Abstract
AIM: To study the modulating effect of GdCl3 and Angelica Sinensis polysaccharides (ASP) on differentially expressed genes in liver of hepatic immunological mice by cDNA microarray.
METHODS: Hepatic immunological injury was induced by lipopolysaccharide (LPS ip, 0.2 mg·kg-1) in bacillus calmetteguerin (BCG ip, 1 mg·kg-1) primed mice; A single dose of 20 mg·kg-1 GdCl3 was simultaneously pretreated and 30 mg·kg-1 ASP (ig, qd × 7 d) was administrated when the BCG+LPS was primed. The mice were sacrificed at the end of the 7th day after ip LPS for 6 h and the liver was removed quickly. The PCR products of 512 genes were spotted onto a chemical material-coated glass plate in array. The DNAs were fixed to the glass plate after series of treatments. The total RNAs were isolated from the liver tissue, and were purified to mRNAs by Oligotex. Both mRNAs from the normal liver tissue and the liver tissue from the mice with hepatic immunological injury or that pretreated with GdCl3 or ASP were reversely transcribed to cDNAs with the incorporation of fluorescent dUTP to prepare the hybridization probes. The mixed probes were hybridized to the cDNA microarray. After high-stringent washing, the cDNA microarray was scanned for fluorescent signals and showed differences between the two tissues.
RESULTS: Among the 512 target genes, 18 differed in liver tissue of hepatic immunological injury mice, and 6 differed in those pretreated by ASP, 7 differed in those pretreated by GdCl3.
CONCLUSION: cDNA microarray technique is effective in screening the differentially expressed genes between two different kinds of tissue. Further analysis of those obtained genes will be helpful to understand the molecular mechanism of hepatic immunological injury and to study the intervention of drug. Both ASP and GdCl3 can decrease the number of the differentially expressed genes in liver tissue of mice with hepatic immunological injury.
INTRODUCTION
The advanced technique of DNA microarray makes it possible to monitor the expression of ten out of thousand genes simultaneously in one hybridization experiment[1,2]. The past year has demonstrated the versatility of microarrays for the analysis of whole model-organism genomes and has seen the development of chips to measure the expression of 40000 human genes. Microarray technology has also become considerably more robust and sensitive[3]. The technology of microarrays has advanced from reverse Northern blots on filters detected using radioactive probes to a highly technical field involving miniaturized synthesis, multi-color fluorescent labeling, and database management. Recently, whole genome has been analyzed[4,5]. Microarray analyses typically follow the steps of gent selection, microarray synthesis, sample preparation, array hybridization, detection, and data analysis with appropriate controls required for each. Functional genomics is the study of gene function through the parallel expression measurements of genomes, most commonly using the technologies of microarrays and serial analysis of gene expression. Microarray could use in some other field also, such as, basic research of drug and target discovery, biomarker determination, pharmacology, toxicogenomics, target selectivity, development of prognostic tests and disease-subclass determination[6]. In this experiment, the DNA segments were spotted on a slide with high density. Then cDNA retro-transcribed from mRNA derived from normal or pathological tissues, which were labeled with Cy3 and Cy5 fluorescence, hybridized with the microarray slide[7]. Through this technique, detection of differentially expressed genes and the construction of differential gene expression profiles are greatly facilitated. The BioDoor 512DNA microarray was used for investigating the changes of gene expression in liver tissue of hepatic immunological mice and studying the effective mechanisms of ASP and GdCl3, which might give important health benefits to understanding, diagnosing and treating the liver injury.
MATERIALS AND METHODS
Animals and treatments
Hepatic immunological injury was induced by lipopolysaccharide (LPS ip, 0.2 mg·kg-1) in bacillus calmetteguerin (BCG ip, 1 mg·kg-1) primed mice (Model); A single dose of 20 mg·kg-1 GdCl3 was pretreated simultaneously (Model+GdCl3) and 30 mg·kg-1 ASP (Model+ASP, ig, qd × 7 d) was administrated when the BCG+LPS was primed. The mice were sacrificed at the end of the 7th day after ip LPS for 6 h and the liver was collected for analysis of cDNA micrarray.
Construction of microarrays[4]
The BioDoor 512 microarray consisted of a total of 512 novel or known genes (provided by United Gene Holdings, Ltd). These genes were amplified through PCR using universal primers and then purified; the purity of PCR production was dissolved in 3 × SSC solution, and these target genes were spotted on silylated slides (Tele Chem, Inc.) by Cartesian 7500 Spotting Robotics (Cartesian, Inc.). After spotting, the slides were hydrated (2 h), dried (0.5 h) at room temperature, UV cross-linked (65 mJ/cm), and then treated with 0.2% SDS (10 min). The slides were dried again and ready for use.
Probe preparation[4]
The method of total RNA extraction was modified from the original single step extraction with Trizol agent. The liver tissue stored in liquid nitrogen were ground completely into tiny granules in 100 mm ceramic mortar (RNase free) and homogenized in Trizol. The pellet of total RNA was dissolved with Milli-Q H2O. The mRNAs were purified using Oligotex mRNA Midi Kit (Qiagen, Inc.). The fluorescent cDNA probes were prepared through reverse transcription and then purified (referring to the protocol of Mark. Schena.). The probes from normal tissues were labeled with Cy3-dUTP, those from the pathological tissues were with Cy5-dUTP.
Hybridization and washing[4]
The probes were mixed and precipitated by ethanol, and resolved in 20 μl hybridization solution (5 × SSC + 0.4% SDS + 50% formamide + 5 × denhardt’s solution). Chip was prehybridized with Hyb sol + 3 μl denatured salmon sperm DNA at 42 °C for 6 h. After denaturing at 95 °C for 5 min, the probe mixture was added on the prehybridized chip and covered with glass. The chips were inocubated at 42 °C for 15-17 h. The slide was then washed in solutions of 2 × SSC + 0.2% SDS, 0.1 × SSC + 0.2% SDS and 0.1 × SSC at 60 °C, respectively for 10 min each, then dried at room temperature.
Detection and analysis[4]
The chip was scanned by ScanArray 5000 laser scanner (General Scanning, Inc) at 2 wavelengths. The acquired image was analyzed by ImaGene 3.0 software (BioDiscovery, Inc.). The intensity of each spot at the 2 wavelengths represented the quantity of Cy3-dUTP and Cy5-dUTP, respectively. The ratio of Cy3 to Cy5 was calculated. The two overall intensity was normalized by a coefficient according to the ratios of the located 40 housekeeping genes.
RESULTS
Differentially expressed gene in liver tissues between mice with or without hepatic immunological injury
Total 18 differentially expressed genes that might be associated with hepatic immnunological injury were selected from microarray. 4 genes showed up-regulation and 14 genes showed down-regulation (Table 1, Figure 1 and Figure 2).
Table 1.
Differentially expressed gene in liver tissues between hepatic immunological injury mice and normal mice
| Genbank-ID | Ratio(Cy5/Cy3) | Classification | Definition |
| Humipasa | 0.253 | Metabolism | Human mRNA for ATP synthase alpha subunit, complete cds. |
| Hsngmrna | 0.285 | H. sapiens hng mRNA for uracil DNA glycosylase. | |
| Humoat | 0.290 | Modulators/Effectors/intracellular transduction | Human ornithine aminotransferase mRNA, complete cds. |
| Hsncadhe | 0.294 | Cell Surface Antigens & Adhesion | Human mRNA for N-cadherin |
| Humpparp | 0.307 | 0 | Human acidic ribosomal phosphoprotein P0 mRNA, complete cds. |
| Humorf06 | 0.319 | Other | Human mRNA for KIAA0106 gene, complete cds. |
| Hsgcsab | 0.373 | Modulators/Effectors/intracellular Transduction | H. sapiens soluble guanylate cyclase small subunit mRNA. |
| Humhspa2 | 0.379 | Stress Response Proteins | Human heat shock protein HSPA2 gene, complete cds. |
| Hspbx3 | 0.439 | Other | Human PBX3 mRNA |
| Hsef1gmr | 0.452 | H. sapiens mRNA for elongation factor-1-gamma | |
| Hsgagmr | 0.460 | Other | Human mRNA for GARS-AIRS-GART |
| Humhhr23 | 0.467 | DNA Synthesis, Repair & Recombination F | Human mRNA for XP-C repair com;lamenting protein(p58/HHR23B), complete cds. |
| Hscatr | 0.470 | Metabolism | Human kidney mRNA for catalase. |
| Humscp2a | 0.5 | Ion Channel & Transport Proteins | Human sterol carrier protein X.sterol carrier protein 2 mRNA, complete cds. |
| Humnpm | 2.062 | Other | Human nucleophosmin mRNA, complete cds. |
| Hsrpl32 | 2.074 | DNA Binding/Transcription/transcription F | Human mRNA for ribosomal protein L32. |
| Hsu32944 | 2.244 | Apoptosis-Related proteins | Human cytoplasmin dynein light chain1(hdlc1) mRNA, complete cds. |
| Hsu32944 | 3.239 | Oncogenes & Tumor Suppressors mRNA, com;oete cds. | Human cytoplasmic dynein light chain 1 (hdlc1) |
Ratio is Cy5/Cy3: High expression (ratio > 2.0), low expression (ratio < 0.5).
Figure 1.
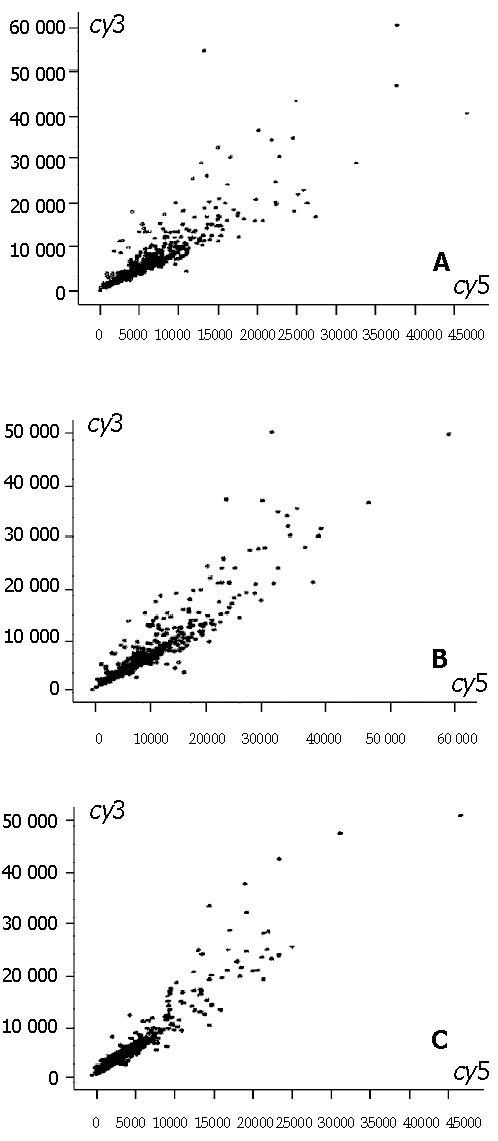
The scatter plots of gene expression pattern by cDNA microarray. A: Model liver(Cy5)/normal liver(Cy3); B: Model+GdCl3 liver (Cy5)/normal liver; C: Mode+ASP liver (Cy5)/normal liver (Cy3). Ratio is Cy5/Cy3: high expression (ratio > 2.0), low expression (ratio < 0.5), no change in expression (0.5 < ratio < 2.0).
Figure 2.
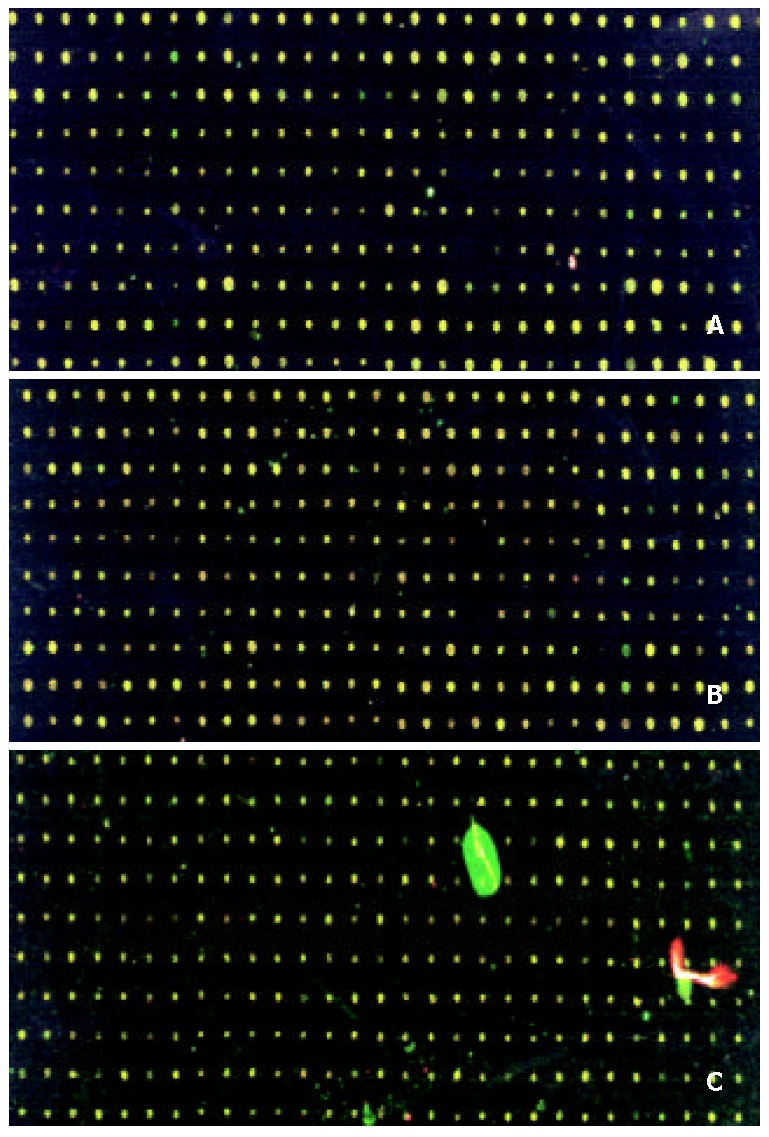
Scanning results of hybridizing signals by cDNA microarray. A: Model liver(Cy5)/normal liver(Cy3); B: Model+GdCl3 liver (Cy5)/normal liver; C: Mode+ASP liver (Cy5)/normal liver (Cy3). Color of spots in the merged image of Cy3 image and Cy5 image: high expression (red), low ex-pression (green), no change in expression (yellow).
Differentially expressed gene in liver tissues between mice with hepatic immunological injury and those pretreated byGdCl3
6 differentially expressed genes associated with the effect of GdCl3 on hepatic immnunological injury were selected from the microarray. 1 gene showed up-regulation and 5 genes showed down-regulation (Table 2, Figure 1 and Figure 2).
Table 2.
Differentially expressed gene in liver tissues between mice with hepatic immunological injury and those pretreated by GdCl3
| Genbank-ID | Ratio(Cy5/Cy3) | Classification | Definition |
| Hsung | 0.313 | DNA Binding/Transcription/Tra nscription Factors | Human cDNA for uracil-DNA glycosylase. |
| Hsu09564 | 0.343 | Modulators/Effectors/intracellular Transduction | Human serine kinase mRNA, complete cds. |
| Hsu83843 | 0.374 | Cell Surface Antigens & Adhesion | Human HIV-1 Nef interacting protein (Nip7-1)mRNA, Partial cds. |
| Hsmyc1 | 0.441 | Oncogenes & Tumor Suppressors | Human mRNA encoding the c-myc oncogene. |
| Humoat | 0.454 | Modulators/Effectors/intracellular transduction | Human ornithine aminotransferase mRNA, complete cds. |
| Hsu70323 | 2.200 | Extracellular Cell signaling & Communication proteins | Human ataxin-2(SCA2) mRNA, complete cds. |
Ratio is Cy5/Cy3: High expression (ratio > 2.0), low expression (ratio < 0.5).
Differentially expressed gene in liver tissues between mice with hepatic immunological injury and those pretreated by ASP
seven differentially expressed genes associated with the effect of ASP on hepatic immnunological injury were selected from the microarray. 1 gene showed up-regulation and 6 genes showed down-regulation (Table 3, Figure 1 and Figure 2).
Table 3.
Differentially expressed gene in liver tissues between mice with hepatic immunological injury and those pretreated by ASP
| Genbank-ID | Ratio(Cy5/Cy3) | Classification | Definition |
| Humscp2a | 0.283 | Ion channel & transport proteins | Human sterol carrier protein X.sterol carrier protein 2 mRNA, complete cds. |
| Humoat | 0.421 | Modulators/Effectors/intracellular transduction | Human ornithine aminotransferase mRNA, complete cds. |
| Hsngmrna | 0.431 | H. sapiens hng mRNA for uracil DNA glycosylase. | |
| humpafaa | 0.461 | Modulators/Effectors/intracellular transduction | Human mRNA for platelet activating factor acetylhydrolase IB gamma-subunit, complete cds. |
| humhbgfb | 0.481 | Extracellular cell signaling & communication proteins | Human heparin-binding growth factor receptor (HBGF-R-alpha-a2) mRNA, complete ceds. |
| Hsu83843 | 0.493 | Cell surface antigens and adhesion | Human HIV-1 Nef interacting protein(Nip7-1)mRNA, Partial cds. |
| Humnlk | 2.353 | Modulators/Effectors/intracellular transduction | Human neroleukin mRNA, complete cds. |
Ratio is Cy5/Cy3: High expression (ratio > 2.0), low expression (ratio < 0.5).
DISCUSSION
Hepatic immunological injury is a result of altering multiple g en es ex p ressio n. cDNA micro array meth o d can simultaneously determine a number of genes altered in the different pathological and physiological status, so gene chip is a useful tool for studying the target cause of disease and the effect of drug[10]. The DNA microarray hybridization applications reviewed include the important areas of gene expression analysis and genotyping for point mutations, single nucleotide polymorphisms (SNPs), and short tandem repeats (STRs)[11,12]. In addition to the many molecular biological and genomic research uses, systems for pharmacogenomic research and drug discovery[6,14], infectious and genetic disease and cancer diagnostics, and forensic and genetic identification purposes[14-16]. Additionally, microarray technology being developed and applied to new areas of proteomic and cellular analysis are reviewed[17,18]. Now, microarray has been developed many kinds, such as, protein microarray, GEM microarray, tissue microarray, they also have different kinds uses[19-21].
The present study showed that 18 differentially expressed genes that might be associated with hepatic immunological injury were found, 4 genes showed up-regulation and 14 genes showed down-regulation. These genes expression could recover to normal level by GdCl3 except the HIV-1 Nef interacting protein (Nip7-1) mRNA (Hsu83843). And by ASP except the HIV-1 Nef interacting protein (Nip7-1) mRNA (Hsu83843), sterol carrier protein X/sterol carrier protein 2 Mrna (Humscp2a), and H. sapiens hug mRNA for uracil DNA glycosylase (Hsngmrna).
Furthermore, cDNA or uracil-DNA glycosylase, serine kinase mRNA, encoding the c-myc oncogene expression (Hsmycl) showed down-regulation in the mice pretreated with GdCl3. In this study we find that using GdCl3 and ASP to pretreat mice with hepatic immunological liver both would cause down-regulation of Hsung and Hsngmrna. Hsung and Hsngmrna both related to encoding uracil-DNA glycosylase. In humans, uracil appears in DNA at the rate of several hundred bases per cell each day as a result of misincorporation of deoxyuridine (dU) or deamination of cytosine. The human UNG-gene at position 12q24.1 encodes nuclear (UNG2) and mitochondrial (UNG1) two forms of uracil-DNA glycosylase[22]. Repair of uracil-DNA is a base-excision pathway initiated by a uracil-DNA glycosylase (UDG) enzyme of which have four families[23]. The most efficient and well characterized of these uracil-DNA glycosylases is UDG which releases uracil from DNA, restores the correct DNA sequence, excises U from single- or double-stranded DNA and is associated with DNA replication forks[24,25]. Uracil DNA glycosylase (UDG) and DNA polymerase beta (beta pol) are the two enzymes of base excision repair (BER)[10]. Krokan et al[22] propose that BER is important both in the prevention of cancer and for preserving the integrity of germ cell DNA during evolution. But Kvaløy et al[25,26] find mutations affecting the function of human UNG gene are seemingly infrequent in human tumor cell lines. So we infer down-regulation of these genes may affect cell proliferation, and GdCl3 and ASP protect liver depend on this mechanism in some degree. Although ornithine aminotransferase mRNA (Humoat) also showed down-regulation, its expression showed a little improvement (Cy5/Cy3 increased from 0.290 to 0.454). Ataxin-2 Mrna (Hsu70323) expression showed up-regulation in the mice pretreated with GdCl3. As for the mice pretreated with ASP, Humoat expression also showed some improvement (Cy5/Cy3 increased from 0.290 to 0.421). TNF alpha can cause liver cell apoptosis through the TNF-alpha receptor or Fas/CD95, which is expressed by liver cells. The TNF-alpha induced expression of the nuclear oncogene c-myc in intact hepatocytes has been studied[27]. The expression of Hsmycl is down-regulation, which may be the reason of preventing hepatic cell from apoptosis. The liver plays a central role in nitrogen metabolism. Nitrogen enters the liver as free ammonia and as amino acids of which glutamine and alanine are the most important precursors. Detoxification of ammonia to urea involves deamination and transamination. Half of the extra NH3 removed by the liver was, ap parently, u tilized by perip ortal glutamate dehydrogenase and aspartate aminotransferase for sequential glutamate and aspartate synthesis and converted to urea as the 2-amino moiety of aspartate[28]. Ornithine aminotransferase is the most important enzyme to metabolize free ammonia and amino acids. The gene exists as a single copy in the malarial genome and is located on chromosomes 6/7/8[29]. Ornithine-delta-aminotransferase (OAT) (EC 2.6.1.13) is a pyridoxal-5’ phosphate dependent mitochondrial matrix enzyme. It controls the L-ornithine (Orn) level in tissues by catalysing the transfer of the delta-amino group of Orn to 2-oxoglutarate. The products of this reaction are L-glutamate-gamma-semialdehyde and L-glutamate[30]. Boon L proves that high protein diet would cause ornithine aminotransferase increased[31]. NOX significantly reduced serum levels of ornithine carbamoyltransferase and aspartate aminotransferase as hepatic injury[32]. The present study showed ornithine aminotransferase mRNA (Humoat) showed down-regulation, but its expression showed a little improvement (Cy5/Cy3 increased from 0.290 to 0.454) and (Cy5/Cy3 increased from 0.290 to 0.421) with GdCl3 and ASP. So we think ornithine aminotransferase is important aspect in hepatic injury. Protective effect of GdCl3 and ASP on the hepatic immunological injury depends on regulating the expression of ornithine aminotransferase. While the heparin-binding growth factor receptor (HBGF-R-alpha-a2) mRNA (humhbgfb) expression showed down-regulation and neuroleukin mRNA(Humnlk) expression showed up-regulation in the mice. Neuroleukin (NLK) is a multifunctional protein, involved in neuronal growth, glucose metabolism, cell motility, and differentiation. Neuroleukin (NLK), autocrine motility factor (AMF) and phosphohexose isomerase (PHI) have been identified same structure, so they have many kinds on functions and expressed in many tissues and organs, such as, brain, bone, liver, etc. NLK could mediate leukemia cell[33]. Romagnoli et al[34] show that the block of NLK commits PC12 cells to caspase-dependent apoptosis, and suggest a general protective role of NLK in the control of cell death in neuronal cells. NLK may be modulating inflammation and is probably involved in protecting CN and the cerebellums against apoptosis[35]. When ASP pretreat mice with hepatic immunological injury, the expression of Humnlk up-regulation may show ASP prevent cell apoptosis. It remains to be demonstrated whether the altered genes are target of protective effect of GdCl3 or ASP on the immunological liver injury, and further analysis the function of these genes is needed.
Footnotes
Edited by Wu XN
References
- 1.Aharoni A, Keizer LC, Bouwmeester HJ, Sun Z, Alvarez-Huerta M, Verhoeven HA, Blaas J, van Houwelingen AM, De Vos RC, van der Voet H, et al. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell. 2000;12:647–662. doi: 10.1105/tpc.12.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schena M, Mulatero P, Schiavone D, Mengozzi G, Tesio L, Chiandussi L, Veglio F. Vasoactive hormones induce nitric oxide synthase mRNA expression and nitric oxide production in human endothelial cells and monocytes. Am J Hypertens. 1999;12:388–397. [PubMed] [Google Scholar]
- 3.Leethanakul C, Knezevic V, Patel V, Amornphimoltham P, Gillespie J, Shillitoe EJ, Emko P, Park MH, Emmert-Buck MR, Strausberg RL, et al. Gene discovery in oral squamous cell carcinoma through the Head and Neck Cancer Genome Anatomy Project: confirmation by microarray analysis. Oral Oncol. 2003;39:248–258. doi: 10.1016/s1368-8375(02)00107-0. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Y, Harlocker SL, Molesh DA, Dillon DC, Stolk JA, Houghton RL, Repasky EA, Badaro R, Reed SG, Xu J. Discovery of differentially expressed genes in human breast cancer using subtracted cDNA libraries and cDNA microarrays. Oncogene. 2002;21:2270–2282. doi: 10.1038/sj.onc.1205278. [DOI] [PubMed] [Google Scholar]
- 5.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 6.Butte A. The use and analysis of microarray data. Nat Rev Drug Discov. 2002;1:951–960. doi: 10.1038/nrd961. [DOI] [PubMed] [Google Scholar]
- 7.Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 8.Welford SM, Gregg J, Chen E. Detection of differentially expressed genesin primary tumor tissues using representational differences analysis coupled to microarray hybridization. Nucleic Acids Res. 1998;26:3059–3065. doi: 10.1093/nar/26.12.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heller MJ. DNA microarray technology: devices, systems, and applications. Annu Rev Biomed Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 10.Armour JA, Barton DE, Cockburn DJ, Taylor GR. The detection of large deletions or duplications in genomic DNA. Hum Mutat. 2002;20:325–337. doi: 10.1002/humu.10133. [DOI] [PubMed] [Google Scholar]
- 11.Stickney HL, Schmutz J, Woods IG, Holtzer CC, Dickson MC, Kelly PD, Myers RM, Talbot WS. Rapid mapping of zebrafish mutations with SNPs and oligonucleotide microarrays. Genome Res. 2002;12:1929–1934. doi: 10.1101/gr.777302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santacroce R, Ratti A, Caroli F, Foglieni B, Ferraris A, Cremonesi L, Margaglione M, Seri M, Ravazzolo R, Restagno G, et al. Analysis of clinically relevant single-nucleotide polymorphisms by use of microelectronic array technology. Clin Chem. 2002;48:2124–2130. [PubMed] [Google Scholar]
- 13.Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics: translating benchside promise into bedside reality. Nat Rev Drug Discov. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman EP, Rao D, Pachman LM. Clarifying the boundaries between the inflammatory and dystrophic myopathies: insights from molecular diagnostics and microarrays. Rheum Dis Clin North Am. 2002;28:743–757. doi: 10.1016/s0889-857x(02)00031-5. [DOI] [PubMed] [Google Scholar]
- 15.Maier S, Olek A. Diabetes: a candidate disease for efficient DNA methylation profiling. J Nutr. 2002;132:2440S–2443S. doi: 10.1093/jn/132.8.2440S. [DOI] [PubMed] [Google Scholar]
- 16.Macgregor PF, Squire JA. Application of microarrays to the analysis of gene expression in cancer. Clin Chem. 2002;48:1170–1177. [PubMed] [Google Scholar]
- 17.Martinez MJ, Aragon AD, Rodriguez AL, Weber JM, Timlin JA, Sinclair MB, Haaland DM, Werner-Washburne M. Identification and removal of contaminating fluorescence from commercial and in-house printed DNA microarrays. Nucleic Acids Res. 2003;31:e18. doi: 10.1093/nar/gng018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam JH, Yu CH, Hwang KA, Kim S, Ahn SH, Shin JY, Choi WY, Joo YR, Park KY. Application of cDNA microarray technique to detection of gene expression in host cells infected with viruses. Acta Virol. 2002;46:141–146. [PubMed] [Google Scholar]
- 19.Reynolds MA. Microarray technology GEM microarrays and drug discovery. J Ind Microbiol Biotechnol. 2002;28:180–185. doi: 10.1038/sj/jim/7000136. [DOI] [PubMed] [Google Scholar]
- 20.Templin MF, Stoll D, Schrenk M, Traub PC, Vöhringer CF, Joos TO. Protein microarray technology. Drug Discov Today. 2002;7:815–822. doi: 10.1016/s1359-6446(00)01910-2. [DOI] [PubMed] [Google Scholar]
- 21.Rimm DL, Camp RL, Charette LA, Costa J, Olsen DA, Reiss M. Tissue microarray: a new technology for amplification of tissue resources. Cancer J. 2001;7:24–31. [PubMed] [Google Scholar]
- 22.Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, Skjelbred C, Akbari M, Aas PA, Slupphaug G. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog Nucleic Acid Res Mol Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- 23.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 24.Dinner AR, Blackburn GM, Karplus M. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature. 2001;413:752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 25.Kvaløy K, Nilsen H, Steinsbekk KS, Nedal A, Monterotti B, Akbari M, Krokan HE. Sequence variation in the human uracil-DNA glycosylase (UNG) gene. Mutat Res. 2001;461:325–338. doi: 10.1016/s0921-8777(00)00063-x. [DOI] [PubMed] [Google Scholar]
- 26.Mendez F, Sandigursky M, Franklin WA, Kenny MK, Kureekattil R, Bases R. Heat-shock proteins associated with base excision repair enzymes in HeLa cells. Radiat Res. 2000;153:186–195. doi: 10.1667/0033-7587(2000)153[0186:hspawb]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 27.Globa AG, Solovyev AS, Terentyev AA, Demidova VS, Shulgina MF, Alesenko AV, Karelin AA. TNF alpha-induced aerobic synthesis of ATP on plasma membranes of target cells. The relation to the expression of the nuclear oncogene c-myc. Biochem Mol Biol Int. 1998;45:1169–1178. doi: 10.1080/15216549800203402. [DOI] [PubMed] [Google Scholar]
- 28.Milano GD, Hotston-Moore A, Lobley GE. Influence of hepatic ammonia removal on ureagenesis, amino acid utilization and energy metabolism in the ovine liver. Br J Nutr. 2000;83:307–315. doi: 10.1017/s0007114500000386. [DOI] [PubMed] [Google Scholar]
- 29.Gafan C, Wilson J, Berger LC, Berger BJ. Characterization of the ornithine aminotransferase from Plasmodium falciparum. Mol Biochem Parasitol. 2001;118:1–10. doi: 10.1016/s0166-6851(01)00357-7. [DOI] [PubMed] [Google Scholar]
- 30.Seiler N. Ornithine aminotransferase, a potential target for the treatment of hyperammonemias. Curr Drug Targets. 2000;1:119–153. doi: 10.2174/1389450003349254. [DOI] [PubMed] [Google Scholar]
- 31.Boon L, Geerts WJ, Jonker A, Lamers WH, Van Noorden CJ. High protein diet induces pericentral glutamate dehydrogenase and ornithine aminotransferase to provide sufficient glutamate for pericentral detoxification of ammonia in rat liver lobules. Histochem Cell Biol. 1999;111:445–452. doi: 10.1007/s004180050380. [DOI] [PubMed] [Google Scholar]
- 32.Nadler EP, Dickinson EC, Beer-Stolz D, Alber SM, Watkins SC, Pratt DW, Ford HR. Scavenging nitric oxide reduces hepatocellular injury after endotoxin challenge. Am J Physiol Gastrointest Liver Physiol. 2001;281:G173–G181. doi: 10.1152/ajpgi.2001.281.1.G173. [DOI] [PubMed] [Google Scholar]
- 33.Chiao JW, Xu W, Seiter K, Feldman E, Ahmed T. Neuroleukin mediated differentiation induction of myelogenous leukemia cells. Leuk Res. 1999;23:13–18. doi: 10.1016/s0145-2126(98)00129-5. [DOI] [PubMed] [Google Scholar]
- 34.Romagnoli A, Oliverio S, Evangelisti C, Iannicola C, Ippolito G, Piacentini M. Neuroleukin inhibition sensitises neuronal cells to caspase-dependent apoptosis. Biochem Biophys Res Commun. 2003;302:448–453. doi: 10.1016/s0006-291x(03)00188-8. [DOI] [PubMed] [Google Scholar]
- 35.Stiles JK, Meade JC, Kucerova Z, Lyn D, Thompson W, Zakeri Z, Whittaker J. Trypanosoma brucei infection induces apoptosis and up-regulates neuroleukin expression in the cerebellum. Ann Trop Med Parasitol. 2001;95:797–810. doi: 10.1080/00034980120111145. [DOI] [PubMed] [Google Scholar]


