Abstract
Lysophosphatidic acid (LPA), a blood-borne lipid mediator, is present in elevated concentrations in ascites of ovarian cancer patients and other malignant effusions. LPA is a potent mitogen in cancer cells. The mechanism linking LPA signal to cancer cell proliferation is not well understood. Little is known about whether LPA affects glucose metabolism to accommodate rapid proliferation of cancer cells. Here we describe that in ovarian cancer cells, LPA enhances glycolytic rate and lactate efflux. A real time PCR-based miniarray showed that hexokinase II (HK2) was the most dramatically induced glycolytic gene to promote glycolysis in LPA-treated cells. Analysis of the human HK2 gene promoter identified the sterol regulatory element-binding protein as the primary mediator of LPA-induced HK2 transcription. The effects of LPA on HK2 and glycolysis rely on LPA2, an LPA receptor subtype overexpressed in ovarian cancer and many other malignancies. We further examined the general role of growth factor-induced glycolysis in cell proliferation. Like LPA, epidermal growth factor (EGF) elicited robust glycolytic and proliferative responses in ovarian cancer cells. Insulin-like growth factor 1 (IGF-1) and insulin, however, potently stimulated cell proliferation but only modestly induced glycolysis. Consistent with their differential effects on glycolysis, LPA and EGF-dependent cell proliferation was highly sensitive to glycolytic inhibition while the growth-promoting effect of IGF-1 or insulin was more resistant. These results indicate that LPA- and EGF-induced cell proliferation selectively involves up-regulation of HK2 and glycolytic metabolism. The work is the first to implicate LPA signaling in promotion of glucose metabolism in cancer cells.
Introduction
Lysophosphatidic acid (LPA), a lysophospholipid mediator, is present in elevated concentrations in ascites of ovarian cancer patients and other malignant effusions [1–3]. LPA regulates diverse biological processes including proliferation, survival, migration and invasion of tumor cells [4–6]. These effects of LPA are mediated via LPA binding to its cognate G-protein coupled receptors (GPCRs) [4–6]. The LPA1, LPA2 and LPA3 receptors are members of the endothelial gene (Edg) subfamily of GPCRs. The purinergic family receptor LPA4 and related LPA5, LPA6 and LPA7 receptors constitute the non-Edg subgroup of LPA receptors, which are structurally distant from the Edg LPA receptors [4–6]. The LPA receptors are expressed differentially in adult tissues. Accumulating evidence suggests that LPA receptors are functionally redundant or opposite. The cellular responses to LPA are determined by the combination of various LPA receptors and G proteins present in a cell [4,6–8].
Among LPA receptors, LPA2 has been the most consistently up-regulated in diverse human malignancies including cancers of ovary [9], breast [10], stomach [11], colorectum [12] and thyroid [13]. Overexpression of LPA2 has been linked to proliferation of ovarian and colon cancer cells, and mesothelioma cells [14–16]. However, molecular mechanisms connecting LPA to proliferative and other oncogenic processes remain poorly defined. We have recently demonstrated that LPA stimulated de novo lipid synthesis in ovarian cancer cells via LPA2-mediated up-regulation of lipogenic enzymes fatty acid synthase and acetyl-CoA carboxylase [17]. The pro-lipogenic action of LPA is essential for LPA-induced cell proliferation, likely through contributing to membrane biogenesis. In addition to its role in lipogenic metabolism, a number of previous studies suggest that LPA may also regulate glucose uptake and metabolism. For examples, LPA stimulated glucose transport in Xenopus oocytes [18], in ethanol-treated rat astrocytes [19], mouse L6 myotubes and 3 T3-L1 adipocytes [20]. In the C13NJ microglia cell line, a proteome analysis indicated LPA increased expression of several glycolytic enzymes such as α-enolase and L-lactate dehydrogenase B [21]. In murine mesangial cells, LPA was found to stimulate hexokinase II (HK2) expression and glycolytic activity although the physiological relevance to LPA-coupled biological functions was not explored [22]. It is generally believed that a primary function of growth factors is to regulate glucose uptake and catabolism, thus enabling anabolic pathways required for mitogenesis [23]. This particularly applies to non-transformed cells where steady-state aerobic glycolysis is generally low in the absence of growth stimuli. Malignant cells are, however, metabolically transformed to sustain a high basal glycolytic rate [24–26]. Hyperactive glycolysis not only provides quick ATP but also serves as a primary route for carbon influx, which is required for biosynthesis of complex macromolecules and formation of organelles in actively proliferating cancer cells [24–26]. Substantial evidence suggests that the glycolytic phenotype of cancer cells is driven by overexpression or hyperactivity of key glycolytic enzymes as a result of activation of oncogene and/or inactivation of tumor suppressors [27–30]. Mutations of mitochondrial DNA that impair functions of the respiratory complexes may also underlie high glycolytic rate seen in cancers [31]. However, the contribution of growth factors to cancer-associated glycolysis is unclear. It remains to be determined whether cancer cell proliferation requires stimulation of additional glycolysis over the high constitutive background.
No studies have thus far connected LPA to glucose metabolism in the context of malignant cells. In the present study, we show that LPA up-regulates glycolytic metabolism to promote proliferation of ovarian cancer cells. The effect of LPA is mediated through LPA2-dependent activation of the sterol regulatory element-binding protein 1 (SREBP-1) and HK2 transcription. We further examined the general significance of growth factor-induced glycolysis in cell proliferation and found that LPA or epidermal growth factor (EGF), but not insulin-like growth factor 1 (IGF-1) or insulin, relies on robust induction of glycolysis to support proliferation of cancer cells. These findings revealed a previously unrecognized role and mechanism for LPA in regulation of glucose metabolism in cancer cells.
Materials and Methods
Reagents
LPA (1-oleoly, 18:1) was obtained from Avanti Polar Lipids, Inc. (Alabaster, AL). Prior to use, LPA was dissolved in PBS containing 0.5% fatty acid-free bovine serum albumin (BSA) from Roche (Indianapolis, IN). d-[5-3H(N)]-glucose was purchased from Perkin Elmer (Boston, MA). EGF, 2-deoxy-d-glucose (2-DG), and AG 1478 were obtained from Sigma Aldrich (St. Louis, MO). IGF-1 and insulin were from Invitrogen (Gaithersburg, MD). Plasmid DNAs were purified using the endo-free purification kit from Qiagen (Valencia, CA). Dharmafect 1 was obtained from Dharmacon, Inc. (Lafayette, CO) and TransIT-TKO was obtained from Mirus Bio (Madison, WI). Luciferase assay reagents were obtained from Promega (Madison, WI). Anti-HK2 antibody was obtained from Cell Signaling (Danvers, MA). The TaqMan Universal PCR Master Mix and qPCR probes for HK2 and GAPDH were obtained from Applied Biosystems (Carlsbad, CA).
Cell Culture
The sources of ovarian cancer cell lines were described previously [16]. These cells were cultured in RPMI medium 1640 supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cell lines were frozen at early passages and used for less than a month in continuous culture.
Gene Knockdown
Lentiviruses carrying short hairpin RNA (shRNA) for LPA1–3 receptors were kind gifts from Dr. S. Huang (Georgia Regents University) [32]. The siRNA oligos for LPA1, LPA2 LPA3, HK2 and SREBP-1 were obtained from Applied Biosystems. Cells were plated in 6-well plates. At around 50% confluence, cells were transfected with target specific siRNA or non-targeting control siRNA (150 pM) with Dharmafect 1 (4 μL, 12 to 16 hours) following the manufacturer's protocol. Approximately 48 hours post transfection, the cells were serum starved overnight before LPA treatment.
Western Blotting
Cells were lysed as previously described [33]. Total cellular proteins were resolved by SDS-PAGE, transferred to immunoblot membrane (polyvinylidene difluoride) (BIO-RAD, Hercules, CA), and immunoblotted with antibodies following the protocols of manufacturers. Immunocomplexes were visualized with an enhanced chemiluminescence detection kit from Amersham (Piscataway, NJ).
Quantitative PCR (qPCR)
Total cellular RNA was isolated from cultured cells using Trizol (Invitrogen). Complementary DNA (cDNA) was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The relative levels of LPA1, LPA2, LPA3, HK2 and GAPDH were determined by reverse transcription (RT) followed by qPCR using gene specific probes, the TaqMan Universal PCR Master Mix, and the Applied Biosystems 7900HT Real-Time PCR System.
Luciferase Assays
The human HK2 promoter sequence (− 1476 to + 73) [34] was PCR amplified from human genomic DNA and cloned into the pGL2-Basic-Luc at XhoI and HindIII sites to construct the luciferase reporter pGL2-1476-HK2-Luc. The truncated forms (− 478 to + 73, − 273 to + 73) were generated by PCR amplification of the corresponding fragments from pGL2-1476-HK2-Luc and re-inserted into the pGL2-Basic-Luc at the XhoI and HindIII sites. The promoter sequences in these plasmids were verified by automatic sequencing. Two potential consensus sites for SREBP (CCAGTCGCCCACACC and CACGCTCCCCCCACCA) in pGL2-478-HK2-Luc were converted into inactive sequences (CCAGGTGTCTTACACC and CACGCGTCTCTTACCA) by site-directed mutagenesis using Lightning Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer's protocol. Primers used for these mutant constructs were listed in Table 1. Caov-3 and OVCA-432 cells were transfected with the luciferase vectors using TransIT-TKO according to the manufacturer. About 48 hours after transfection, the cells were starved overnight and treated with LPA or vehicle (BSA) for 12 to 16 hours. Cell extracts were prepared and assayed for luciferase activity using a luciferase assay kit from Promega.
Table 1.
Oligos Used in the Work
| -1476 | 5′-GCACTCGAGGGATTATGATTTTTGTTTATTTTTCCT-3′ (forward) |
| -478 | 5′-GCACTCGAGCCGGCCGTGCTACAATAG-3′ (forward) |
| -273 | 5′ -GCACTCGAGCTCATGCGCCTTTCCGTC-3′ (forward) |
| + 73 | 5′-GCAAAGCTTCGGATTTTCTTAGCTGGGTG-3′ (reverse) |
| SRE1 Mut | 5′-CAGAGGCCCGTTTTTCCAGGTGTCTTACACCCCGGGTCC GCGAT-3′ (forward) |
| 5′-ATCGCGGACCCGGGGTGTAAGACACCTGGA AAA ACG GGC CTC TG-3′ (reverse) | |
| SRE2 Mut | 5′-GGGTCCGCGATCACGCGTCTCTTACCCATAGCCGAGCCTG-3′ (forward) |
| 5′-CAGGCTCGGCTATGGGTAAGAGACGCGTGATCGCGGACCCG-3′ (reverse) |
PCR-Based Gene Expression Array
The human glucose metabolism RT2 profiler PCR array (PAHS-006ZA) was obtained from Qiagen (Valencia, CA). Caov-3 cells were treated with LPA or vehicle for 12 hours before RNA isolation using RNeasy mini kit (Qiagen). Complementary DNA (cDNA) was synthesized using RT2 First Strand Kit (Qiagen) and qPCR was carried out using RT2 SYBR®Green qPCR Mastermix according to the manufacturer (Qiagen). The CT values were normalized to the levels of GAPDH. Fold changes relative to vehicle-treated control cells were calculated and presented in bar graphs.
Glycolysis
Glycolysis in cultured cells was measured as describes [28] with some modifications. Briefly, cells were plated in 6-well or 12-well plates, serum starved and treated with vehicle, LPA or other growth factors for 12 to 16 hours. Radioactive glucose [5-3H (N)] was added to the medium at a concentration of 1 μCi/ml and incubated for 24 hours. Hydrochloric acid was then added to medium at a final concentration of 0.2 N to terminate all biological reactions. The acidified medium (0.6 ml) was collected into 15 ml high-clarity polypropylene conical tubes (Falcon). A 0.5 ml micro centrifuge tube containing 0.25 ml distilled water was uncapped and inserted into the 15 ml tube. Precautions were taken to prevent direct contact between two solutions. The 15-ml tubes were tightly capped to allow diffusion between two liquid phases for more than 48 hours. Radioactivities in water and medium were determined by liquid scintillation counting. The glycolytic rate was calculated with the formula 3.4a/(a + b) where a is radioactivity present in water and b is activity present in the medium [19].
Lactate Measurement
The lactate contents in culture supernatants were determined using a lactate assay kit (Eton Bioscience, San Diego, CA) following the protocol of the manufacture.
Hexokinase Activity Assay
Cells were lysed with a lysis buffer containing 15 mM Tris pH 7.8, 0.25 mM sucrose, 0.5 mM dithiothreitol (DTT), 1 mM aminohexanoic acid, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 2 μg/ml leupeptin. The lysates were then sonicated (5 times, 30 seconds each) in a water bath, followed by centrifugation at 2000 g at 4°C for 5 minutes. The cell extracts (50 μl) were added to 950 μl of reaction buffer (100 mM Tris-HCl, pH 7.8, 5 mM ATP, 10 mM MgCl2, 10 mM glucose, 0.4 mM NADP, and 0.15 U/ml of G6PD (Sigma-Aldrich) and incubated at 37°C. The HK enzymatic activity was monitored by measuring glucose-6-phosphate dehydrogenase (G6PD)-dependent conversion of NADP to NADPH spectrophotometrically at 340 nm. The activity was presented as nanomoles of NADPH generated from one milligram of protein per minute at 37°C.
Quantification of Cellular ATP
After washing twice with PBS, cells in 6-well plates were lysed on ice in H2O containing 0.75% NP-40 and vertexed at full speed for 10 seconds, and incubated on ice for 10 minutes. After centrifugation (16,000g, 3 minutes), the supernatants were collected and diluted 100 times with H2O for ATP measurement with an ATP bioluminescence assay kit (Sigma-Aldrich). ATP concentrations were calculated from the standard curve and presented as nMol of ATP per 106 cells.
Statistics
All numerical data were presented as mean ± SD of triplicate assays, representative of three independent experiments. The statistical significances were analyzed using Student's t test unless otherwise stated, where P < .05 was considered statistically significant. In all figures, the statistical significances were indicated with * if P < .05 or ** if P < .01.
Results
LPA Activates Glycolysis in Ovarian Cancer Cells
LPA is an important mitogen in ovarian cancer and other neoplastic cells. To understand the molecular mechanism of LPA-mediated cancer cell proliferation, we examined whether LPA treatment stimulates glucose metabolism using ovarian cancer cells as a model. During glycolysis, one molecule of water is released when 2-phosphoglycerate is converted to phosphoenolpyruvate. By labeling cells with 5-3H glucose, we were able to quantitate the glycolytic rate by measuring generation of 3H water [28]. We treated a panel of ovarian cancer cell lines with LPA (10 μM) and labeled the cells with 5-3H glucose. LPA stimulated one- to four-fold increases in glycolysis in ovarian cancer cell lines Caov-3 and OVCA-432 cells (Figure 1A). The LPA-induced increases in the glycolytic flux were concurrent with significant rises in lactate present in culture supernatants of LPA-treated cells (Figure 1A). Similar effects were observed in LPA-treated OVCAR-3 and OVCAR-8 cells (data not shown).
Figure 1.
LPA activates glycolysis in ovarian cancer cells. (A) Ovarian cancer cell lines in 6-well plates were treated with LPA (10 μM) or vehicle for 12 to 16 hours before addition of 5-3H glucose (1 μCi/ml). The cells were then incubated for 24 hours before measuring conversion of 5-3H glucose to 3H-H2O. The results were presented as % conversion of glucose (upper). Lactate concentrations in culture supernatants were quantified with a lactate assay kit (lower). (B) Caov-3 and OVCA-432 cells in 12 well plates were treated with increasing concentrations of LPA and glycolytic rate quantified as in (A). Cell numbers were determined with a Coulter counter. For this and all following figures, data are mean ± SD of triplicates, representative of three independent experiments. Statistical significances of data were determined by Student's t test and indicted by * if P < 0.05 or by ** if P < 0.01.
LPA stimulated glycolysis in a dose-dependent manner similar to that of the mitogenic response to LPA in these cells (Figure 1B). The LD50 values were approximately 0.3 to 0.6 μM in Caov-3 and 2.7 to 4.1 μM in OVCA-432 cells. The activities reached plateau from 10 to 50 μM. We subsequently used 10 μM LPA for the remainder of the study.
LPA Up-Regulates Expression of HK2 to Promote Glycolysis
LPA is known to up-regulate expression of a variety of pro-oncogenic protein factors such as VEGF, COX-2, IL-6, IL-8, cyclin D1 and kruppel-like factor 5 [35–41]. We determined whether LPA stimulated glycolysis via regulation of any rate-limiting enzymes of the glycolytic pathway. To this end, we treated Caov-3 cells with LPA for 12 hours and isolated RNA to perform a PCR-based glucose metabolism array to identify gene expression signatures involved in glycolysis. While LPA treatment led to modest up- or down-regulation (less than three-fold) of a number of glycolytic genes (Figure 2A), the most robust change was close to 20-fold induction of HK2 by LPA. This pattern of changes in gene expression induced by LPA was reproducible in an independent array experiment (Supplemental Figure S1). Quantitative PCR (qPCR) analysis confirmed that LPA induced HK2 mRNA expression in Caov-3 in a time-dependent manner with maximal level reached at around 12 hours (Figure 2B). Similarly, LPA stimulated potent and sustained induction of HK2 mRNA in OVCA-432 cells (Figure 2B).
Figure 2.
LPA stimulates expression of HK2. (A) Caov-3 cells were treated with LPA (10 μM) or vehicle (BSA) for 12 hours. The human glucose metabolism RT2 profiler PCR array was performed on cDNAs prepared from total cellular RNA as described in Experimental Procedures. Expression levels of glycolytic genes were normalized on GAPDH and then compared between LPA-treated and control cells and presented as fold changes over vehicle controls (defined as arbitrary 1). (B, C) Caov-3 and OVCA-432 cells were treated with LPA (10 μM) for the indicated periods of time (hours). Induction of HK2 mRNA (B) and protein (C) was analyzed by RT-qPCR and immunoblotting, respectively. HK2 protein levels were quantified by densitometry and presented as fold changes relative to control cells. (D) Hexokinase activity was determined in Caov-3 and OVCA-432 cells treated for 16 hours with LPA or vehicle and presented as nanomoles/mg protein/min at 37 °C. (E) HK-2 expression was down-regulated by HK2 specific siRNA in Caov-3 and OVCA-432 cells. LPA-induced glycolysis in HK-2 knockdown and control siRNA-treated cells were determined as described in Figure 1.
Western blotting analysis confirmed that LPA induced expression of HK2 protein in ovarian cancer cell lines (Figure 2C). The kinetics of HK2 protein expression was essentially consistent with the time course of LPA induction of HK2 mRNA. While HK2 mRNA peaked around 12 hours, the protein levels reached a plateau between 12 and 16 hours. Compared to the multi-fold induction of HK2 mRNA, the increases in the full-length HK2 protein did not seem to be proportional, suggesting that post-transcriptional or post-translational modifications of HK2 expression occurred. Indeed, on Western blotting analysis of LPA-treated cells, prolonged exposure revealed a novel band of approximately 70 kDa specifically present in LPA-treated cells, likely representing a product cleaved from the full-length HK2 following its induction.
The majority of HK2 protein in cells is attached to the mitochondria. The mitochondria-associated HK2 is considered to be the active form of the enzyme, contributing significantly to the cellular glycolytic activity [42,43]. LPA stimulation increased both mitochondrial and cytosolic fractions of HK2 (data not shown). In line with this, LPA-treated cells showed significant increases in HK enzymatic activity (Figure 2D). To gain molecular evidence for involvement of HK2 in LPA-driven glycolysis, we used siRNA to knockdown HK2 expression. As shown in Figure 2E, down-regulation of HK2 in these cells attenuated LPA-driven glycolysis, indicating that HK2 mediates the glycolysis-promoting effect of LPA.
LPA Stimulates HK2 Expression Through the SREBP-1 Transcription Factor
We next explored the mechanism underlying LPA up-regulation of HK2. A fragment (− 1476 to + 73) of the human HK2 gene promoter [34] was cloned from human genomic DNA and inserted into the pGL2-Basic-Luc reporter. Serial deletion of 5′ sequences generated two truncation mutants containing − 478 to + 73, and − 273 to + 73 fragments of the promoter. These constructs were transfected into Caov-3 and OVCA-432 cells, and LPA-induced luciferase activity was determined by luciferase assays. As illustrated in Figure 3, LPA treatment stimulated a multi-fold increases in luciferase activity in cells transfected with pGL2-1476 to + 73-Luc. Comparable stimulation of luciferase activity by LPA was observed in cells transfected with pGL2-478 to + 73-Luc. However, further deletion to − 273 to + 73 resulted in a drastic decrease in luciferase activity induced by LPA (Figure 3), suggesting that major responsive element(s) required for LPA stimulation resided within the sequence between − 478 and − 273. The Insilico analysis revealed several potential cis regulatory elements within this region, including sites for cAMP-responsive element binding protein, nuclear factor 1, specificity protein 1 (Sp1) and SREBP. We have previously shown that LPA activates Sp1 and SREBP in ovarian cancer cells [17,36]. However, Sp1 is a transcription factor activated rapidly following LPA treatment [36], which does not echo the delayed HK2 induction in LPA-treated cells. HK2 has been previously shown to be transcriptionally induced by SREBP-1 in human myocytes [44] and in rat adipose tissue and skeleton muscles [45]. The existence of two closely positioned sterol response elements (SREs) within the responsive region and strong activation of SREBP by LPA in ovarian cancer cell lines [17] prompted us to examine the potential role of SREBP in transcriptional activation of HK2. As shown in Figure 3A, mutation of either SRE sites (SRE1 or SRE2) significantly reduced LPA-driven luciferase activity. Simultaneous mutation of both SRE sites, however, did not further eliminate LPA-induced activity. The remaining response of the double mutant was similar to that of individual mutation. The results indicated that the two adjacent SREs act in concert to drive HK2 transcription. Interestingly, the SREBP-1 regulation of the fatty acid synthase promoter has been also reported to rely on two tandem SRE sites for full activation [46]. In further support of the role of SREBP, siRNA knockdown of SREBP-1abrogated LPA-dependent up-regulation of HK2 in Caov-3 cells (Figure 3B). SREBP siRNA also consistently reduced LPA-induced HK-2 protein expression in OVCA-432 cells. The minor remaining activity in OVCA-432 cells (Figure 3B) could be due to incomplete knockdown or the presence of SREBP1-independent input.
Figure 3.
LPA stimulates HK-2 expression through transcriptional activation mediated by SREBP. (A) Caov-3 and OVCA-432 cells were transfected with the indicated luciferase constructs. After 48 hours, transfected cells were starved for 16 to 24 hours and then treated for 16 hours with LPA (10 μM) or vehicle before measurement of luciferase activity with a luminometer. LPA-induced luciferase activities were presented as fold increases over vehicle-treated control cells. (B) LPA-induced HK2 expression in SREBP-1 siRNA knockdown cells and control cells (Ctrl-si) was analyzed by immunoblotting and densitometry. The cells were treated for 16 hours with 10 μM LPA or vehicle.
LPA2 is the Major LPA Receptor That Mediates HK2 Expression and Glycolysis
Caov-3, OVCA-432 and other ovarian cancer cell lines express the Edg LPA receptors LPA1, LPA2, and LPA3 while the non-Edg receptors are either absent or are expressed inconsistently among ovarian cancer cell lines [38,47]. To identify the LPA receptor subtype responsible for the pro-glycolytic action of LPA, we focused on the Edg LPA receptors. LPA receptor siRNA was used to knockdown expression of LPA1, LPA2, or LPA3 in Caov-3 cells. Knockdown of LPA2 but not other Edg LPA receptors, led to suppression of LPA-induced HK2 expression (Figure 4). A similar result was observed in OVCA-432 cells where LPA receptors were stably knocked down using lentivirus-mediated shRNA (Figure 4). Consistent with the requirement of HK2 for LPA-induced glycolysis, knockdown of LPA2 in both cell lines inhibited LPA-induced glycolysis (Figure 4). Therefore LPA2 is the major LPA receptor subtype accounting for LPA up-regulation of HK2 and glycolysis in ovarian cancer cells. The conclusion is also consistent with the essential role of LPA2 in LPA-dependent activation of SREBP as we demonstrated recently [17]. In addition to ovarian cancer cells, we also examined the effects of LPA in breast and colon cancer cell lines that have been reported to overexpress LPA2 [10,15,48]. As shown in Figure 4B, LPA stimulated HK2 expression, glycolysis and proliferation of the MDA-231 breast cancer and HCT116 colon cancer cell lines.
Figure 4.
LPA2 is the major LPA receptor subtype responsible for HK2 induction and glycolysis. Each of LPA1 -3 receptors was knocked down by siRNA in Caov-3 cells or by lentivirus-transduced shRNA in OVCA-432 cells. (A) The cells were serum starved and treated with LPA (10 μM) or vehicle for 16 hours before immunoblotting analysis of HK2 protein. (B) Glycolysis of these cells was determined as in Figure 1 and the results were presented as fold increases relative to vehicle-treated control cells (defined as one fold). (C) LPA-induced increases in HK2 expression and glycolysis were reproduced in LPA2-expressing MDA-231 breast cancer and HCT116 colon cancer cell lines.
Growth Factors Differ in Their Ability to Activate HK2 Expression and Glycolysis
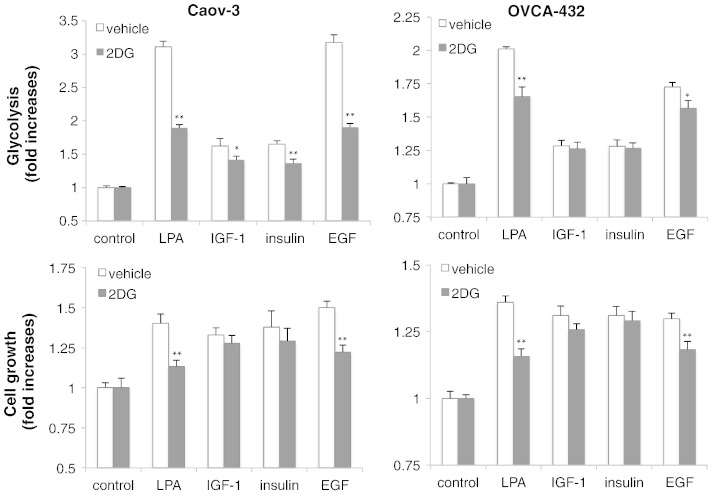
Although active glycolysis is a general phenomenon of rapidly growing cancer cells, few studies have addressed potential regulatory role of growth factors present in tumor microenvironments. To understand the general role of glycolytic metabolism in cancer cell proliferation, we compared LPA with a number of growth factors involved in ovarian cancer pathogenicity including EGF, IGF-1, and insulin. Interestingly, when their proliferative responses were adjusted to be at similar levels by using appropriate concentrations of growth factors, their glycolytic activities varied dramatically (Figure 5A). LPA and EGF strongly activated glycolysis while IGF-1 and insulin showed only modest stimulatory effects on glycolysis, not proportional to their mitogenic activities. Likewise, IGF-1 and insulin only slightly increased HK2 expression in contrast to the strong induction of HK2 by LPA or EGF in these cells (Figure 5B). In addition, all these growth factors increased cellular ATP levels in Caov-3 and OVCA-432 cells (Figure 5A).
Figure 5.
LPA, EGF, insulin and IGF-1 display differential effects on glycolysis and HK2 expression. (A) Caov-3 and OVCA-432 cells in 6-well plates were serum starved and treated with LPA (10 μM), IGF-1 (20 ng/ml in Caov-3, 25 ng/ml in OVCA-432), insulin (10 μg/ml in Caov-3, 0.125 μg/ml in OVCA-432), or EGF (40 ng/ml in Caov-3, 2 ng/ml in OVCA-432). Cell numbers, glycolysis, and lactate levels were determined as in Figure 1. ATP contents in the cells treated for 20 hours with each of these growth factors were measured with an ATP bioluminescence assay kit and the data presented as nMol ATP per 106 cells. (B) Immunoblotting analysis of HK2 induction was performed in Caov-3 and OVCA-432 cells stimulated for 16 hours with LPA, EGF, IGF-1 or insulin as described in (A). (C) The effects of LPA on glycolysis and HK2 expression were examined as in (A) and (B) in the absence or presence of the EGFR inhibitor AG 1478 (0.75 μM). AG 1478 was added to culture 0.5 hour before LPA.
LPA has been shown to transactivate EGFR, which is involved in diverse biological responses to LPA [38]. The similarity between LPA and EGF in up-regulation of HK2 and glycolysis suggests that LPA may exert its effects via EGFR transactivation. We examined the possibility by treatment of Caov-3 and OVCA-432 cells with LPA in the presence of the EGFR specific inhibitor AG 1478. As demonstrated in Figure 5C, inhibition of EGFR prevented LPA from inducing HK2 expression and glycolysis in these cells.
LPA and EGF Stimulates Glycolysis to Promote Cell Proliferation
The differential effects of LPA, EGF, IGF-1 and insulin suggest that robust induction of glycolysis by growth factors may not be necessarily required to support active cell proliferation. A modest increase such as that seen in IGF-1 or insulin-stimulated cells may be sufficient to supply cells with quick energy and biosynthetic intermediates. The excess glycolysis seen in the context of LPA or EGF stimulation may be redundant and indispensable for cell proliferation. Hyperactive glycolysis and/or lactate production in vivo have been linked to other non-autonomous cellular functions in cancer such as acidification of extracellular environment and evasion of immune surveillance [49,50]. To address whether up-regulation of glycolysis is indeed required for LPA-induced cell proliferation, we used 2-DG, a well-defined glycolytic inhibitor that targets primarily HK2 to prevent glucose from entering the glycolytic pathway [51,52]. To this end, Caov-3 and OVCA-432 cells were stimulated with LPA, EGF, IGF-1 or insulin in the presence or absence of 2-DG. We chose nontoxic concentrations of 2-DG so that LPA-driven glycolysis was significantly reduced but remained to be higher than the levels in IGF- or insulin-stimulated cells. As shown in Figure 6, this partial inhibition of glycolysis was sufficient to suppress LPA-induced proliferation in Caov-3 and OVCA-432 cells. EGF-induced growth of these cells was also sensitive to the partial inhibition of glycolysis. However, in Caov-3 cells, 2-DG only slightly inhibited IGF-1- or insulin-induced glycolysis. Consistent with this, 2-DG did not significantly affect mitogenic activity of insulin or IGF-1. In OVCA-432 cells, 2-DG had little, if any, effects on either glycolytic or proliferative effects of IGF-1 or insulin (Figure 6). The results indicate that enhancement of glycolysis is specifically required as an integral component of the complex proliferative response to LPA or EGF.
Figure 6.
Up-regulation of glycolysis is required for LPA and EGF-induced cell proliferation. Caov-3 and OVCA-432 cells were treated in the same conditions as in Figure 5A in the presence of 2-DG (1.25 mM in Caov-3 and 3 mM in OVCA-432) or vehicle. 2-DG was added 12 hours after the growth factors. Cell numbers (upper) and glycolysis (lower) stimulated by these growth factors were quantified as in Figure 1. The data were presented as fold increases over the corresponding controls that are defined as one fold.
Discussion
Previous studies in Xenopus oocytes, primary mammalian cells such as rat astrocytes and murine mesangial cells, and murine cell lines hinted at the possibility that LPA up-regulates glucose metabolism in these non-transformed cells although the related physical significance is unknown [18–22]. Such a role of LPA in glucose metabolism, however, has not been extended to malignant cells which generally maintain high basal levels of glycolysis [24–26]. We have recently reported that LPA enhances de novo lipogenesis in ovarian cancer cells [17]. Since the increase in lipid biosynthesis requires not only activation of lipogenic enzymes but also the availability of the lipid synthesis precursor acetyl-CoA, an important intermediate of glucose metabolism. We thus speculated that LPA might play a more general role in cancer cell metabolism. We initially monitored the effect of LPA on expression of glucose transporters (GLUT1, GLUT4) but did observe any significant changes in response to LPA. Consistently, LPA did not increase glucose transport into ovarian cancer cells as measured with 2-deoxy-d-[2, 6-3H] glucose (data not shown). However, LPA strongly stimulated glycolytic flux and lactate production in cells of Caov-3, OVCA-432 and other ovarian cancer lines. A glucose metabolism PCR array led to the identification of HK2, the enzyme catalyzing the irreversible first rate-limiting step of glycolysis, as most markedly induced by LPA. We further demonstrated that LPA stimulated HK2 expression through LPA2-dependent activation of SREBP. To understand biological significance of LPA-regulated glycolysis, we compared LPA with EGF, IGF-1 and insulin. Interestingly, their glycolysis-promoting activities varied considerably. LPA and EGF strongly activated glycolysis while IGF-1 and insulin exhibited only modest effects, not proportional to their potent mitogenic activities. In line with the differential effects, LPA- or EGF-driven cell proliferation depends on the strong up-regulation of glycolysis.
Malignantly transformed cells exhibit an altered metabolic profile exemplified by a heightened glycolytic rate in normoxic conditions or the Warburg effect [24–26]. Tumors at advanced stages often experience hypoxia, leading to stabilization of HIF-1α protein, an important regulator of many glycolytic enzymes [53]. However, hypoxia is not the causal factor of the glycolytic phenotype that occurs in both hypoxic and oxygenated regions of a tumor. Tumor cells in vitro also glycolyse when cultured in normoxic conditions. Ras, Akt, and c-Myc have been reported to activate expression of various glycolytic enzymes [27–29]. In contrast, loss of the p53 tumor suppressor inhibits the mitochondrial respiratory chain by suppression of SCO2 (the synthesis of cytochrome c oxidase protein) and promotes glycolysis via TIGAR, a p53-inducible regulator of glycolysis and apoptosis [30]. However, these defects present only in some tumors do not explain the generally altered pattern of glucose metabolism in a wide spectrum of cancers. Other unrecognized mechanisms are likely important in the development and maintenance of the glycolytic phenotype of cancer cells. The results of the present study suggest that growth factors such as LPA and EGF could contribute significantly to the high glycolytic levels seen in cancer cells. Moreover, up-regulation of glycolysis is essential for LPA- or EGF-dependent cell proliferation. The requirement suggests the importance of glycolysis to meet the bioenergetic and biosynthetic demands of proliferatively active cancer cells.
In contrast to LPA and EGF, IGF-1 and insulin are relatively weaker stimuli of glycolysis in ovarian cancer cells although they exhibited as strong mitogenic activities. IGF-1 or insulin-driven growth of ovarian cancer cells was not much affected by 2-DG. This is not surprising as the glycolytic inhibitor did not decrease glycolytic levels in IGF-1- or insulin-treated cells as dramatically as in LPA- or EGF-stimulated cells. The result also suggests that a modest increase in glycolysis was adequate to meet requirements of bioenergy and biomasses for proliferation of IGF-1 or insulin-stimulated cells. Alternatively, as physiological endocrine factors, IGF-1 and insulin could modulate other metabolic processes such as lipid oxidation and glutamate metabolism to compensate for relatively restrained glycolysis [54–56].
A mechanistic finding of the current study is that LPA induces glycolysis through LPA2 activation of SREBP to enhance HK2 expression. We have previously shown that LPA2-dependent activation of SREBP is also involved in LPA induction of the lipogenic enzyme FAS to promote de novo lipid synthesis [17]. The results of the current study suggest that SREBP acts as a transcription factor that up-regulates expression of HK2 as well. Therefore SREBP lies at a convergence point to coordinately regulate both lipid anabolism and glucose catabolism in LPA-stimulated cells.
Compared to other LPA receptors, LPA2 is most commonly overexpressed in various cancers. A number of recent studies have provided strong evidence that LPA2 is causally linked to tumorigenesis in animals. Liu et al reported that transgenic expression of LPA receptors driven by the MMTV promoter leads to development of mammary tumors including estrogen receptor-positive and -negative breast carcinomas [57]. LPA2 transgenic mice showed a more aggressive tumorigenic phenotype compared to the LPA1 or LPA3 transgenic animals [57]. The most direct evidence to implicate LPA2 in cancer stems from the recent studies by Yun's group using the LPA2 null mice [58,59]. Compared to wild type mice, LPA2-deficient mice were more resistant to intestinal tumorigenesis induced by colitis or by ApcMin mutation [58,59]. The oncogenic mechanism of LPA2 remains elusive. Most studies have focused on the ability of LPA2 to stimulate expression of oncogenic mediators [35–41]. The link from LPA to activation of HK2 expression and glycolytic metabolism as revealed by the present study reflects another oncogenic process mediated primarily by LPA2. Given the prominent pro-oncogenic function of LPA2 and its non-essential physiological role in mice [60], LPA2 could be an ideal target for therapeutic intervention of cancer.
Conclusions
In sum, LPA stimulates glycolysis through induction of HK2. The effect of LPA is mediated by the LPA2 receptor-dependent activation of the SREBP transcription factor. Further functional analysis indicates that growth factors involved in ovarian oncogenesis exhibit differential effects on glycolysis. Like LPA, EGF strongly activates glycolysis while IGF-1 and insulin induce glycolysis at a modest level, not proportional to their strong mitogenic activity. Consistently, LPA or EGF relies on robustly enhanced glycolysis to support cell proliferation while the mitogenic activity of IGF-1 or insulin is largely resistant to glycolytic inhibition. The work represents the first to link LPA signaling to glucose metabolism and the glycolytic phenotype of cancer cells.
Author Contributions
AM and XF were responsible for experimental design and preparation of the manuscript. AM, YM, FY, YG, ZF, EMM, EB and HS generated reagents, performed experiments and analyzed the data. All authors read and approved the final version of the manuscript.
The following is the supplementary data related to this article.
Quantitative PCR-based array of glycolytic genes in Caov-3 cells treated with LPA for 12 hours. Experimental conditions and data presentation were as described in Figure 2A.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2015.09.003.
Acknowledgements
The work was supported in part by the NIH/NCI grant R21 CA161478 (XF), the Department of Defense Ovarian Cancer Research Program grant W81XWH-11-1-0541 (XF), the Jeffress Memorial Fund award (XF), and the NIH grant P30 CA16059 to Massey Cancer Center of Virginia Commonwealth University (VCU) School of Medicine.
Footnotes
Conflict of Interest: The authors declare that there are no conflicts of interest.
References
- 1.Xiao YJ, Schwartz B, Washington M, Kennedy A, Webster K, Belinson J, Xu Y. Electrospray ionization mass spectrometry analysis of lysophospholipids in human ascitic fluids: comparison of the lysophospholipid contents in malignant vs nonmalignant ascitic fluids. Anal Biochem. 2001;290:302–313. doi: 10.1006/abio.2001.5000. [DOI] [PubMed] [Google Scholar]
- 2.Westermann AM, Havik E, Postma FR, Beijnen JH, Dalesio O, Moolenaar WH, Rodenhuis S. Malignant effusions contain lysophosphatidic acid (LPA)-like activity. Ann Oncol. 1998;9:437–442. doi: 10.1023/a:1008217129273. [DOI] [PubMed] [Google Scholar]
- 3.Aoki J, Inoue A, Okudaira S. Two pathways for lysophosphatidic acid production. Biochim Biophys Acta. 2008;1781:513–518. doi: 10.1016/j.bbalip.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, Mutoh T, Lin ME, Teo ST, Park KE, Mosley AN. LPA receptors: subtypes and biological actions. Annu Rev Pharmacol Toxicol. 2010;50:157–186. doi: 10.1146/annurev.pharmtox.010909.105753. [DOI] [PubMed] [Google Scholar]
- 5.Houben AJ, Moolenaar WH. Autotaxin and LPA receptor signaling in cancer. Cancer Metastasis Rev. 2011;30:557–565. doi: 10.1007/s10555-011-9319-7. [DOI] [PubMed] [Google Scholar]
- 6.Lee SC, Fujiwara Y, Tigyi GJ. Uncovering unique roles of LPA receptors in the tumor microenvironment. Receptors Clin Investig. 2015;2 doi: 10.14800/rci.440. [pii: e440] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Z, Cheng CT, Zhang H, Subler MA, Wu J, Mukherjee A, Windle JJ, Chen CK, Fang X. Role of LPA4/p2y9/GPR23 in negative regulation of cell motility. Mol Biol Cell. 2008;19:5435–5445. doi: 10.1091/mbc.E08-03-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jongsma M, Matas-Rico E, Rzadkowski A, Jalink K, Moolenaar WH. LPA is a chemorepellent for B16 melanoma cells: action through the cAMP-elevating LPA5 receptor. PLoS One. 2011;6:e29260. doi: 10.1371/journal.pone.0029260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang X, Schummer M, Mao M, Yu S, Tabassam FH, Swaby R, Hasegawa Y, Tanyi JL, LaPushin R, Eder A. Lysophosphatidic acid is a bioactive mediator of ovarian cancer. Biochim Biophys Acta. 2002;1582:257–264. doi: 10.1016/s1388-1981(02)00179-8. [DOI] [PubMed] [Google Scholar]
- 10.Kitayama J, Shida D, Sako A, Ishikawa M, Hama K, Aoki J, Arai H, Nagawa H. Over-expression of lysophosphatidic acid receptor-2 in human invasive ductal carcinoma. Breast Cancer Res. 2004;6:R640–R646. doi: 10.1186/bcr935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita H, Kitayama J, Shida D, Ishikawa M, Hama K, Aoki J, Arai H, Nagawa H. Differential expression of lysophosphatidic acid receptor-2 in intestinal and diffuse type gastric cancer. J Surg Oncol. 2006;93:30–35. doi: 10.1002/jso.20397. [DOI] [PubMed] [Google Scholar]
- 12.Shida D, Watanabe T, Aoki J, Hama K, Kitayama J, Sonoda H, Kishi Y, Yamaguchi H, Sasaki S, Sako A. Aberrant expression of lysophosphatidic acid (LPA) receptors in human colorectal cancer. Lab Invest. 2004;84:1352–1362. doi: 10.1038/labinvest.3700146. [DOI] [PubMed] [Google Scholar]
- 13.Schulte KM, Beyer A, Kohrer K, Oberhauser S, Roher HD. Lysophosphatidic acid, a novel lipid growth factor for human thyroid cells: over-expression of the high-affinity receptor edg4 in differentiated thyroid cancer. Int J Cancer. 2001;92:249–256. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1166>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 14.Yu S, Murph MM, Lu Y, Liu S, Hall HS, Liu J, Stephens C, Fang X, Mills GB. Lysophosphatidic acid receptors determine tumorigenicity and aggressiveness of ovarian cancer cells. J Natl Cancer Inst. 2008;100:1630–1642. doi: 10.1093/jnci/djn378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun CC, Sun H, Wang D, Rusovici R, Castleberry A, Hall RA, Shim H. LPA2 receptor mediates mitogenic signals in human colon cancer cells. Am J Physiol Cell Physiol. 2005;289:C2–C11. doi: 10.1152/ajpcell.00610.2004. [DOI] [PubMed] [Google Scholar]
- 16.Yamada T, Yano S, Ogino H, Ikuta K, Kakiuchi S, Hanibuchi M, Kanematsu T, Taniguchi T, Sekido Y, Sone S. Lysophosphatidic acid stimulates the proliferation and motility of malignant pleural mesothelioma cells through lysophosphatidic acid receptors, LPA1 and LPA2. Cancer Sci. 2008;99:1603–1610. doi: 10.1111/j.1349-7006.2008.00848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee A, Wu J, Barbour S, Fang X. Lysophosphatidic acid activates lipogenic pathways and de novo lipid synthesis in ovarian cancer cells. J Biol Chem. 2012;287:24990–25000. doi: 10.1074/jbc.M112.340083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson FJ, Moyes C, Scott PH, Plevin R, Gould GW. Lysophosphatidic acid stimulates glucose transport in Xenopus oocytes via a phosphatidylinositol 3′-kinase with distinct properties. Biochem J. 1996;316:161–166. doi: 10.1042/bj3160161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomas M, Lazaro-Dieguez F, Duran JM, Marin P, Renau-Piqueras J, Egea G. Protective effects of lysophosphatidic acid (LPA) on chronic ethanol-induced injuries to the cytoskeleton and on glucose uptake in rat astrocytes. J Neurochem. 2003;87:220–229. doi: 10.1046/j.1471-4159.2003.01993.x. [DOI] [PubMed] [Google Scholar]
- 20.Yea K, Kim J, Lim S, Park HS, Park KS, Suh PG, Ryu SH. Lysophosphatidic acid regulates blood glucose by stimulating myotube and adipocyte glucose uptake. J Mol Med. 2008;86:211–220. doi: 10.1007/s00109-007-0269-z. [DOI] [PubMed] [Google Scholar]
- 21.Bernhart E, Kollroser M, Rechberger G, Reicher H, Heinemann A, Schratl P, Hallstrom S, Wintersperger A, Nusshold C, DeVaney T. Lysophosphatidic acid receptor activation affects the C13NJ microglia cell line proteome leading to alterations in glycolysis, motility, and cytoskeletal architecture. Proteomics. 2010;10:141–158. doi: 10.1002/pmic.200900195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coy PE, Taneja N, Lee I, Hecquet C, Bryson JM, Robey RB. LPA is a novel lipid regulator of mesangial cell hexokinase activity and HKII isoform expression. Am J Physiol Renal Physiol. 2002;283:F271–F279. doi: 10.1152/ajprenal.00093.2001. [DOI] [PubMed] [Google Scholar]
- 23.Vander Heiden MG, Plas DR, Rathmell JC, Fox CJ, Harris MH, Thompson CB. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol. 2001;21:5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 25.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 27.Racker E, Resnick RJ, Feldman R. Glycolysis and methylaminoisobutyrate uptake in rat-1 cells transfected with ras or myc oncogenes. Proc Natl Acad Sci U S A. 1985;82:3535–3538. doi: 10.1073/pnas.82.11.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 29.Chesney J, Telang S. Regulation of Glycolytic and Mitochondrial Metabolism by Ras. Curr Pharm Biotechnol. 2013;14:251–260. doi: 10.2174/1389201011314030002. [DOI] [PubMed] [Google Scholar]
- 30.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;14:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 31.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Wu X, Pan ZK, Huang S. Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer metastasis. Cancer Res. 2010;70:9979–9990. doi: 10.1158/0008-5472.CAN-10-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Z, Swaby RF, Liang Y, Yu S, Liu S, Lu KH, Bast RC, Jr., Mills GB, Fang X. Lysophosphatidic acid is a major regulator of growth-regulated oncogene alpha in ovarian cancer. Cancer Res. 2006;66:2740–2748. doi: 10.1158/0008-5472.CAN-05-2947. [DOI] [PubMed] [Google Scholar]
- 34.Malkki M, Laakso M, Deeb SS. The human hexokinase II gene promoter: functional characterization and detection of variants among patients with NIDDM. Diabetologia. 1997;40:1461–1469. doi: 10.1007/s001250050850. [DOI] [PubMed] [Google Scholar]
- 35.Hu YL, Tee MK, Goetzl EJ, Auersperg N, Mills GB, Ferrara N, Jaffe RB. Lysophosphatidic acid induction of vascular endothelial growth factor expression in human ovarian cancer cells. J Natl Cancer Inst. 2001;93:762–768. doi: 10.1093/jnci/93.10.762. [DOI] [PubMed] [Google Scholar]
- 36.Song Y, Wu J, Oyesanya RA, Lee Z, Mukherjee A, Fang X. Sp-1 and c-Myc mediate lysophosphatidic acid-induced expression of vascular endothelial growth factor in ovarian cancer cells via a hypoxia-inducible factor-1-independent mechanism. Clin Cancer Res. 2009;15:492–501. doi: 10.1158/1078-0432.CCR-08-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Symowicz J, Adley BP, Woo MM, Auersperg N, Hudson LG, Stack MS. Cyclooxygenase-2 functions as a downstream mediator of lysophosphatidic acid to promote aggressive behavior in ovarian carcinoma cells. Cancer Res. 2005;65:2234–2242. doi: 10.1158/0008.5472.CAN-04-2781. [DOI] [PubMed] [Google Scholar]
- 38.Oyesanya RA, Lee ZP, Wu J, Chen J, Song Y, Mukherjee A, Dent P, Kordula T, Zhou H, Fang X. Transcriptional and post-transcriptional mechanisms for lysophosphatidic acid-induced cyclooxygenase-2 expression in ovarian cancer cells. FASEB J. 2008;22:2639–2651. doi: 10.1096/fj.07-101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang X, Yu S, Bast RC, Liu S, Xu HJ, Hu SX, LaPushin R, Claret FX, Aggarwal BB, Lu Y. Mechanisms for lysophosphatidic acid-induced cytokine production in ovarian cancer cells. J Biol Chem. 2004;279:9653–9661. doi: 10.1074/jbc.M306662200. [DOI] [PubMed] [Google Scholar]
- 40.Yang M, Zhong WW, Srivastava N, Slavin A, Yang J, Hoey T, An S. G protein-coupled lysophosphatidic acid receptors stimulate proliferation of colon cancer cells through the {beta}-catenin pathway. Proc Natl Acad Sci U S A. 2005;102:6027–6032. doi: 10.1073/pnas.0501535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Bialkowska A, Rusovici R, Chanchevalap S, Shim H, Katz JP, Yang VW, Yun CC. Lysophosphatidic acid facilitates proliferation of colon cancer cells via induction of Krüppel-like factor 5. J Biol Chem. 2007;282:15541–15549. doi: 10.1074/jbc.M700702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakashima RA, Mangan PS, Colombini M, Pedersen PL. Hexokinase receptor complex in hepatoma mitochondria: evidence from N, N′-dicyclohexylcarbodiimide-labeling studies for the involvement of the pore-forming protein VDAC. Biochemistry. 1986;25:1015–1021. doi: 10.1021/bi00353a010. [DOI] [PubMed] [Google Scholar]
- 43.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gosmain Y, Lefai E, Ryser S, Roques M, Vidal H. Sterol regulatory element-binding protein-1 mediates the effect of insulin on hexokinase II gene expression in human muscle cells. Diabetes. 2004;53:321–329. doi: 10.2337/diabetes.53.2.321. [DOI] [PubMed] [Google Scholar]
- 45.Gosmain Y, Dif N, Berbe V, Loizon E, Rieusset J, Vidal H, Lefai E. Regulation of SREBP-1 expression and transcriptional action on HKII and FAS genes during fasting and refeeding in rat tissues. J Lipid Res. 2005;46:697–705. doi: 10.1194/jlr.M400261-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Magaña MM, Osborne TF. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 47.Kishi Y, Okudaira S, Tanaka M, Hama K, Shida D, Kitayama J, Yamori T, Aoki J, Fujimaki T, Arai H. Autotaxin is overexpressed in glioblastoma multiforme and contributes to cell motility of glioblastoma by converting lysophosphatidylcholine to lysophosphatidic acid. J Biol Chem. 2006;281:17492–17500. doi: 10.1074/jbc.M601803200. [DOI] [PubMed] [Google Scholar]
- 48.Wu J, Mukherjee A, Lebman D, Fang X. Gene expression of the lysophosphatidic acid receptor 1 is target of transforming growth factor beta. Oncogene. 2013;32:3198–3206. doi: 10.1038/onc.2012.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 50.Choi SY, Collins CC, Gout PW, Wang Y. Cancer-generated lactic acid: a regulatory, immunosuppressive metabolite? J Pathol. 2013;230:350–355. doi: 10.1002/path.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen W, Gueron M. The inhibition of bovine heart hexokinase by 2-deoxy-d-glucose-6-phosphate: Characterization by 31P NMR and metabolic implications. Biochimie. 1992;74:867–873. doi: 10.1016/0300-9084(92)90070-u. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z, Lu W, Garcia-Prieto C, Huang P. The Warburg effect and its cancer therapeutic implications. J Bioenerg Biomembr. 2007;39:267–274. doi: 10.1007/s10863-007-9086-x. [DOI] [PubMed] [Google Scholar]
- 53.Marin-Hernandez A, Gallardo-Perez JC, Ralph SJ, Rodriguez-Enriquez S, Moreno-Sanchez R. HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Rev Med Chem. 2009;9:1084–1101. doi: 10.2174/138955709788922610. [DOI] [PubMed] [Google Scholar]
- 54.Xu X, Gopalacharyulu P, Seppanen-Laakso T, Ruskeepaa AL, Aye CC, Carson BP, Mora S, Oresic M, Teleman AA. Insulin signaling regulates fatty acid catabolism at the level of CoA activation. PLoS Genet. 2012;8:e1002478. doi: 10.1371/journal.pgen.1002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wasa M, Wang HS, Tazuke Y, Okada A. Insulin-like growth factor-I stimulates amino acid transport in a glutamine-deprived human neuroblastoma cell line. Biochim Biophys Acta. 2001;1525:118–124. doi: 10.1016/s0304-4165(00)00178-1. [DOI] [PubMed] [Google Scholar]
- 56.Gallagher EJ, LeRoith D. Minireview: IGF, Insulin, and Cancer. Endocrinology. 2011;152:2546–2551. doi: 10.1210/en.2011-0231. [DOI] [PubMed] [Google Scholar]
- 57.Liu S, Umezu-Goto M, Murph M, Lu Y, Liu W, Zhang F, Yu S, Stephens LC, Cui X, Murrow G. Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell. 2009;15:539–550. doi: 10.1016/j.ccr.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin S, Wang D, Iyer S, Ghaleb AM, Shim H, Yang VW, Chun J, Yun CC. The absence of LPA2 attenuates tumor formation in an experimental model of colitis-associated cancer. Gastroenterology. 2009;136:1711–1720. doi: 10.1053/j.gastro.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin S, Lee SJ, Shim H, Chun J, Yun CC. The absence of LPA receptor 2 reduces the tumorigenesis by ApcMin mutation in the intestine. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1128–G1138. doi: 10.1152/ajpgi.00321.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Contos JJ, Ishii I, Fukushima N, Kingsbury MA, Ye X, Kawamura S, Brown JH, Chun J. Characterization of lpa(2) (Edg4) and lpa(1)/lpa(2) (Edg2/Edg4) lysophosphatidic acid receptor knockout mice: signaling deficits without obvious phenotypic abnormality attributable to lpa(2) Mol Cell Biol. 2002;22:6921–6929. doi: 10.1128/MCB.22.19.6921-6929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative PCR-based array of glycolytic genes in Caov-3 cells treated with LPA for 12 hours. Experimental conditions and data presentation were as described in Figure 2A.