC | OS improved 4.7 to 6.7 mo | 2004 | ||
| Dasatinib | Multityosine kinase inhibitor | Chronic-phase CML, Philadelphia chromosome positive (Ph +) | Complete cytogenic response improved 66.2% to 76.8% | 2010 |
| Refractory CML and ALL, Ph + | No phase III data | 2006 | ||
| Panitumumab | EGFR | EGFR-expressing metastatic colorectal cancer | PFS improved 60 to 96 d | 2006 |
| Sunitinib | Multikinase (VEGFR, PDGFR, KIT, FLT3, RET) | Pancreatic neuroendocrine tumor | PFS improved 5.4 to 10.2 mo | 2011 |
| Renal cell carcinoma | ORR 25.5%-36.5% | 2006 | ||
| GIST | TTP improved 6 to 27 wk | 2006 | ||
| Lapatinib | HER-2 | ER/PR +, HER2 + breast cancer | PFS improved 13 to 35 wk | 2010 |
| HER2 + breast cancer | TTP improved 18 to 24 wk | 2007 | ||
| Pazopanib | VEGFR | Advanced soft tissue sarcoma | PFS improved 1.6 to 4.6 mo | 2012 |
| Advanced renal cell carcinoma | PFS improved 4.2 to 9.2 mo | 2009 | ||
| Vandetanib | VEGFR, EGFR | Medullary thyroid cancer | ORR improved 1% to 44% | 2011 |
| Crizotinib | c-Met, anaplastic lymphoma kinase (ALK) | ALK-positive NSCLC | PFS improved 3.0 to 7.7 mo | 2011/2013 |
| Axitinib | VEGFR | Renal cell carcinoma | Improved PFS 4.7 to 6.7 mo | 2012 |
| Bosutinib | Bcr-Abl, Src-family kinases | CML/ALL Ph + | No phase III data | 2012 |
| Cabozantinib | Pan-tyrosine kinase inhibitor | Metastatic medullary thyroid cancer | PFS improved 4.0 to 11.2 mo | 2012 |
| Ponatinib | Multikinase inhibitor | CML/ALL Ph + | Phase III trial stopped | 2012 |
| Regorafenib | Multikinase inhibitor | GIST | PFS improved 0.9 to 4.8 mo | 2013 |
| Refractory metastatic colorectal cancer | Improved OS 5.0 to 6.4 mo | 2012 | ||
| Afatinib | EGFR, HER2, HER4 | Metastatic NSCLC with mutant EGFR | PFS improved 6.9 to 11.1 mo | 2013 |
| Ibrutinib | Burton’s tyrosine kinase | CML | ORR 58.3% | 2014 |
| Mantle cell lymphoma | ORR 69% | 2013 | ||
| Ceritinib | ALK | ALK-positive metastatic NSCLC | ORR 54.6% | 2014 |
| Ramucirumab | VEGFR | Gastric cancer | OS improved 3.8 to 5.2 mo | 2014 |
| Metastatic NSCLC | OS improved 9.1 to 10.6 mo | 2014 | ||
| Serine/threonine kinase inhibitors | ||||
| Vemurafenib | BRAFV600E | Melanoma V600E mutant | PFS improved 1.6 to 5.3 mo | 2011 |
| Trametinib | MEK1, MEK2 | Melanoma BRAF V600E/V600K mutant | PFS improved 1.5 to 4.8 mo | 2013 |
| Dabrafenib | BRAF, CRAF | Melanoma BRAF V600E mutant | PFS improved 2.7 to 5.1 mo | 2013 |
| Other kinase inhibitors | ||||
| Sorafenib | Multikinase inhibitor (BRAF, VEGFR, PDGFR, FLT3, KIT) | Differentiated thyroid cancer | PFS improved 5.8 to 10.8 mo | 2013 |
| Hepatocellular carcinoma | OS improved 7.9 to 10.7 mo | 2007 | ||
| Renal cell carcinoma | PFS improved 84 to 167 d | 2005 | ||
| Temsirolimus | mTOR | Renal cell carcinoma | Improved PFS 3.1 to 5.5 mo | 2007 |
| Everolimus | mTOR | HER2-negative breast cancer | PFS improved 3.2 to 7.8 mo | 2012 |
| Pancreatic neuroendocrine tumor | PFS improved 4.6 to 11.0 mo | 2011 | ||
| Renal cell carcinoma | PFS improved 1.9 to 4.9 mo | 2009 | ||
| Idelalisib | Phosphoinositide-3 kinase | Relapsed CLL | PFS HR 0.18 | 2014 |
| SLL | ORR 58% | |||
| Follicular NHL | ORR 54% | |||
| HDAC inhibitors | ||||
| Vorinostat | HDAC | Cutaneous T-cell lymphoma | ORR 30% | 2006 |
| Romidepsin | HDAC | Cutaneous T-cell lymphoma | ORR 34-35% | 2009 |
| Belinostat | HDAC | Refractory peripheral T-cell lymphoma | ORR 25.8% | 2014 |
| Panobinostat | HDAC | Refractory multiple myeloma | PFS improved 5.8 to 10.6 mo | 2015 |
| Proteasome inhibitors | ||||
| Bortezomib | Proteasome | Mantle cell lymphoma | PFS improved 14 to 25 mo | 2014 |
| Multiple myeloma | ORR 28% | 2003 | ||
| Carfilzomib | Proteasome inhibitor | Multiple myeloma | ORR 23% | 2012 |
| PARP inhibitors | ||||
| Olaparib | PARP | Ovarian cancer with germline BRCA mutation | ORR 34% | 2014 |
DOR: duration of response; ORR: overall response rate; PFS: progression-free survival; TTP: time to progression.
HR = Hazard Ratio DFS = Disease Free Survival OS = Overall Survival.
The concern with selective targeted agents is that parallel signaling pathways can compensate for the inhibition of a single kinase and resistant mutations can quickly develop. Conversely, with multitargeted kinase inhibitors, there are issues with off-target effects leading to toxicity. Also, because multiple pathways are affected, there is a poor understanding of the true mechanism of these agents in a particular tumor, making it difficult to develop further improvements in specificity or activity.
Besides phosphorylation, other posttranslational protein modifications have been identified as targets for cancer therapy. One of the earliest attempts at this strategy was inhibition of RAS isoprenylation by farnesyltransferase inhibitors [5]. More than 30 years ago, the attachment of farnesyl side chains to RAS proteins (including H-, N-, and K-RAS) was found to be critical for wild-type and mutant oncogenic protein localization and function. Farnesyltransferase inhibitors showed promise in preclinical tumor models; however, they have failed in the clinic because of the presence of alternate isoprenylation pathways through the geranyl side chain attachment. Given the presence of multiple isoprenylation modifications, dual inhibitors targeting both farnesylation and geranylation were tested; however, this strategy showed increased toxicity as approximately 30 mammalian proteins are known to undergo similar posttranslational modifications. A recent review [6] describes the many unsuccessful attempts at targeting RAS. Given its important role in several types of cancer, novel strategies are needed to target KRAS mutant-driven tumors.
Similar to phosphorylation, protein acetylation has been extensively explored as a strategy to target cancer. Histone deacetylase (HDAC) inhibitors such as vorinostat (Zolinza) and romidepsin (Istodax) are FDA approved for the treatment of cutaneous T-cell lymphoma [7]. Recently, LBH589 (Panobinostat, Novartis) in combination with dexamethasone and bortezomib was shown to slow progression of multiple myeloma in a phase III trial. The mechanism behind HDAC inhibitor cytotoxicity is complex and poorly understood. One proposed mechanism is that HDAC inhibition leads to increased expression of tumor suppressors, although several other cellular processes are altered as well.
Chaperone proteins represent a unique class of therapeutic targets. After translation, the chaperone machinery plays an important role in protein homeostasis by facilitating nascent protein maturation and folding. HSP90 is critical for the maintenance of more than 600 proteins including several oncoproteins. Even though highly potent HSP90 inhibitors such as geldanamycin and its derivatives (17-AAG or 17-DMAG) (reviewed in [8]) have been extensively studied, no HSP90 inhibitors are FDA approved because of their toxicity. Next-generation HSP90 inhibitors, such as ganetespib [9], have shown promise for improved tumor selectivity in preclinical models. Preliminary results from an ongoing clinical trial suggest that the single agent ganetespib has activity in a subset of advanced-stage non–small cell lung carcinoma (NSCLC) patients [10]. Furthermore, a phase IIB/III study evaluating the safety and therapeutic activity of ganetespib in combination with docetaxel in NSCLC found that this combination improved survival [11]. It is unclear why ganetespib is less toxic than first-generation HSP90 inhibitors. It may be due to a more tolerable dosing schedule or relatively selective degradation of oncoproteins. Future studies will improve our understanding of how to effectively use HSP90 inhibitors in patients.
Inducing Protein Degradation as an Attractive Alternative to Inhibition
Resistance to kinase inhibitors is a major challenge limiting the clinical effectiveness of these drugs. Single amino acid substitutions can make a kinase inhibitor ineffective. Given that a tumor typically contains a billion cells per cubic centimeter and the fact that tumors cells are genetically unstable, it is likely that a resistance mutation will be present. Prolonged exposure to a drug will select for the resistant population. This phenomenon has been clearly demonstrated in non–small cell lung cancer expressing EGFR with a threonine-790-methionine (T790M) point mutation [12]. Careful analysis has indicated that the T790M mutation is a de novo mutation, inferring that TKI treatment selects for these cells. A drug that selectively degrades a target should theoretically be effective in kinase inhibitor–resistant and –sensitive cells.
One approach to eliminating a target rather than simply inhibiting it is to deliver small interfering RNA (siRNA). Indeed, several studies suggest that the reduction of a protein by an siRNA can be effective for treating cancer. Reducing the amount of a specific protein has certain advantages over inhibition of its activity as a protein’s physical presence can serve critical functions beyond its catalytic activity. One of the first examples of this concept came from the yeast (Saccharomyces cerevisiae) protein Pbs2p. Both the kinase activity of Pbs2p and its function as scaffolding were found to be equally important in maintaining cellular osmolarity [13]. A recent review article by Rauch et al. [14] identifies more than 70 kinases whose catalytic activity and physical presence (mainly as scaffolding proteins) are critical in disease pathogenesis. Among these kinases, some of the most relevant to cancer are in the ErbB family which includes EGFR and Her2. ERBB3 is a very interesting member of this family as it has no kinase activity but can facilitate signal transduction by forming a heterodimer with another family member [15]. Furthermore, a kinase-defective mutant of EGFR (K721M) can activate downstream signaling with assistance of ErbB2 (Her2) after EGF stimulation [16]. These findings demonstrate that ErbB family members and other receptor tyrosine kinases have important cellular functions that are independent of kinase activity.
In addition to its scaffolding role, EGFR has many other functions beyond kinase activity. EGFR was recently found to protect cells from autophagy by a kinase-independent mechanism [17]. Also, the EGFRvIII mutant can sequester the proapoptotic protein PUMA in a kinase independent manner, promoting drug resistance [18]. Furthermore, in genetically engineered mouse models, EGFR knockout is embryonically lethal [19], whereas transgenic mice expressing a kinase-dead form of EGFR are viable with minimal defects [20]. Also, mice that express a kinase-inactive form of EGFR (V765G-EGFR) are fertile and show a significant reduction in intestinal polyps when crossed with APCMin mice compared with APCMin mice carrying wild-type EGFR. These findings suggest that EGFR kinase activity is important for tumorigenesis, but its physical presence is essential for cell survival. Along these lines, in patients with colorectal cancer, EGFR expression correlates with prognosis but not with response to EGFR inhibitors such as cetuximab. We recently reported our finding that degradation of EGFR is more efficacious than treatment with small molecule inhibitors and that this strategy can overcome resistance from an acquired EGFR mutation (T790M) [21]. We discuss this strategy in more detail later in this review.
Proteosome Inhibitors as Anticancer Agent
So far, the most successful drug targeting protein degradation is bortezomib (Velcade, Millennium Pharmaceuticals). It is approved for use in patients with multiple myeloma and non-Hodgkin’s lymphoma. Bortezomib binds to the catalytic site of the 26S proteasome, ultimately inhibiting the degradation of proteins. Because the proteasome degrades most cellular proteins, this drug class has many side effects including peripheral neuropathy and myelosuppression [22]. A proposed mechanism of bortezomib’s anticancer activity is the attenuation of IκB degradation which promotes inactivation of NF-κB. However, the true mechanism behind this drug’s actions is poorly understood as countless proteins are affected by proteasome inhibition.
Given the lack of target specificity seen with proteasome inhibitors, more selective approaches that target the degradation of a specific oncoprotein or tumor suppressor are needed. One way to accomplish this goal is to target protein ubiquitination or neddylation processes. A novel inhibitor of neddylation, pevonedistat (MLN4924), is currently in clinical trials for hematologic malignancies and melanoma. Similarly, the E3 ubiquitin ligase MDM2 has been targeted in patients using the drug RO5045337A. Several recent reviews discuss these efforts in more detail. Below, we discuss novel approaches of targeting protein-protein interaction to target oncogenes and tumor suppressors.
Targeting Ubiquitination-Mediated Protein Degradation
Ubiquitination-mediated protein degradation is an exciting target for cancer therapy. The ubiquitination cascade consists of three enzyme groups: E1 is a group of ubiquitin-activating enzymes, E2 is a group of ubiquitin-conjugating enzymes, and E3 is a group of ubiquitin ligases. This cascade regulates the addition of ubiquitin moieties to specific proteins within a cell, leading to protein degradation. By altering these pathways, one can potentially manipulate the degradation of a protein. An example of this strategy is the use of 5-deazaflavin derivatives to inhibit the ubiquitin ligase MDM2, resulting in decreased degradation of p53, a tumor suppressor [23]. Although 5-deazaflavin derivatives inhibit MDM2-mediated p53 ubiquitination, they have poor substrate selectivity as they target a number of other kinases. An alternative strategy being studied is the use of Nutlins (particularly Nutlin-3) to disrupt the interaction between p53 and MDM2 [24]. Nutlins are currently in clinical trials for pediatric cancers containing wild-type p53.
Like p53, a reduction in p27 levels has been correlated with a poor prognosis in many types of cancer. Mechanistic studies indicate that the loss of p27 depends on SKP2 E3 ligase-mediated proteasomal degradation. A recent report describes a small molecule capable of disrupting a critical interaction between SKP2 with its partner SKP1 [25]. Similarly, the interaction of SKP2 with phosphorylated p27 requires the adapter protein CKS1 for degradation. Small molecule inhibitors capable of disrupting the SKP2:CKS1 interaction have been developed [26]. Another example of this strategy is targeting PML, a known tumor suppressor. PML is downregulated in many human malignancies because of enhanced ubiquitination-mediated proteasomal degradation. Agents that manipulate PML ubiquitination are in development.
Similar to the downregulation of tumor suppressors, the overexpression of oncoproteins is critical for cancer development and progression. The amount of a specific protein in a cell depends on the rate it is synthesized and its stability or half-life. So far, efforts to target an oncoprotein’s stability have been limited. As discussed above, we believe that the physical presence of an oncoprotein, independent of its activity, can promote cancer progression. Therefore, inducing degradation of an oncogenic protein, as opposed to mere inhibition of its activity, is a promising strategy. A better understanding of the factors responsible for the stability of oncoproteins is needed to accomplish this.
A New Approach of Targeted Oncoprotein Degradation
EGFR Degradation: Disruption Of Homo- And Heterodimerization
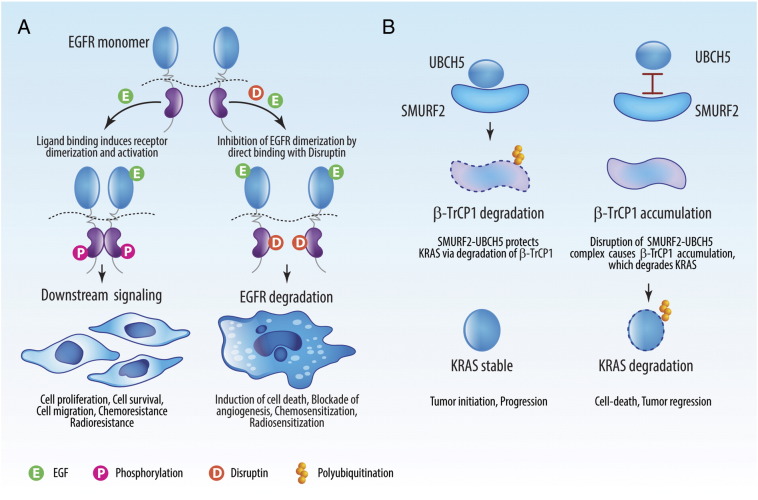
We have studied the selective degradation of EGFR as a novel strategy to target EGFR-driven tumors. Under physiological conditions, EGFR is stable with a half-life greater than 12 hours. Its stability depends on the ability to form homo- or heterodimers as EGFR is highly unstable in its monomer state. When inactive, EGFR can form a dynamic heterocomplex with the chaperone protein HSP90, promoting stability [27]. Upon stimulation by various growth factors, EGFR is phosphorylated and dissociates with HSP90 to form a homodimer with another EGFR molecule or a heterodimer with another EGFR family member (such as ErbB-2, -3, or -4) or related tyrosine kinase (such as c-Met). Based on these findings, we identified a critical eight–amino acid stretch (amino acids 768-SVDNPHVC-775) that lies within the αC-β4 loop region of the EGFR kinase domain corresponding to the binding region of HSP90 [28]. We found that mutating the first six amino acids in this sequence disrupts EGFR homo- and heterodimerization, making the protein highly unstable (half-life < 3 hours). Using this knowledge, we developed a peptide named Disruptin (Figure 1A), which inhibits dimerization of EGFR, leading to rapid degradation. As cancer cells have higher levels of activated EGFR than normal tissue, Disruptin preferentially targets tumor cells expressing EGFR. Furthermore, Disruptin has therapeutic efficacy in preclinical xenograft models carrying the T790M mutation which makes tumors resistant to erlotinib. This demonstration of selective EGFR degradation with the peptide Disruptin represents a novel therapy for EGFR-driven tumors, including those that are TKI resistant [28].
Figure 1.
Disruption of protein-protein interactions to induce oncoproteins degradation. (A) Disruptin (D), a peptide containing an eight–amino acid stretch of human EGFR, is capable of disrupting the protein:protein interactions between EGFR:HSP90 and EGFR homodimers. These interactions are critical for maintaining EGFR stability. In the presence of high levels of EGF, as seen in the tumor microenvironment, treatment with Disruptin causes rapid degradation of EGFR. In preclinical models, treatment with Disruptin induces regression of EGFR-dependent tumors including those resistant to TKIs. (B) Interaction between the SMURF2:UBCH5 protein complex and β-TrCP1 plays a critical role in the maintenance of KRAS protein stability. The SMURF2:UBCH5 complex polyubiquitinates and degrades β-TrCP1, which indirectly protects KRAS from degradation. We found that the disruption of the SMURF2:UBCH5 complex can promote β-TrCP1 accumulation, leading to rapid KRAS degradation.
EGFR Degradation: Targeting Receptor Trafficking
Receptor trafficking also plays a critical role in determining the fate of EGFR molecules. Monoubiquitination and polyubiquitination of EGFR are involved in its stability, localization, and functionality. In addition to its ability to signal from the cell membrane, EGFR is also involved in signal transduction after it is endocytosed. The endocytosed receptor can be recycled back to the cell membrane, or it can be trafficked to the multivesicular bodies for lysosomal degradation. Factors involved in EGFR trafficking can be categorized into two groups: 1) factors that promote receptor recycling and 2) factors that promote EGFR endocytosis and degradation. Among the trafficking factors that promote receptor stability, Vav proteins (guanine nucleotide exchange factors) have been shown to positively regulate EGFR signaling by attenuating receptor internalization and degradation. Similarly, SGEF and DYRK1A can protect EGFR from degradation by delaying lysosomal sorting. Factors like Odin/ANKS1A also interact with EGFR, increasing receptor recycling. In contrast, Ephrin A5 promotes EGFR degradation by cooperating with c-CBL, an E3 ligase known to polyubiquitinate this protein. Also, Presenilin, a component of gamma-secretase protease complex, causes EGFR degradation indirectly by negatively regulating the expression and activity of the RING-type ubiquitin ligase Fbw7. All of these molecules represent novel molecular targets for EGFR-driven tumors.
Because ubiquitination plays an important role in EGFR endocytosis, trafficking, and lysosomal degradation, various ubiquitin ligases and deubiquitinating enzymes have been shown to regulate EGFR stability. The most extensively studied E3 ubiquitin ligase responsible for EGF-mediated EGFR polyubiquitination is c-CBL. In addition, AIP4/ITCH, pVHL, and UBE4B play roles in the polyubiquitination and degradation of EGFR [29,30]. We have recently identified that the HECT-type E3 ubiquitin ligase SMURF2 directly interacts with EGFR. SMURF2’s ubiquitin ligase activity is critical for maintaining EGFR protein stability [31]. Similar to SMURF2, deubiquitinating enzymes such as USP18 and USP2A protect EGFR from degradation. USP18 negatively regulates the expression of a microRNA (miR-7) which blocks EGFR translation by binding to the 3’-UTR of the EGFR transcript. USP2A antagonizes EGFR endocytosis via deubiquitination. Knowledge of the factors responsible for maintaining EGFR protein stability will allow for the development of novel drugs targeting this important receptor.
Downregulation of Mutant KRAS Protein: siRNA
Mutant KRAS has a high prevalence in pancreatic (90% KRAS mutant), colorectal (50% mutant), and lung (30% mutant) cancers. Effective treatment strategies are greatly needed for patients with KRAS-driven tumors. Direct inhibition of this protein is not feasible because of high intracellular GTP concentrations (micromolar) compared with the picomolar affinity between KRAS and GTP. Because KRAS acts as an on/off switch for multiple signaling pathways, the predominant strategy for targeting mutant KRAS tumors has focused on inhibiting downstream pathways including RAF-MEK-ERK and PI3K-AKT-mTOR. These approaches have had limited success because of the activation of compensatory signaling. In contrast, various studies using mutant KRAS–specific siRNA-mediated knockdown have shown promise.
Early attempts using either an anti-KRAS ribozyme [32] or shRNA-mediated knockdown of KRAS [33] demonstrated tumor growth delay of mutant KRAS-driven NSCLC. However, in these studies, compensatory activation of STAT3 and EGFR signaling was noted, suggesting that targeting mutant KRAS alone may not be sufficient. Along this point, a recent report demonstrated better in vivo tumor control when mutant KRAS siRNA was combined with either RAF or PI3K siRNAs [34]. These studies confirm the proof-of-principle that siRNA-mediated physical ablation of mutant KRAS is an attractive therapeutic strategy; however, we believe that the clinical success of these therapies depends on the long-term and tumor-specific delivery of siRNA payloads, which remains a challenge. Recently, one such siRNA delivery system has been developed, called Local Drug Eluter (LODER). LODER is able to protect siRNA from degradation and promotes localized prolonged siRNA release, an essential requirement for clinical success [35]. Using this delivery system, mutant KRAS siRNA (siG12D LODER) was delivered in pancreatic tumor xenograft models successfully. Such a system is currently being tested in a phase I (NCT01188785) and a phase II (NCT01676259) clinical trial for unresectable locally advanced pancreatic cancer patients.
Downregulation of Mutant KRAS Protein: Targeting Ubiquitination Machinery
Targeting posttranslational modifications of KRAS is a promising alternative to siRNA-based methods. Supporting this idea is a recent report that identifies a small molecule capable of inhibiting KRAS through its interaction with prenyl-binding protein PDEδ, a step critical for KRAS localization to the endomembrane [36]. Our work related to this strategy has focused on targeting the ubiquitination machinery that regulates KRAS stability. Monoubiquitination of mutant KRAS is known to enhance its activity, amplifying downstream signaling [37]. Based on the mechanistic studies of other proteins, we speculated that monoubiquitination competes with β-TrCP1–mediated polyubiquitination to maintain RAS protein levels. Interestingly, we observed that, under physiological conditions, the GTP-bound active form of mutant KRAS has a similar half-life as the GDP-bound inactive form of wild-type KRAS. Our observation is in direct contrast with previous reports which show that an active protein has a shorter half-life than its inactive form. We hypothesized that this difference is due to enhanced monoubiquitination of the mutant form of KRAS. Based on our prior work, we hypothesized that the E3 ubiquitin ligase SMURF2 regulates the stability of GTP-bound mutant KRAS. As expected, we found that the loss of SMURF2 caused mutant KRAS to become highly unstable.
Although we discovered that SMURF2 ubiquitin ligase activity is critical in maintaining mutant KRAS protein stability, we found that SMURF2 does not monoubiquitinate mutant KRAS as hypothesized. Instead, it indirectly protects mutant KRAS by polyubiquitinating and degrading β-TrCP1 [38]. As a proof-of-principle study, we showed that si/sh-RNA targeting of SMURF2 attenuates the growth of mutant KRAS–driven tumors. Although the finding that SMURF2 silencing can degrade mutant KRAS and block the growth of mutant KRAS–dependent tumors is encouraging, direct inhibition of SMURF2 ubiquitin ligase activity is not a viable strategy because of the critical importance of SMURF2 in mitosis [39]. We further found that SMURF2 can monoubiquitinate its partner ubiquitin conjugating enzyme (E2) UBCH5 to form an E3:E2 complex required for β-TrCP1 degradation. These findings suggest that the SMURF2:UBCH5 complex is critical in maintaining mutant KRAS protein stability and could be explored to develop the novel anti-KRAS strategy proposed in Figure 1B.
Future Directions
Inhibition of oncogenic kinase activity has led to the discovery and development of novel therapies that benefit patients. However, kinase inhibition has significant limitations including the emergence of drug resistance leading to limited therapeutic durability. Targeting protein stability is an interesting alternative strategy that is potentially applicable to any oncoprotein or tumor suppressor. Strategies such as disrupting protein-protein interactions to destabilize an oncoprotein or to stabilize a tumor suppressor protein are in the early phases of development. Although targeting protein-protein interactions using synthetic agents is challenging, recent advancements in structural and computational proteomics allow us to identify novel synthetic mimetics capable of disrupting specific protein-protein interaction to induce oncoprotein degradation. Such an approach can abolish the presence of an oncoprotein, which, in theory, can overcome resistance to kinase inhibitors. Future studies will determine if this approach can improve the outcomes of patients.
Acknowledgements
We thank Wayne Klohs for helpful discussion, Mary Davis for assistance in manuscript preparation, and Steven Kronenberg for assistance with graphics. This work was supported, in whole or in part, by National Institutes of Health grants R01CA131290 (to M.K.N.), R01CA160981 (to D.R.), P50CA097248 (project IV to M.K.N.), as well as R01 CA193690 and P01 CA085878 (A.R.). This work was also supported by the Alfred Taubman Scholarship (to T.S.L.). This project is a component of the University of Michigan Medical School's Fast Forward Strategic Research Initiative.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health grants R01CA131290 (to M.K.N.), R01CA160981 (to D.R.), P50CA097248 (project IV to M.K.N.), as well as R01 CA193690 and P01 CA085878 (A.R.). This work was also supported by the Alfred Taubman Scholarship (to T.S.L.).
References
- 1.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 2.Raymond E, Faivre S, Armand JP. Epidermal growth factor receptor tyrosine kinase as a target for anticancer therapy. Drugs. 2000;60(Suppl. 1):15–23. doi: 10.2165/00003495-200060001-00002. [discussion 41–12] [DOI] [PubMed] [Google Scholar]
- 3.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.Kohl NE, Conner MW, Gibbs JB, Graham SL, Hartman GD, Oliff A. Development of inhibitors of protein farnesylation as potential chemotherapeutic agents. J Cell Biochem Suppl. 1995;22:145–150. doi: 10.1002/jcb.240590819. [DOI] [PubMed] [Google Scholar]
- 6.Vasan N, Boyer JL, Herbst RS. A RAS renaissance: emerging targeted therapies for KRAS-mutated non–small cell lung cancer. Clin Cancer Res. 2014;20:3921–3930. doi: 10.1158/1078-0432.CCR-13-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemoine M, Younes A. Histone deacetylase inhibitors in the treatment of lymphoma. Discov Med. 2010;10:462–470. [PubMed] [Google Scholar]
- 8.Neckers L. Hsp90 inhibitors as novel cancer chemotherapeutic agents. Trends Mol Med. 2002;8:S55–S61. doi: 10.1016/s1471-4914(02)02316-x. [DOI] [PubMed] [Google Scholar]
- 9.Shimamura T, Perera SA, Foley KP, Sang J, Rodig SJ, Inoue T, Chen L, Li D, Carretero J, Li YC. Ganetespib (STA-9090), a nongeldanamycin HSP90 inhibitor, has potent antitumor activity in in vitro and in vivo models of non–small cell lung cancer. Clin Cancer Res. 2012;18:4973–4985. doi: 10.1158/1078-0432.CCR-11-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Socinski MA, Goldman J, El-Hariry I, Koczywas M, Vukovic V, Horn L, Paschold E, Salgia R, West H, Sequist LV. A multicenter phase II study of ganetespib monotherapy in patients with genotypically defined advanced non–small cell lung cancer. Clin Cancer Res. 2013;19:3068–3077. doi: 10.1158/1078-0432.CCR-12-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramalingam SS, Owonikoko TK, Behera M, Subramanian J, Saba NF, Kono SA, Gal AA, Sica G, Harvey RD, Chen Z. Phase II study of docetaxel in combination with everolimus for second- or third-line therapy of advanced non–small-cell lung cancer. J Thorac Oncol. 2013;8:369–372. doi: 10.1097/JTO.0b013e318282709c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 14.Rauch J, Volinsky N, Romano D, Kolch W. The secret life of kinases: functions beyond catalysis. Cell Commun Signal. 2011;9 doi: 10.1186/1478-811X-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 16.Deb TB, Su L, Wong L, Bonvini E, Wells A, David M, Johnson GR. Epidermal growth factor (EGF) receptor kinase-independent signaling by EGF. J Biol Chem. 2001;276:15554–15560. doi: 10.1074/jbc.M100928200. [DOI] [PubMed] [Google Scholar]
- 17.Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, Hung MC. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu H, Cao X, Ali-Osman F, Keir S, Lo HW. EGFR and EGFRvIII interact with PUMA to inhibit mitochondrial translocalization of PUMA and PUMA-mediated apoptosis independent of EGFR kinase activity. Cancer Lett. 2010;294:101–110. doi: 10.1016/j.canlet.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- 20.Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, Lee DC. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- 21.Ahsan A, Ramanand SG, Bergin IL, Zhao L, Whitehead CE, Rehemtulla A, Ray D, Pratt WB, Lawrence TS, Nyati MK. Efficacy of an EGFR-specific peptide against EGFR-dependent cancer cell lines and tumor xenografts. Neoplasia. 2014;16:105–114. doi: 10.1593/neo.14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (bortezomib) Cancer Invest. 2004;22:304–311. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 24.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res. 2008;14:5318–5324. doi: 10.1158/1078-0432.CCR-07-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ungermannova D, Lee J, Zhang G, Dallmann HG, McHenry CS, Liu X. High-throughput screening AlphaScreen assay for identification of small-molecule inhibitors of ubiquitin E3 ligase SCFSkp2-Cks1. J Biomol Screen. 2013;18:910–920. doi: 10.1177/1087057113485789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahsan A, Ramanand SG, Whitehead C, Hiniker SM, Rehemtulla A, Pratt WB, Jolly S, Gouveia C, Truong K, Van Waes C. Wild-type EGFR is stabilized by direct interaction with HSP90 in cancer cells and tumors. Neoplasia. 2012;14:670–677. doi: 10.1593/neo.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahsan A, Ray D, Ramanand SG, Hegde A, Whitehead C, Rehemtulla A, Morishima Y, Pratt WB, Osawa Y, Lawrence TS. estabilization of the epidermal growth factor receptor (EGFR) by a peptide that inhibits EGFR binding to heat shock protein 90 and receptor dimerization. J Biol Chem. 2013;288:26879–26886. doi: 10.1074/jbc.M113.492280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L, Yang H. The von Hippel-Lindau tumor suppressor protein promotes c-Cbl-independent poly-ubiquitylation and degradation of the activated EGFR. PLoS One. 2011;6:e23936. doi: 10.1371/journal.pone.0023936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirisaengtaksin N, Gireud M, Yan Q, Kubota Y, Meza D, Waymire JC, Zage PE, Bean AJ. UBE4B protein couples ubiquitination and sorting machineries to enable epidermal growth factor receptor (EGFR) degradation. J Biol Chem. 2014;289:3026–3039. doi: 10.1074/jbc.M113.495671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray D, Ahsan A, Helman A, Chen G, Hegde A, Gurjar SR, Zhao L, Kiyokawa H, Beer DG, Lawrence TS. Regulation of EGFR protein stability by the HECT-type ubiquitin ligase SMURF2. Neoplasia. 2011;13:570–578. doi: 10.1593/neo.11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YA, Nemunaitis J, Scanlon KJ, Tong AW. Anti-tumorigenic effect of a K-ras ribozyme against human lung cancer cell line heterotransplants in nude mice. Gene Ther. 2000;7:2041–2050. doi: 10.1038/sj.gt.3301331. [DOI] [PubMed] [Google Scholar]
- 33.Sunaga N, Shames DS, Girard L, Peyton M, Larsen JE, Imai H, Soh J, Sato M, Yanagitani N, Kaira K. Knockdown of oncogenic KRAS in non–small cell lung cancers suppresses tumor growth and sensitizes tumor cells to targeted therapy. Mol Cancer Ther. 2011;10:336–346. doi: 10.1158/1535-7163.MCT-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan TL, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, Hsu DJ, Grace D, Carver JO, Zuber J. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov. 2014;4:1182–1197. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zorde Khvalevsky E, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, Shemi A, Golan T, Domb AJ, Yavin E. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20723–20728. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 37.Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, Haviv S, Asara JM, Pandolfi PP, Cantley LC. Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci Signal. 2011;4:ra13. doi: 10.1126/scisignal.2001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shukla S, Allam US, Ahsan A, Chen G, Krishnamurthy PM, Marsh K, Rumschlag M, Shankar S, Whitehead C, Schipper M. KRAS protein stability is regulated through SMURF2: UBCH5 complex-mediated beta-TrCP1 degradation. Neoplasia. 2014;16:115–128. doi: 10.1593/neo.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osmundson EC, Ray D, Moore FE, Gao Q, Thomsen GH, Kiyokawa H. The HECT E3 ligase Smurf2 is required for Mad2-dependent spindle assembly checkpoint. J Cell Biol. 2008;183:267–277. doi: 10.1083/jcb.200801049. [DOI] [PMC free article] [PubMed] [Google Scholar]