Abstract
Viral infections dynamically alter the composition and metabolic potential of marine microbial communities and the evolutionary trajectories of host populations with resulting feedback on biogeochemical cycles. It is quite possible that all microbial populations in the ocean are impacted by viral infections. Our knowledge of virus–host relationships, however, has been limited to a minute fraction of cultivated host groups. Here, we utilized single-cell sequencing to obtain genomic blueprints of viruses inside or attached to individual bacterial and archaeal cells captured in their native environment, circumventing the need for host and virus cultivation. A combination of comparative genomics, metagenomic fragment recruitment, sequence anomalies and irregularities in sequence coverage depth and genome recovery were utilized to detect viruses and to decipher modes of virus–host interactions. Members of all three tailed phage families were identified in 20 out of 58 phylogenetically and geographically diverse single amplified genomes (SAGs) of marine bacteria and archaea. At least four phage–host interactions had the characteristics of late lytic infections, all of which were found in metabolically active cells. One virus had genetic potential for lysogeny. Our findings include first known viruses of Thaumarchaeota, Marinimicrobia, Verrucomicrobia and Gammaproteobacteria clusters SAR86 and SAR92. Viruses were also found in SAGs of Alphaproteobacteria and Bacteroidetes. A high fragment recruitment of viral metagenomic reads confirmed that most of the SAG-associated viruses are abundant in the ocean. Our study demonstrates that single-cell genomics, in conjunction with sequence-based computational tools, enable in situ, cultivation-independent insights into host–virus interactions in complex microbial communities.
Introduction
Viruses are the most abundant biological entities on Earth, surpassing the number of their potential host cells by at least one order of magnitude (Suttle, 2005). In the ocean, viral infections kill ~10–20% of planktonic biomass each day (Suttle, 2007; Evans and Brussaard, 2012). These infections are believed to have a major impact on microbial community composition, evolution and global geochemical cycles (Jover et al., 2014). Most of the current knowledge of viral–host interactions is based on laboratory experimentation with pure cultures. Although cultivation has yielded phages from some groups of marine bacterioplankton, including SAR11 (Zhao et al., 2013), SAR116 (Kang et al., 2013), cyanobacteria (Sullivan et al., 2003) and Bacteroidetes (Holmfeldt et al., 2013), this approach has been limited to the 0.1–1% of host cells that are amenable to cultivation and is biased towards certain taxonomic groups and life strategies (Rappé and Giovannoni, 2003; Swan et al., 2013). Furthermore, viral types detected using plaque assays on pure cultures are biased toward virulent phages causing complete lysis (Brüssow and Hendrix, 2002). As a result of these methodological limitations, the overarching question of ‘who infects whom and how in situ, has been challenging to address.
Several novel methods have reduced culture-dependence in the study of phages. For example, metagenomics offers a glimpse at the global viral diversity, but rarely allows linking a viral sequence to its host. In simple systems, it has been possible to infer putative host linkages by comparing spacers in clusters of regularly interspaced short palindromic repeats to viral metagenomic sequences (Anderson et al., 2011; Berg Miller et al., 2012). Another way to putatively assign hosts is by similarity to tRNA genes (Mizuno et al., 2013), a common site of integration for temperate phages (Fouts, 2006). Methods have been developed for more complex communities, such as phageFISH (fluorescence in situ hybridization) targeting specific viral genes (Allers et al., 2013), infection of cultured microbial strains with tagged viral communities to identify uncultured viruses (Deng et al., 2012, 2014), or the use of microfluidic digital PCR to associate single cells with environmental viruses (Tadmor et al., 2011).
Over the past decade, single-cell genomics (SCG) has emerged as a powerful tool for the recovery of genomic information from uncultured, individual cells of environmental microorganisms (Lasken, 2012; Stepanauskas, 2012). Importantly, SCG recovers sequences of all DNA molecules in a cell, including host chromosomes and extrachromosomal genetic elements, thus offering an opportunity for cultivation-independent, cell-specific analysis of organismal interactions, such as infections, symbioses and predation. Previous application of this approach revealed a complete genome of a novel nanovirus from a putatively infected, uncultivated Picozoan protist cell (Yoon et al., 2011). In another study, 127 single amplified genomes (SAGs) of the uncultured gammaproteobacterial clade SUP05 revealed 69 viral genomic sequences, suggesting that a third of the cells were infected. These new viral sequences were then used as references to mine viromes and microbial metagenomes to show that these phages are endemic and persistent over time (Roux et al., 2014).
The enormous opportunity to study in situ virus–host interactions using SCG is accompanied by novel computational challenges, which cannot be fully addressed by existing bioinformatics tools. Although many software packages have been developed for the identification of prophages (that is, Phage_Finder (Fouts, 2006), Prophinder (Lima-Mendez et al., 2008) or PhiSpy (Akhter et al., 2012)), pathogenicity islands (that is, PIPS (Soares et al., 2012)) and horizontal gene transfer events (that is, Alien Hunter (Vernikos and Parkhill, 2006)), they may be poorly suited when analyzing novel viruses in uncultured microorganisms, because of the reliance of these tools on genomic sequences of viral isolates in public databases. Further challenges are presented by the incomplete and fragmented nature of SCG assemblies. Finally, SCG captures microbial cells during various types of interactions with viruses, including lytic infections, lysogeny, chronic infections, as well as non-infectious attachment, with highly divergent biological consequences. Ideally, these interaction types should be discriminated in SCG sequence analyses.
To realize the potential for SCG to uncover viral–host interactions within new microbial groups, using new bioinformatics approaches, we searched for viruses in 58 previously sequenced SAGs of surface ocean bacteria and archaea, originating from four locations: the Gulf of Maine, the Mediterranean Sea, and the North Pacific and South Atlantic subtropical gyres (Swan et al., 2013). This enabled cultivation-independent detection and characterization of in situ cell interactions with viruses that are highly divergent from previously sequenced genomes. Our results revealed complete or near-complete genomes of phages belonging to all three major tailed groups (Podoviridae, Myoviridae and Siphoviridae), infecting phylogenetically diverse hosts. We detected and uncovered genomic sequences of first known viruses infecting several ubiquitous, yet uncultured marine bacterioplankton groups, including Marinimicrobia (formerly Marine Group A, SAR406), Verrucomicrobia, Gammaproteobacteria SAR92 and SAR86 and Thaumarchaeota (Marine Group I).
Materials and methods
Sources of SAG sequences
We analyzed 57 previously published SAGs from surface ocean (Swan et al., 2013). These SAGs are from the Gulf of Maine (36 SAGs), the Hawaii Ocean Time Series (HOT) (10 SAGs), the Mediterranean Sea (7 SAGs) and Southern Atlantic (4 SAGs). The original assemblies were performed on Illumina 2 × 150 bp reads (Illumina, San Diego, CA, USA) using a combination of Velvet and Allpaths assemblers (Swan et al., 2013), and only contigs ⩾2000 bp were kept for further analyses. In an attempt to improve genome recoveries, we re-sequenced 10 virus-containing SAGs using the PacBio technology (Pacific Biosciences Inc., Menlo Park, CA, USA). We created linear terminal deoxynucleotidyl transferase libraries for 10 of the SAGs (AAA076-E06, AAA160-C11, AAA160-J20, AAA164-A08, AAA164-A21, AAA164-B23, AAA164-I21, AAA164-M04, AAA164-P11, AAA168-E21). Single-cell MDA products were randomly sheared to ~3 kb target size using G-tubes (Covaris, Inc., Woburn, MA, USA). Poly(dA) tails were then added to the 3'ends using terminal deoxynucleotidyl transferase and the poly(dA)-tailed library annealed with poly(dT) sequencing primer for sequencing on the PacBio RS instrument (Pacific Biosciences). These PacBio sequences were co-assembled with Illumina reads with SPAdes 3.1 using —careful, —sc and —pacbio arguments (Nurk et al., 2013). In addition, we obtained new Illumina-only assemblies using SPAdes 3.1. Alternative SPAdes assemblies were performed with and without k-mer pre-normalization of Illumina reads (Swan et al., 2013). k-mer normalization was performed with the open source software kmernorm (http://sourceforge.net/projects/kmernorm/) using settings k=19, t=80, c=2. The SPAdes assembler improved assembly contiguity, as compared with the original, Velvet-Allpath assemblies, in 8 of the 10 re-analyzed SAGs (Supplementary Tables 1 and 2). The k-mer pre-normalization further improved the contiguity of SPAdes assemblies of eight SAGs. Significantly, this enabled the assembly into single contigs, likely representing complete genomes, of all viral sequences in Verrucomicrobia SAGs AAA164-A21 (four viral contigs in Velvet-Allpaths assemblies) and AAA168-E21 (three viral contigs in Velvet-Allpaths assemblies). The addition of long PacBio reads had minimal impact on overall assemblies (see Supplementary Material).
In addition to the 57 SAGs listed above, we also sequenced one more Roseobacter-clade SAG from the Gulf of Maine, AAA160-J18. This SAG was generated from the same sample and using the same methods as other Gulf of Maine SAGs (Martinez-Garcia et al., 2012). This SAG was sequenced at the Bigelow Laboratory Single Cell Genomics Center (scgc.bigelow.org). A total of 11 464 853 '2 × 150' reads were generated with a NextSeq 500 (Illumina). Read digital normalization and assembly were performed in the same way as for the remaining 57 SAGs (Supplementary Table S1), producing 2.21 Mbp in 47 contigs larger than 2 kbp, with an average GC content of 40.5%.
DNA segments 2.4–11.8 kb in length that were 100% identical to the contig 00001 of Verrucomicrobia SAG AAA164-B23 were found in seven taxonomically diverse SAGs (Supplementary Table 3), in which sequence coverage depth was about 1000-fold lower than in AAA164-B23. To avoid the risk of false discovery, we assumed that these sequences were Illumina library cross-contaminants, originating from the SAG with the longest and highest-coverage fragment. We therefore eliminated these contigs from the seven assemblies (Supplementary Table 3). Similarly, a 7.0 kb segment of contig 00005 from Roseobacter SAG AAA076-E06 was also found in Roseobacter SAG AAA015-O19, with the likely contaminating sequence having a 1500 × lower sequence coverage depth than in AAA076-E06. This AAA015-O19 contig was also removed from further analysis.
Assessing the risks of SAG contamination with free viral particles
We assessed the risk of a cell being contaminated with a free viral particle during cell sorting, which would obscure single cell genomics data interpretation. Single-drop sort mode on the MoFlo flow cytometer (Beckman Coulter, Indianapolis, IN, USA) was used to separate individual cells in this study, which prevents sorting a droplet that contains other detectable particles than the cell of interest in the sorted and neighboring droplets. We used light side scatter as the trigger and maximized the voltage on side scatter and green fluorescence, which is used for the detection of SYTO-9-stained nucleic acids. This renders most viral particles visible to the instrument and enables their effective exclusion from sorted droplets, the only exception being extremely small particles, such as the ssDNA viruses (Tomaru and Nagasaki, 2007; Holmfeldt et al., 2012). There is a possibility of an accidental co-sort of a cell and a large, free viral particle, if the particle is in the shade of a cell while passing through the cytometer's laser beam, and we estimated the frequency of such events. The diameter of the sample flow stream, using our cell-sorting conditions, is about 5 μm (Gerrit van den Engh, personal communication). The most abundant bacterioplankton cells in the ocean are under 1 μm in diameter: Prochlorococcus is ~0.6 × 0.6 μm (Partensky et al., 1999), Pelagibacter ubique is 0.1 × 0.9 μm (Rappé et al., 2002). Using a conservative assumption of an average diameter being 1 μm among the sorted cells, we estimated the average shaded volume to be <4 × 10−12 ml. Assuming the presence of 109 viral particles per ml of the analyzed sample, which is typical for surface ocean (Suttle, 2005), and accounting for the 10 × dilution of the sample prior to cell sorting, we estimate the frequency of free viral particles being in cells' shade to be less than 1 in 2500. This indicates high probability that viruses detected in our study sample of 58 cells were either inside or attached to the analyzed cells during cell sorting.
Identification of viral sequences
Because multiple displacement amplification (MDA) amplifies only dsDNA and ssDNA, our study did not target RNA viruses. To identify SAG contigs originating from DNA viruses, we used a combination of five criteria, listed below.
Marker genes
Gene prediction of sequenced and assembled SAGs was performed using Prodigal (Hyatt et al., 2010). The translated protein sequences were then used as queries in BLASTp (Altschul et al., 1990) searches (e-value <0.001, max. 10 hits) of the GenBank nr database (Updated 12 July 2013). We identified homologous sequences containing words within the sequence description indicative of viral genes (*phage, *virus, virion, prophage, terminase, capsid, head, tail, fiber, baseplate, portal, lysis, structural, T4, lambda, mu, lambdoid, podo*, myovir*, siphovir*, integrase, transposase). Query sequences homologous to hypothetical proteins were also identified, because viral genomes are generally enriched in them. We also searched for tRNA genes, which are common sites for prophage integration into host genomes, using tRNAscan (Lowe and Eddy, 1997).
DNA sequence anomalies
In bacterial genomes, a GC skew is associated with the origin of replication (Karlin and Campbell, 1998). GC skew can also be associated with the insertion of foreign DNA, including prophages (Grigoriev, 1998). Local anomalies in GC content and codon usage may also aid in the detection of prophages and other laterally acquired genetic elements within bacterial genomes (Akhter et al., 2012; Soares et al., 2012). For each contig, we calculated GC content and GC skew with custom scripts using a sliding window of 1600 bp. Tetramer frequencies have been used to detect contaminating sequences in SAG assemblies (Woyke et al., 2009; Swan et al., 2011; Martinez-Garcia et al., 2012). Here, we extracted tetramer frequencies using a sliding window of 1600 bp and 200 bp step size to have a minimum of three windows for each contig. Principal component analysis was used on a matrix of N × 136 where N is the number of windows in the genome and 136 is the number of unique reverse-complimentary tetramers, as in the study by Swan et al. (2013).
Metagenomic fragment recruitment
To further improve the detection of viral sequences within SAGs, sequence data from cellular metagenomes and viral metagenomes (viromes) were recruited to SAG contigs. SAG contigs originating from host chromosomes were expected to recruit predominantly cellular metagenomic sequences, while contigs originating from viruses were expected to recruit virome sequences. In this analysis, we used the Line P prokaryote metagenome (IMG/MER GOLD Project ID Gm00303) (Swan et al., 2013; Wright et al., 2014) and the Pacific Ocean Virome metagenome (Hurwitz and Sullivan, 2013). These metagenomes integrate coastal and off-shore samples from multiple depths. Importantly, these virome libraries did not have a strong bacterial recruitment signal (Hurwitz and Sullivan, 2013; Hurwitz, Brum, et al., 2014; Hurwitz, Westveld, et al., 2014). Both of these metagenomic datasets have an average read length of ~300–350 bp, facilitating the comparison of their fragment recruitment. The Line P metagenome was sequenced using a combination of Illumina paired-end and Sanger technologies, with a mean read length of 387 bp, whereas the POV metagenome was sequenced using 454 technology with a mean read length of 316 bp. Metagenomic fragment recruitment was done using BLASTx (e-value <0.001, 1 hit per read) on predicted SAG proteins. Fragment recruitment estimates were performed for viral and bacterial contigs with 50, 70 and 90% sequence identity thresholds. The 70% threshold produced the best contrast between viral and bacterial recruitment and therefore was used in subsequent analyses.
Sequence coverage depth
During a lytic infection, many copies of the virus genome are present in the host cell. Therefore, when performing shotgun sequencing of an infected cell, viral contigs are expected to have significantly higher sequence coverage depth, as compared with contigs of the host genome. In contrast, only one viral genome copy typically is present in other types of virus–cell interactions (lysogeny, unspecific attachment of the virus to the cell), leading to a similar sequence coverage depth between host and virus contigs. To discriminate between active infections and other forms of bacteria–phage associations, the sequence coverage depth between putative bacterial and viral contigs was compared. The depth of coverage was calculated by mapping reads on assembled contigs using Bowtie (Langmead et al., 2009) and BEDTools (Quinlan and Hall, 2010). We relied on the average coverage for each contig rather than utilizing a sliding window approach, to limit the impact of uneven MDA (Raghunathan et al., 2005; Woyke et al., 2009).
Confirmation of viral sequences
Contigs that contained multiple viral marker genes, DNA sequence anomalies, high metagenomic viral recruitment or low metagenomic bacterial recruitment were manually inspected using diverse comparative genomics tools (see below), to confirm the presence or absence of viral sequences. Such manual verification was performed on 29 SAGs, and viruses were confirmed in 20 of them. See Supplementary Material for more information on tests of available bioinformatics tools to search for viral sequences in SAGs.
Phylogenetic and comparative genomic analyses
All alignments were constructed with MUSCLE (Edgar, 2004) implemented in Geneious V6.1.8 and evaluated for optimal amino acid substitution models using ProtTest 3 (Darriba et al., 2011). For the Podoviridae DNA pol A, the initial alignment was provided by Schmidt et al. (2014) and phylogeny was performed using phyML V3.1 (Guindon et al., 2010) with 100 bootstrap replicates and the LG model with a gamma distribution (+G), estimated rates of variation among sites and a proportion of invariable sites (+I). For the phylogenetic analysis of the T4-like major capsid protein gp23, all amino acid Myoviridae gp23 sequences ⩾100 amino acids were downloaded from GenBank (2199 sequences as of 15 September 2014), and then clustered at 90% similarity using CD-hit (Li and Godzik, 2006) to reduce the number of sequences. Sequences that did not align properly or created long branches in the tree were removed. Phylogeny was performed in Geneious V6.1.8 using neighbor joining with the Jukes Cantor model with 1000 bootstrap replicates. For the whole-genome phylogeny of Myoviridae, initial alignments were obtained from Zhao et al. (2013), and more Myoviridae genomes were added to reflect the gp23 phylogeny (Sinorhizobium phage phiM12 (NC_004735), Rhodothermus phage RM378 (NC_013697), Deftia phage phiW-14 (NC_013697), Cronobacter phage vB_CsaM_GAP32 (NC_019401)), as well as the viral SAG genomes from AAA164-P11 and AAA160-J20. The alignments were stripped to keep only the 13 proteins for which homologs were found in the SAG sequences (Supplementary Table 4). Alignments of each protein were edited with Gblocks (Talavera and Castresana, 2007) using the following settings: −b1=(N/2)+1; −b2=(N/2)+1; −b3=(N/2); −b4=2; −b5=h, with N=number of taxa (Sassera et al., 2011). Phylogeny was performed using phyML V3.1 (Stamatakis et al., 2008) under the LG model following a gamma distribution and empirical base frequencies with 100 bootstrap replicates.
Taxonomic families were tentatively assigned to each virus, based on the best blast hits (Supplementary Table 5). To do so, we compared each protein of each viral contig to a database composed of all available viral genomes in GenBank (5453 genomes⩾1000 nucleotide long) using tBLASTn. Only the top hits were kept and the viral family with the most hits was tentatively assigned to the virus. To obtain additional evidence, whole-genome synteny comparisons were performed with EasyFig for Mac version 2.1 (Sullivan et al., 2011) using tBLASTx and filtering of small hits and annotations. The viral genomes used for comparisons were selected based on their similarity to the SAG viruses, as determined by BLASTp searches (Supplementary Table 6). The average nucleotide identity was calculated with the JSpecies software under the ANIm parameters (Richter and Rosselló-Móra, 2009). Because viral taxonomy is often based on morphology rather than genomic sequence, the assigned families should be viewed as tentative.
Experimental infection of Roseobacter strains with cyanophages
A pool containing 25 μl from each of 374 cyanophage lysates (Supplementary Table 7) was filtered through a 0.22 μm filter and 0.1 ml of the filtered phage mix was used to inoculate 0.1 ml of 34 Roseobacter isolates (Supplementary Table 8) in logarithmic phase. Isolates were grown either on half-strength YTSS (2.5 g of yeast extract, 4 g of tryptone, 15 g of sea salts per liter) at 30 °C or ZoBell (5 g peptone, 1 g yeast extract and 0.01 g FeC6H5O7 added per liter of 80%, 0.45 μm-filtered seawater) at 21 °C. The virus-bacteria mix was incubated for 1 h at 25 °C before transferring 0.3 ml of the culture to 3.5 ml of 4% top agar and poured onto a bottom plate. Plates were incubated at 25 °C for 24 h and then 21 °C until growth was observed. Plates were checked daily for plaque formation to record infections.
Data availability
Viral contig sequences, phylogenetic trees and alignments are available in Supplementary Materials of this publication. SAG genomic sequences are available on the D.O.E. Joint Genome Institutes IMG portal, http://img.jgi.doe.gov/.
Results and discussion
Detection of viral sequences in SAGs
The combination of searches for viral marker genes, DNA sequence anomalies and contrasts in metagenomic fragment recruitment detected phage-like sequences in 20 of the 58 analyzed SAGs (33%) (Table 1). All but one of these putative virus-containing SAGs were collected from the Gulf of Maine. The one additional putative virus was found in a Roseobacter SAG AAA300-J04 from the North Pacific subtropical gyre. For the first time, viruses were found in the following, ubiquitous groups of marine bacterioplankton: Marinimicrobia (formerly called Marine Group A and SAR406; 2 SAGs), Verrucomicrobia (10 SAGs), Gammaproteobacteria lineages SAR86 (1 SAG) and SAR92 (1 SAG), and Thaumarchaeota (1 SAG). Viruses were also found in Bacteroidetes (1 SAG) and Alphaproteobacteria (3 SAGs). Of the 20 SAG-associated viruses, most shared some genes with cultivated phages belonging to the Podoviridae (10 SAGs), Myoviridae (6 SAGs) and Siphoviridae (3 SAGs) (Table 1 and Supplementary Table 5). The observed frequency of SAG-associated viruses, their higher abundance in the most productive study site and the predominant viral types are consistent with prior observations of marine phage–host interactions (Fuhrman and Suttle, 1993; Wommack and Colwell, 2000; Weinbauer, 2004; Suttle, 2005; Zhao et al., 2013). Significantly, the use of SCG allowed the specific matching of genomic sequences of each virus and its host, without cultivation.
Table 1. SAG-associated viral sequences: SAG ID, predicted viral taxonomic group, length of viral contigs, host taxonomic group, estimated fraction of host genome recovered, the ratio of viral versus bacterial contig sequence coverage depth, the multiple displacement amplification critical point value and the predicted viral–host interaction.
| SAG | Viral group | Viral contigs | Viral bp | Host group | Host genome recovery | Ratio viral/bacterial depth of coverage | MDA Cp, hours | Inferred interaction |
|---|---|---|---|---|---|---|---|---|
| AAA300-J04* | Podoviridae | 1 | 42 985 | Roseobacter | 22% | 0.015 | 8.4 | Unclear |
| AAA160-D02 | Podoviridae | 2 | 44 447 | SAR92 | 64% | 3.75 | 7.4 | Unclear |
| AAA160-C11 | Podoviridae | 2 | 66 562 | Marinimicrobia | 91% | 0.044 | 5.7 | Unclear |
| AAA160-P02 | Podoviridae | 3 | 51 410 | Bacteroidetes | 85% | 0.137 | 5.1 | Unclear |
| AAA164-I21 | Podoviridae | 3 | 65 751 | Verrucomicrobia | 30% | 31.28 | 7.0 | Unclear |
| AAA164-M04 | Podoviridae | 4 | 75 811 | Verrucomicrobia | 53% | 2.92 | 6.7 | Unclear |
| AAA168-E21* | Podoviridae | 3 | 38 179 | Verrucomicrobia | 58% | 141 | 5.8 | Unclear |
| AAA164-O14 | Podoviridae | 1 | 39 559 | Verrucomicrobia | 62% | 4.40 | 6.4 | Unclear |
| AAA164-A21 | Podoviridae | 4 | 50 683 | Verrucomicrobia | 24% | 105 | 5.2 | Unclear |
| AAA164-B23 | Podoviridae | 2 | 60 351 | Verrucomicrobia | 0% | 734 | 5.7 | Late lytic |
| AAA164-P11 | Myoviridae | 4 | 135 147 | Verrucomicrobia | 5% | 888 | 6.3 | Late lytic |
| AAA076-E06 | Myoviridae | 13 | 203 548 | Roseobacter | 2% | 1111 | 6.6 | Late lytic |
| AAA160-J18 | Myoviridae | 4 | 66 442 | Roseobacter | 74% | 0.047 | 6.8 | Unclear |
| AAA160-I06 | Myoviridae | 1 | 6458 | Marinimicrobia | 96% | 0.003 | 5.3 | Unclear |
| AAA168-P09 | Myoviridae | 2 | 10 003 | SAR86 | 96% | 0.007 | 5.4 | Unclear |
| AAA160-J20 | Myoviridae | 4 | 159 127 | Thaumarchaeota | 0% | 2.45 | 7.3 | Unclear |
| AAA160-J14 | Siphoviridae | 1 | 19 019 | SAR116 | 38% | 0.122 | 8.7 | Potentially temperate |
| AAA164-A08 | Siphoviridae | 3 | 82 090 | Verrucomicrobia | 1% | 41 499 | 6.6 | Late lytic |
| AAA164-L15 | Siphoviridae | 1 | 28 638 | Verrucomicrobia | 50% | 5.76 | 6.4 | Unclear |
| AAA164-N20 | Phycodnaviridae | 5 | 62 064 | Verrucomicrobia | 37% | 794 | 6.7 | Unspecific attachment |
Abbreviation: MDA, multiple displacement amplification; SAG, single amplified genome.
Complete circular viral genome assemblies are marked with an asterisk. Additional information about the host component of these SAGs is available in Swan et al., 2013.
Modes of viral–host interactions
The presence of viruses inside of or attached to cells may be the result of several types of interactions with divergent biological consequences, including lysogeny, lytic infections, chronic infections and unspecific attachment. In an effort to discriminate between these modes of interaction, we examined the SAGs containing viral sequences for: (i) sequence-based evidence for virus integration into the host genome; (ii) the fraction of viral and host genome recovered, in relation to the speed of the single-cell MDA reaction; and (iii) difference in sequence coverage depth between phage and host contigs.
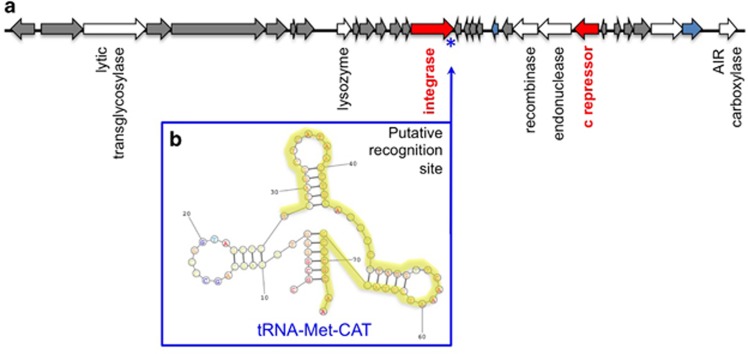
To be considered a prophage, we expected viral sequences to be flanked by host genes and to encode for an integrase. We found no contigs containing both viral and bacterial genes in the analyzed data set. Lysogeny predominantly occurs in environments of low bacterial abundance and productivity (McDaniel et al., 2002; Williamson et al., 2002; McDaniel et al., 2008). As most viruses detected in this study are from the productive Gulf of Maine, the apparent absence of lysogens among the 20 viruses may not be surprising. Prophages usually integrate in hypervariable regions containing multiple repeats, which are difficult to assemble from short fragment sequence reads (Zerbino and Birney, 2008). Thus, the failure to detect prophages because of the fragmented nature of SAG assemblies cannot be excluded. Nevertheless, we detected a viral contig in the SAR116 SAG AAA160-J14 that harbors an integrase, suggesting the potential of this phage for lysogeny (Figure 1). For a phage to integrate into its host's genome, it must contain a phage attachment site (attP), which is an exact match to the bacterial attachment site (attB), often located in tRNA genes (Campbell, 2003; Sullivan et al., 2009; Mizuno et al., 2013). We compared the tRNA sequences of all 58 SAGs to the putative viral sequences and found one 48 bp exact match between the attP site of the phage found in SAR116 AAA160-J14 and a bacterial tRNA-Met-CAT gene found in contig 00007 of the same SAG, confirming the association of this particular phage with its host and suggesting potential for phage integration. This potentially lysogenic phage also encodes a c repressor, which may confer immunity of lysogenic cells against similar phages, in this way protecting the host from other, potentially lytic infections (Canchaya et al., 2003). Lysogeny may permit phage's survival at low host abundance and protect from host defense mechanisms, UV radiation and, in some cases, grazing (Paul, 2008). The SAR116 cluster is one of the most ubiquitous and abundant groups of Alphaproteobacteria in the ocean, although only two laboratory cultures are currently available and sequenced (Oh et al., 2010; Grote et al., 2011). The new, potentially temperate phage does not share any homology with the two prophages found in the genome of the SAR116 isolate Candidatus Puniceispirillum marinum (Oh et al., 2010) nor the only lytic phage that has been isolated from SAR116 so far (Kang et al., 2013), suggesting that the diversity of lytic and temperate phages capable of infecting SAR116 remains far from saturation.
Figure 1.
Phage associated with the SAR116 SAG AAA160-J14 and its genetic potential for integration at the bacterial tRNA-Met-CAT site. (a) Phage genome organization with the attP site represented by an asterisk. (b) Bacterial tRNA-Met-CAT, with the attB region that is identical to phage's recognition site highlighted in yellow.
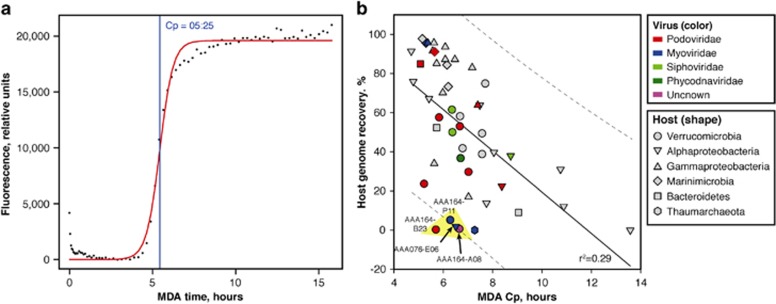
We considered the following criteria to determine whether a cell was undergoing a late lytic infection: (i) high sequence coverage depth of putative viral, as compared with host contigs; (ii) low host genome recovery, indicating infection-induced DNA degradation; and (iii) complete or near-complete recovery of viral genome, indicative of the availability of multiple genome copies for MDA. However, factors other than viral infections can cause poor genome recovery and uneven sequence coverage depth in SCG data, such as incomplete cell lysis, varied DNA packaging in the cell, as well as MDA artifacts, complicating the application of these criteria (Stepanauskas, 2012). To circumvent these issues, we analyzed the relationship between the speed of MDA reaction and the recovery of host genome. The availability of a longer DNA template is expected to result in a faster MDA reaction (Zhang et al., 2006). Accordingly, we observed a negative correlation between the MDA critical point and the fraction of genome recovered in a SAG assembly (Figure 2). Four SAGs were at or outside of the 95% confidence intervals for the model prediction and had low MDA critical point values (fast MDA) and low host genome recovery (yellow-shaded region in Figure 2b). In all four SAGs, complete or near-complete viral genomes were recovered, and the ratio of sequence coverage depth of viral versus host contigs exceeded 500 (Table 1). The collective evidence suggests that these four cells were undergoing late lytic infections during their collection. Interestingly, all of these putative lytic infections were identified in metabolically active cells, including three Verrucomicrobia cells that were sorted based on their uptake of polysaccharide laminarin and one Alphaproteobacteria cell that was sorted based on electron transport system activity (Martinez-Garcia et al., 2012) (Table 1). This is consistent with prior observations that the most metabolically active cells in a given environment are most susceptible to phage infections (Fuhrman and Suttle, 1993; Zhao et al., 2013).
Figure 2.
Single-cell whole-genome MDA kinetics. (a) SAG reaction kinetics, where dots indicate actual measurements, the red curve indicates a logistic model fit, and the blue line indicates the critical point (Cp). The Cp is the time required to reach half of the maximal fluorescence of DNA labeled with SYTO-9. (b) Correlation between the MDA Cp and the estimated genome recovery. Each data point represents a SAG. The solid line indicates a linear regression fit. Dotted lines represent 95% confidence intervals for the linear regression model prediction. Yellow shading indicates SAGs inferred to undergo late lytic infections. The r2 of this regression increases to 0.48 when six outlier SAGs are excluded from the analysis.
Modes of phage–host interactions in the remaining virus-containing SAGs could not be determined with confidence and may include lytic infections, lysogeny or chronic infection. Among these scenarios, chronic infections (Weinbauer, 2004) are especially poorly understood, owing to detection difficulties in traditional plaque assays, and SCG may offer a novel way for their study. Chronic infections and lysogeny would result in a phage with a similar depth of coverage to its host, which is the case for most of the assembled viral sequences in the studied SAGs. Non-infectious and non-specific attachment of viral particles to cell's surface is also a possibility, although existing evidence suggests this to be uncommon among marine microorganisms (Deng et al., 2012). We expect that the ability to categorize phage–host interactions using SCG will continue to improve as more refined laboratory and computational tools are developed. Moreover, an increased availability of SCG data may lead to the observation of consistent infectivity patterns for given microbial species.
Diversity and abundance of SAG-associated viruses
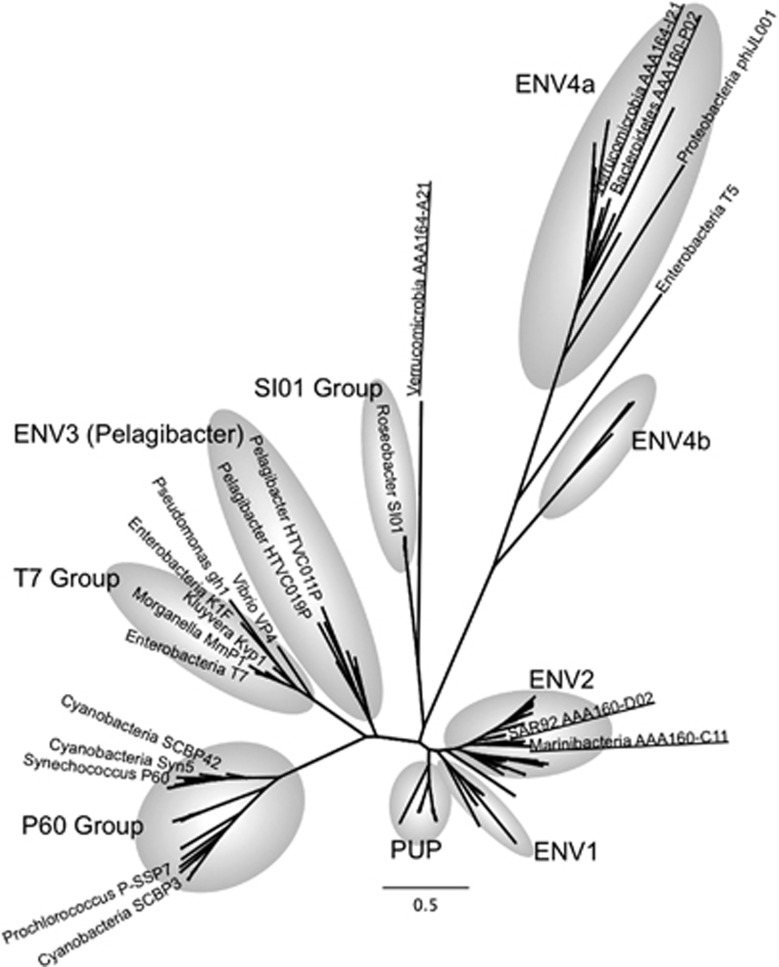
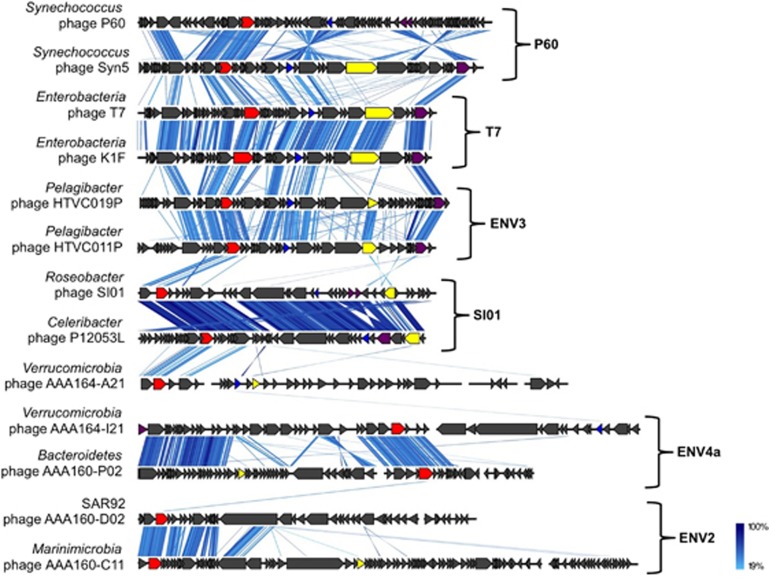
On the basis of genomic sequence similarity, 9 of the 20 SAG-associated viruses were putatively assigned to Podoviridae, a large and diverse family of phages within the order Caudovirales that is characterized by icosahedral capsids and short, non-contractile tails (Table 1 and Supplementary Table 5). These viruses were identified in phylogenetically diverse SAGs, including five Verrucomicrobia, two Proteobacteria, one Marinimicrobia and one Bacteroidetes. Podoviridae genome sizes typically range ~40–42 kb, whereas our obtained assemblies vary between 44 and 75 kb. Complete, circular viral genome assemblies were obtained from the Roseobacter SAG AAA300-J04 and Verrucomicrobia AAA168-E21. Five of the discovered T7-like phages contain viral DNA polymerase A genes, which cluster with environmental sequences previously amplified with degenerate primers or metagenomic sequences (Labonté et al., 2009; Schmidt et al., 2014) and are distant from cultured isolates (Figure 3). On the basis of the DNA polymerase A phylogeny, the newly sequenced genomes of phages AAA164-I21 (Verrucomicrobia) and AAA160-P02 (Bacteroidetes) clustered in group ENV4a, phages AAA160-C11 (Marinimicrobia) and AAA160-D02 (SAR92) clustered in group ENV2, and phage AAA164-A21 clustered distantly with group SI01. Prior to our study, PCR amplicon sequences of polymerase A were the only information available about ENV4a and ENV2. We looked at position Phe762 of the DNA pol A, a well-studied site where mutations affect dideoxynucleotide (ddNTP) incorporation, polymerase activity and fidelity. Sequences in the ENV4a group had a leucine (L) at that position, which has been suggested for phages with a broad host range, temperate phages or phages infecting slow growing bacteria. In contrast, phages in the ENV2 group and phage AAA164-A21 had a tyrosine (Y) at position Phe762, which is associated with lytic phages (Schmidt et al., 2014). Unfortunately, other indicators, such as integration into host genome, percent genome recovery and sequence coverage depth could not confirm these predictions for the four phages (Table 1). The general genome organization of the DNA polymerase A gene harboring phages differs from previously sequenced phage genomes (Figure 4). Cultured viral isolates, even those obtained from distant hosts (for example, E. coli and Synechococcus), were more closely related to each other than to the uncultured marine phages from this study.
Figure 3.
Phylogenetic analysis of the Podoviridae-like DNA polymerase A from SAGs (underlined), isolates (labeled) and metagenomes (not labeled). The grey shading indicates the pre-defined groups from Schmidt et al. (2014) and Labonté et al. (2009). The tree was generated using maximum likelihood, with 100 bootstrap replicates, using the LG model with a gamma distribution (+G), estimated rates of variation among sites and a proportion of invariable sites (+I). Bootstrap replicates are not shown for clarity. Scale bar represents the number of amino acid substitution per site.
Figure 4.
Synteny between Podoviridae genomes that are most closely related to SAG-associated Podoviridae, based on DNA pol A phylogeny. Each arrow represents a gene: DNA pol A (red), major capsid protein (blue), tail fibers (yellow), terminase (purple) and others (black). tBLASTx was used to identify homologous regions. Color legend indicates DNA sequence identity.
Three SAG-associated genomes are most similar to the recently discovered Podoviridae phages infecting Cellulophaga (Holmfeldt et al., 2013). Phages in AAA300-J04 (Roseobacter) and AAA164-B23 (Verrucomicrobia) have a genome organization similar to the Cellulophaga phage 3:2 (Supplementary Figure 1A). The phage in AAA164-M04 (Verrucomicrobia) is most similar to the Cellulophaga phage 40:1 (Supplementary Figure 1B). The last putative Podoviridae genome, which is a complete circular genome (AAA168-E21; Verrucomicrobia), shares a similar genome organization with Pelagibacter HTVC010P (Supplementary Figure 1C), a phage with a small capsid that is highly prevalent in the ocean, but is the only isolate from its group available at the moment. Our results show that marine Podoviridae are highly diverse in their genome content, are widely distributed in marine environments (Figure 5) and infect a wide taxonomic range of marine bacterioplankton groups. Some of the putative Podoviridae genomes, such as AAA164-B23 (60.4 kb) and AAA164-M04 (75.8 kb), have larger genomes than the Podoviridae average of 40–42 kb. Podoviridae phages with very large genomes (70–75 kb) infecting Cellulophaga were shown to be generalist phages, and were lytic on their host of isolation, but lysogenic or inefficient on other hosts (Holmfeldt et al., 2014). Inefficient infections could be quite common in marine environments, but are rarely isolated in the laboratory owing to a bias against virulent phages. SCG has the promise to resolve this methodological limitation.
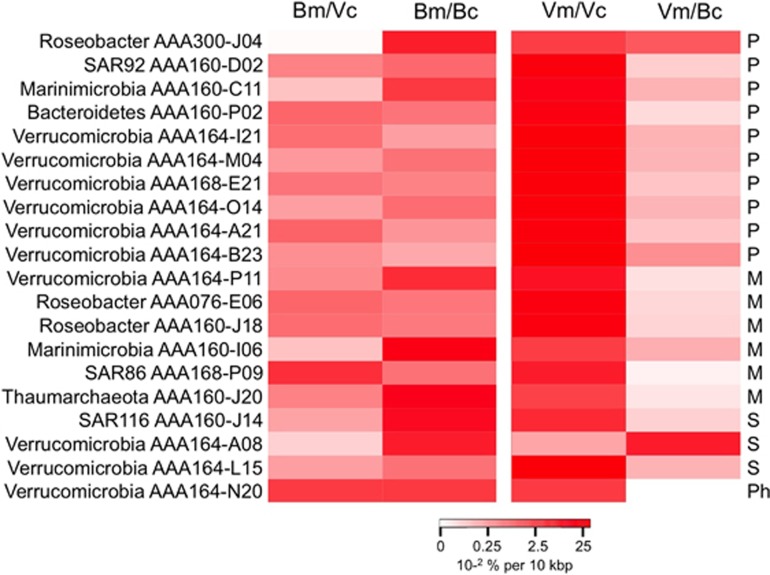
Figure 5.
Fragment recruitment of microbial metagenomic reads from the Line P metagenome (Bm) and viral metagenomic reads from the Pacific Ocean Virome (Vm) on the viral (Vc) and bacterial contigs (Bc). The scale bar indicates the fraction of metagenomic reads aligning to each reference with ⩾70% nucleotide identity, normalized by the length of the genome. The right column indicates whether the virus belongs to the Podoviridae (P), Myoviridae (M), Siphoviridae (S) or Phycodnaviridae (Ph) families.
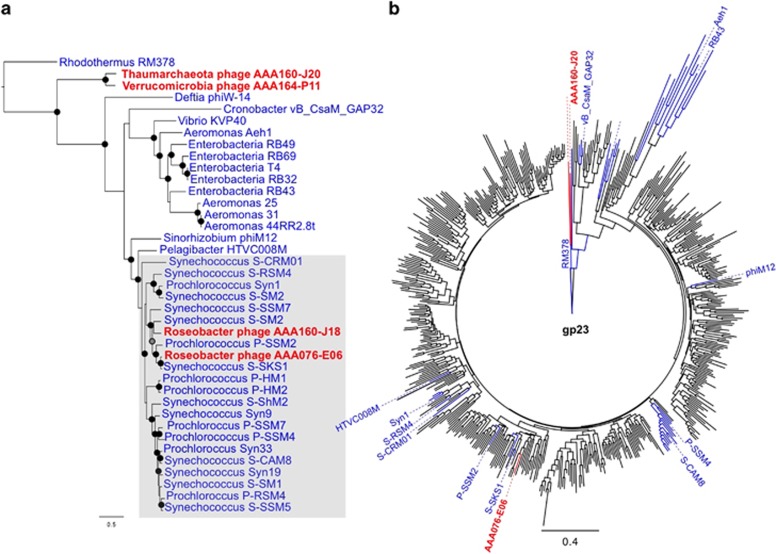
The Myoviridae family consists of phages with a contractile tail and genome sizes varying from 33 to 170 kb (King et al., 2012). We found Myoviridae-like sequences in five SAGs, with three of these assemblies likely near-complete. Two of them, found in AAA164-P11 (135 kb; Verrucomicrobia) and AAA160-J20 (159 kb; Thaumarchaeota), encode the conserved T4 core genes (Supplementary Table 5) but are evolutionarily distant from existing viral isolates and may represent a novel group within the large Myoviridae family (Figure 6). Although these two phages share similar gene organization and core genes, their plasticity regions are highly divergent, consistent with the infection of different hosts. Phylogeny of the gp23 major capsid protein gene also shows that these phages are distant to other known Myoviridae sequences, as they cluster close to the root (Rhodothermus RM378 phage). Metagenomic fragment recruitment analysis suggested that relatives of these two phages are abundant in the ocean (Figure 5). Interestingly, similar phages were not observed in the environment when samples were surveyed with degenerate PCR primers, designed using sequences from cultured isolates (Filée et al., 2005), which may be explained by primer sequence mismatches. Myoviridae infections of halophilic and methanogenic Euryarchaeota have been reported previously (Prangishvili et al., 2006). To the best of our knowledge, this is the first report of viral infections in Thaumarchaeota.
Figure 6.
Phylogenetic analysis of Myoviridae phages recovered from SAGs, isolates and environmental sequences. (a) Phylogenetic tree of 13 concatenated conserved proteins within the large Myoviridae family, showing that phages found in Verrucomicrobia SAG AAA164-P11 and Thaumarchaeota SAG AAA160-J20 form a novel phylogenetic group, and that Roseobacter phages AAA076-E06 and AAA160-J18 are most similar to cyanophages. The following parameters were used: maximum likelihood, 100 bootstrap replicates, LG model with a gamma distribution (+G), estimated rates of variation among sites and a proportion of invariable sites (+I). Bootstrap replicates >95% and 75–95% are represented by black and gray circles, respectively. (b) Phylogenetic tree of the major capsid protein gp23 of Myoviridae phages from SAGs (red), isolates (blue) and PCR amplicons from environmental samples (black). The following parameters were used: Neighbor Joining, Jukes Cantor model, 1000 bootstrap replicates. Both trees are rooted with the divergent Rhodothermus phage RM378. Scale bar represents the number of amino acid substitution per site.
Surprisingly, the Myoviridae-like phage associated with the Roseobacter SAG AAA076-E06 shares 82.9% and 75.8% average nucleotide identity with phages P-SSM2 (103/330 genes) (Sullivan et al., 2005) and S-SKS1 (218/281 genes), respectively, which infect cyanobacteria (Figure 6 and Supplementary Figure 2). Although non-specific phage attachment to the sorted cell cannot be fully excluded, several lines of evidence suggest a lytic infection, including a fast MDA reaction, a very limited host genome recovery and a high sequence coverage depth of viral as compared with bacterial contigs. To the best of our knowledge, there are no prior reports of cyanophages infecting Proteobacteria, although phage cross-infections of Beta- and Gamma-proteobacteria provide some evidence for such possibility (Jensen et al., 1998). Interestingly, another Roseobacter SAG (AAA160-J18) from the same lineage also displayed viral contigs (66 442 bp over 4 contigs) similar to the Roseobacter phage AAA076-A06 and other cyanophages (Figure 6). These results provide additional evidence that cyanophage-like viruses infect cells from the Roseobacter lineage. To test the hypothesis that a cyanophage may infect a Roseobacter, we performed a plaque assay using a mixture of 374 cyanophage isolates on 34 Roseobacter strains, but no plaques were observed. Although these results failed to provide experimental evidence for cross-infections of cyanobacteria and Roseobacter, they do not prove the absence of such events in the environment, where the genetic diversity of phages and hosts is much greater than what could be captured in our laboratory experiment. Experimental studies of phage host range are very tedious and few. Arguably, the most extensive study was performed by Moebus and Nattkemper (1981), where 286 bacterial hosts and 215 phages from multiple sites in the Atlantic Ocean were tested. That study demonstrated a wide spectrum of host range and modularity in phage–host specificity (Flores et al., 2011), although none of the hosts were identified. It is generally assumed that the true host range of most environmental phages remains unknown (Breitbart, 2012), and SCG has the potential to directly bridge this knowledge gap.
Three SAG-associated viruses had a weak homology to other, previously sequenced phages and prophages, with predominant blast hits to uncharacterized Siphoviridae. We tentatively assigned these phages to the Siphoviridae family, a large family of phages with a long non-contractile tail. Siphoviridae phages evolve rapidly by horizontal gene transfer and genome reorganization (Hatfull et al., 2008; Ignacio-Espinoza and Sullivan, 2012). Consequently, they have very mosaic genomes. Very few marine Siphoviridae phages are cultured and their genomic content greatly differs from each other (Huang et al., 2012), which is consistent with our findings. The evidence for these sequences belonging to viruses includes similarity of most genes to phage genes (Supplementary Table 5) and high recruitment of viral metagenomes, as compared with bacterial metagenomes (Figure 5). This observation further reinforces the diversity of marine viral communities, including the presence of many enigmatic lineages that can be illuminated using SCG.
Viral contigs in the Verrucomicrobia SAG AAA164-N20 were most similar to Bathycoccus sp. RCC1105 virus BpV1 (Supplementary Figure 3), with overall 88.4% average nucleotide identity. Virus BpV1 belongs to the Phycodnaviridae, a family of large double-stranded DNA viruses (160-560 kb) known to infect marine and freshwater eukaryotic algae. A total of 62 kb of the viral genome were recovered from the SAG, corresponding to 31.2% of the 198 519-bp-long BpV1 genome. The recovery of the bacterial genome was not anomalous, in relation to the speed of the MDA reaction (Figure 2), providing no support for a lytic infection. We are not aware of prior reports of eukaryote viruses infecting bacteria and therefore view these results with caution. Bathycoccus and other members of the Prasinophyceae class, such as Micromonas and Ostreococcus, are abundant in the Gulf of Maine, and the presence of viruses infecting these phytoplankton groups in the analyzed sample is likely. We therefore speculate that this finding is a result of a non-specific attachment of a viral particle on the bacterial cell.
Conclusions
A combination of comparative genomics, metagenomic fragment recruitment, sequence anomalies and irregularities in sequence coverage depth and genome recovery identified members of all three-tailed phage families in 20 of 58 surface ocean SAGs of phylogenetically diverse Bacteria and Archaea. At least five phage–host interactions had the characteristics of late lytic infections, all of which were found in cells of high metabolic activity. One virus had the genetic potential for lysogeny. Our findings include first known viruses of Thaumarchaeota, Marinimicrobia, Verrucomicrobia and Gammaproteobacteria clusters SAR86 and SAR92, presenting SCG as a valuable tool to link uncultured phages and hosts. Viruses were also found in SAGs of Alphaproteobacteria and Bacteroidetes. Near-complete genomes of multiple, novel viruses were recovered, along with the genomic sequences of uncultured microbial cells with which each of the viruses were associated. Fragment recruitment of viral metagenomic reads confirmed that most of the SAG-associated viruses are abundant in the ocean. These results suggest that continued technological developments in microbial SCG, such as improved bioinformatics tools, scaled-up SAG sequencing and single-cell gene expression analyses have the potential to revolutionize studies of viral–cell interactions in complex microbial communities.
Acknowledgments
We thank Chris Harris, Ben Tupper and Joe Brown for their assistance in computational tool development, as well as Joaquin Martinez-Martinez and Willie Wilson for valuable comments. This work was supported by the National Science Foundation grants OCE-1148017 (to RS and KEW), OCE-1136488 (to RS) and OCE-1232982 (to RS and BKS), the United States Department of Energy Joint Genome Institute (DOE JGI) Community Science Program grant 2011-387 (to RS and BKS) and the Natural Sciences and Engineering Research Council (NSERC) of Canada, Canada Foundation for Innovation (CFI), and the Canadian Institute for Advanced Research (CIFAR; SJH). The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231. The contributions of SK were funded under Agreement No. HSHQDC-07-C-00020 awarded by the Department of Homeland Security Science and Technology Directorate (DHS/S&T) for the management and operation of the National Biodefense Analysis and Countermeasures Center (NBACC), a Federally Funded Research and Development Center. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Homeland Security. In no event shall the DHS, NBACC or Battelle National Biodefense Institute (BNBI) have any responsibility or liability for any use, misuse, inability to use or reliance upon the information contained herein. The Department of Homeland Security does not endorse any products or commercial services mentioned in this publication.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akhter S, Aziz RK, Edwards RA. (2012). PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 40: e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers E, Moraru C, Duhaime MB, Beneze E, Solonenko N, Barrero-Canosa J et al. (2013). Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ Microbiol 15: 2306–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson RE, Brazelton WJ, Baross JA. (2011). Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol Ecol 77: 120–133. [DOI] [PubMed] [Google Scholar]
- Berg Miller ME, Yeoman CJ, Chia N, Tringe SG, Angly FE, Edwards RA et al. (2012). Phage-bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ Microbiol 14: 207–227. [DOI] [PubMed] [Google Scholar]
- Breitbart M. (2012). Marine viruses: truth or dare. Ann Rev Mar Sci 4: 425–448. [DOI] [PubMed] [Google Scholar]
- Brüssow H, Hendrix RW. (2002). Phage genomics: small is beautiful. Cell 108: 13–16. [DOI] [PubMed] [Google Scholar]
- Campbell A. (2003). Prophage insertion sites. Res Microbiol 154: 277–282. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H. (2003). Prophage genomics. Microbiol Mol Biol Rev 67: 238–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Gregory A, Yilmaz S, Poulos B, Hugenholtz P, Sullivan MB. (2012). Contrasting life strategies of viruses that infect photo-and heterotrophic bacteria, as revealed by viral tagging. MBio 3: e00373–00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P et al. (2014). Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513: 242–245. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C, Brussaard CPD. (2012). Regional variation in lytic and lysogenic viral infection in the Southern Ocean and its contribution to biogeochemical cycling. Appl Environ Microbiol 78: 6741–6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée J, Tétart F, Suttle CA, Krisch HM. (2005). Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc Natl Acad Sci USA 102: 12471–12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. (2011). Statistical structure of host-phage interactions. Proc Natl Acad Sci USA 108: E288–E297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE. (2006). Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34: 5839–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Suttle CA. (1993). Viruses in marine planktonic systems. Oceanography 6: 51–63. [Google Scholar]
- Grigoriev A. (1998). Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res 26: 2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Bayindirli C, Bergauer K, Carpintero de Moraes P, Chen H, D'Ambrosio L et al. (2011). Draft genome sequence of strain HIMB100, a cultured representative of the SAR116 clade of marine Alphaproteobacteria. Stand Genomic Sci 5: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hatfull GFG, Cresawn S, Hendrix R. (2008). Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res Microbiol 159: 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt K, Howard-Varona C, Solonenko N, Sullivan MB. (2014). Contrasting genomic patterns and infection strategies of two co-existing Bacteroidetes podovirus genera. Environ Microbiol 16: 2501–2513; http://www.ncbi.nlm.nih.gov/pubmed/24428166 (accessed 8 September 2014. [DOI] [PubMed] [Google Scholar]
- Holmfeldt K, Odić D, Sullivan MB, Middelboe M, Riemann L. (2012). Cultivated single-stranded DNA phages that infect marine Bacteroidetes prove difficult to detect with DNA-binding stains. Appl Env Microb 78: 892–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt K, Solonenko N, Shah M, Corrier K, Riemann L, Verberkmoes NC et al. (2013). Twelve previously unknown phage genera are ubiquitous in global oceans. Proc Natl Acad Sci USA 110: 12798–12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang K, Jiao N, Chen F. (2012). Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage-host genetic exchanges. Environ Microbiol 14: 540–558. [DOI] [PubMed] [Google Scholar]
- Hurwitz BL, Brum JR, Sullivan MB. (2014). Depth-stratified functional and taxonomic niche specialization in the ‘core' and ‘flexible' Pacific Ocean Virome. ISME J 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz BL, Sullivan MB. (2013). The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One 8: e57355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz BL, Westveld AH, Brum JR, Sullivan MB. (2014). Modeling ecological drivers in marine viral communities using comparative metagenomics and network analyses. Proc Natl Acad Sci USA 111: 10714–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignacio-Espinoza JC, Sullivan MB. (2012). Phylogenomics of T4 cyanophages: lateral gene transfer in the ‘core' and origins of host genes. Env Microbiol 14: 2113–2136. [DOI] [PubMed] [Google Scholar]
- Jensen E, Schrader H, Rieland B, Thompson TL, Lee KW, Nickerson KW et al. (1998). Prevalence of Broad-Host-Range Lytic Bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl Environ Microbiol 64: 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover LF, Effler TC, Buchan A, Wilhelm SW, Weitz JS. (2014). The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat Rev Microbiol 12: 519–528. [DOI] [PubMed] [Google Scholar]
- Kang I, Oh H-M, Kang D, Cho J-C. (2013). Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc Natl Acad Sci USA 110: 12343–12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Campbell AM. (1998). Comparative DNA analysis. Annu Rev Genet 32: 185–225. [DOI] [PubMed] [Google Scholar]
- King A, Adams M, Carstens E, Lefkowitz E. (2012) Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses 2nd Ed. Elsevier Academic Press: San Diego, California. [Google Scholar]
- Labonté JM, Reid KE, Suttle CA. (2009). Phylogenetic analysis indicates evolutionary diversity and environmental segregation of marine podovirus DNA polymerase gene sequences. Appl Environ Microbiol 75: 3634–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS. (2012). Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol 10: 631–640. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. (2008). Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 24: 863–865. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia M, Brazel DM, Swan BK, Arnosti C, PSG Chain, Reitenga KG et al. (2012). Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One 7: e35314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel L, Breitbart M, Mobberley J, Long A, Haynes M, Rohwer F et al. (2008). Metagenomic analysis of lysogeny in Tampa Bay: implications for prophage gene expression. PLoS One 3: e3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel L, Houchin LA, Williamson SJ, Paul JH. (2002). Lysogeny in marine Synechococcus. Nature 415: 496. [DOI] [PubMed] [Google Scholar]
- Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R. (2013). Expanding the marine virosphere using metagenomics. PLoS Genet 9: e1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moebus K, Nattkemper H. (1981). Bacteriophage sensitivity patterns among bacteria isolated from marine waters. Helgolander Meeresuntersuchungen 34: 375–385. [Google Scholar]
- Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Computational Biol 20: 714–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H-M, Kwon KK, Kang I, Kang SG, Lee J-H, Kim S-J et al. (2010). Complete genome sequence of ‘Candidatus Puniceispirillum marinum' IMCC1322, a representative of the SAR116 clade in the Alphaproteobacteria. J Bacteriol 192: 3240–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partensky F, Hess WR, Vaulot D. (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol R 63: 106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul JH. (2008). Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J 2: 579–589. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Forterre P, Garrett RA. (2006). Viruses of the Archaea: a unifying view. Nat Rev Microbiol 4: 837–848. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan A Jr, Ferguson HR Jr, Bornarth CJ, Song W, Driscoll M, Lasken RS et al. (2005). Genomic DNA amplification from a single bacterium. Appl Environ Microbiol 71: 3342–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418: 630–633. [DOI] [PubMed] [Google Scholar]
- Rappé MS, Giovannoni SJ. (2003). The uncultured microbial majority. Annu Rev Microbiol 57: 369–394. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Hawley AK, Torres Beltran M, Scofield M, Schwientek P, Stepanauskas R et al. (2014). Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta- genomics. Elife 3: e03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassera D, Lo N, Epis S, D'Auria G, Montagna M, Comandatore F et al. (2011). Phylogenomic evidence for the presence of a flagellum and cbb(3) oxidase in the free-living mitochondrial ancestor. Mol Biol Evol 28: 3285–8296. [DOI] [PubMed] [Google Scholar]
- Schmidt HF, Sakowski EG, Williamson SJ, Polson SW, Wommack KE. (2014). Shotgun metagenomics indicates novel family A DNA polymerases predominate within marine virioplankton. ISME J 8: 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares SC, Abreu VAC, Ramos RTJ, Cerdeira L, Silva A, Baumbach J et al. (2012). PIPS: pathogenicity island prediction software. PLoS One 7: e30848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stepanauskas R. (2012). Single cell genomics: an individual look at microbes. Curr Opin Microbiol 15: 613–620. [DOI] [PubMed] [Google Scholar]
- Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. (2005). Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, Sarracino D et al. (2009). The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial ‘mobilome'. Environ Microbiol 11: 2935–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Waterbury JB, Chisholm SW. (2003). Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424: 1047–1051. [DOI] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27: 1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. (2007). Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5: 801–812. [DOI] [PubMed] [Google Scholar]
- Suttle CA. (2005). Viruses in the sea. Nature 437: 356–361. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, Gonzalez JM et al. (2013). Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA 110: 11463–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmor A, Ottesen E, Leadbetter J, Phillips R. (2011). Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science 333: 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577. [DOI] [PubMed] [Google Scholar]
- Tomaru Y, Nagasaki K. (2007). Flow cytometric detection and enumeration of DNA and RNA viruses infecting marine eukaryotic microalgae. J Oceanogr 63: 215–221. [Google Scholar]
- Vernikos GS, Parkhill J. (2006). Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22: 2196–2203. [DOI] [PubMed] [Google Scholar]
- Weinbauer MG. (2004). Ecology of prokaryotic viruses. FEMS Microbiol Rev 28: 127–181. [DOI] [PubMed] [Google Scholar]
- Williamson SJ, Houchin LA, Mcdaniel L, Paul JH. (2002). Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl Environ Microbiol 68: 4307–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol R 64: 69–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Xie G, Copeland A, González JM, Han C, Kiss H et al. (2009). Assembling the marine metagenome, one cell at a time. PLoS One 4: e5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JJ, Mewis K, Hanson NW, Konwar KM, Maas KR, Hallam SJ. (2014). Genomic properties of Marine Group A bacteria indicate a role in the marine sulfur cycle. ISME J 8: 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH et al. (2011). Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332: 714–717. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW et al. (2006). Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol 24: 680–686. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC et al. (2013). Abundant SAR11 viruses in the ocean. Nature 494: 357–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Viral contig sequences, phylogenetic trees and alignments are available in Supplementary Materials of this publication. SAG genomic sequences are available on the D.O.E. Joint Genome Institutes IMG portal, http://img.jgi.doe.gov/.