Abstract
To investigate the genetic basis of microbial evolutionary adaptation to salt (NaCl) stress, populations of Desulfovibrio vulgaris Hildenborough (DvH), a sulfate-reducing bacterium important for the biogeochemical cycling of sulfur, carbon and nitrogen, and potentially the bioremediation of toxic heavy metals and radionuclides, were propagated under salt stress or non-stress conditions for 1200 generations. Whole-genome sequencing revealed 11 mutations in salt stress-evolved clone ES9-11 and 14 mutations in non-stress-evolved clone EC3-10. Whole-population sequencing data suggested the rapid selective sweep of the pre-existing polymorphisms under salt stress within the first 100 generations and the slow fixation of new mutations. Population genotyping data demonstrated that the rapid selective sweep of pre-existing polymorphisms was common in salt stress-evolved populations. In contrast, the selection of pre-existing polymorphisms was largely random in EC populations. Consistently, at 100 generations, stress-evolved population ES9 showed improved salt tolerance, namely increased growth rate (2.0-fold), higher biomass yield (1.8-fold) and shorter lag phase (0.7-fold) under higher salinity conditions. The beneficial nature of several mutations was confirmed by site-directed mutagenesis. All four tested mutations contributed to the shortened lag phases under higher salinity condition. In particular, compared with the salt tolerance improvement in ES9-11, a mutation in a histidine kinase protein gene lytS contributed 27% of the growth rate increase and 23% of the biomass yield increase while a mutation in hypothetical gene DVU2472 contributed 24% of the biomass yield increase. Our results suggested that a few beneficial mutations could lead to dramatic improvements in salt tolerance.
Introduction
In the post-genomic era, one of the major challenges in microbiology is the assessment of gene function and the linkage between phenotypes and genotypes (Rodrigues et al., 2011; Amaral et al., 2014). However, a phenotype is often controlled by multiple genes and correlated to environmental conditions, making it difficult to link a phenotype (for example, functional trait) to the associated gene(s). High-throughput genome-wide mutagenesis strategies, such as transposon saturation mutagenesis or homologous recombination, allow us to map genes to their functional traits on a genome-wide scale (Garst et al., 2013). Despite the widespread application of these approaches, only non-essential genes can be correlated with phenotypes, and extensive follow-up sequencing and characterization of mixed populations are required to assess genotype–phenotype relationships. Microbial experimental evolution combined with whole-genome sequencing has the potential to bypass these limitations and provide direct evidence of functions for mutations in essential genes, non-essential genes and non-coding regions (Elena and Lenski, 2003; Brockhurst et al., 2011; Dettman et al., 2012; Barrick and Lenski, 2013). In this approach, populations are allowed to evolve under controlled laboratory conditions and the genetic changes are identified by sequence comparisons between the evolved strains or populations and the ancestral strain. In this way, genotypic changes can be assoicated with biological responses to applied constraints.
Salinity (for example, elevated NaCl) is a key environmental factor affecting many organisms, and salt tolerance is a complex trait involving multiple cellular pathways (Warringer et al., 2003). To ameliorate the effects of excessive salinity, physiological responses/changes, such as intracellular accumulation of organic solutes, membrane lipid composition changes and efflux of Na+, have been observed in diverse microorganisms (Roberts, 2005; Krämer, 2010). Histidine kinases have been observed important in receiving and transducing salt stress signals in microorganisms (Marin et al., 2003; Wang et al., 2012). However, key functional genes involved in microbial evolutionary adaptation to salt stress are largely unknown. Recent experimental evolution of E. coli or yeast under salt stress revealed limited numbers of mutations. For instance, in E. coli, mutations have been identified in the proline ABC transporter gene proV (Dragosits et al., 2013; Winkler et al., 2014), and in the osmoprotectant biosynthesis genes otsBA (Stoebel et al., 2009). In yeast, mutations were found in the proton efflux pump gene pma1, the global transcriptional repressor gene cyc8 (Anderson et al., 2010) and the mot2 gene that has an unknown role in salt tolerance (Dhar et al., 2011). Here, we employ experimental evolution coupled with whole-genome whole-population sequencing and site-directed mutagenesis to discover and verify the functional genes conferring salt tolerance in an anaerobic environmental bacterium Desulfovibrio vulgaris.
Desulfovibrio vulgaris is widely distributed in anaerobic, sulfate-rich environments such as gas pipelines, subterranean tank environments, off-shore hydrocarbon production facilities, marine sediments and freshwater sediments that can also have high salt concentrations (Postgate, 1984). The Hildenborough strain (DvH) has been extensively studied as a model organism of sulfate-reducing bacteria due to its potential for bioremediation (Lovley and Phillips, 1994) and biocorrosion (Postgate, 1984; Zhou et al., 2011). The complexity of, and differences between, the cellular responses to short-term salt shock or salt adaptation in DvH have been reported (Mukhopadhyay et al., 2006; He et al., 2010). Taking advantages of the available knowledge, we initiated an experimental evolution of DvH under salt stress to investigate the molecular mechanisms of evolutionary adaptation to salt stress. Analysis of the representative evolved strains demonstrated significant changes in salt tolerance via gene transcription, metabolite abundances (for example, organic solutes) and cellular composition (for example, phospholipid fatty acids) (Zhou et al., 2013), but the genetic basis of salt adaptation remains elusive (that is, evolution).
In this study, we aimed to determine the genetic basis of evolutionary adaptation to salt stress in DvH by addressing the following questions: (i) What mutations were underlying the improved salt tolerance in evolved DvH? (ii) How rapid did the mutations arise under salt stress and how did mutation frequencies change over time? (iii) How quickly did the salt tolerance improvement occur? (iv) What individual mutations were associated with salt tolerance? To answer these questions, we sequenced the representative stress-evolved strain ES9-11 and non-stress-evolved strain EC3-10, monitored the temporal changes of mutation frequencies and salt tolerance in representative populations ES9 and EC3 over 1200 generations evolution, examined mutation selections in six independently salt stress-evolved ES populations and six non-stress-evolved EC populations, and evaluated the functions of individual mutations with site-directed mutants (SDMs). Our results indicated the rapid genetic and phenotypic adaptation in as few as 100 generations, and that a few mutations could explain significant improvement in salt tolerance. The results underscore the observation that biological systems can achieve significant phenotypic adjustments with minor genotypic changes.
Materials and methods
Bacteria strains and growth conditions
Twelve populations were founded from colonal isolates of Desulfovibrio vulgaris Hildenborough (DvH, ATCC 29579). Six populations (EC1 to EC6, non-stress-evolved) were cultured in standard growth condition (defined medium LS4D with 60 mM lactate as electron donor and 50 mM sulfate as electron acceptor) and six populations (ES7 to ES12, stress evolved) were cultured in salt stress condition (LS4D+100 mM NaCl) (Zhou et al., 2013) for 1200 generations. Briefly, the cultures were maintained at 37 °C and serially transferred every 48 h with a 1-to-100 dilution (10 ml culture volume in 18 × 150 mm anaerobic culture tubes). Population samples were archived at 100-generation interval and stored at −80 °C. Single colony isolates ES9-11 and EC3-10 were obtained from plates of population ES9 or EC3, respectively.
Whole-genome and whole-population DNA sequencing
Genomic DNA was isolated with CTAB (hexadecyltrimethylammonium bromide) method and purified with phenol/chloroform (Zhou et al., 1996). Illumina sequencing of genomic DNA from evolved colony isolates ES9-11, EC3-10 and the ancestral isolate An-1 was conducted at the DOE Joint Genome Institute (Walnut Creek, CA, USA) with 1 × 35 cycling format. Illumina sequencing of genomic DNA samples from stress-evolved population ES9, non-stress-evolved population EC3 archived at 100, 300, 800 or 1200 generations, and the ancestral population was conducted at Los Alamos National Laboratory with 1 × 100 cycling format. Alignments to the DvH reference sequence in NCBI (NC_002937.3) and single-nucleotide polymorphism (SNP) calls were performed with maq-0.7.1 (Li et al., 2008). The insertions/deletions (InDel) were identified with an in-house tool to locate regions that did not have a reading start and manually sorted out.
Mutation validation and PCR genotyping
The regions harboring mutations were PCR amplified from genomic DNA templates isolated from the colony isolates (ES9-11, EC3-10, An-1 from the ancestral population), and ancestral population to verify the mutations identified with Illumina sequencing. Similarly, PCR genotyping of individual clones in populations ES9 or EC3 was performed with genomic DNA templates. PCR genotyping of the evolving population samples at different time points (time series) was performed with DNA prepared from glycerol stocks by boiling (2 min) and precipitation with 1/10 volume of sodium acetate (3 M, pH 5.2) and 2 volumes of ethanol. Phusion high-fidelity DNA polymerase (New England Biolabs, Inc., Ipswich, MA, USA; Cat: F530L) was used for PCR with primers listed in Supplementary Table S3. PCR fragments were purified with the QIAquick PCR purification kit (QIAGEN, Germantown, MD, USA; Cat: 28104) and sequenced with ABI 3730 capillary sequencer (Applied Biosystems/Hitachi, Foster City, CA, USA).
Salt tolerance phenotype test
Growth phenotypes of the ancestral DvH, populations ES9 and EC3 evolved for different numbers of generations (100, 200, 300, 800, 1000 or 1200 generations) were examined in non-stress (LS4D) or salt stress (LS4D+250 mM NaCl) conditions with three replicates for each population. Growth rate, yield and lag phase values were obtained independently from the growth curve of each replicate (Zhou et al., 2013). The changes of growth rate, yield and lag phase along the evolution were obtained by dividing the measurements in evolved populations by the measurements in the ancestral population.
Site-directed mutagenesis and growth phenotype test of SDMs
DvH strain JW710 (Keller et al., 2009) with a deletion of the upp gene was used for generation of SDMs. upp encodes the pyrimidine salvage enzyme uracil phosphoribosyl transferase, which allows the recycling of free pyrimidines and the incorporation of base analogs into nucleoside monophosphates. Incorporation of the pyrimidine analog 5-fluorouracil (5-FU) is lethal in D. vulgaris. Deletion of upp makes DvH resistant to 5-FU and re-introduction of upp restores sensitivity, which provides a selection marker for a two-step integration and excision in mutagenesis (Keller et al., 2009). SNPs were introduced into JW710 with a strategy similar to that of E. coli (Warming et al., 2005). Briefly, with first targeting, the ancestral nucleotide was replaced by the kanamycin resistance marker gene and upp gene. With second targeting, the SNP replaced the kanamycin resistance marker gene and upp gene and the mutants were selected as 5-FUR. Details about the site-directed mutagenesis and the gene specific primers are in Supplementary Figure S5 and Supplementary Table S5. Growth phenotypes of the SDMs and JW710 were tested in LS4D or LS4D+250 mM NaCl as described above.
Results
Mutations in evolved DvH strains
To investigate the evolutionary adaptations to salt stress, 12 populations were grown under standard growth conditions (LS4D, non-stress-evolved, populations EC1~EC6) or salt stress conditions (LS4D+100 mM NaCl, stress-evolved, populations ES7~ES12) for 1200 generations (1200 g), respectively (Supplementary Figure S1, the experimental flowchart). Dramatically improved salt tolerance has been observed in stress-evolved strain ES9-11 compared with the non-stress-evolved strain EC3-10 and ancestral DvH (Zhou et al., 2013). As a first step in identifying the genetic bases of salt adaptation, the genomes of ES9-11, EC3-10 and the ancestral DvH were sequenced.
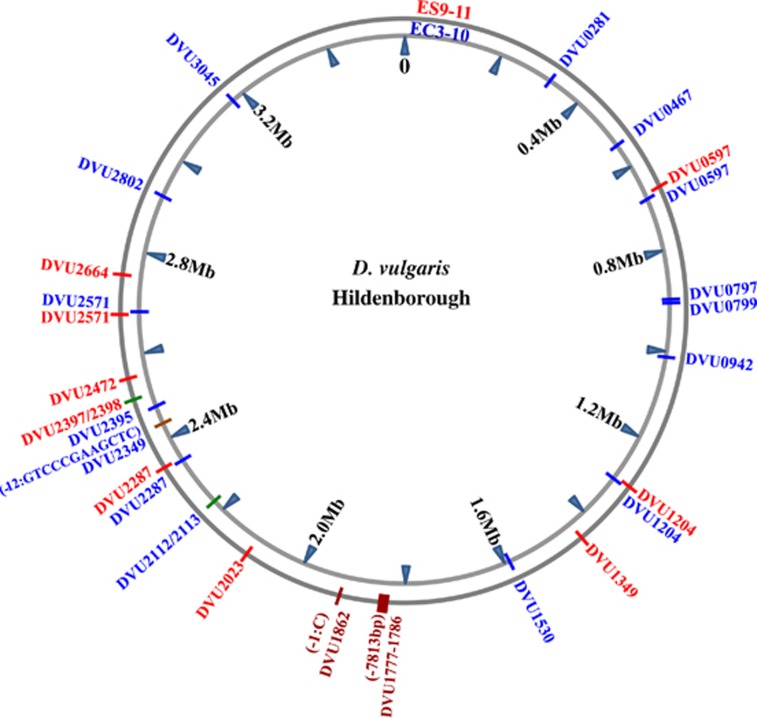
Sequences of these genomes covered 99% of the NCBI Reference DvH Sequence NC_002937.3 with average sequencing depths of 70x for ES9-11, 91x for EC3-10 and 59x for the ancestor. As shown in Figure 1 and Table 1, nine SNPs specific to ES9-11 including seven missense mutations, one silent mutation and one mutation in a non-coding region were identified. Fourteen SNPs including ten missense mutations, three silent mutations and three mutations in a non-coding region were identified in EC3-10. In addition, two deletions in ES9-11 and one deletion in EC3-10 were identified. Besides these specific mutations, 22 mutations were common in the ancestor, ES9-11 and EC3-10 (Supplementary Table S1), and these results indicated the genetic variation between the DvH strain used in this study and the type strain genome originally deposited in the NCBI database. Eleven polymorphic loci in the ancestral DvH but homogenous alleles in ES9-11 and EC3-10 were identified (Supplementary Table S2,Supplementary Figure S3a), suggesting the existence of polymorphisms in the ancestor and that these pre-existing mutations were under selection during evolution. Interestingly, different SNPs in four genes (DVU0597, DVU1204, DVU2287 and DVU2571) were identified in ES9-11 and EC3-10 (gray highlighted in Table 1). Among these, SNPs in DVU2287 were from the pre-existing polymorphisms in ancestral DvH and the others were new mutations.
Figure 1.
Maps of mutations identified in D. vulgaris Hildenborough strains evolved for 1200 generations under salt stress (ES9-11, outer circle) or non-stress condition (EC3-10, inner circle). Red: specific mutations in ES9-11. Blue: specific mutations in EC3-10. Green: SNPs in intergenic regions. Dark brown: deletions.
Table 1. Mutations identified in stress-evolved strain ES9-11 or non-stress-evolved strain EC3-10.
| Strain | Mutation type | Mutation position | Affected gene(s) | Mutation position | Nucleotide change | Amino-acid change |
|---|---|---|---|---|---|---|
| ES9-11 | SNP | Gene | DVU0597a, lytS, regulatory protein, putative regulator of cell autolysis | 666077 | C → T | Thr → Ile |
| SNP | Gene | DVU2571a, feoB, ferrous iron transport protein B | 2685757 | G → A | Ala → Val | |
| SNP | Gene | DVU1204a, fabF, 3-oxoacyl-(acyl-carrier-protein) synthase II | 1296562 | C → T | Gly → Ser | |
| SNP | Gene | DVU2287a, cooK, hydrogenase, CooK subunit, selenocysteine-containing, putative | 2381877 | G → C | stop → Ser | |
| SNP | Gene | DVU2472, conserved hypothetical protein | 2581002 | G → T | Arg → Leu | |
| SNP | Gene | DVU2664, pstB-2, phosphate ABC transporter, ATP-binding protein | 2775672 | C → G | Ala → Pro | |
| SNP | Gene | DVU1349, selGGPS, geranylgeranyl diphosphate synthase | 1426830 | T → G | Val → Gly | |
| SNP | Gene | DVU2023, hypothetical protein | 2104739 | G → C | Val = Val | |
| SNP | Intergenic | DVU2397, hypothetical protein | 2502193 | G → C | non-coding (-199) | |
| Deletion | Gene | DVU1777–DVU1786 | 1842155 to 1849967 | −7813 nt | −10 genes | |
| Deletion | Gene | DVU1862, GGDEF domain protein | 1930359 | −1: C | 27 amino acids changed | |
| EC3-10 | SNP | Gene | DVU0597b, lytS, regulatory protein, putative regulator of cell autolysis | 666481 | C → T | Pro → Ser |
| SNP | Gene | DVU2571b, feoB, ferrous iron transport protein B | 2685839 | A → G | Phe → Leu | |
| SNP | Gene | DVU1204b, fabF, 3-oxoacyl-(acyl-carrier-protein) synthase II | 1296677 | C → T | Met → Ile | |
| SNP | Gene | DVU2287b, cooK, hydrogenase, CooK subunit, selenocysteine-containing, putative | 2381876 | T → G | stop → Gly | |
| SNP | Gene | DVU0942, fur, ferric uptake regulator | 1034709 | G → A | Glu → Lys | |
| SNP | Gene | DVU2395, sensor histidine kinase | 2499343 | G → A | Gln → stop | |
| SNP | Gene | DVU0797, conserved hypothetical protein | 883424 | C → T | Glu → Lys | |
| SNP | Gene | DVU0799, conserved hypothetical protein | 885112 | G → A | Ser → Phe | |
| SNP | Gene | DVU2802, transcriptional regulator, GntR family | 2905203 | G → A | Ala → Thr | |
| SNP | Gene | DVU3045, fexB, sensory box histidine kinase/response regulator | 3169435 | G → C | Gly → Arg | |
| SNP | Gene | DVU1530, metallo-beta-lactamase family protein | 1599469 | C → T | Asp = Asp | |
| SNP | Gene | DVU0281, exopolysaccharide biosynthesis protein, putative | 326403 | G → A | Asp = Asp | |
| SNP | Gene | DVU0467, trpD, anthranilate phosphoribosyltransferase | 535249 | G→C | Leu = Leu | |
| SNP | Intergeneric | DVU2112, hypothetical protein | 2207959 | G→C | non-coding (+199) | |
| Deletion | Gene | DVU2349, carbohydrate phosphorylase family protein | 2442657 | −12: GTCCCGAAGCTC | −4 amino acids |
Abbreviation: SNP, single-nucleotide polymorphism.
Gray highlight: genes with different SNPs in ES9-11 or EC3-10.
To determine whether these mutations were specific and fixed within the populations and responsible for salt tolerance, PCR genotyping and growth phenotype tests of randomly selected clones from the 1200 g populations ES9 (ES9-1 to ES9-15) or EC3 (EC3-1 to EC3-15) were conducted. As shown in Supplementary Table S4, the mutations were fixed or nearly fixed (allele frequency>0.93) and specific to the respective populations. ES9-11 and EC3-10 had the best salt tolerance (for example, shortest lag phase and highest growth rate) within the populations (Supplementary Figure S2). The clone ES9-12 had the longest lag phase compared with other ES9 strains. Coincidently, SNPs DVU0597a (C707T) and DVU2472 (G974T) were not observed in ES9-12 (Supplementary Table S4), suggesting the possible correlations between these two mutations and salt tolerance.
Dynamics of mutation frequencies over evolution
It is expected that mutations selected only under salt stress conditions are the most likely targets for salt tolerance. To examine how rapid the pre-existing mutations were selected and new mutations arose under salt stress and their frequency changes over time, allele frequencies over the course of 1200 g were monitored in stress-evolved populations ES9 with non-stress-evolved EC3 as a control. An average sequence coverage of 62x was achieved with Illumina sequencing. A total of 13 mutations with frequencies between 0.2 and 0.7 were identified in the ancestral population (Figure 2), and 11 have been observed as polymorphic loci in the genome of the ancestral clone (Supplementary Table S2).
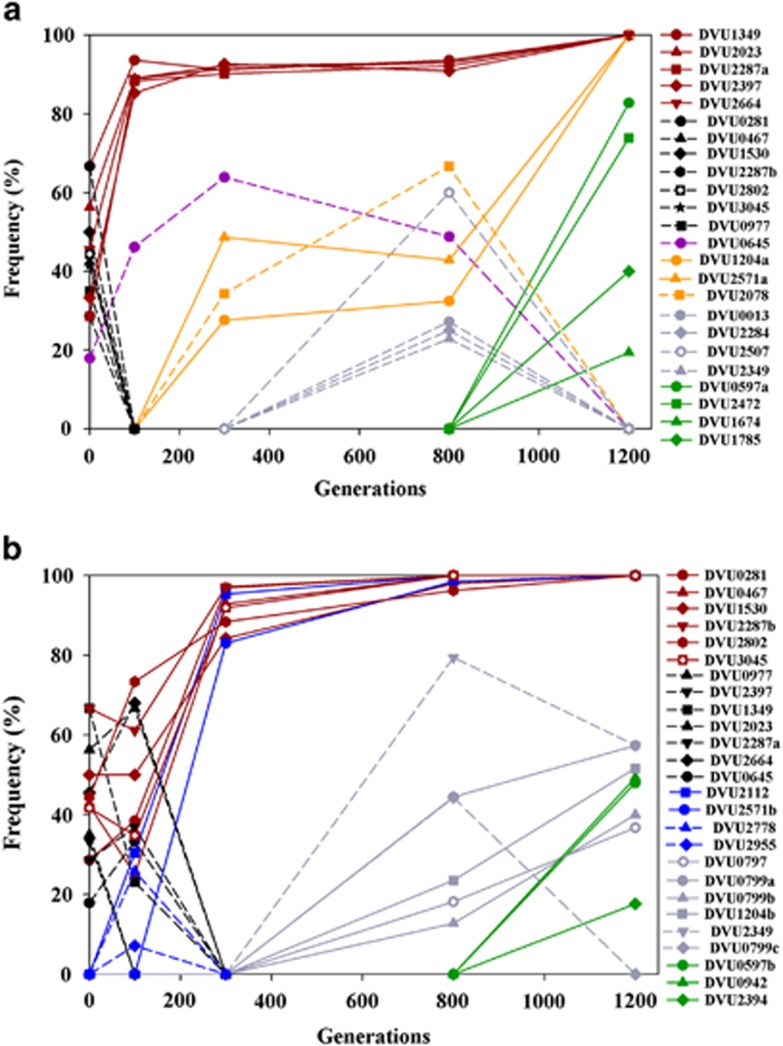
Figure 2.
Dynamics of mutation (SNPs) frequencies in stress-evolved population ES9 (a) or non-stress-evolved population EC3 (b) during 1200 generations of evolution. Solid line: fixed mutations or new mutations with increasing frequencies; dashed line, extinct mutations or mutations with decreasing frequencies. Mutational cohorts were marked as different colors: dark red, selected pre-existing mutations at 100 generations; black, extinct pre-existing mutations at 100–300 generations; dark pink, extinct pre-existing mutations at later generations; orange, new mutations appeared before 300 generations in ES9; blue, new mutations appeared before 300 generations in EC3; dark grey, new mutations appeared after 300 generations; dark green, new mutations appeared after 800 generations.
The initial sorting of pre-existing polymorphisms was remarkably different in ES9 compared with EC3. It is likely that there were multiple dominant clones in the ancestral population according to the parallel changes of allele frequencies over time (Lang et al., 2013). Each population selected different dominant ancestral clone(s) that contained several mutations within the first few hundred generations. Under salt stress-evolving conditions in ES9, one or two clones bearing five mutations in DVU1349, DVU2023, DVU2287a, DVU2397 and DVU2664 were selected at 100 g and another clone bearing six mutations in DVU0281, DVU0467, DVU1530, DVU2287b, DVU2802 and DVU3045 went extinct (Figure 2a). In contrast, under the non-stress-evolving condition in EC3, the clone bearing the latter six mutations was selected, and clone(s) bearing the former five mutations went extinct within 300 g (Figure 2b). Overall, the initial selective sweep of the pre-existing polymorphisms was quicker in ES9 than in EC3.
After the rapid selective sweep of the pre-existing mutations, different new mutations arose, selected or went extinct in ES9 or EC3. In stress-evolved ES9, new mutations DVU1204a and DVU2571a became common by 300 g, rapidly sweeping through the population after 800 g, and were fixed at 1200 g, suggesting one or both of them were highly beneficial (Figure 2a). A clone bearing a pre-existing mutation in DVU0645 became common within 300 g. Five new mutations arose presumably in this clone around 300 g. However, this clone was outcompeted after 800 g by other clone(s) carrying four new mutations in DVU0597a, DVU2472, DVU1674 and DVU1785. Different from what was observed in ES9, in non-stress-evolved EC3, new mutations in DVU2112 and DVU2571b arose and were nearly fixed within 300 g (Figure 2b). Six new mutations appeared after 300 g and five of them remained common in the population with frequencies lower than 0.6 till 1200 g except one (SNP DVU0799 went extinct at 1200 g). After 800 g, three new mutations arose and the frequencies increased between 0.2 and 0.5. During the 1200 g of evolution, numbers of new mutations were 11 in ES9 and 13 in EC3. Among these, 5 in ES9 and 3 in EC3 went extinct, suggesting that new genetic variations during evolution were less in ES9 than in EC3.
At 1200 g, the mutation frequencies were generally higher in ES9 than in EC3. Mutation in DVU2472 was unique in ES9, suggesting that it might be beneficial for salt tolerance. Although different mutations in four genes (DVU0597, DVU2571, DVU1204 and DVU2287) were found in ES9 or EC3, the allele frequency dynamics suggested the distinctiveness of these mutations in these populations. Different DVU2287 alleles probably existed in different ancestral clones that were selected within the first 100 g in ES9 or EC3. DVU2571a was fixed at 1200 g in ES9, while DVU2571b was fixed at 800 g in EC3, although both mutations arose within 300 g. Alleles DVU1204a and DVU0597a were fixed or nearly fixed in ES9. In contrast, frequencies of DVU1204b and DVU0597b were about 0.5 in EC3. Due to the relatively high Na+ concentration (~210 mM) in LS4D (the evolution condition for EC3), and improved salt tolerance in EC3-10 and the best growth performances of EC3 among EC populations (Zhou et al., 2013), mutations in DVU2287, DVU2571, DVU1204 and DVU0597 might also contribute to salt tolerance.
Allele frequencies in independently evolved ES or EC populations
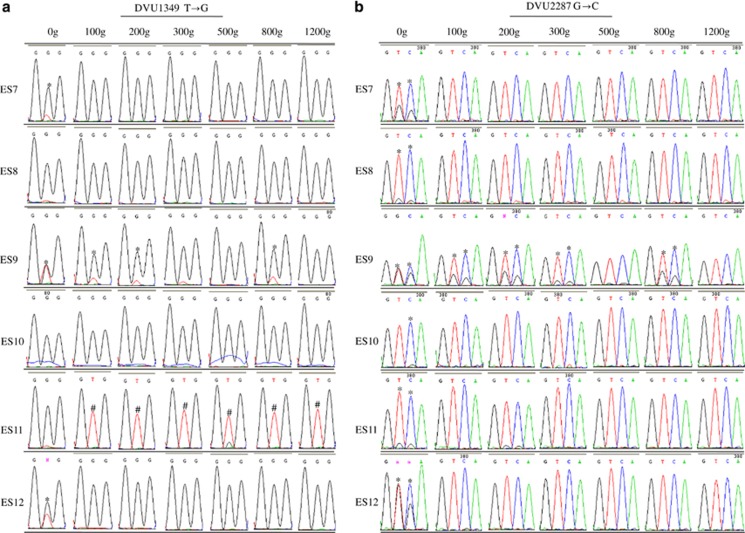
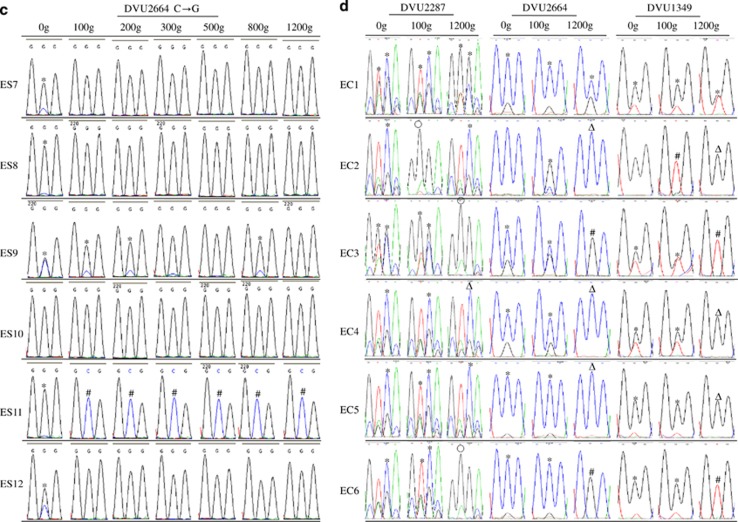
To determine whether the selection of pre-existing mutations and arising of new mutations were common in salt stress evolution condition, PCR genotyping of six independently evolved ES or EC populations was conducted. As shown in Figure 3, the numbers of the founding ancestral clones (0 g) carrying polymorphic loci of DVU1349, DVU2664 or DVU2287 were 8, 10 or 12, respectively. At 100 g, ES9 was the only ES population that had polymorphisms and all other ES populations (except ES11) had homogenous mutations at these loci. At 1200 g, all ES populations had the same mutations as ES9 except ES11 did not have mutations in DVU1349 or DVU2664. In contrast, five or six EC populations had these pre-existing polymorphic loci at 100 g. At 1200 g, either polymorphic loci, ES-type mutations as seen in ES9, or EC-type mutations as seen in EC3 were found in EC populations. These data suggested that the rapid selective sweep of pre-existing polymorphisms was common in ES populations, yet appeared to be largely random in EC populations.
Figure 3.
Selection of pre-existing polymorphisms in six salt stress-evolved ES populations (a–c) or six non-stress-evolved EC populations (d). Chromatograms of the sequences of the nucleotide(s) with mutations and the adjacent nucleotides are shown. *: polymorphisms; Δ: ES-type mutation as seen in ES9; ○: EC-type mutation as seen in EC3; #: no mutation.
Population genotyping data of the four new mutations in ES9 (Supplementary Figure S3c) confirmed the allele frequency dynamics obtained from sequencing data (Figure 2). EC3 was the only EC population carrying new mutations in DVU1204b, DVU2571b and DVU0597b (Supplementary Figure S3d). In addition, PCR genotyping of randomly selected clones from six ES populations (15 clones per population, 1200 g) showed that new mutations DVU0597a, DVU1204a and DVU2472, which arose de novo in ES9, were not present in other ES populations. However, an alternative DVU2571 allele with a deletion of 15 bp at position 1641 was identified in population ES8 (Supplementary Figure S3b). We previously reported that ES9 had the highest salt tolerance (Zhou et al., 2013); therefore, both pre-existing mutations and new mutations might be targets of salt adaptation.
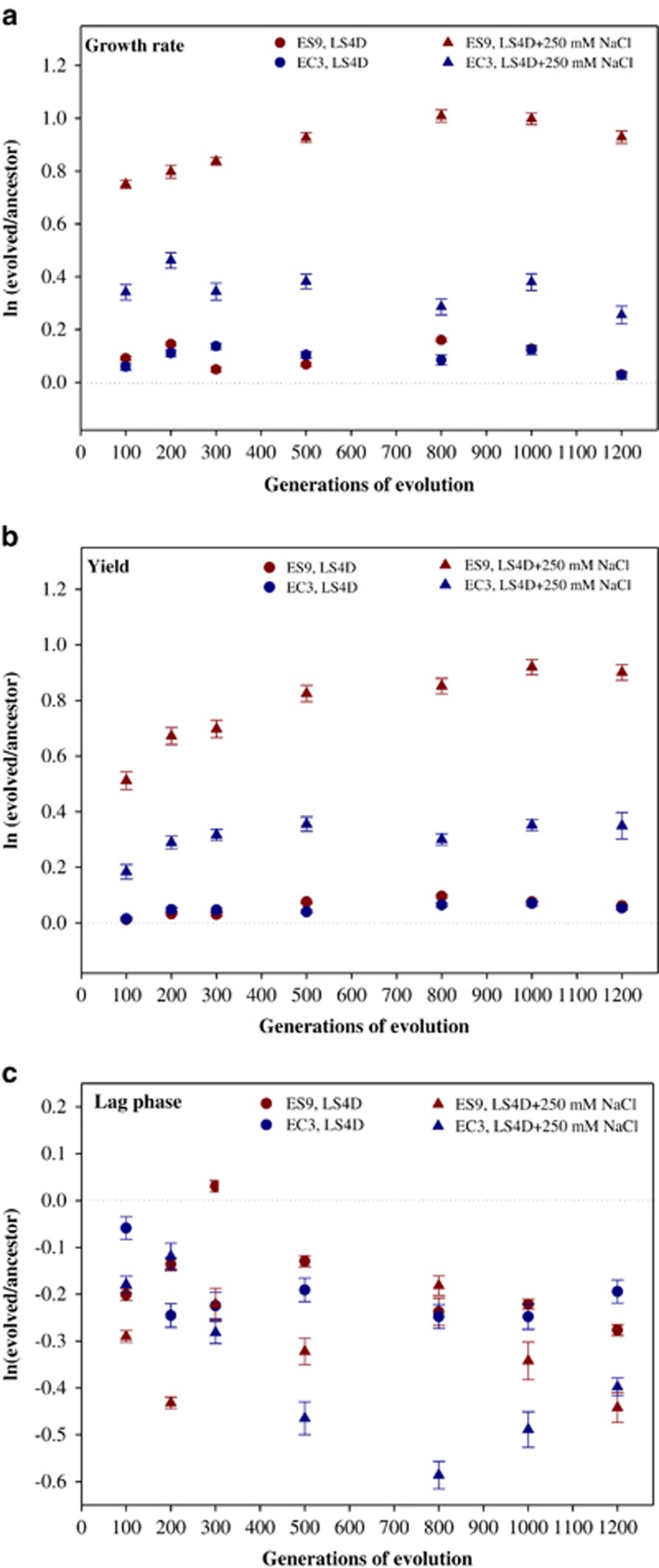
Rapid phenotypic adaptation to salt stress
To uncover how rapid the salt tolerance was improved, growth phenotypes of population ES9 evolved for different generations were tested with EC3 and the ancestral population as controls. Consistent with the rapid selective sweep of pre-existing mutations within 100 g, significant increases of salt tolerance were observed in ES9 at 100 g with increased growth rates (2.0-fold), increased biomass yields (1.8-fold), and a shortened lag-phase (0.7-fold) under salt stress conditions (LS4D+250 mM NaCl) (Figure 4). As the number of generations increased, growth rate and biomass yield gradually increased in ES9. In contrast, significantly less improvement of growth rate or biomass yield was observed in EC3 (Figures 4a and b). Under the non-stress condition (LS4D), there were no apparent differences between ES9 and EC3 as both showed improved growth rate or biomass yield to some extent relative to the ancestor. Decreased lag phases were observed in both ES9 and EC3 over the course of 1200 g but with large fluctuations (Figure 4c).
Figure 4.
Rapid adaptation of D. vulgaris Hildenborough to salt stress measured by (a) growth rate, (b) biomass yield and (c) lag phase. Error bars indicate standard errors.
The superior salt tolerance of ES9 indicated that beneficial mutations for salt tolerance were selected in ES9 and the improved growth of ES9 and EC3 relative to the ancestor on LS4D implied the presence of mutations that provided general growth advantages in both populations. Some mutations in EC3 might have pleiotropic effects on salt tolerance, or mutations DVU2287b, DVU2571b, DVU1204b and DVU0597b might provide EC3 improved salt tolerance.
Contribution of individual mutations to salt tolerance
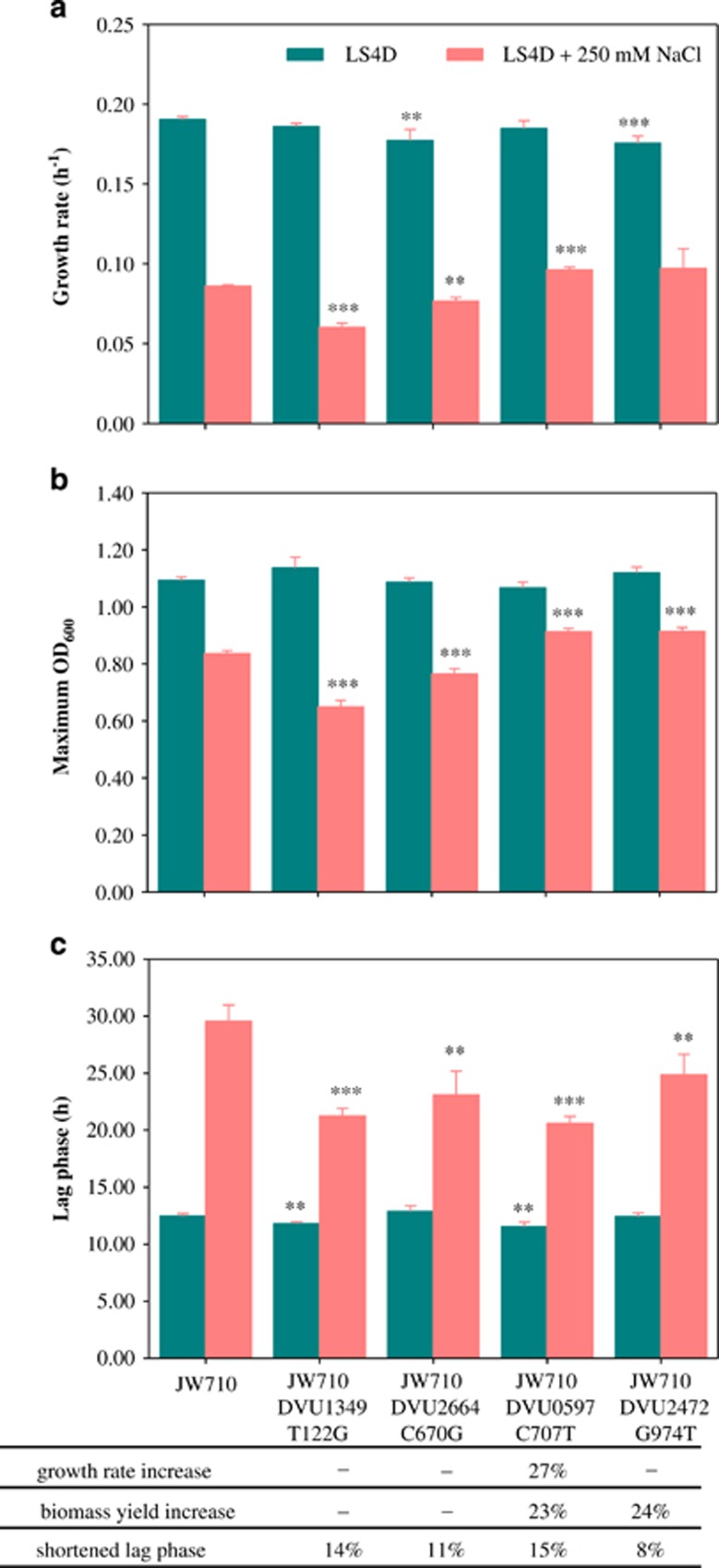
To determine the contributions of individual mutations to improved salt tolerance, individual SNPs were introduced into DvH strain JW710 (Δupp) with a two-step integration and excision strategy (Supplementary Figure S4) and growth phenotypes of the SDMs were tested. Three early-fixed pre-existing mutations in DVU1349, DVU2664 and DVU2287 (except the synonymous mutation in DVU2023 and mutation in the upstream of DVU2397) and four new mutations (DVU1204a, DVU2571a, DVU0597a and DVU2472) identified in ES9 were tested. No SDMs for DVU2287a, DVU1204a or DVU2571a could be obtained. The first step integration in DVU1204 or DVU2571 was not successful, suggesting these two genes might be essential. The second step integration in DVU2287 failed although the first integration succeeded. When grown in LS4D, no significant differences were observed in SDMs compared with JW710. In contrast, improvements of one or more growth parameters were found for each SDM when grown in high salinity medium (LS4D+250 mM NaCl) (Figure 5, Supplementary Figure S5).
Figure 5.
Effects of SNP mutations on (a) growth rate, (b), biomass yield and (c) lag phase under salt stress condition (LS4D+250 mM NaCl). Growth parameters of each SDM were compared with D. vulgaris strain JW710 (t test). ***P<0.01. **P<0.05. Error bars indicate standard deviations. Contribution of individual SDMs to salt tolerance was shown as percentages (changes in SDM vs JW710/changes in ES9-11 vs the ancestor).
Under the high salinity condition, the SDMs containing individual SNPs contributed to shorter lag times. However, pre-existing mutations in a geranylgeranyl diphosphate synthase gene DVU1349 (SelGGPS), and a phosphate ABC transporter gene DVU2664 (pstB-2), did not contribute to growth rate or yield increases. New mutations in DVU0597a (lytS, regulatory protein) and DVU2472 (conserved hypothetical protein) contributed to increased yields. DVU0597a contributed to increased growth rate as well. Compared with the salt tolerance improvement in ES9-11 (Zhou et al., 2013), about 8–15% of the shortened lag phase was conferred by each SNP (Figure 5). DVU0597a conferred about 27% of the growth rate increase and 23% of the yield increase. DVU2472 mutation conferred about 24% of the yield increase. The results demonstrated that these SNPs were beneficial for salt tolerance and SNPs in DVU0597a and DVU2472 contributed more to salt tolerance. The function of SNPs in DVU1204, DVU2287 and DVU2571 needs to be tested with different site-directed mutagenesis strategies in the future.
Discussion
Despite our increased knowledge of physiological acclimation, the genetic basis of microbial evolutionary adaptation to physiological constraint (for example, salinity) is largely unknown. In this study, several alleles that confer salt tolerance in the anaerobic sulfate-reducing bacterium D. vulgaris Hildenborough were identified with whole-genome sequencing of experimentally evolved DvH and site-directed mutagenesis. Rapid genetic adaptation involved sorting of pre-existing genetic variations within 100 g. As the number of generations increased, newly arising mutations were selected or went extinct. Consistently, salt tolerance rapidly increased by 100 g followed by a more gradual increase. The results indicated that a few mutations made substantial contributions to improved salt tolerance, a trait that is controlled by many genes and cellular pathways.
Whether evolutionary changes occur through many mutations with small fitness effects or few mutations with larger effects is one of the oldest questions in biology (Dettman et al., 2012). Association between a few mutations/genes and dramatic phenotypic changes or fitness improvement have been reported in prokaryotes, eukaryotes and viruses (Taubenberger et al., 2005; Wang et al., 2005; Zeyl 2005; Herring et al., 2006; Schoustra et al., 2009). Results from this study indicated the linkage between significantly increased salt tolerance and a few mutations. Site-directed mutagenesis is one of the ‘gold' standards to verify whether a particular mutation is beneficial; however, it remains difficult for microorganisms other than E. coli (Herring et al., 2006). Here, a site-directed mutagenesis approach was developed (Supplementary Figure S4). Growth phenotype tests of the SDMs demonstrated that these mutations were beneficial for salt adaptation. Single nucleotide changes in DVU0597 or DVU2472 accounted for up to 27% of observed increases in growth rate or yield in salt stress condition, with little to no effect on these features in the absence of salt stress (Figure 5). Such a large contribution of a single nucleotide change to phenotypic changes was also observed in other evolved microorganisms (Herring et al., 2006; Summers et al., 2012). However, early selected pre-existing mutations in DVU1349 or DVU2664 did not contribute to salt tolerance as much as later arising new mutations in DVU0597 and DVU2472 that was different from what was observed in other adaptive evolution experiments (Barrick and Lenski, 2013). Through the process of selection, it seems intuitive to select against pre-existing mutations that may not be the most beneficial mutations.
By what mechanism(s) did the SNPs in DVU0597 or DVU2472 increase salt tolerance? According to the annotation, DVU0597 encodes a regulatory protein LytS, a transmembrane protein with an N-terminal sensing domain and an intracellular C-terminal histidine kinase domain. The predicted protein structure suggests that LytS may function in signal sensing and transduction. Roles of lytS/lytR two-component systems in carbon starvation in DvH (Rajeev et al., 2011) or controlling autolysis rate in Staphylococcus aureus (Brunskill and Bayles, 1996) have been reported. The cessation of stationary phase and entrance into death phase of an SDM harboring the SNP in lytS was much shorter than control strain JW710 and other SDMs when grown in either the non-stress or high salinity medium (Supplementary Figure S5). These results are consistent with an effect on altered control of carbon storage and cell lysis. However, how LytS functions in increasing salt tolerance remains unknown. Functional significance of the hypothetical gene DVU2472 in salt tolerance was suggested by growth phenotypes of the SDM, but further study is needed to determine the underlying mechanism. Deletion mutant of DVU0597 had increased salt tolerance while a deletion mutant of DVU2472 had decreased salt tolerance (Supplementary Figure S6), suggesting the SNP in DVU0597a was a possible loss-of-function mutation while the SNP in DVU2472 was a possible gain-of-function mutation.
In addition to the beneficial mutations demonstrated by SDMs, mutations likely contributing via general adaptive benefits were identified. Mutations that conferred general adaptive benefits were also reported in other microbial experimental evolution studies (Dettman et al., 2012). Different alleles of DVU2571 or DVU2287 were identified in stress-evolved ES8 and ES9 and non-stress-evolved EC3 (Figure 1, Table 1, Supplementary Figure S3). DVU2571 encodes a ferrous iron transport protein FeoB that functions as an Fe2+ permease (Marlovits et al., 2002; Cartron et al., 2006). Mutations in iron acquisition-related genes were identified in salt-adapted or butanol-adapted E. coli (Dragosits et al., 2013) as well as being up-expressed genes when exposed to various stresses, in statoinary-phase cells or biofilm populations in DvH (Clark et al., 2006, 2012; Zhou et al., 2011). Although the role of iron transportation in stress tolerance is not clear, mutations in iron transport-related genes might be common in evolutionary adaptation to stress conditions. DVU2287 encodes a selenocysteine-containing hydrogenase CooK, a component of the membrane-localized Coo complex. The Coo complex is involved in energy metabolism through cycling hydrogen equivalents generated by oxidation of organic compounds to the periplasm and providing electrons for sulfate reduction (Voordouw, 2002). SNPs in DVU2287 (G824C in ES9 and T823G in EC3) changed the selenocysteine codon to serine in ES9 or glycine in EC3, which might lead to the alteration of the selenocysteine-containing CooK to become seleno independent. Therefore, the selection of these mutations was possibly beneficial for a general growth advantage.
Mutations in DVU1204 might be beneficial for salt tolerance. DVU1204 encodes 3-oxoacyl-(acyl-carrier-protein) synthase II FabF (also named as beta-ketoacyl-ACP synthase II), which has been shown to function in fatty acid biosynthesis in Synechocystis (Moche et al., 2001) as well as the synthesis of both unsaturated fatty acids and saturated fatty acids in Clostridium (Zhu et al., 2009). The DvH genome lacks unsaturated fatty acids biosynthesis genes fabA and fabB (Campbell and Cronan, 2001); but increased percentages of unsaturated fatty acids have been observed in evolved strains ES9-11 and EC3-10 (Zhou et al., 2013). DVU1204 might contribute to increased salt tolerance via functioning in biosynthesis of unsaturated fatty acids to alter overall membrane characteristics.
The sequencing results demonstrated that the ancestral DvH was a population that contained multiple clones (or ecotypes). Compared with the NCBI database, over 20 mutations and polymorphic loci were found in the starting materials, although only a few culture cycles were involved before isolation of the founder clones for experimental evolution. These results suggested possible rapid ecological diversification as seen in other microbial laboratory evolution experiments (Treves et al., 1998; Rainey et al., 2000; Koeppel et al., 2013). Interestingly, clone(s) carrying the pre-existing genetic variations in the ancestral population were rapidly sorted within 100 g in six stress-evolved populations. In contrast, the selection of pre-existing mutations was random in non-stress-evolved populations. After the initial rapid sorting of pre-existing mutations, in the following several hundreds of evolution, multiple new mutations arose and remained common without sweeping through the population, indicating significant clonal interference. Hundreds of generations of clonal interference have been also observed in other evolving microbial populations (Barrick and Lenski, 2009; Lang et al., 2013). At 1200 g, more fixed or nearly fixed mutations were observed in stress-evolved ES9 than in non-stress-evolved EC3. Consistently, ES9 had dramatic salt tolerance improvement while only a certain level of salt tolerance improvement was observed in EC3 (Figure 4), and these results suggested stronger selection pressure under salt stress conditions. The results provided evidence for the chance effects and deterministic selection in evolutionary outcomes.
In summary, mutations beneficial for salt tolerance were identified with experimental evolution and whole-genome whole-population sequencing. Mutations in a few genes such as DVU0597 (lytS, a regulatory protein potentially involved in signal sensing) and DVU2472 (a hypothetical gene) contributed substantially to the dramatic salt tolerance increase. Rapid selective sweep of pre-existing mutations was followed by slow fixation of new mutations. Mutations in both essential genes and non-essential genes, which led to loss- or gain-of-function of the genes, beneficial for salt tolerance or general growth advantages under high-salt conditions were observed. Future studies include covering the mutation spectra by sequencing all of the independently stress-evolved populations, to verify functions of mutations in DVU1204, DVU2571 and DVU2287 with alternative mutagenesis approaches, to test the interactions among mutations, to investigate the molecular mechanisms of how individual mutations contribute to salt tolerance, and to explore the possible ecological distinctions within the starting materials.
Acknowledgments
This material by ENIGMA- Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory is based upon work supported by the US Department of Energy, Office of Science, Office of Biological & Environmental Research under contract number DE-AC02-05CH11231. We thank Adam Deutschbauer for his comments on the manuscript, Joel Martin and Anna Lipzen for assistance with sequence data analysis, Kimberly L Keller for providing cloning vectors.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amaral GRS, Dias GM, Wellington-Oguri M, Chimetto L, Campeão ME, Thompson FL et al. (2014). Genotype to phenotype: identification of diagnostic vibrio phenotypes using whole genome sequences. Int J Syst Evol Microbiol 64: 357–365. [DOI] [PubMed] [Google Scholar]
- Anderson JB, Funt J, Thompson DA, Prabhu S, Socha A, Sirjusingh C et al. (2010). Determinants of divergent adaptation and dobzhansky-muller interaction in experimental yeast populations. Curr Biol 20: 1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J, Lenski R. (2009). Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol 74: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. (2013). Genome dynamics during experimental evolution. Nat Rev Genet 14: 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Colegrave N, Rozen DE. (2011). Next-generation sequencing as a tool to study microbial evolution. Mol Ecol 20: 972–980. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Bayles KW. (1996). Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol 178: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JW, Cronan JE. (2001). Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Ann Rev Microbiol 55: 305–332. [DOI] [PubMed] [Google Scholar]
- Cartron M, Maddocks S, Gillingham P, Craven CJ, Andrews S. (2006). Feo – transport of ferrous iron into bacteria. Biometals 19: 143–157. [DOI] [PubMed] [Google Scholar]
- Clark M, He Q, He Z, Huang K, Alm E, Wan X et al. (2006). Temporal transcriptomic analysis as Desulfovibrio vulgaris Hildenborough transitions into stationary phase during electron donor depletion. Appl Environ Microbiol 72: 5578–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M, He Z, Redding A, Joachimiak MP, Keasling JD, Zhou JZ et al. (2012). Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: Carbon and energy flow contribute to the distinct biofilm growth state. BMC Genomics 13: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, Rodrigue N, Melnyk AH, Wong A, Bailey SF, Kassen R. (2012). Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Mol Ecol 21: 2058–2077. [DOI] [PubMed] [Google Scholar]
- Dhar R, SÄGesser R, Weikert C, Yuan J, Wagner A. (2011). Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J Evol Biol 24: 1135–1153. [DOI] [PubMed] [Google Scholar]
- Dragosits M, Mozhayskiy V, Quinones-Soto S, Park J, Tagkopoulos I. (2013). Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol Syst Biol 9: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. (2003). Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4: 457–469. [DOI] [PubMed] [Google Scholar]
- Garst A, Lynch M, Evans R, Gill R. (2013). Strategies for the multiplex mapping of genes to traits. Microb Cell Fact 12: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhou A, Baidoo E, He Q, Joachimiak MP, Benke P et al. (2010). Global transcriptional, physiological, and metabolite analyses of the responses of Desulfovibrio vulgaris Hildenborough to salt adaptation. Appl Environ Microbiol 76: 1574–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring C, Raghunathan A, Honisch C, Patel T, Applebee M, Joyce A et al. (2006). Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet 38: 1406–1412. [DOI] [PubMed] [Google Scholar]
- Keller KL, Bender KS, Wall JD. (2009). Development of a markerless genetic exchange system for Desulfovibrio vulgaris Hildenborough and its use in generating a strain with increased transformation efficiency. Ann Rev Microbiol 75: 7682–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel AF, Wertheim JO, Barone L, Gentile N, Krizanc D, Cohan FM. (2013). Speedy speciation in a bacterial microcosm: new species can arise as frequently as adaptations within a species. ISME J 7: 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer R. (2010). Bacterial stimulus perception and signal transduction: response to osmotic stress. Chem Record 10: 217–229. [DOI] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D et al. (2013). Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500: 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Phillips EJP. (1994). Reduction of chromate by Desulfovibrio vulgaris and its c3 cytochrome. Appl Environ Microbiol 60: 726–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin K, Suzuki I, Yamaguchi K, Ribbeck K, Yamamoto H, Kanesaki Y et al. (2003). Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA 100: 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. (2002). The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc Natl Acad Sci USA 99: 16243–16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moche M, Dehesh K, Edwards P, Lindqvist Y. (2001). The crystal structure of beta-ketoacyl-acyl carrier protein synthase II from Synechocystis sp. at 1.54A resolution and its relationship to other condensing enzymes. J Mol Biol 305: 491–503. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, He Z, Alm E, Arkin A, Baidoo E, Borglin S et al. (2006). Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J Bacteriol 188: 4068–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postgate JR. (1984) The Sulfate-Reducing Bacteria. Cambridge University Press: Cambridge. [Google Scholar]
- Rainey PB, Buckling A, Kassen R, Travisano M. (2000). The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol Evol 15: 243–247. [DOI] [PubMed] [Google Scholar]
- Rajeev L, Luning E, Dehal P, Price M, Arkin A, Mukhopadhyay A. (2011). Systematic mapping of two component response regulators to gene targets in a model sulfate reducing bacterium. Genome Biol 12: R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JLM, Serres MH, Tiedje JM. (2011). Large-scale comparative phenotypic and genomic analyses reveal ecological preferences of Shewanella species and identify metabolic pathways conserved at the genus Level. Appl Environl Microbiol 77: 5352–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoustra SE, Bataillon T, Gifford DR, Kassen R. (2009). The properties of adaptive walks in evolving populations of fungus. PLoS Biol 7: e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebel DM, Hokamp K, Last MS, Dorman CJ. (2009). Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet 5: e1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers ZM, Ueki T, Ismail W, Haveman SA, Lovley DR. (2012). Laboratory evolution of Geobacter sulfurreducens for enhanced growth on lactate via a single-base-pair substitution in a transcriptional regulator. ISME J 6: 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. (2005). Characterization of the 1918 influenza virus polymerase genes. Nature 437: 889–893. [DOI] [PubMed] [Google Scholar]
- Treves DS, Manning S, Adams J. (1998). Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol 15: 789–797. [DOI] [PubMed] [Google Scholar]
- Voordouw G. (2002). Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J Bacteriol 184: 5903–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M et al. (2005). The origin of the naked grains of maize. Nature 436: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. (2012). The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J 31: 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J, Ericson E, Fernandez L, Nerman O, Blomberg A. (2003). High-resolution yeast phenomics resolves different physiological features in the saline response. Proc Natl Acad Sci USA 100: 15724–15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler JD, Garcia C, Olson M, Callaway E, Kao KC. (2014). Evolved osmotolerant Escherichia coli mutants frequently exhibit defective N-Acetylglucosamine catabolism and point mutations in cell shape-regulating protein MreB. Appl Environl Microbiol 80: 3729–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl C. (2005). The number of mutations selected during adaptation in a laboratory population of Saccharomyces cerevisiae. Genetics 169: 1825–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Baidoo E, He Z, Mukhopadhyay A, Baumohl JK, Benke P et al. (2013). Characterization of NaCl tolerance in Desulfovibrio vulgaris Hildenborough through experimental evolution. ISME J 7: 1790–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. (1996). DNA recovery from soils of diverse composition. Appl Environ Microbiol 62: 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou A et al. (2011). How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat Rev Microbiol 9: 452–466. [DOI] [PubMed] [Google Scholar]
- Zhu L, Cheng J, Luo B, Feng S, Lin J, Wang S et al. (2009). Functions of the Clostridium acetobutylicium FabF and FabZ proteins in unsaturated fatty acid biosynthesis. BMC Microbiol 9: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.