Abstract
AIM: To evaluate the level of sperm chromosome aberrations in male patients with hepatitis B, and to directly detect whether there are HBV DNA integrations in sperm chromosomes of hepatitis B patients.
METHODS: Sperm chromosomes of 14 tested subjects (5 healthy controls, 9 patients with HBV infection, including 1 with acute hepatitis B, 2 with chronic active hepatitis B, 4 with chronic persistent hepatitis B, 2 chronic HBsAg carriers with no clinical symptoms) were prepared using interspecific in vitro fertilization between zona-free golden hamster ova and human spermatozoa, and the frequencies of aberration spermatozoa were compared between subjects of HBV infection and controls. Fluorescence in situ hybridization (FISH) to sperm chromosome spreads was carried out with biotin-labeled full length HBV DNA probe to detect the specific HBV DNA sequences in the sperm chromosomes.
RESULTS: The total frequency of sperm chromosome aberrations in HBV infection group (14.8%, 33/223) was significantly higher than that in the control group (4.3%, 5/116). Moreover, the sperm chromosomes in HBV infection patients commonly presented stickiness, clumping, failure to staining, etc, which would affect the analysis of sperm chromosomes. Specific fluorescent signal spots for HBV DNA were seen in sperm chromosomes of one patient with chronic persistent hepatitis. In 9 (9/42) sperm chromosome complements containing fluorescent signal spots, one presented 5 obvious FISH spots, others presented 2 to 4 signals. There was significant difference of fluorescence intensity among the signal spots. The distribution of signal sites among chromosomes was random.
CONCLUSION: HBV infection can bring about mutagenic effects on sperm chromosomes. Integrations of viral DNA into sperm chromosomes which are multisites and nonspecific, can further increase the instability of sperm chromosomes. This study suggested that HBV infection can create extensively hereditary effects by alteration genetic constituent and/or induction chromosome aberrations, as well as the possibility of vertical transmission of HBV via the germ line to the next generation.
INTRODUCTION
Hepatitis B virus (HBV) infection is a serious public health problem worldwide[1-3], especially in Far-East Asia such as China. During HBV infection, HBV can be found in saliva, vaginal secretions, semen and other tissues beyond the liver and blood[4-6]. It is well established that HBV DNA could integrate not only into the host hepatocytes but also into sperm cells[7-19]. Extensive studies have confirmed that HBV DNA integrated into the hepatocytes will increase chromosome instability and cause genetic recombination and hepatocarcinogenesis[7-17]. In this study, in order to clarify the inheritable influence of HBV on sperm chromosomes, we used interspecific in vitro fertilization with zona-free hamster eggs to prepare sperm chromosomes and compared the aberration frequencies of sperm chromosomes between patients with hepatitis B and the controls. In addition, FISH technique with HBV DNA as a probe was used to detect HBV sequences in sperm chromosomes and to analyze features of sperm chromosomes integrated HBV DNA. The genetic significance of the results from this investigation was discussed.
MATERIALS AND METHODS
Subjects
Nine men with HBV infection, including 1 subject with acute hepatitis, 6 with chronic hepatitis (2 with chronic active hepatitis, 4 with chronic persistent hepatitis), 2 chronic HBsAg carriers with no clinical symptoms, and 5 healthy men as controls were studied. The age range was from 22 to 38 years old (mean 27), without the history of exposure of radiation and use of mutagenic agent. The status of the markers of HBV infection of the subjects tested was listed in Table 1.
Table 1.
Markers of HBV infection of the tested subjects
| Subjects | Age |
Serum |
Seminal fluid |
||||||
| HBsAg | Anti-HBc | HBeAg | Anti-HBs | Anti-HBe | HBV-DNA | HBsAg | HBV-DNA | ||
| Hepatitisa | |||||||||
| 1 | 38 | + | + | - | - | - | + | + | - |
| 2 | 32 | + | + | + | - | - | + | - | + |
| 3 | 25 | + | + | - | - | + | + | - | - |
| 4 | 22 | + | + | + | - | - | - | + | + |
| 5 | 26 | + | + | - | - | + | - | + | + |
| 6 | 23 | + | + | - | - | + | - | - | + |
| 7 | 27 | + | + | + | - | - | + | + | - |
| 8 | 22 | + | + | - | - | + | - | - | - |
| 9 | 25 | + | + | - | - | - | + | - | + |
| Controls | |||||||||
| 1 | 32 | - | - | - | - | - | - | - | - |
| 2 | 29 | - | - | - | - | - | - | - | - |
| 3 | 23 | - | - | - | - | - | - | - | - |
| 4 | 25 | - | - | - | - | - | - | - | - |
| 5 | 26 | - | - | - | - | - | - | - | - |
1: Acute hepatitis; 2, 3: Chronic active hepatitis; 4, 5, 6, 7: Chronic persistent hepatitis; 8, 9: Chronic HBsAg carriers.
Preparation of sperm chromosomes
The technique of interspecific in vitro fertilization of zona-free golden hamster was used to prepare the sperm chromosomes. Those procedures included the treatment of semen samples from the tested subjects, superovulation of golden hamsters and egg processing, insemination and postinsemination culture and preparation of chromosome slides[20,21].
Analysis of aberrations of sperm chromosome
Sperm chromosome slides were stained with 10-fold diluted Giemsa in 50 ml Sorensen’s buffer, pH6.8, for 20-30 min and observed under the light microscope. After the observation, the slides were stored at -70 °C for FISH test. Frequency distributions of spermatozoa with chromosome aberrations were evaluated by using Chi-square test.
Fluorescence in situ hybridization of sperm chromosomes
Labeling HBV DNA probe with biotin Recombinant plasmid, pHBV-1 containing 3.2 kb HBV genomic DNA, was taken to amplify according to the routine method[22]. The 3.2 kb HBV DNA probe with its vector (altogether 6.2 kb) was labeled with biotin-14-dATP by nick translation (GIBCOBRL No. 18247-015). Unincorporated nucleotides were separated by cold ethanol precipitation method.
In situ hybridization Sperm chromosome slides were orderly treated with RNase (Sigma) at 100 mg/L for 60 min at 37 °C, then at 50 mg/L pepsin (Sigma) in 0.01N HCl for 10 min at 37 °C, at last at 1% polyformaldehyde in PBS for 10 min at room temperature. Chromosomes were denatured at 75 °C for 4 min in 70% formamide in 2 × SSC. In situ hybridization with denatured DNA probe mentioned above was performed with a modification of the procedure described by our previous paper[23]. Briefly, 10 µl hybridization buffer (50% deionized formamide, 10% dextran sulfate, and 2 × SSC) containing 40 ng/µl biotin-labeled HBV DNA probe, 500 ng/µl sheared salmon sperm DNA was placed on each slide. A coverslip (18 × 18 mm) was then applied and sealed with rubber cement, followed by overnight incubation in a humidified chamber at 37 °C.
Detection of hybridization signals Post-hybridization washing was carried out, referring to Korenberg’s methods for mapping small DNA probes[24], once in 40% formamide, 2 × SSC for 10 min at 40 °C, then twice, each for 5 min, in 2 × SSC at room temperature. Hybridization signals were detected with FITC-conjugated avidin and amplified with goat biotinylated anti-avidin antibody followed by another layer of FITC-avdin (avidin and anti-avidin, Vector Laboratories). To increase the intensity of the hybridization signal, a second round of amplification was applied by sandwich method as above. In order to reduce the nonspecific binding, the slides were preincubated in 4 × SSC with 15% nonfat dry milk for 10 min at 37 °C prior to each step of immunological reactions. The chromosomes were counterstained with propidium iodide (PI, Sigma) and 4, 6-diamidino-2-phenylindole (DAPI, Sigma), 2 µg/mL each in PBS/glycerol (1:9, v/v) containing 0.2% (1,4)-diazobicyclo-(2, 2, 2) octane (DABCO, Sigma) as an anti-fade agent. Photograghy was taken under a fluorescence microscope (BH-2, Olympus) with the G excitation filter, EY-455 help excitation filter and Y-475 barrier filter, using Fuji ASA 400 color film.
RESULTS
Analysis of chromosomal quality
Under the same conditions of the experiment, sperm of the subjects with HBV infection as well as healthy controls appeared to be able to penetrate into zona-free hamster oocytes and to develop to the first-cleavage metaphase. However, the quality of metaphase spreads of sperm chromosomes had significant difference between the above two groups (P < 0.005, χ2-test). The quality of a number of sperm chromosome plates of the subjects with HBV infection was low, showing generally hard to be stained, stickiness of chromosome, clumping, tortuosity and so on, making the analyzable metaphase spreads greatly decreased. Whereas, few of these phenomena occurred in the controls although a small proportion of sperm chromosome plates of the controls could not be analyzed because of poor separation of chromosomes (Table 2).
Table 2.
Analytical results of sperm chromosomes of the tested subjects
| Sub-jects | Number of metaphase spreads observed | Number (%)of analyzable metaphase spreads | Number(%) of normal sperm karyotypes |
Number of abnormal sperm |
Rates (%) of anormal sperm | FISH Re-sults | |||||||
| With aneu-ploidy |
With structural aberrations |
||||||||||||
| g | r | tr | dic | pul | ace | del | |||||||
| Hepatitis | |||||||||||||
| 1 | 73 | 23 (31.5) | 20 (87.0) | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 13.0 | - |
| 2 | 69 | 20 (29.0) | 15 (75.0) | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 25.0 | - |
| 3 | 72 | 24 (33.3) | 22 (91.7) | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 8.3 | - |
| 4 | 68 | 29 (42.6) | 25 (86.2) | 0 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 13.8 | - |
| 5 | 86 | 30 (34.9) | 28 (93.3) | 1 (1)a | 0 | 1 | 1a | 0 | 0 | 0 | 0 | 6.7 | - |
| 6 | 113 | 43 (38.1) | 30 (69.8) | 5 (2)a | 4 (1)b | 2 (1)a | 2 (1)a | 0 | 2 | 1b | 0 | 30.2 | + |
| 7 | 82 | 19 (23.2) | 19 (100 ) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | - |
| 8 | 61 | 23 (37.7) | 20 (87.0) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 13.0 | - |
| 9 | 48 | 12 (25.0) | 11 (91.7) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.3 | - |
| Total | 672 | 223 (33.2) | 190 (85.2) | 9 (3)a | 6 (1)b | 5 | 5 | 4 | 4 | 3 | 1 | 14.8 | - |
| Controls | |||||||||||||
| 1 | 57 | 31 (54.3) | 29 (93.5) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 6.5 | - |
| 2 | 61 | 21 (34.4) | 21 (100 ) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | - |
| 3 | 50 | 19 (38.0) | 19 (100 ) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | - |
| 4 | 36 | 17 (47.2) | 16 (94.1) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.9 | - |
| 5 | 42 | 28 (66.7) | 26 (92.9) | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 7.1 | - |
| Total | 246 | 116 (47.2) | 111 (95.7) | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 4.3 | - |
g = Gap; r = Ring; tr = Triradial; dic = Dicentric chromosome; pul = Pulverization; ace = Acentric fragment; del = Deletion
For containing both numerically and structurally chromosomal aberrations simultaneously
For containing gap and acentric fragment chromosome aberrations simultaneously
Incidence of chromosome aberrations
Among the analyzable metaphase spreads, chromosome aberrations observed included numerical anomaly (aneuploidy), gap, ring chromosome, triradial, dicentric chromosome, pulverization, acentric fragment and deletion, orderly. Of the 233 analyzable sperm metaphase spreads in the hepatitis group, 33 (14.8%) complements contained chromosome aberrations, being significantly higher than 5 (4.3%) chromosome aberrations in the control group (P < 0.005, χ2-test).
Signals of in situ hybridization
Using specific whole length HBV DNA as probe to perform in situ hibridization on sperm chromosome slides, the sperm metaphases of one patient with chronic persistent hepatitis (subject 6) presented positive signals, all other tested subjects’ spermatozoa (50 per subject), as well as all hamster oocytes (more than 200) did not have FISH signal (Table 1). Of 42 chromosome complements of subject 6 with chronic persistent hepatitis, 9 complements showed clearly twin yellow spots on some chromosomes. In 9 sperm metaphase spreads with positive signals, one had 5 signals on different chromosomes, the rest ranged from 2 to 4 spots involving unrelated chromosomes. The intensity of signal presented distinct difference among the spots. Positive signals detected were not identical in the chromosomes and in distribution (Figure 1).
Figure 1.
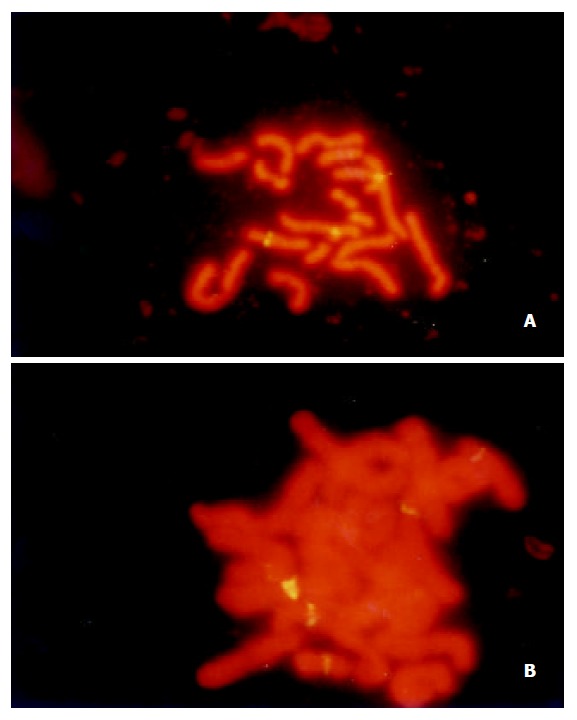
Detection of HBV DNA sequences in sperm chromo-somes by FISH with biotinylated whole length HBV DNA probe: (A) a well separated chromosome plate from one pa-tient (subject 6) with chronic persistent hepatitis, with three fluorescent signals on different chromosomes; (B) a poorly sepa-rated chromosome plate from the same patient with five fluo-rescent signals.
DISCUSSION
Our studies showed that the frequency of sperm chromosomal aberrations was greatly elevated after the infection of hepatitis B virus, and FISH could directly visualize the integration of HBV DNA sequences into sperm chromosomes. These results suggested that HBV infection could produce inheritable biological effects by carrying genetic materials damaged by virus or carrying altered genetic constituent due to the insertions of virus DNA in germ cells. As we know, this is the first report about the influence of biological factors on human sperm chromosomes.
In our study, we unexpectedly found that the sperm chromosomes frequently presented stickiness, faint stain and extreme tortuosity in the tested subjects with HBV infection and analyzable metaphases were low. Nevertheless, these changes were not found in the healthy controls under the strictly identical conditions. A previous report demonstrated that this manifestation had occurred in the chromosomes of somatic cells of HB patients with positive HBsAg[25]. The reasons of causing chromosomal stickiness in the HBV infection patients were unclear. The possibilities we considered might include the following events: First, antigen components of HBV such as core protein might interfere with the assembly of chromosome from chromatin due to its interaction with histones, it was confirmed that the core protein of HBV had participated in the organization of nucleosomes with histones in the hepatitis B virus minichromosome[26,27], so the condensation degree of chromosome was decreased and staining of chromosome became more difficult. Second, local despiralizations in the chromosomes occurred due to virus or its components. Third, premature chromosome condensation (PCC) might be induced by HBV, because virus-induced PCC was a common phenomenon[28]. Chromosome stickiness itself may be a manifestation of chromosomal pulverizations.
Previous reports have indicated that HBV infection can induce genetic alterations in somatic cells including hepatocytes and blood cells etc, showing increase of the rate of chromosomal aberrations or SCEs[29-35]. It was reported that influenza in spermatocytes of mice could greatly induce the increase of chromosomal anomalies[36]. Our studies revealed that the incidence of chromosomal aberrations in the HBV infection group was significantly higher than that in the controls. Although in the present study the individual sample sizes and analyzable chromosome numbers were not large enough and it was difficult to compare the differences of rates and types of aberrations among the different clinical and serological states of HBV infection, there were some features worthy to be considered: First, the incidence of chromosome aberrations did not appear to be closely correlated with the seminal fluid status of HBV infection; Second, the chromosome pulverizations were not observed in the controls but were present in 4 sperm cells in the HBV infection group; Third, subject 6 with proven HBV integrations in the sperm chromosomes had prominently higher level of chromosomal aberration than the other HBV infection subjects; Fourth, the chromosome breakages were more frequent in the HBV infection group than that in the controls.
FISH onto sperm chromosomes by specific HBV DNA as a probe had showed positive signals in an HBV patient. The results directly indicated that HBV could penetrate blood-testis barrier and enter male germ line and integrate into their genome. So the possibility of vertical transmission of HBV via the germ cells was further confirmed[18,19]. Our preliminary observations showed that HBV integrations into sperm chromosomes had the feature of multi-site integrations and that the predilection site of HBV insertion into chromosomes seemed to be unlikely present. These results were similar with other previous reports about HBV-related hepatocellular carcinoma either in HCC-derived cell lines or in liver tumor samples[7-17,32,33]. During spermatogeneis, the stem cells sequentially undergo proliferation by mitosis and differentiation into spermatocytes and formation into spermatozoa by meiosis. The occurred important events of meiosis were the pairing and recombination of chromosomes during the prophase of meiosis I[37]. Thus it was possible that relocation of viral sequences and multi-site integrations could occur via genetic recombination when HBV was inserted into male reproductive cells, but the genetic effect of initial HBV insertion into various spermatogenic stages must have been greatly various. If the insertion occurred in stem cells, their all daughter cells would carry the integrated form of viral DNA and the effects would be long-term. In our studies, we were surprised by the fact that FISH showed strong signals in a patient using single small DNA probe. The reasonable explanations for this result included that the copies of HBV strand-invasion may be multiple, HBV sequence generated duplications in situ after the invasion into host genome, during the processes of making spermatozoa, unequal cross-over or DNA rearrangements occurred repeatedly due to HBV insertion into DNA of stem cells. According to the sperm chromosome aberration analysis (Table 2), such a subject with HBV integrations had relatively higher incidence of sperm chromosome aberration than that in other subjects without HBV integration. This correlation implied that viral DNA integrations into germ line significantly increased the instability of gametic genome. Thus, HBV integration into sperm cells would create extensively inheritable effects including offspring from such sperm fusion with ovum presenting HBV infection status in newborn infant and producing congenital or hereditary disease (for example, embryonal tumor) by altering genetic constituent and/or inducing mutations[38-40].
We conclude that H BV infection can bring about mutagenic effects on sperm chromosomes which show that the incidence of sperm chromosome aberrations is significantly elevated after the infection of HBV. Integrations of viral DNA into sperm chromosomes with feature of multisites and nonspecific can further increase the instability of sperm chromosomes. Our study suggests that HBV infection can create extensively inheritable effects not only by altering genetic constituent and/or inducing mutations but also possibly by vertical transmission of HBV via the germ line to the next generation.
Footnotes
Supported by the Natural Science Foundation of Guangdong Province, No. 940567; and by the National Natural Science Foundation of China, No. 39970374
Edited by Xu XQ and Wu XN
References
- 1.Fang JN, Jin CJ, Cui LH, Quan ZY, Choi BY, Ki M, Park HB. A comparative study on serologic profiles of virus hepatitis B. World J Gastroenterol. 2001;7:107–110. doi: 10.3748/wjg.v7.i1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan JC, Ma JY, Pan BR, Ma LS. Studies on viral hepatitis B in China. Shijie Huaren Xiaohua Zazhi. 2001;9:611–616. [Google Scholar]
- 3.Rabe C, Pilz T, Klostermann C, Berna M, Schild HH, Sauerbruch T, Caselmann WH. Clinical characteristics and outcome of a cohort of 101 patients with hepatocellular carcinoma. World J Gastroenterol. 2001;7:208–215. doi: 10.3748/wjg.v7.i2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heathcote J, Cameron CH, Dane DS. Hepatitis-B antigen in saliva and semen. Lancet. 1974;1:71–73. doi: 10.1016/s0140-6736(74)92289-2. [DOI] [PubMed] [Google Scholar]
- 5.Darani M, Gerber M. Letter: Hepatitis-B antigen in vaginal secretions. Lancet. 1974;2:1008. doi: 10.1016/s0140-6736(74)92094-7. [DOI] [PubMed] [Google Scholar]
- 6.Dejean A, Lugassy C, Zafrani S, Tiollais P, Brechot C. Detection of hepatitis B virus DNA in pancreas, kidney and skin of two human carriers of the virus. J Gen Virol. 1984;65(Pt 3):651–655. doi: 10.1099/0022-1317-65-3-651. [DOI] [PubMed] [Google Scholar]
- 7.Rogler CE, Sherman M, Su CY, Shafritz DA, Summers J, Shows TB, Henderson A, Kew M. Deletion in chromosome 11p associated with a hepatitis B integration site in hepatocellular carcinoma. Science. 1985;230:319–322. doi: 10.1126/science.2996131. [DOI] [PubMed] [Google Scholar]
- 8.Hino O, Shows TB, Rogler CE. Hepatitis B virus integration site in hepatocellular carcinoma at chromosome 17; 18 translocation. Proc Natl Acad Sci USA. 1986;83:8338–8342. doi: 10.1073/pnas.83.21.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tokino T, Fukushige S, Nakamura T, Nagaya T, Murotsu T, Shiga K, Aoki N, Matsubara K. Chromosomal translocation and inverted duplication associated with integrated hepatitis B virus in hepatocellular carcinomas. J Virol. 1987;61:3848–3854. doi: 10.1128/jvi.61.12.3848-3854.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou YZ, Slagle BL, Donehower LA, vanTuinen P, Ledbetter DH, Butel JS. Structural analysis of a hepatitis B virus genome integrated into chromosome 17p of a human hepatocellular carcinoma. J Virol. 1988;62:4224–4231. doi: 10.1128/jvi.62.11.4224-4231.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urashima T, Saigo K, Kobayashi S, Imaseki H, Matsubara H, Koide Y, Asano T, Kondo Y, Koike K, Isono K. Identification of hepatitis B virus integration in hepatitis C virus-infected hepatocellular carcinoma tissues. J Hepatol. 1997;26:771–778. doi: 10.1016/s0168-8278(97)80241-3. [DOI] [PubMed] [Google Scholar]
- 12.Pineau P, Marchio A, Terris B, Mattei MG, Tu ZX, Tiollais P, Dejean A. A t(3; 8) chromosomal translocation associated with hepatitis B virus intergration involves the carboxypeptidase N locus. J Virol. 1996;70:7280–7284. doi: 10.1128/jvi.70.10.7280-7284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hino O, Tabata S, Hotta Y. Evidence for increased in vitro recombination with insertion of human hepatitis B virus DNA. Proc Natl Acad Sci USA. 1991;88:9248–9252. doi: 10.1073/pnas.88.20.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hino O, Ohtake K, Rogler CE. Features of two hepatitis B virus (HBV) DNA integrations suggest mechanisms of HBV integration. J Virol. 1989;63:2638–2643. doi: 10.1128/jvi.63.6.2638-2643.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokino T, Matsubara K. Chromosomal sites for hepatitis B virus integration in human hepatocellular carcinoma. J Virol. 1991;65:6761–6764. doi: 10.1128/jvi.65.12.6761-6764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su TS, Hwang WL, Yauk YK. Characterization of hepatitis B virus integrant that results in chromosomal rearrangement. DNA Cell Biol. 1998;17:415–425. doi: 10.1089/dna.1998.17.415. [DOI] [PubMed] [Google Scholar]
- 17.Shamay M, Agami R, Shaul Y. HBV integrants of hepatocellular carcinoma cell lines contain an active enhancer. Oncogene. 2001;20:6811–6819. doi: 10.1038/sj.onc.1204879. [DOI] [PubMed] [Google Scholar]
- 18.Hadchouel M, Scotto J, Huret JL, Molinie C, Villa E, Degos F, Brechot C. Presence of HBV DNA in spermatozoa: a possible vertical transmission of HBV via the germ line. J Med Virol. 1985;16:61–66. doi: 10.1002/jmv.1890160109. [DOI] [PubMed] [Google Scholar]
- 19.Peng HW, Su TS, Han SH, Ho CK, Ho CH, Ching KN, Chiang BN. Assessment of HBV persistent infection in an adult population in Taiwan. J Med Virol. 1988;24:405–412. doi: 10.1002/jmv.1890240407. [DOI] [PubMed] [Google Scholar]
- 20.Huang T, Liu X, Cai M, Huang J. [Chromosomal aberrations induced in human spermatozoa by mitomycin C] Yichuan Xuebao. 1997;24:291–295. [PubMed] [Google Scholar]
- 21.Kamiguchi Y, Mikamo K. An improved, efficient method for analyzing human sperm chromosomes using zona-free hamster ova. Am J Hum Genet. 1986;38:724–740. [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis . Molecular cloning-a labora-tory manual. 3rd edition, New york: Clod Spring Habor Laboratory Press; 2001. pp. 1.21–1.83. [Google Scholar]
- 23.Huang JM, Huang TH, Fang XW, Zhuang TG, Liu HX. Detec-tion of aneuploidy in human sperm by FISH. Zhonghua Yixue Yichuanxue Zazhi. 1997;14:248–249. [Google Scholar]
- 24.Korenberg JR, Chen XN. Human cDNA mapping using a high-resolution R-banding technique and fluorescence in situ hybridization. Cytogenet Cell Genet. 1995;69:196–200. doi: 10.1159/000133962. [DOI] [PubMed] [Google Scholar]
- 25.Vianello MG, Fasce LC, Mura MS, Andreoni G, Scalise G. [Cytogenic analysis in HBsAg positive subjects, healthy carriers and patients with acute-phase hepatitis] Boll Ist Sieroter Milan. 1979;58:266–272. [PubMed] [Google Scholar]
- 26.Bock CT, Schranz P, Schröder CH, Zentgraf H. Hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Genes. 1994;8:215–229. doi: 10.1007/BF01703079. [DOI] [PubMed] [Google Scholar]
- 27.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 28.Aula P. Virus-induced premature chromosome condensation (PCC) in single cells and G-bands of PCC-chromatin. Hereditas. 1973;74:81–87. doi: 10.1111/j.1601-5223.1973.tb01106.x. [DOI] [PubMed] [Google Scholar]
- 29.Chen HL, Chiu TS, Chen PJ, Chen DS. Cytogenetic studies on human liver cancer cell lines. Cancer Genet Cytogenet. 1993;65:161–166. doi: 10.1016/0165-4608(93)90227-d. [DOI] [PubMed] [Google Scholar]
- 30.Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A. Recurrent chromosomal abnormalities in hepato-cellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 1997;18:59–65. [PubMed] [Google Scholar]
- 31.Pang E, Wong N, Lai PB, To KF, Lau JW, Johnson PJ. A compre-hensive karyotypic analysis on a newly developed hepatocellu-lar carcinoma cell line, HKCI-1, by spectral karyotyping and com-parative genomic hybridization. Cancer Genet cytogenet. 2000;121:9–16. doi: 10.1016/s0165-4608(99)00247-2. [DOI] [PubMed] [Google Scholar]
- 32.Livezey KW, Negorev D, Simon D. Hepatitis B virus-transfected Hep G2 cells demonstrate genetic alterations and de novo viral integration in cells replicating HBV. Mutat Res. 2000;452:163–178. doi: 10.1016/s0027-5107(00)00072-5. [DOI] [PubMed] [Google Scholar]
- 33.Livezey KW, Negorev D, Simon D. Increased chromosomal alterations and micronuclei formation in human hepatoma HepG2 cells transfected with the hepatitis B virus HBX gene. Mutat Res. 2002;505:63–74. doi: 10.1016/s0027-5107(02)00140-9. [DOI] [PubMed] [Google Scholar]
- 34.Wang PL, Liu SF, Shen SL, Dong XY. A study of sister-chromatid exchange in peripheral lymphocytes of hepatitis B patients with HBsAg positive and their children. Mutat Res. 1986;175:249–254. doi: 10.1016/0165-7992(86)90062-x. [DOI] [PubMed] [Google Scholar]
- 35.Chatterjee B, Ghosh PK. Constitutive heterochromatin polymorphism and chromosome damage in viral hepatitis. Mutat Res. 1989;210:49–57. doi: 10.1016/0027-5107(89)90043-2. [DOI] [PubMed] [Google Scholar]
- 36.Sharma G, Polasa H. Cytogenetic effects of influenza virus infection on male germ cells of mice. Hum Genet. 1978;45:179–187. doi: 10.1007/BF00286960. [DOI] [PubMed] [Google Scholar]
- 37.Hecht NB. The making of a spermatozoon: a molecular perspective. Dev Genet. 1995;16:95–103. doi: 10.1002/dvg.1020160202. [DOI] [PubMed] [Google Scholar]
- 38.Naumova AK, Kisselev LL. Biological consequences of interactions between hepatitis B virus and human nonhepatic cellular genomes. Biomed Sci. 1990;1:233–238. [PubMed] [Google Scholar]
- 39.Kim H, Lee MJ, Kim MR, Chung IP, Kim YM, Lee JY, Jang JJ. Expression of cyclin D1, cyclin E, cdk4 and loss of heterozygosity of 8p, 13q, 17p in hepatocellular carcinoma: comparison study of childhood and adult hepatocellular carcinoma. Liver. 2000;20:173–178. doi: 10.1034/j.1600-0676.2000.020002173.x. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T, Miyamoto H, Hino O, Kitagawa T, Koike M, Iizuka T, Sakamoto H, Ooshima A. Primary hepatocellular carcinoma with hepatitis B virus-DNA integration in a 4-year-old boy. Hum Pathol. 1986;17:202–204. doi: 10.1016/s0046-8177(86)80296-9. [DOI] [PubMed] [Google Scholar]


