Abstract
Dimethylsulphide (DMS) has an important role in the global sulphur cycle and atmospheric chemistry. Microorganisms using DMS as sole carbon, sulphur or energy source, contribute to the cycling of DMS in a wide variety of ecosystems. The diversity of microbial populations degrading DMS in terrestrial environments is poorly understood. Based on cultivation studies, a wide range of bacteria isolated from terrestrial ecosystems were shown to be able to degrade DMS, yet it remains unknown whether any of these have important roles in situ. In this study, we identified bacteria using DMS as a carbon and energy source in terrestrial environments, an agricultural soil and a lake sediment, by DNA stable isotope probing (SIP). Microbial communities involved in DMS degradation were analysed by denaturing gradient gel electrophoresis, high-throughput sequencing of SIP gradient fractions and metagenomic sequencing of phi29-amplified community DNA. Labelling patterns of time course SIP experiments identified members of the Methylophilaceae family, not previously implicated in DMS degradation, as dominant DMS-degrading populations in soil and lake sediment. Thiobacillus spp. were also detected in 13C-DNA from SIP incubations. Metagenomic sequencing also suggested involvement of Methylophilaceae in DMS degradation and further indicated shifts in the functional profile of the DMS-assimilating communities in line with methylotrophy and oxidation of inorganic sulphur compounds. Overall, these data suggest that unlike in the marine environment where gammaproteobacterial populations were identified by SIP as DMS degraders, betaproteobacterial Methylophilaceae may have a key role in DMS cycling in terrestrial environments.
Introduction
Methylated sulphur compounds are significant intermediates in the global biogeochemical sulphur cycle owing to their role in transferring sulphur between the atmosphere, terrestrial and marine ecosystems (Lomans et al., 1997; Bentley and Chasteen, 2004; Schäfer et al., 2010). Dimethylsulphide (DMS), a volatile organic sulphur compound, provides the largest input of biogenic sulphur from the oceans into the atmosphere with approximately 21 Tg per year (Watts, 2000). Its oxidation products affect atmospheric chemistry and contribute to the atmospheric aerosol burden (Andreae, 1990). It has been suggested that DMS contributes to regulation of global climate by acting as a climate-cooling gas (Charlson et al., 1987); although, whether this is indeed the case remains controversial (Quinn and Bates, 2011).
In the marine environment, breakdown of dimethylsulphoniopropionate (DMSP) is the major source for DMS (Johnston et al., 2008). In contrast, the sources of DMS in terrestrial environments, including freshwater bodies, are diverse, including reduction of dimethylsulphoxide (Zinder and Brock, 1978), degradation of sulphur containing amino acids (Kadota and Ishida, 1972; Finster et al., 1990; Kiene and Hines, 1995) and anaerobic degradation of methoxylated aromatic compounds (Bak et al., 1992; Lomans et al., 2001). Lomans et al. (1997) reported concentrations of DMS in freshwater sediments in the range of 1–44 nM, similar to, if not higher than typical DMS concentrations in the upper marine water column (Lana et al., 2011). DMS is also emitted from a range of plant species including trees, cruciferous vegetables, lichens and wheat (Bending and Lincoln, 1999; Watts, 2000; Crespo et al., 2012). Anthropogenic sources of DMS include the pulp and paper, dairy and brewery industries for instance (Keenan and Lindsay, 1968; Scarlata and Ebeler, 1999; Zhang et al., 2006).
Annual DMS emission to the atmosphere from terrestrial ecosystems has been estimated at 3.8 Tg (Watts, 2000). As in marine systems, microbial metabolism of DMS is likely to have a major role in controlling DMS emissions by acting as a sink for DMS. Utilisation of DMS as a carbon and energy source by microorganisms has been well documented and a wide range of methylotrophic DMS degrading bacteria have been isolated from soils, biofilters, sewage and other terrestrial environments. A large number of DMS degrading isolates of Hyphomicrobium and Thiobacillus spp. have been described, but whether these represent the dominant active DMS-degrading populations in terrestrial environments remains unknown (reviewed in Schäfer et al., 2010).
DNA stable isotope probing (SIP) allows relating the function of uncultivated microorganisms to their identities through incorporation of stable isotope labels (for example, 13C or 15N) into their DNA (Radajewski et al., 2000). SIP has been applied with a range of labelled substrates to identify functionally active microorganisms in the environment (Neufeld et al., 2007b). SIP experiments with 13C-labelled methanol, methylamine, formaldehyde and formate have been carried out to identify active methylotrophs in Lake Washington sediments (Nercessian et al., 2005; Kalyuzhnaya et al., 2008). These studies revealed the dominance of Methylophilaceae in the SIP enrichments. [13C2]-DMS was used previously to investigate the active marine methylotrophs during a phytoplankton bloom in the English Channel (Neufeld et al., 2008a), suggesting that DMS degrading bacteria in seawater were Gammaproteobacteria, related to but distinct from previously isolated DMS-degrading Methylophaga strains (Schäfer, 2007).
Previous studies have demonstrated the potential of SIP combined with metagenomics for the analysis of functionally defined, yet uncultivated microorganisms (Dumont et al., 2006; Kalyuzhnaya et al., 2008; Neufeld et al., 2008b). In the current study, we applied a SIP-metagenomics approach to identify aerobic bacterial populations utilising DMS as a carbon and energy source in a soil and a lake sediment and to gain insight into functional aspects of microbial populations degrading DMS.
Materials and methods
Sampling
Soil samples were collected from the top 10 cm of Hunts Mill field with a crop of Brassica oleraceae (Wellesbourne, Warwickshire, UK) using an ethanol-sterilised trowel on 20 January 2010. Hunts Mill is an organic sandy loam soil with 73% sand, 12% silt and 14% clay (Whitfield, 1974). A field growing Brassica oleraceae was chosen for soil sampling since brassicas have a high content of sulphur containing glucosinolates, which may be released to soil via the roots and be subject to microbial degradation that releases volatile sulphur compounds (Crespo et al., 2012). Samples were obtained from five discrete locations in a 2.5 × 1.5 m area and a composite sample was derived. Soil was transferred to the laboratory immediately and kept at 4 °C until SIP incubations were set up. Lake sediment was collected on 08 June 2011 from Tocil Lake, a small eutrophic lake on the campus of the University of Warwick (Coventry, UK). Samples were taken from close to the shore to a depth of 6.5 cm using an acrylic corer and divided into four layers (0–1 cm, 1–2.5 cm, 2.5–4.5 cm and 4.5–6.5 cm) to measure DMS uptake rates. Sediment samples for SIP analysis were collected on 28 June 2011 as above and kept at 4 °C for 1 day until the surface layer was sliced off to a depth of 2.5 cm and homogenised with a spatula.
DMS uptake
[12C2]- or [13C2]-DMS uptake rates of soil and lake sediment samples were measured before time course SIP experiments. [12C2]-DMS uptake rates of lake sediment samples were measured in triplicate. Two grams of homogenised samples from each layer were placed in 125-ml crimp-top serum vials, sealed with butyl rubber stoppers and amended with 100 nanomoles of [12C2]-DMS. A gas chromatograph (Agilent Technologies, Cheshire, UK) fitted with a 30 m × 0.32 mm column (DB-1) and a flame ionisation detector was used to monitor the concentration of DMS in the headspace of the incubation vials. Helium was used as the carrier gas at a temperature of 200 °C. Similarly, an experiment was carried out to assess [12C2]- and [13C2]-DMS uptake by the soil samples. Triplicate microcosms were set up using 1 g of soil sample and amended with [13C2]- or [12C2]-DMS as described above. 13C-labelled DMS was synthesised as described previously (Neufeld et al., 2008a). DNA extraction, fractionation and PCR/denaturing gradient gel electrophoresis (DGGE) analysis were carried out as described below and in Supplementary Information to confirm that DMS uptake and label incorporation occurred.
Stable isotope probing
Stable isotope probing (SIP) of DMS-degrading bacteria was carried out according to Neufeld et al. (2007b). Three sets of triplicate microcosms were prepared using 1 g of wet soil or lake sediment sample per microcosm and amended with 450 nmoles per gram of DMS initially. GC measurements were taken twice a day and more DMS was provided to the incubation bottles when it was depleted. [12C2]-DMS incubations were set up as controls to assess potential differences in the rate of DMS oxidation by microorganisms during SIP incubations using the heavy isotope-labelled [13C2]-DMS.
DNA extraction and gradient fractionation
DNA was extracted from approximately 500 mg of sample from the microcosms using the Fast DNA soil extraction kit (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer's instructions. DNA concentrations were estimated using a spectrophotometer (NanoDrop ND-1000, Nanodrop products, Wilmington, DE, USA) and found to be in a range of 150–200 ng μl−1. In all, 15–20 μl aliquots of DNA (3 μg of DNA) were subjected to ultracentrifugation and DNA was recovered via gradient fractionation as described previously (Neufeld et al., 2007c).
Pyrosequencing of 16S rRNA genes
Pyrosequencing of the hypervariable regions (V1 and V2) of bacterial 16S rRNA genes was performed using a Roche 454 GS Junior (Roche 454 Life Sciences, Branford, CT, USA) at Micropathology Ltd. (Coventry, UK). The manufacturer's Universal Tailed Amplicon Sequencing protocol was applied with primers 27 F (Lane, 1991) and 519 R (Muyzer et al., 1993). Analyses of the pyrosequencing data were performed using a pipeline through the Quantitative Insights Into Microbial Ecology (QIIME) software, version 1.6.0 (Caporaso et al., 2010). Details can be found in Supplementary Material. Data have been submitted to the NCBI read archive under bioproject number PRJNA247775.
Metagenomics
Multiple displacement amplification was carried out to obtain micrograms of DNA from the ‘heavy' (fraction 7) and ‘light' (fraction 12) DNA of the second time-point SIP incubations. For this purpose, 100 times diluted templates were amplified using the Illustra GenomiPhi V2 DNA amplification kit (GE Life Sciences, Little Chalfont, UK) according to the manufacturer's instructions. PCR-DGGE analysis of phi29 amplified DNA did not reveal major changes in community fingerprints due to multiple displacement amplification (Supplementary Figure 1). Metagenomic libraries of the Genomiphi-amplified 13C-labelled DNA from ‘heavy' and ‘light' fractions were sequenced using the Illumina HiSeq2000 technology (GATC Biotech, Konstanz, Germany) which generated 8–12 Gb of 100 bp paired-end sequence data per sample. Read data have been submitted to the EBI short read archive under study accession PRJEB6310.
Assembly, annotation and analysis of metagenomes
Raw metagenomic reads from all samples were assembled into contigs using Ray Meta, an open source, de novo metagenome assembler (Boisvert et al., 2012). Assembled sequences were then annotated using Metagenomics Rapid Annotation using Subsystem Technology (MG-RAST) server version 3.2 (Meyer et al., 2008) and compared against the M5NR and Subsystems databases for taxonomic and functional distributions, respectively. In both cases, the maximum e-value cutoff was 1e−5, minimum identity cutoff was 60% and minimum alignment length cutoff was 15. The assembled contigs obtained by Ray Meta were also screened for functional genes involved in methylotrophy and sulphur metabolism using the BLAST search programme (Altschul et al., 1990) (see Supplementary Information). Assembled contigs are available on MGRAST (IDs 4528621.3, 4528622.3, 4528623.3 and 4528624.3). Taxonomic profiling based on unassembled metagenomic read data was carried out with Kraken (Wood and Salzberg, 2014). Sample reads were searched against a custom-made MiniKraken database that includes Archaea, Bacteria, Virus, Protozoa, Fungi and Plant RefSeq (NCBI Reference Sequence) data sets. Additional detail of Kraken analysis is given in the Supplementary Information.
Statistical analysis
Details of statistical analyses of 454 amplicon sequencing and metagenomic assemblies are given in the Supplementary Information. Briefly, principal components analysis (PCA) was applied to determine the operational taxonomical units (OTUs) primarily contributing to the differences in pyrosequencing data sets. This was followed by canonical variates analysis to identify the OTUs contributing to the discrimination of the samples by both time and treatment (12C or 13C). Analysis of variance and least significant difference (LSD) tests were further applied to the key OTUs identified by PCA to find significant differences at the 5% level. Metagenomics profiles obtained by MG-RAST (Meyer et al., 2008) were analysed using Statistical Analysis of Metagenomic Profiles (STAMP) software version 2.0 (Parks and Beiko, 2010). Two-sided Fisher's exact test was applied to compare the family-level proportions in the ‘heavy' and ‘light' DNA fractions.
Results
Assessment of DMS uptake potentials in lake sediment and soil samples
The top two layers of the lake sediment were found to be the most active in terms of [12C2]-DMS uptake with a rate up to 53.6±0.91 nmol DMS g−1 (wet weight) sediment day−1. Therefore, SIP incubations were set up using the samples from the top layers (0–2.5 cm) of the lake sediment.
Uptake of [12C2]- and [13C2]-DMS into soil was also assessed using a total amount of 15 μmoles of DMS amended to the incubations in 20 days. DGGE fingerprinting of the fractions (Supplementary Figure 2) showed that a restricted bacterial community consumed both 12C- and 13C- labelled DMS. A distinct pattern was observed in the ‘heavy' part of the [13C2]-DMS fractions (fractions 7 and 8) when compared with the ‘light' part of the same microcosm (fractions 11 and 12) representing the rest of the bacterial community that did not assimilate the [13C2]-DMS. Three dominant bands (Supplementary Figure 2, bands 1–3) were common in both ‘heavy' DNA fraction of the [13C2]-DMS incubation and ‘light' DNA fraction of the [12C2]-DMS incubation indicating the potential DMS degraders in the soil sample as these bands disappeared on the ‘light' fractions of the [13C2]-DMS incubation. Two of these bands had high identity to 16S rRNA genes of species from the Methylophilaceae family while the third represented a species of Thiobacillus (Supplementary Table 1).
Time course SIP experiments
The soil incubations were terminated after the samples had consumed approximately a total of 2.7, 8 and 15 μmoles of DMS per gram of soil (days 11, 17 and 20, respectively; Supplementary Figure 3a). The lake sediment incubations were terminated after approximately 5, 10 and 20 μmoles of DMS per gram of sediment had been consumed (days 10, 12 and 16, respectively; Supplementary Figure 3b). Initial microbial diversity analysis of DNA from lake sediment SIP gradients was carried out using DGGE (Supplementary Figure 4) and sequencing of dominant bands suggested proliferation of Methylophilus and Thiobacillus spp. (Supplementary Table 1).
454-analysis of bacterial diversity in SIP gradient fractions
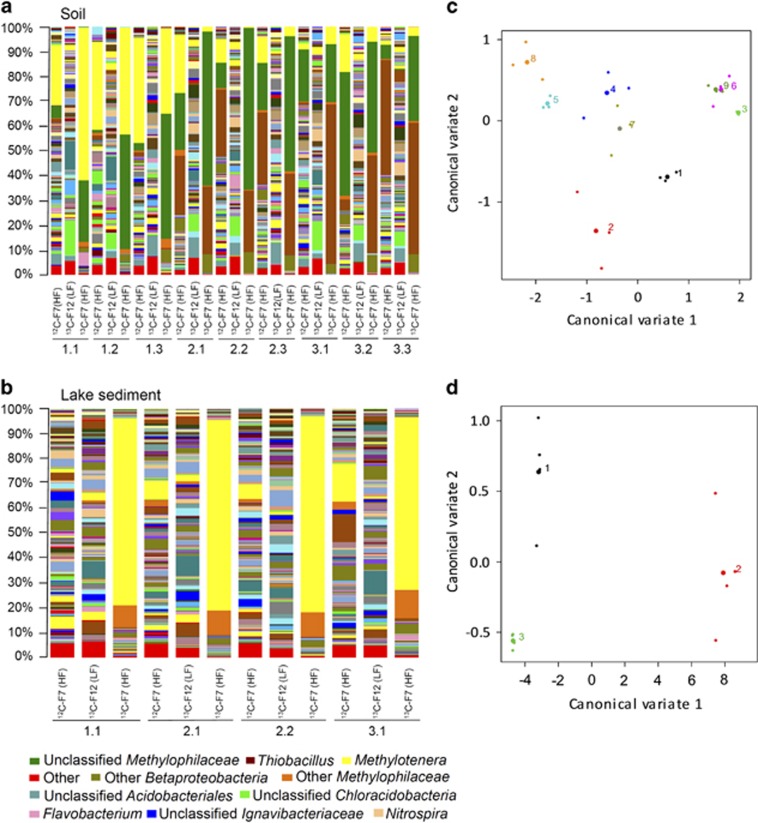
The ‘heavy' and ‘light' SIP gradient fractions from the three time points were further analysed by pyrosequencing of the 16S rRNA genes. A total of 31 401 quality-filtered, chimera-free sequences were obtained from 39 samples. These sequences were assigned to 9080 distinct OTUs at 97% identity (average 805±327 reads per sample with an average length of 481 bp). Taxonomic analysis revealed distinct community profiles in the ‘heavy' and ‘light' DNA fractions of the soil and lake sediment SIP samples (Figures 1a and b; Supplementary Table 2). The changes in the read abundance for particular OTUs in different DNA fractions and across the time course of the incubations suggested the degradation and assimilation of 13C-labelled DMS by specific bacterial populations. PCA supported the difference between the bacterial communities in DMS-SIP samples (Supplementary Figure 5). The first four principal components explained 45.86% and 59.88% of the total variability in the soil and lake sediment samples, respectively. Canonical variates analysis of the data clearly separated 13C-labelled ‘heavy' DNA from 13C-labelled ‘light' DNA fractions from different time points (Figures 1c and d). Betaproteobacteria dominated the 13C-labelled ‘heavy' fractions (F7) of all samples with percentages between 87.5 and 99.5. At the family level, members of the Methylophilaceae were prevalent across both soil and lake sediment 13C-labelled ‘heavy' DNA samples.
Figure 1.
Genus-level taxonomic profiling of the pyrosequencing data sets of 16S rRNA genes of heavy (fraction 7; F7) and light fractions (fraction 12, F12) from [12C2]-DMS or [13C2]-DMS incubated microcosms using (a) soil sample or (b) lake sediment sample. (c, d) Ordination of canonical variates (CV1 and CV2) produced from canonical variate analysis of pyrosequencing data at family level. Sample labels include the fraction number (F7 (HF) or F12 (LF)) and type of carbon used (12C or 13C). Numbers under each plot refer to the SIP experiment time point and replicate number. i.e. 2.1: second time point, replicate 1. Numbers in (c) represent nine different SIP fractions from the soil microcosm samples. (1) Time point 1, 12C ‘heavy' fraction (F7), (2) Time point 1, 13C ‘heavy' fraction (F7), (3) Time point 1, 13C ‘light' fraction (F12), (4) Time point 2, 12C ‘heavy' fraction (F7), (5) Time- point 2, 13C ‘heavy' fraction (F7), (6) Time point 2, 13C ‘light' fraction (F12), (7) Time point 3, 12C ‘heavy' fraction (F7), (8) Time point 3, 13C ‘heavy' fraction (F7), (9) Time point 3, 13C ‘light' fraction (F12). Numbers in (d) represent three different treatments in the lake sediment microcosm samples. (1) 12C ‘heavy' fraction (F7), (2) 13C ‘heavy' fraction (F7), (3) 13C ‘light' fraction (F12). Big dots represent the means and small dots represent the individual observations. Note that only the most predominant taxa in each fraction are colour-annotated, including Methylotenera (yellow), unclassified Methylophilaceae (green), Thiobacillus (brown), other Methylophilaceae (amber) and other Betaproteobacteria (dark green). The relative abundances of the 15 most abundant genera found in the ‘heavy' DNA fractions identified in the pyrosequencing data are shown in the Supplementary Table 2.
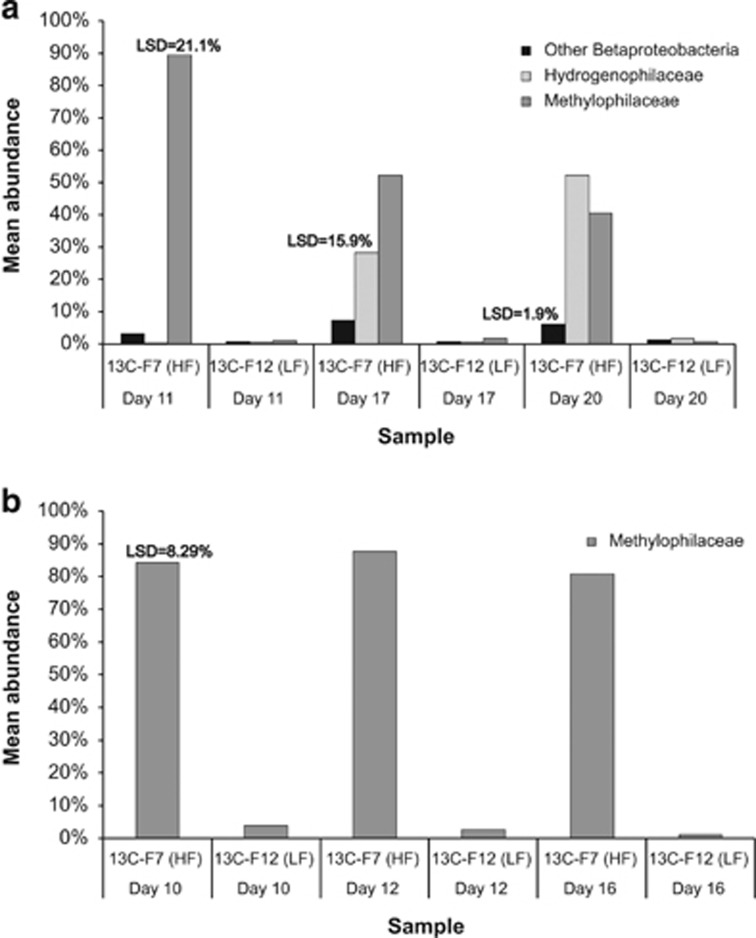
In the soil DMS-SIP, Methylophilaceae comprised an average of 89.4% (±1.5%) of the sequences from the 13C-labelled ‘heavy' fractions in the first time-point samples (day 11; 2.7 μmoles of DMS), decreasing to 63.2% (±3.7%) and 40.5% (±10.4%) in the second (day 17; 8 μmoles of DMS) and third time-point samples (day 20; 15 μmoles of DMS), respectively. The mean Methylophilaceae read abundances in the 13C-labelled ‘heavy' fractions were significantly higher than those in the 13C-labelled ‘light' fractions at each time point (LSD=21%, P=0.028; Figure 1a and Figure 2a). When the data sets were grouped according to the treatment only (12C or 13C-labelled DMS), the pairwise difference between the Methylophilaceae read abundances was even more pronounced (64% and 1% for 13C-labelled ‘heavy' and ‘light' fractions, respectively; LSD=12%, P=0.00). There was a concomitant increase in the mean read abundance of Hydrogenophilaceae in the 13C-labelled ‘heavy' fractions from 0.2% in the first time point (day 11) to 28.4% (±4.1%) and 52.2% (±12.4%) in the second (day 17) and third time points (day 20), respectively. Similarly, the mean read abundances of Hydrogenophilaceae in the 13C-labelled ‘heavy' fractions from the second and third time points were significantly higher than that of the 13C-labelled ‘light' fractions (LSD=15.9%, P=0.0018; Figures 1a and 2a). Hydrogenophilaceae sequence abundances contributed significantly (P=0.0001) to the discrimination of the samples by treatment only (26.9% and 1% for 13C-labelled ‘heavy' fraction and 13C-labelled ‘light' fraction; LSD=9.2%). Other Betaproteobacteria also increased in the second and third time point ‘heavy' fractions (7.4±1.1% and 6.1±2.1%, respectively) compared with the first time point (3.2±0.8%). Likewise, there was a significant pairwise difference between 13C-labelled ‘heavy' and ‘light' fractions (LSD=1.9%, P=0.047; Figure 2a). At the genus level, the Methylophilaceae were separated into Methylotenera and unclassified Methylophilaceae sequences in the first time-point (day 11) soil samples. Their relative abundances decreased while Thiobacillus (Hydrogenophilaceae) abundance increased in the second and third time-point (days 17 and 20, respectively) samples (Figure 1a; Supplementary Table 2a). The ‘light' fractions of the [13C2]-DMS microcosms were not enriched with Methylophilaceae or Hydrogenophilaceae sequences, but comprised highly diverse bacterial communities representing the soil bacterial community. Interestingly, the ‘heavy' fraction of the 12C-control microcosm of the soil sample had an increase in read abundance for Methylophilaceae and Thiobacillus species in addition to the background microbial community, suggesting the proliferation of these organisms on DMS in the 12C-microcosm that lead to their dominance in the microbial community.
Figure 2.
Mean abundances and LSD values of Methylophilaceae (dark grey), Hydrogenophilaceae (light grey) and other Betaproteobacteria (black) in 13C-labelled ‘heavy' and ‘light' fractions from each time point. Note that only families which had significantly higher abundances in the ‘heavy' fractions were shown. F7: ‘heavy' fraction F12: ‘light' fraction. (a) Soil, (b) Lake sediment.
The lake sediment DMS-SIP 13C-labelled ‘heavy' fractions were similarly dominated by members of the Methylophilaceae, comprising 84.2% of the sequences from the first time-point sample (day 10; 5 μmoles of DMS). Their read abundance increased to an average of 87.6% in the second time-point replicates (day 12; 10 μmoles of DMS) and decreased to 80.9% in the third time-point sample (day 16; 20 μmoles of DMS). The pairwise differences between the Methylophilaceae read abundances in the 13C-labelled ‘heavy' and ‘light' fractions at each time point were significant (LSD=8.2%, P=0.036; Figures 1b and 2b). This was also the case when the data sets were grouped by treatment only (85% and 2.5% for 13C-labelled ‘heavy' and ‘light' fractions, respectively; LSD=4.1%, P=0.00). Methylotenera remained the most dominant genus in the lake sediment samples from the three time points (Figure 1b; Supplementary Table 2b). While Thiobacillus spp. detected by DGGE band sequencing in the lake sediment ‘heavy' DNA were also detected by 454 amplicon sequencing, the read abundance remained fairly constant and they were not found as one of the OTUs contributing to the differences between the data sets according to the PCA which suggests that these did not proliferate significantly. As for the soil DMS-SIP experiment, the ‘light' DNA fractions comprised diverse microbial communities with only minor increase in read abundance of Methylotenera related bacteria from the second to third time point (days 12 and 16, respectively).
Taxonomic profiling of the metagenomes
Average contig lengths were 1279 and 1728 bp for the ‘light' and ‘heavy' fractions of the [13C2]-DMS soil incubation, respectively (Table 1). Based on all the protein annotation sources (M5NR database) used by MG-RAST, the ‘heavy' and ‘light' metagenomes were dominated by bacteria (74.9 and 76.9%). The proportions of Archaea or Eukarya were not >6.4% in either fraction while unassigned sequences comprised 13.4% and 14.8% of the ‘heavy' and ‘light' metagenomes, respectively. Kraken analysis showed that of the classified reads in the ‘heavy' fraction of soil SIP, 85.9% were bacterial, 0.17% were archaeal and 0.87% viral DNA, while in the ‘light' fraction of soil SIP 17.2% were bacterial, 0.61% were archaeal and 1.48% were of viral origin. Eukaryotic sequences (fungi, protozoa, plant and algae) comprised 13% and 80.6% of the soil ‘heavy' and ‘light' fractions, respectively.
Table 1. Assembly results of metagenomic reads from the soil and lake sediment SIP fractions.
|
Soil |
Lake Sediment |
|||
|---|---|---|---|---|
| Light | Heavy | Light | Heavy | |
| Total number of contigs | 80 987 | 28 382 | 53 331 | 22 094 |
| Total contig length (bp) | 103 612 778 | 49 050 429 | 65 621 201 | 28 430 434 |
| Average contig length (bp) | 1279 | 1728 | 1230 | 1286 |
| N50 (bp) | 1567 | 2 708 | 1449 | 1628 |
| Median (bp) | 769 | 934 | 767 | 753 |
Raw sequences obtained by Illumina sequencing were assembled into contigs using Ray Meta (Boisvert et al., 2012).
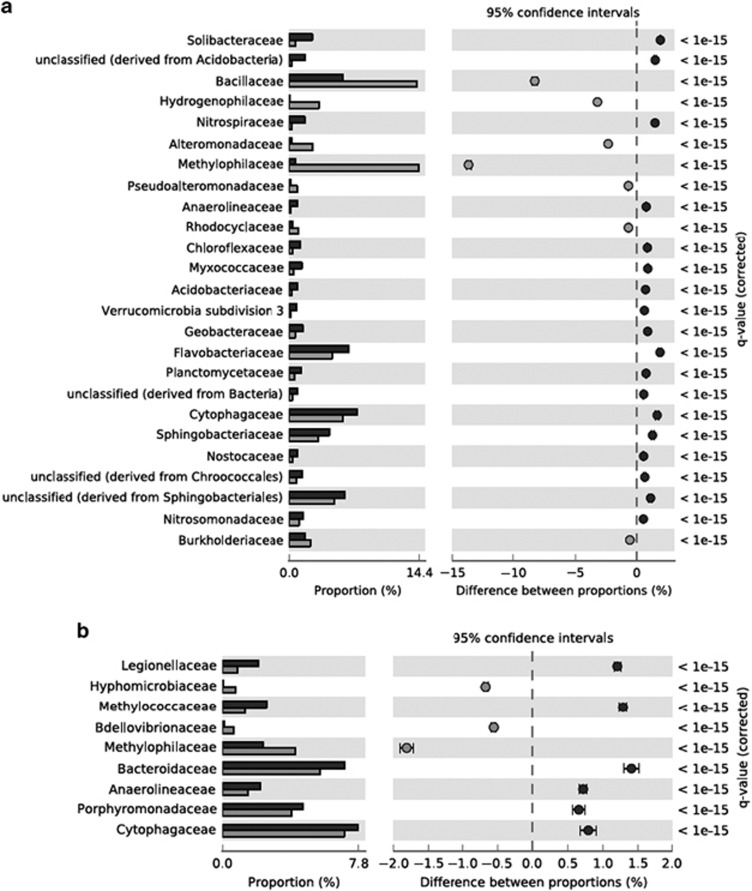
Proteobacteria and Bacteroidetes predominated the soil ‘heavy' and ‘light' fractions with 45.8% and 32.8%, respectively. Sphingobacteria dominated the sequences among the Bacteroidetes in the ‘light' fraction (34.7%) while Betaproteobacteria dominated in the ‘heavy' fraction (60.9%). With increasing taxonomic resolution at the family level, unclassified Sphingobacteriales was the most dominant group in the ‘light' metagenome with 5.0% abundance. However, in the ‘heavy' metagenome, Methylophilaceae were the most abundant family (11.1%). According to STAMP comparison, the Methylophilaceae family was also indicated as the most overrepresented family in the ‘heavy' fraction (Figure 3a). STAMP comparisons of the ‘heavy' and ‘light' fractions at the genus level revealed that Methylovorus, Methylotenera and Methylobacillus were the most overrepresented genera within the Methylophilaceae family in the ‘heavy' metagenome (Supplementary Figure 6a). Bacillus was the most abundant genus in the ‘heavy' fraction (11%).
Figure 3.
Family-level taxonomic analysis of metagenomic sequences obtained from ‘light' and ‘heavy' DNA samples from the SIP incubations using the STAMP software. Dark grey bars represent the ‘light' DNA and light grey bars represent the ‘heavy' DNA. Methylophilaceae family had the greatest proportional difference in the both ‘heavy' metagenomes. (a) Soil, (b) Lake sediment.
For the lake sediment metagenomes, the average contig lengths were 1230 and 1286 bp for the ‘light' and ‘heavy' fractions, respectively (Table 1). Similar to the soil metagenomes, bacteria dominated the sequences in both fractions with 81.8% and 81.4%. Unassigned sequences, Archaea and Eukarya comprised 12.8, 2.4 and 2.4% of the ‘heavy' metagenome, and 12.8%, 3.9% and 1.0%, in the ‘light' metagenome, respectively. Kraken analysis showed that of the classified reads in the ‘heavy' fraction of the lake sediment SIP, 71.7% were bacterial, 1.68% were archaeal and 3.3% viral DNA, while in the ‘light' fraction of lake sediment SIP 46.13% were bacterial, 6.6% were archaeal and 4.2% were of viral origin. In all, 23.2% and 43% were Eukaryotic sequences (fungi, protozoa, plant and algae) in the ‘heavy' and ‘light' fractions, respectively.
Taxonomic assignments of the lake metagenomic sequences showed differences compared with the soil metagenome. In both fractions, Bacteroidetes was the most dominant phylum (37.8% and 38.4% of the ‘light' and ‘heavy' fractions, respectively) followed by Proteobacteria which increased from 24.5% in the ‘light' fraction to 38.4% in the ‘heavy' fraction. Within Proteobacteria, the most abundant class in the ‘light' fraction was Gammaproteobacteria (39.8%) whereas Betaproteobacteria was the most abundant in the ‘heavy' fraction with 30.3%.
Comparison of the ‘heavy' and ‘light' fractions from the lake sediment SIP incubations by STAMP software indicated Methylophilaceae, Hyphomicrobiaceae and Bdellovibrionaceae as the most overrepresented taxa (Figure 3b). At the genus level, the fractional abundances of Methylobacillus, Methylotenera, Hyphomicrobium and Bdellovibrio sequences significantly increased in the ‘heavy' fraction (Supplementary Figure 6b).
Analysis of unassembled reads using Kraken also suggested that Methylophilaceae and Hydrogenophilaceae increased in abundance in the soil ‘heavy' SIP fractions, although the estimate of the fractional abundances (15.4% and 17.1%, respectively) was lower than that derived by amplicon sequencing. Similarly, Kraken analysis estimated the Methylophilaceae fractional abundance to be 45.5% and no marked increase of Hydrogenophilaceae in the lake sediment ‘heavy' fraction (Supplementary Figure 7). Overall, Kraken analysis had low fraction of matching kmers in the unassembled read data sets ranging between 1.4% and 6.9% suggesting that the coverage of the uncultivated microorganisms in the database was poor (Supplementary Table 3).
Functional profiling of the metagenomes
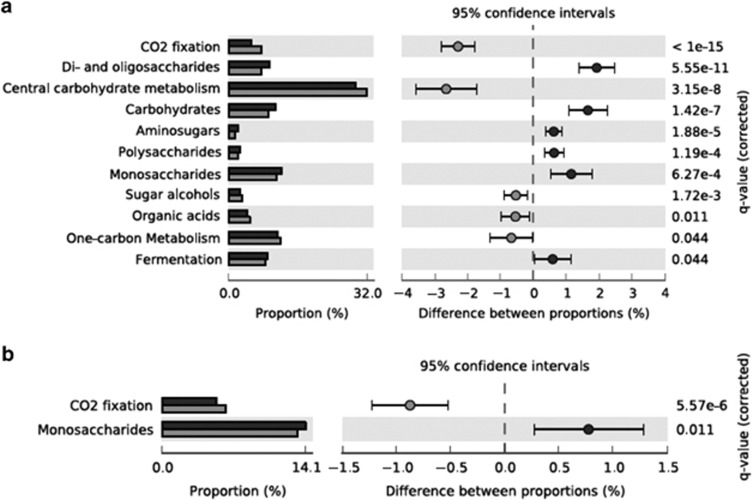
Analysis of the MG-RAST subsystems category ‘carbohydrate metabolism' in soil fractions revealed the over-representation of ‘central carbohydrate metabolism', ‘CO2 fixation', ‘one-carbon metabolism', ‘sugar alcohols' and ‘organic acids' in the ‘heavy' metagenome (Figure 4a). Within the category ‘one-carbon metabolism', the percentage of the sequences from the serine-glyoxylate cycle decreased from 91.9% in the ‘light' to 87.3% in the ‘heavy' fractions (corrected P-value=1.24e−7). However, the abundance of the sequences representing the ribulose-monophosphate pathway significantly increased from 0.30% to 1.3% in the ‘heavy' fraction (corrected P-value=1.09e−5; Supplementary Figure 8a). Sequences matching to ‘CO2 fixation' genes occurred significantly more in the ‘heavy' metagenome than in the ‘light' metagenome (corrected P-value=<1e−15). Among the CO2 fixation genes, sequences from the ‘Calvin-Benson cycle' were the most dominant in both fractions (36.8% and 43.9%). However, the number of genes for ‘CO2 uptake' and ‘carboxysome' increased significantly in the ‘heavy' fraction (corrected P-value=7.56e−10 and 3.25e−4, respectively). The abundance of genes related to sulphur metabolism was also investigated (Supplementary Figure 9a). In comparison with the 12C-DNA fraction, the most overrepresented genes in the ‘heavy' metagenome were involved in ‘inorganic sulphur assimilation', ‘sulphur oxidation' and ‘sulphate reduction-associated complexes' (corrected P-value=2.70e−5, 3.29e−10 and <1e−15, respectively).
Figure 4.
Functional analysis of metagenomic sequences from the carbohydrate metabolism category (MG-RAST) obtained from ‘light' and ‘heavy' DNA samples from the SIP incubations using the STAMP software. Dark grey bars represent the ‘light' DNA and light grey bars represent the ‘heavy' DNA. (a) Soil, (b) Lake sediment.
For the lake sediment metagenomes, within MG-RAST subsystems category ‘carbohydrate metabolism', only genes related to ‘CO2 fixation' were significantly more abundant in the ‘heavy' metagenome (corrected P-value=5.57e−6; Figure 4b). Within the one-carbon metabolism, only the number of sequences related to the ‘serine-glyoxylate cycle' was found significantly more abundant according to the STAMP analysis (corrected P-value=1.07e−3, Supplementary Figure 8b). The abundance of ‘sulphur oxidation' genes within ‘sulphur metabolism' category was significantly higher in the ‘heavy' fraction (corrected P-value=2.77e−4; Supplementary Figure 9b).
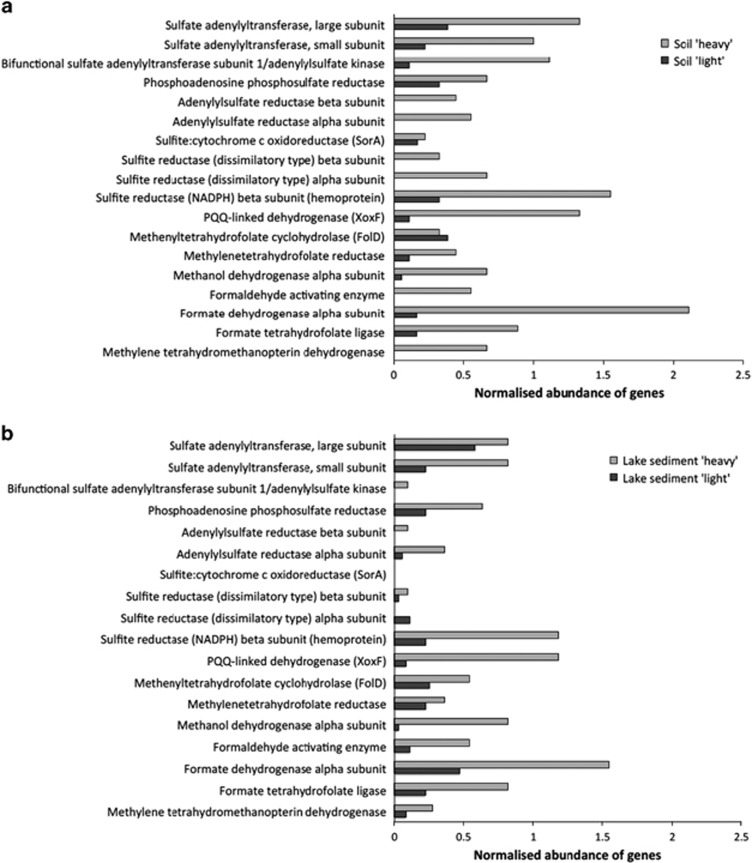
Additionally, genes encoding for enzymes involved in methylotrophy and oxidation of sulphide and sulphite were searched within the metagenomes using BLAST to identify the affiliation of the relevant genes and to compare their relative abundance to the recA gene, which is known to exist as a single copy in bacterial genomes (Eisen, 1995; Wu et al., 2011) (Supplementary Information). A higher relative abundance of genes encoding enzymes of tetrahydrofolate and tetrahydromethanopterin linked C1 oxidation, formaldehyde activating enzyme, formate oxidation as well as methanol dehydrogenase and PQQ linked dehydrogenase was found in ‘heavy' metagenomes (Figure 5a). The gene encoding the large subunit of dimethylsulphide monooxygenase (dmoA) was not found in metagenomes. Similarly, the relative abundance per recA copy of several genes encoding enzymes involved in inorganic sulphur compound oxidation/assimilation was higher in the ‘heavy' fraction, including flavocytochrome c sulphide dehydrogenase, dissimilatory sulphite reductase, adenylylsulphate reductase and sulphate adenylyltransferase. Similar changes in gene frequency were detected in the metagenome of the ‘heavy' fraction of the lake sediment microcosm (Figure 5b).
Figure 5.
Relative abundance of methylotrophy- and sulphur-related functional genes relative to recA (single copy control gene) in ‘heavy' and ‘light' metagenomes. Dark grey bars represent the ‘light' DNA and light grey bars represent the ‘heavy' DNA. (a) Soil, (b) Lake sediment. Numbers are based on features identified in assembled reads of the metagenomes as shown in Supplementary Table 4.
The methylotrophy and sulphur compound oxidation genes in the ‘heavy' metagenomes were mainly related to Methylophilaceae and Thiobacillus species, while those detected in the ‘light' fractions were affiliated with more diverse bacteria including members of the Bacteroidetes group that was abundant based on 454 read data. Similarly, the recA genes (e-value <1e−30) in the ‘heavy' soil metagenome were exclusively affiliated with Methylophilaceae and Thiobacillus, but originated from a wide diversity of bacteria in the ‘light' soil metagenome. In the lake sediment ‘heavy' metagenome, recA sequences were mainly originating from Methylotenera, Bdellovibrio and other bacteria while the diversity of recA in the ‘light' fraction reflected a more diverse community (Supplementary Table 4).
Discussion
The fate of DMS in terrestrial ecosystems and the microbial populations involved in its degradation have remained poorly characterised due to a lack of adequate cultivation-independent tools to study active DMS degrading bacteria. While microbial communities from complex ecosystems such as soils are now routinely analysed using high-throughput sequencing (Tringe et al., 2005; Kurokawa et al., 2007; Sul et al., 2009; Delmont et al., 2012), the polyphyletic nature of DMS degrading microbial taxa means that populations degrading DMS cannot be identified using ribosomal sequence analysis alone.
The analysis of DMS-SIP experiments suggested that DMS degradation in microcosms induced the proliferation of a limited number of populations in the community. DGGE analysis, 454 amplicon sequencing of gradient fractions and taxonomic profiling of metagenomic reads were in agreement, suggesting that the DMS-assimilating bacterial populations were dominated by Betaproteobacteria. 454-pyrosequencing data analysis revealed the dominance of species from the Methylophilaceae family in all 13C-labelled ‘heavy' DNA. While in soil DMS-SIP microcosms, Thiobacillus species also had increasing read abundance during the incubation, their lower read abundance in lake sediment SIP microcosms suggested that they were potentially less important in DMS assimilation in the lake sediment samples compared with soils. Metagenomic analysis also suggested that Bacillaceae were abundant in both ‘light' and ‘heavy' soil metagenomes, however, this was not supported by 454 amplicon sequencing or analysis of functional genes. While the analyses of the metagenomes showed the dominance of Methylobacillus and Methylotenera in both 13C-labelled ‘heavy' DNA fractions from soil and lake sediment, there were also differences between the experiments with soil and lake sediment, consisting of higher representation of Methylovorus and Thiobacillus in soil SIP microcosms, while analysis of the lake sediment ‘heavy' metagenome also indicated increased abundance of Hyphomicrobium and Bdellovibrionaceae. There were differences between the relative abundances of key microbial taxa estimated based on amplicon pyrosequencing, assembled metagenomic data and unassembled read data which are likely reflecting the intrinsic differences of these approaches. While ribosomal RNA gene databases would arguably be the most robust tool to match sequences of uncultivated organisms to taxonomic identifiers, kmer-based approaches, such as in Kraken require 100% matches of kmers and are based on databases that are less representative of uncultivated organisms than is possible with matching of amplicon sequences against rRNA databases. The latter is reflected in the low matching of kmers of raw read data to the Kraken database that only classified between 1.3% and 6.9% of reads in all samples (see Supplementary Information). In addition, estimates based on assembled features against the M5NR database would only assess a subset of sequence information (only the assembled part) while also potentially underestimating abundant species due to the fact that different coverage of such contigs would not be considered further. Despite these differences, each of these approaches agreed that Methylophilacaea and Thiobacillus were the most important taxa that were markedly enriched in the ‘heavy' fraction of the soil SIP, and that Methylophilaceae but not Thiobacillus were markedly enriched in the ‘heavy' fraction of the lake sediment SIP.
A critical issue in SIP is cross-feeding, which may give false positive identification of substrate assimilating taxa (Neufeld et al., 2007c). The higher abundance of Bdellovibrionaceae in the ‘heavy' metagenomes of the lake microcosms is most likely a consequence of these bacteria predating on the bacteria growing in response to DMS addition rather than an active involvement in methylotrophy or sulphur oxidation. Previous SIP studies have repeatedly shown Bdellovibrio-like organisms in experiments with different 13C-labelled carbon sources (Morris et al., 2002; Hutchens et al., 2004) and Bdellovibrio-like organisms were also successfully investigated using SIP with 13C-labelled biomass (Chauhan et al., 2009). Conversely, based on prior knowledge of DMS metabolism from cultivation-dependent studies, it might be argued that the true DMS degraders in the microcosms were Thiobacillus species, as a number of Thiobacillus spp. have previously been shown to degrade DMS (Sivelä and Sundman, 1975; Smith and Kelly, 1988; Visscher and Taylor, 1993; de Zwart et al., 1997), and that Methylophilaceae were labelled in SIP due to assimilation of intermediates of DMS degradation (for example, formate, formaldehyde and methanethiol) released by Thiobacillus spp. High abundance of Methylophilaceae representatives might be due to the ability of some strains to use formaldehyde as a carbon source (Nercessian et al., 2005). Microcosm experiments carried out with Lake Washington sediment to study the response of the microbial community to methane addition have shown increases of non-methanotrophic Methylophilaceae populations alongside those of bona fide methanotrophs, showing cooperation between Methylococcaceae and Methylophilaceae in methane utilisation (Beck et al., 2013), but the abundance of the primary methane degrader (Methylobacter) was substantial compared with that of the cooperating Methylophilaceae populations (Beck et al., 2013; Oshkin et al., 2014). According to the labelling pattern observed by the 454-pyrosequencing data in our study, the scenario that Methylophilaceae were getting labelled via cross-feeding is unlikely since Thiobacillus spp., the known DMS degrader, were not detected in the first time point of the soil DMS-SIP samples and only became a more dominant part of the community after Methylophilaceae were already established. The proliferation of Thiobacillus species in soil SIP microcosms could have been due to utilisation of intermediates produced by Methylophilaceae, for example, by utilising sulphide/sulphite as energy sources and assimilating 13CO2, but it is equally possible that Thiobacillus populations, which assimilate DMS after its initial oxidation to CO2 (Kanagawa and Kelly, 1986), were degrading DMS, which might also explain the increase of the CO2 fixation genes in the ‘heavy' metagenomes.
The family Methylophilaceae includes obligate and facultative methylotrophs but none of the members of this family has been shown to degrade DMS. We tested several cultures from the Methylophilaceae for their ability to degrade DMS including Methylophilus leisingeri, Methylophilus methylotrophus and Methylotenera versatilis, but none of the strains tested was able to degrade DMS, suggesting the available cultivated strains may not be representative of the SIP-detected populations. Comparative genomic analysis of Methylotenera mobilis JLW8, Methylotenera versatilis 301, Methylovorus glucosetrophus SIP3-4 with two other Methylophilaceae species (Methylobacillus flagellatus KT and unclassified Methylophilales strain HTCC2181) suggested that these methylotrophic Betaproteobacteria are metabolically versatile (Lapidus et al., 2011) and that closely related but metabolically distinct Methylophilaceae strains may co-exist in the same environment (Chistoserdova, 2011a); our data strongly support that suggestion and further indicate that certain Methylophilaceae are able to degrade DMS. An increase in the abundance of the genes from the Ribulose-monophosphate pathway in the soil ‘heavy' fraction was in agreement with these bacteria being known to use the Ribulose-monophosphate pathway for formaldehyde assimilation (Chistoserdova, 2011b). The absence of homologues of the DMS monooxygenase enzyme characterised in Hyphomicrobium sulfonivorans S1 (Boden et al., 2011) suggests that the dominant DMS assimilating microorganisms detected here use an alternative enzyme for DMS degradation, possibly via methanethiol, utilising as yet unidentified methyltransferases that have been suggested to be involved in DMS oxidation in certain Thiobacillus isolates (Visscher and Taylor, 1993) and in Methylophaga thiooxydans (Boden et al., 2011).
The frequency of genes of methylotrophic metabolism markedly increased in ‘heavy' soil and lake sediment metagenomes and their taxonomic classification supported that these uncultivated Methylophilaceae and Thiobacillus populations were involved in methylotrophy and oxidation of reduced inorganic sulphur compounds. DMS monooxygenase is unlikely to be the primary enzyme of DMS oxidation in these populations, and thus further studies of the biochemistry of DMS degradation in these groups of bacteria are essential. The methylotrophic populations detected in the ‘heavy' fractions also had genes encoding methanol dehydrogenase genes and XoxF, which was previously shown to be expressed during growth of Methylophaga thiooxydans on DMS (Schäfer, 2007). Methylotrophy genes detected in the ‘heavy' metagenomes suggest that these bacteria have the potential to degrade methyl groups of DMS via tetrahydrofolate or tetrahydromethanopterin bound intermediates. In the Thiobacillus spp. detected, sulphide and sulphite oxidation could occur via the dissimilatory sulphite reductase, adenylylsulphate reductase and sulphate adenylyltransferase, but it is less clear which enzymes might be involved in sulphite oxidation in Methylophilaceae.
The combination of SIP, 454 sequencing of gradient fractions and metagenomic sequencing proved useful for identifying microbial populations actively involved in DMS cycling in two terrestrial environments. It detected known DMS degraders (Thiobacillus) but also implicated a group of methylotrophs not previously realised as DMS degraders, uncultured Methylophilaceae. These findings reemphasise the significance of members of the Methylophilaceae as widely distributed and versatile methylotrophs (Lapidus et al., 2011; Chistoserdova, 2011a), which have important roles in not only carbon and nitrogen, but also sulphur cycling in terrestrial environments likely using as yet unidentified enzymes to degrade DMS.
Acknowledgments
HS was supported by a UK Natural Environment Research Council Advanced Fellowship NE/E013333/1), ÖE by a postgraduate scholarship from the University of Warwick and an Early Career Fellowship from the Institute of Advanced Study, University of Warwick, UK, respectively. Lawrence Davies is acknowledged for help with QIIME.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andreae MO. (1990). Ocean-atmosphere interactions in the global biogeochemical sulfur cycle. Mar Chem 30: 1–29. [Google Scholar]
- Bak F, Finster K, Rothfuß F. (1992). Formation of dimethylsulfide and methanethiol from methoxylated aromatic compounds and inorganic sulfide by newly isolated anaerobic bacteria. Arch Microbiol 157: 529–534. [Google Scholar]
- Beck DAC, Kalyuzhnaya MG, Malfatti S, Tringe SG, Glavina del Rio T, Ivanova N et al. (2013). A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between Methylococcaceae and the Methylophilaceae. Peer J 1: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bending GD, Lincoln SD. (1999). Characterisation of volatile sulphur-containing compounds produced during decomposition of Brassica juncea tissues in soil. Soil Biol Biochem 31: 695–703. [Google Scholar]
- Bentley R, Chasteen TG. (2004). Environmental VOSCs—formation and degradation of dimethyl sulfide, methanethiol and related materials. Chemosphere 55: 291–317. [DOI] [PubMed] [Google Scholar]
- Boden R, Borodina E, Wood AP, Kelly DP, Murrell JC, Schäfer H. (2011). Purification and characterization of dimethylsulfide monooxygenase from Hyphomicrobium sulfonivorans. J Bacteriol 193: 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. (2012). Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol 13: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson RJ, Lovelock JE, Andreae MO, Warren SG. (1987). Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326: 655–661. [Google Scholar]
- Chauhan A, Cherrier J, Williams HN. (2009). Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. Proc Natl Acad Sci 106: 4301–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. (2011. a). Methylotrophy in a lake: from metagenomics to single-organism physiology. Appl Environ Microbiol 77: 4705–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. (2011. b). Modularity of methylotrophy, revisited. Environ Microbiol 13: 2603–2622. [DOI] [PubMed] [Google Scholar]
- Crespo E, Hordijk CA, de Graaf RM, Samudrala D, Cristescu SM, Harren FJM et al. (2012). On-line detection of root-induced volatiles in Brassica nigra plants infested with Delia radicum L. root fly larvae. Phytochemistry 84: 68–77. [DOI] [PubMed] [Google Scholar]
- Delmont TO, Prestat E, Keegan KP, Faubladier M, Robe P, Clark IM et al. (2012). Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J 6: 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zwart JMM, Sluis JMR, Kuenen JG. (1997). Competition for dimethyl sulfide and hydrogen sulfide by Methylophaga sulfidovorans and Thiobacillus thioparus T5 in continuous cultures. Appl Environ Microbiol 63: 3318–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont MG, Radajewski SM, Miguez CB, McDonald IR, Murrell JC. (2006). Identification of a complete methane monooxygenase operon from soil by combining stable isotope probing and metagenomic analysis. Environ Microbiol 8: 1240–1250. [DOI] [PubMed] [Google Scholar]
- Eisen J. (1995). The RecA protein as a model molecule for molecular systematic studies of bacteria: Comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol 41: 1105–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finster K, King GM, Bak F. (1990). Formation of methylmercaptan and dimethylsulfide from methoxylated aromatic compounds in anoxic marine and fresh water sediments. FEMS Microbiol Lett 74: 295–301. [Google Scholar]
- Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC. (2004). Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol 6: 111–120. [DOI] [PubMed] [Google Scholar]
- Johnston AWB, Todd JD, Sun L, Nikolaidou-Katsaridou MN, Curson ARJ, Rogers R. (2008). Molecular diversity of bacterial production of the climate-changing gas, dimethyl sulphide, a molecule that impinges on local and global symbioses. J Exp Bot 59: 1059–1067. [DOI] [PubMed] [Google Scholar]
- Kadota H, Ishida Y. (1972). Production of volatile sulfur compounds by microorganisms. Annu Rev Microbiol 26: 127–138. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, Szeto E et al. (2008). High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol 26: 1029–1034. [DOI] [PubMed] [Google Scholar]
- Kanagawa T, Kelly DP. (1986). Breakdown of dimethyl sulfide by mixed cultures and by Thiobacillus thioparus. FEMS Microbiol Lett 34: 13–19. [Google Scholar]
- Keenan TW, Lindsay RC. (1968). Evidence for a dimethyl sulfide precursor in milk. J Dairy Sci 51: 112–114. [Google Scholar]
- Kiene RP, Hines ME. (1995). Microbial formation of dimethyl sulfide in anoxic Sphagnum peat. Appl Environ Microbiol 61: 2720–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A et al. (2007). Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lana A, Bell TG, Simó R, Vallina SM, Ballabrera-Poy J, Kettle AJ et al. (2011). An updated climatology of surface dimethlysulfide concentrations and emission fluxes in the global ocean. Global Biogeochem Cycle 25: GB1004–GB1021. [Google Scholar]
- Lane DJ. (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons Ltd.: Chichester, pp 115–175. [Google Scholar]
- Lapidus A, Clum A, LaButti K, Kaluzhnaya MG, Lim S, Beck DAC et al. (2011). Genomes of three methylotrophs from a single niche reveal the genetic and metabolic divergence of the Methylophilaceae. J Bacteriol 193: 3757–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomans BP, AJP Smolders, Intven LM, Pol A, Op Den Camp HJM, Van Der Drift C. (1997). Formation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl Environ Microbiol 63: 4741–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomans BP, Luderer R, Steenbakkers P, Pol A, van der Drift C, Vogels GD et al. (2001). Microbial populations involved in cycling of dimethyl sulfide and methanethiol in freshwater sediments. Appl Environ Microbiol 67: 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass E, Kubal M et al. (2008). The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Radajewski S, Willison TW, Murrell JC. (2002). Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl Environ Microbiol 68: 1446–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, De Waal EC, Uitterlinden AG. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. (2005). Bacterial populations active in metabolism of C1 Compounds in the sediment of Lake Washington, a freshwater lake. Appl Environ Microbiol 71: 6885–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW et al. (2007. a). DNA stable-isotope probing. Nat Protoc 2: 860–866. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Dumont MG, Vohra J, Murrell JC. (2007. b). Methodological considerations for the use of stable isotope probing in microbial ecology. Microb Ecol 53: 435–442. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Boden R, Moussard H, Schäfer H, Murrell JC. (2008. a). Substrate-specific clades of active marine methylotrophs associated with a phytoplankton bloom in a temperate coastal environment. Appl Environ Microbiol 74: 7321–7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld JD, Chen Y, Dumont MG, Murrell JC. (2008. b). Marine methylotrophs revealed by stable-isotope probing, multiple displacement amplification and metagenomics. Environ Microbiol 10: 1526–1535. [DOI] [PubMed] [Google Scholar]
- Oshkin IY, Beck DAC, Lamb AE, Tchesnokova V, Benuska G, MacTaggart TL et al. (2014). Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J doi:10.1038/ismej.2014.203. [DOI] [PMC free article] [PubMed]
- Parks DH, Beiko RG. (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26: 715–721. [DOI] [PubMed] [Google Scholar]
- Quinn PK, Bates TS. (2011). The case against climate regulation via oceanic phytoplankton sulphur emissions. Nature 480: 51–56. [DOI] [PubMed] [Google Scholar]
- Radajewski S, Ineson P, Parekh NR, Murrell JC. (2000). Stable isotope probing as a tool in microbial ecology. Nature 403: 646–649. [DOI] [PubMed] [Google Scholar]
- Scarlata CJ, Ebeler SE. (1999). Headspace solid-phase microextraction for the analysis of dimethylsulfide in beer. J Agric Food Chem 47: 2505–2508. [DOI] [PubMed] [Google Scholar]
- Schäfer H. (2007). Isolation of Methylophaga spp. from marine dimethylsulfide-degrading enrichment cultures and identification of polypeptides induced during growth on dimethylsulfide. Appl Environ Microbiol 73: 2580–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer H, Myronova N, Boden R. (2010). Microbial degradation of dimethylsulphide and related C1-sulphur compounds: organisms and pathways controlling fluxes of sulphur in the biosphere. J Exp Bot 61: 315–334. [DOI] [PubMed] [Google Scholar]
- Sivelä S, Sundman V. (1975). Demonstration of Thiobacillus type bacteria, which utilize methyl sulfides. Arch Microbiol 103: 303–304. [Google Scholar]
- Smith NA, Kelly DP. (1988). Mechanism of oxidation of dimethyl disulphide by Thiobacillus thioparus strain E6. J Gen Microbiol 134: 3031–3039. [Google Scholar]
- Sul WJ, Park J, Quensen JF, Rodrigues JLM, Seliger L, Tsoi TV et al. (2009). DNA-Stable Isotope Probing integrated with metagenomics for retrieval of biphenyl dioxygenase genes from polychlorinated biphenyl-contaminated river sediment. Appl Environ Microbiol 75: 5501–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW et al. (2005). Comparative metagenomics of microbial communities. Science 308: 554–557. [DOI] [PubMed] [Google Scholar]
- Visscher PT, Taylor BF. (1993). A new mechanism for the aerobic catabolism of dimethyl sulfide. Appl Environ Microbiol 59: 3784–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts SF. (2000). The mass budgets of carbonyl sulfide, dimethyl sulfide, carbon disulfide and hydrogen sulfide. Atmos Environ 34: 761–779. [Google Scholar]
- Whitfield WAD. (1974). The soils of the National Vegetable Research Station, Wellesbourne. Report of the National Vegetable Research Station for 1973. The British Society for the Promotion of Vegetable Research: Wellesbourne, UK, pp 21–30. [Google Scholar]
- Wood DE, Salzberg SL. (2014). Kraken: Ultrafast metagenomic sequence classification using exact alignment. Genome Biol 15: R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Wu M, Halpern A, Rusch DB, Yooseph S, Frazier M et al. (2011). Stalking the fourth domain in metagenomic data: Searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS ONE 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liss SN, Allen DG. (2006). The effects of methanol on the biofiltration of dimethyl sulfide in inorganic biofilters. Biotech Bioeng 95: 734–743. [DOI] [PubMed] [Google Scholar]
- Zinder SH, Brock TD. (1978). Dimethyl sulphoxide reduction by micro-organisms. J Gen Microbiol 105: 335–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.