Abstract
Soil is the key resource that must be managed to ensure sustainable forest productivity. Soil microbial communities mediate numerous essential ecosystem functions, and recent studies show that forest harvesting alters soil community composition. From a long-term soil productivity study site in a temperate coniferous forest in British Columbia, 21 forest soil shotgun metagenomes were generated, totaling 187 Gb. A method to analyze unassembled metagenome reads from the complex community was optimized and validated. The subsequent metagenome analysis revealed that, 12 years after forest harvesting, there were 16% and 8% reductions in relative abundances of biomass decomposition genes in the organic and mineral soil layers, respectively. Organic and mineral soil layers differed markedly in genetic potential for biomass degradation, with the organic layer having greater potential and being more strongly affected by harvesting. Gene families were disproportionately affected, and we identified 41 gene families consistently affected by harvesting, including families involved in lignin, cellulose, hemicellulose and pectin degradation. The results strongly suggest that harvesting profoundly altered below-ground cycling of carbon and other nutrients at this site, with potentially important consequences for forest regeneration. Thus, it is important to determine whether these changes foreshadow long-term changes in forest productivity or resilience and whether these changes are broadly characteristic of harvested forests.
Introduction
Forests are important ecosystems that provide cultural, economic and ecological benefits. Globally, forests contribute to almost half of the terrestrial net primary productivity and store 45% of terrestrial carbon (Suzuki et al., 2004). Thus, forests play a critical role in the global carbon cycle, and via fluxes of greenhouse gases, they strongly impact global climate (Foley, 2005; Bonan, 2008; Pan et al., 2011). Economic contributions of forests are also considerable. The forest sector contributes to ∼1% of the global gross domestic product, employs over 10 million people in management and conservation and is a direct source of income to over 1 billion people (FAO, 2012). For these reasons, forests must be responsibly and effectively managed.
Soil is the key resource that must be properly managed to ensure forest sustainability. The importance and complexity of soil management are compounded by global environmental challenges such as climate change, fresh water scarcity and biodiversity loss (Bouma et al., 2012). Soil microbial communities mediate key processes, such as nutrient cycling, transport of water and nutrients as well as maintenance of soil structure. Decomposition of biomass is an essential role of soil communities that enables cycling of carbon and other nutrients sequestered in biomass, making those nutrients available for primary productivity (Moorhead and Sinsabaugh, 2006). Enzymes involved in decomposition are additionally important for the conversion of biomass to commercial products, such as fuels, aromatic compounds and animal feed (Tuck et al., 2012).
Recent studies raise concern that harvesting forests over multiple rotations may deplete soil productivity via poorly understood mechanisms (Egnell, 2011; Thiffault et al., 2011). Several studies have shown immediate effects of harvesting on soil communities (Busse et al., 2006; Smith et al., 2007). However, few studies have examined long-term effects that might impact sustainable forest productivity, and those have shown small or inconsistent effects and lack of differentiation among harvesting regimes (Ponder et al., 2012). Because tree regeneration can take decades, long-term effects of harvesting are critical to evaluating the sustainability of forest management. Recent investigations of Long-Term Soil Productivity Study (LTSP) sites in British Columbia demonstrated that bacterial and fungal community composition remained significantly altered 10 and 15 years after forest harvesting and replanting (Hartmann et al., 2009, 2012). This raises the critical question of whether these changes in community taxonomic composition alter the capacity of the soil to deliver ecosystem functions. Such a change in capacity is not axiomatic, because of the potential for functional redundancy within microbial communities (that is, different taxonomic groups can have a common function). However, the capacity for biomass degradation is not universally distributed among microbial taxa, and lignin degradation in particular is believed to be restricted to a few fungal and bacterial groups (Hatakka, 2005). Thus, we hypothesized that long-term shifts in forest soil community composition because of harvesting alter the capacity of those communities for degradation of biomass, particularly lignocellulose. We further hypothesized that the degree of organic matter removal during harvesting would modulate the magnitude of the impact of harvesting on the capacity for biomass degradation.
To test the above hypotheses, we analyzed 21 shotgun soil metagenomes from one of the above LTSP sites in the Interior Douglas Fir biogeoclimatic zone of British Columbia, Canada. We compared harvesting treatments varying in severity of organic matter (OM) removal. Both the organic and mineral soil layers were investigated. In a focused analysis of the soil metagenomes, we evaluated the effects of harvesting on the occurrence of genes encoding carbohydrate and lignin decomposition.
Materials and methods
Study site
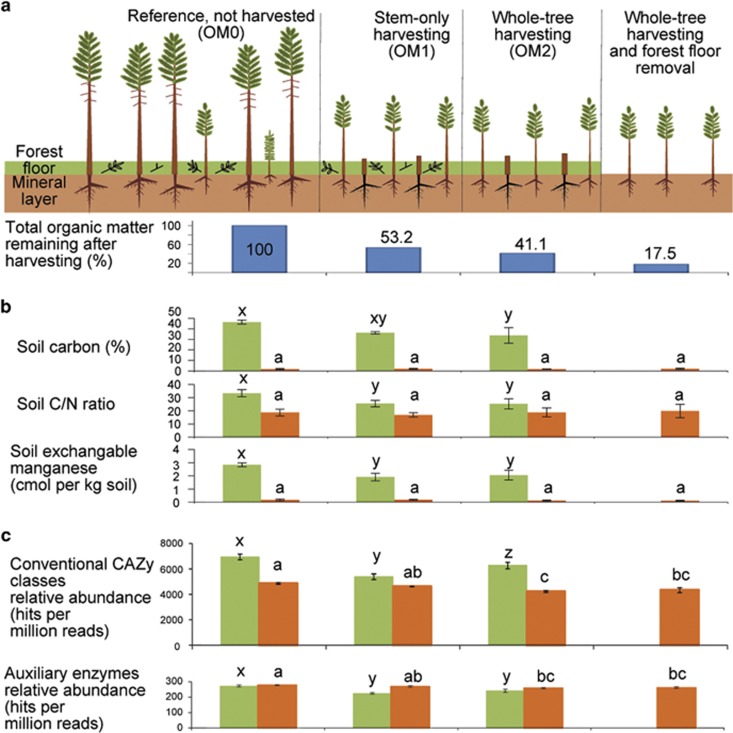
Soil samples were collected from the LTSP Study site at O'Connor Lake, British Columbia, Canada (Hope, 2006). This site is located in the Interior Douglas Fir biogeoclimatic zone characterized by warm, dry summers and cool winters. For this study, we focused on four tree harvesting treatments at this site that varied in levels of OM removal and had no soil compaction (Figure 1a). The treatments included a reference where no harvesting occurred (OM0), stem-only harvesting, leaving behind the crowns and branches (OM1), whole-tree harvesting (OM2) and whole-tree harvesting plus removal of the forest floor (organic soil layer; OM3). The maximum degree of total OM removal, occurring in OM3, was 83% (Figure 1a). Treatment plots were 40 m by 70 m. Harvesting treatments occurred in 2000, followed by replanting with Lodgepole pine (Pinus contorta) seedlings in 2001.
Figure 1.
Forest harvesting treatments, soil chemistry and abundance of CAZy genes. (a) Schematic of forest harvesting treatments that were followed by replanting with lodgepole pine. (b) Soil chemistry; green, organic horizon; orange, mineral horizon (n=3); see Supplementary Table S1 for additional data. (c) Relative abundance of total conventional and AA CAZy genes in soil metagenomes (n=3). Letters indicate significant differences based on analysis of variance (ANOVA) and Tukey's honestly significant difference (HSD) tests (P<0.05).
Soil sampling
Soils were sampled as previously described (Hartmann et al., 2012). Each sample consisted of a composite of three independent samples mixed in the field (as opposed to five in Hartmann et al., 2012), and there were three such composite samples per treatment plot. At each sample point, both the organic layer (forest floor) and top 20 cm of the mineral layer were collected separately. However, OM3 treatments lacked an organic layer.
Soil chemistry
Samples for chemical analysis were collected 11 years after the harvesting treatments on 26 and 27 May 2010. Chemical analyses were done using standard methods, as previously described (Hannam et al., 2008). Total C, N and S were determined on a combustion elemental analyzer. Exchangeable Ca, Mg, K, Na, Al, Mn and Fe were determined by extraction with 0.1 M BaCl2 and inductively coupled plasma spectrophotometric analysis. Mineralizable N was determined by anaerobic incubation; available P by Bray extraction; and pH was measured in 0.1 M CaCl2. All laboratory analyses were completed at the Ministry of Environment Laboratory in Victoria, BC, and results are shown in Supplementary Table S1. Although samples for chemical analysis were collected 1 year before those for DNA analysis (below), the chemistry was relatively stable, with no significant changes from 6 to 11 years after the harvesting treatments in average values in pH, total C, total N or available P for each site (Graeme Hope, unpublished data).
DNA extraction and metagenome generation
Samples for DNA analysis were collected 12 years after the harvesting treatments on 18 June 2011. Soil samples were homogenized by sieving (2 mm) to remove rocks and plant material. DNA was directly extracted from a subsample of the homogenized soil (0.5 g) using the FastDNA Spin kit for soil (MP Biomedicals, Solon, OH, USA). DNA concentration was measured by fluorescence using Picogreen (Life Technologies, Carlsbad, CA, USA) and 1.5 to 2 μg were submitted for shotgun sequencing at the Michael Smith Genome Sciences Centre, Vancouver, Canada. Sets of triplicate samples were barcoded and pooled on a single lane of an Illumina Hiseq 2000 instrument (Illumina, San Diego, CA, USA). Sequencing generated 75-base paired-end reads. Raw reads were filtered with the NGS QC toolkit v2.3 (Patel and Jain, 2012) using a cutoff PHRED quality value of 20 over 70% of the length. Paired ends were then joined when possible using FLASH v1.2.6 (Magoc and Salzberg, 2011). Raw and high-quality reads were assembled using SOAPdenovo (Luo et al., 2012) at a range of kmers (21, 23, 25, 27, 29 and 31). Default settings for all SOAPdenovo assemblies were used (flags: -d 1 and -R). Contigs generated by each assembly were combined by similarity using the SGA program (Simpson and Durbin, 2010) with default parameters. Data were deposited in the European Nucleotide Archive (Study accession PRJEB8420, sample accessions ERS656878 to ERS65689).
Gene identification
To study the diversity of genes involved in biomass degradation, we searched our high-quality unassembled reads against the Carbohydrate Active Enzyme Database (CAZy), as well as the recently added auxiliary redox enzymes (Levasseur et al., 2013), using BLASTX (Altschul et al., 1997) with an optimized E-value threshold of 1E−5. The database included the four conventional CAZy classes (Cantarel et al., 2009), glycoside hydrolase (GH), glycosyl transferase (GT), polysaccharide lyase (PL) and carbohydrate esterase (CE), as well as the recently added auxiliary activity (AA) redox enzymes (Levasseur et al., 2013). Both forward and reverse reads as well as joined paired ends were searched. Hits were assigned to CAZy gene families according to the classification of the closest reference sequence. For each gene family, richness was measured as the number of distinct genes matched in the database. Both abundance and richness were normalized to hits, or unique matches, per million reads to account for differences in metagenome size (and thus sequence coverage). A BLAST database was created by downloading information from the CAZy website from each protein family and then downloading all corresponding sequences from Genbank using custom scripts. The CAZy database was accessed on 13 May 2013 for the conventional CAZy classes and on 6 May 2014 for the AA class. As AA enzymes do not share a common mechanism and mainly target lignin, we analyzed them separately.
Data analysis
Differences in relative abundances among all treatments were measured by analysis of variance. The post hoc pairwise comparisons between treatments were done using Tukey's honestly significant difference test. Differences in richness between treatments were measured using t-tests on square-root transformed data. For the latter test, gene families with no data for at least 50% of the samples were removed from the analysis.
Similarity of samples based on profiles of gene family relative abundance was measured using the Bray–Curtis dissimilarity index. Gene family profiles were compared using principal coordinate analysis. Significant differences among a priori defined groups were tested using permutational multivariate analysis of variance (Anderson, 2001). Within-group multivariate dispersion (variance) was determined using PERMDISP (Anderson, 2006).
Predictive gene families that segregated significantly between soil layers or among harvesting treatments were identified using random forest analysis (Breiman, 2001) with 1000 trees followed by the Boruta algorithm for feature selection (Kursa and Rudnicki, 2010). Gene families were characterized using information from the CAZy website obtained on 15 August 2014 and previous publications on cellulases (Berlemont and Martiny, 2012) and hemicellulases (Shallom and Shoham, 2003; Zhao et al., 2013).
Soil chemistry was analyzed in each layer separately, and differences among treatments were measured with analysis of variance and Tukey's honestly significant difference tests.
For multivariate analysis, relative abundances were square-root transformed. Analyses were done with all samples when testing for soil layer effects, and on subsets of either organic or mineral soil layer samples when testing for harvesting treatment effects. A P-value of 0.05 was the criterion for statistical significance. Permutational multivariate analysis of variance and PERMDISP were done with 999 permutations using PRIMER6+ (Clarke and Gorley, 2006). Principal coordinate analysis was performed in the R platform v2.15 (R Core Team, 2012) using the package Vegan v2.0–10 (Oksanen et al., 2009). Random forest with Boruta feature selection was done in R using packages randomForest v4.6-7 and Boruta v3.0, respectively.
Results
Soil chemistry
At 11 years after harvesting, soil chemistry remained altered in the organic layer of harvested treatment plots (Figure 1 and Supplementary Table S1). As expected, the organic layer showed higher nutrient content for both macronutrients (C, N, Ca, S, K and Mg) and micronutrients (Mn and Fe). In addition, harvesting reduced C, C/N and Mn in the organic layer; and Ca and Al in the mineral layer. In all the cases, the nutrients were decreased in the harvested treatments versus the unharvested references, and no differences were found between the OM1 and OM2 treatments.
Analysis of unassembled reads
Sequencing and quality control produced 2 413 768 803 high-quality reads totaling 187.2 Gbases (average read length=77.5 bases). Combining triplicate metagenomes improved assembly, but it remained limited, with only 2.14% of the total reads forming contigs (Supplementary Table S2). Given the poor assembly of our metagenomes, we instead analyzed unassembled high-quality short reads to evaluate the carbohydrate and lignin degradation capacity of the communities. Before this analysis of the metagenomes, we optimized and validated the identification of CAZy genes among short, unassembled sequence reads, using simulated sequencing data sets. Simulated paired-end reads, including typical sequencing errors, were generated from four well-assembled and annotated genomes (two bacterial and two fungal). These reads were searched by BLASTX for matches to the CAZy Database. This analysis is further described in the Supplementary Text.
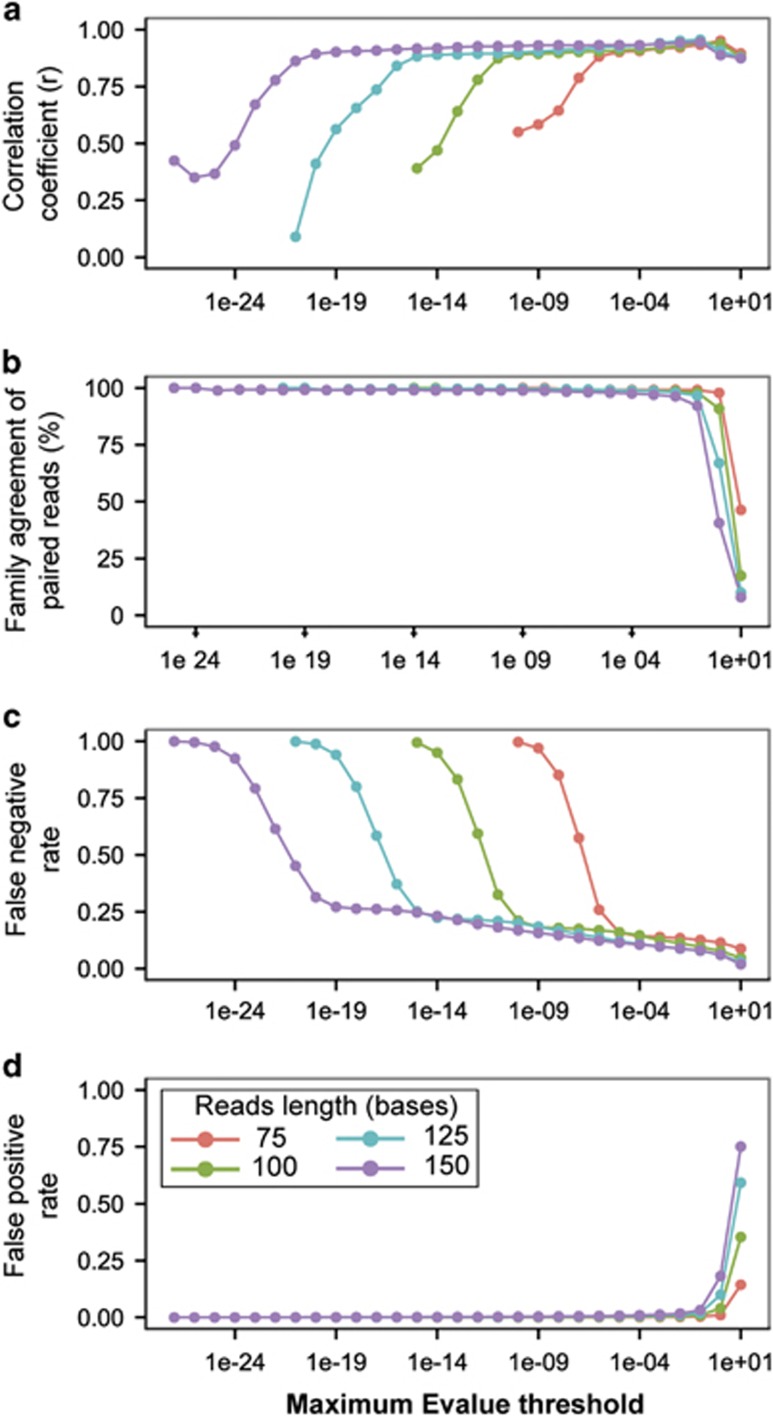
Results of analysis of the Rhodococcus jostii genome (Figure 2) were representative of the results with the three additional genomes (Supplementary Figures S1–S10) and indicated that, for 75-bp reads, an E-value of 10−5 was in the optimal range for maximizing the number of accurately identified hits. As a measure of accuracy of gene identification, we compared predicted and experimental gene family profiles, based on relative abundances of the CAZy families in the genomes. Predicted and experimental profiles agreed well within an E-value range of 10−2 to 10−5, with correlation coefficients almost always >0.90 (Figure 2a and Supplementary Figure S1). As an additional test of accuracy, we compared identification of individual reads in assembled paired ends that, in the majority of cases, should be from the same gene. For subject-level identification (best hit in the database), agreement between paired-end reads was variable (Supplementary Figure S2) and no E-value threshold yielded high agreement. In contrast, for family-level identification, paired ends agreed very strongly, nearly 100%, at E-values of 10−2 and lower (Figure 2b and Supplementary Figure S3). The number of hits (Supplementary Figure S4) was further constrained by false positives and false negatives. With E-values above 10−1, false positives dominated the results (Figure 2c and Supplementary Figure S5), whereas with E-values below 10−6, the false negative rate increased sharply, especially for the auxiliary activities class (Figure 2d and Supplementary Figure S6).
Figure 2.
Effects of read length and E-value threshold on accuracy of CAZy gene analysis using simulated, unassembled sequencing reads from the Rhodococcus jostii complete genome. (a) Correlation between the experimental and expected gene family relative abundance profiles. (b) Agreement between family level identification of paired-end reads. (c) False negative gene identification. (d) False positive gene identification.
Abundance and diversity of carbohydrate and lignin degradation genes
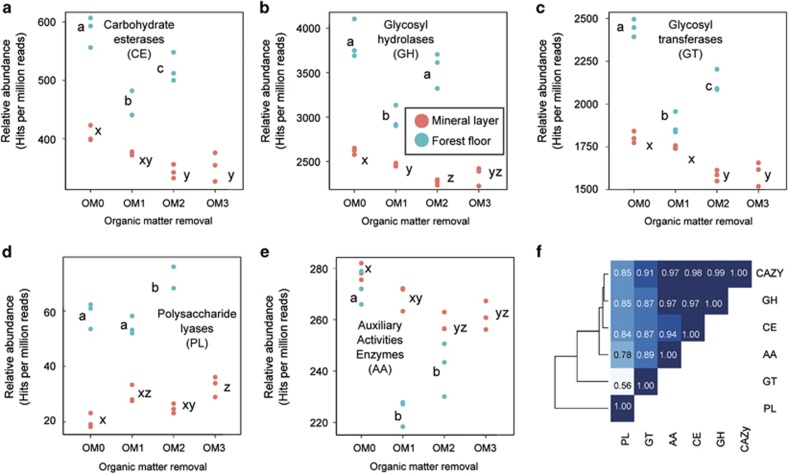
As an indicator of overall genetic potential for carbohydrate and lignin degradation, we examined the relative abundances of 231 CAZy gene families. The optimized BLASTX analysis of unassembled forest soil metagenomes yielded a total of 12 537 586 hits to the four conventional CAZy classes and 6 22 805 hits to the AA class. Relative abundances of CAZy genes were greater in the organic versus mineral soil layer (Figure 1c) with the exception of AA enzymes that had similar abundances in both layers. Harvesting generally reduced the relative abundances of total CAZy genes and CAZy classes (Figure 3). These reductions in relative abundances were much greater in the organic versus mineral layer. In the mineral layer, abundances of CAZy genes did not differ greatly among the three harvested treatments, but in the organic layer, they were consistently higher in the whole-tree harvesting (OM2) versus stem-only harvesting (OM1) treatment. This was true for all conventional CAZy classes, and although the difference was not significant, AA exhibited the same trend. Notably, these harvesting effects on gene abundances were evident 12 years after harvesting and replanting the experimental plots.
Figure 3.
Effects of harvesting treatments on CAZy gene classes. (a–e) Relative abundances of gene classes in the four harvested treatments. Letters indicate significant differences based on analysis of variance (ANOVA) and Tukey's honestly significant difference (HSD) tests (P<0.05). (f) Mantel correlations between classes of patterns of similarity among harvesting treatments, based on gene family relative abundance profiles. Hierarchical clustering indicates pairwise similarity of patterns among classes.
Generally, the presence of CAZy gene families was similar among the metagenomes. The number of CAZy families was slightly (~2%) higher in the organic versus mineral layer. The presence of families did not differ among harvested treatments, and the same gene families tended to dominate each CAZy class in a particular soil layer (Supplementary Table S3). In both layers, the dominant GH was GH13 and the dominant GT was GT2 that accounted for 17% and 10%, respectively, of all detected CAZy genes. In the organic layer, the dominant CE was CE1 and the dominant PL was PL1, whereas in the mineral layer, they were CE4 and PL4. Finally, the dominant AA was AA1 in all metagenomes, except the organic layer of the reference plot where it was AA3.
On the other hand, numbers of distinct genes within gene families (richness) varied substantially among metagenomes. Richness of CAZy families was higher in the organic versus mineral layer, on average 22% higher for the conventional classes and 18% for the AA gene families (heteroscedastic t-tests, both significant at P<0.001). Harvesting affected the richness of a small number of gene families, 26, all but one in the conventional CAZy classes (Supplementary Table S4). For those affected families, harvesting mainly increased richness in the mineral layer, whereas it mainly decreased the richness in the organic layer.
Gene family profiles
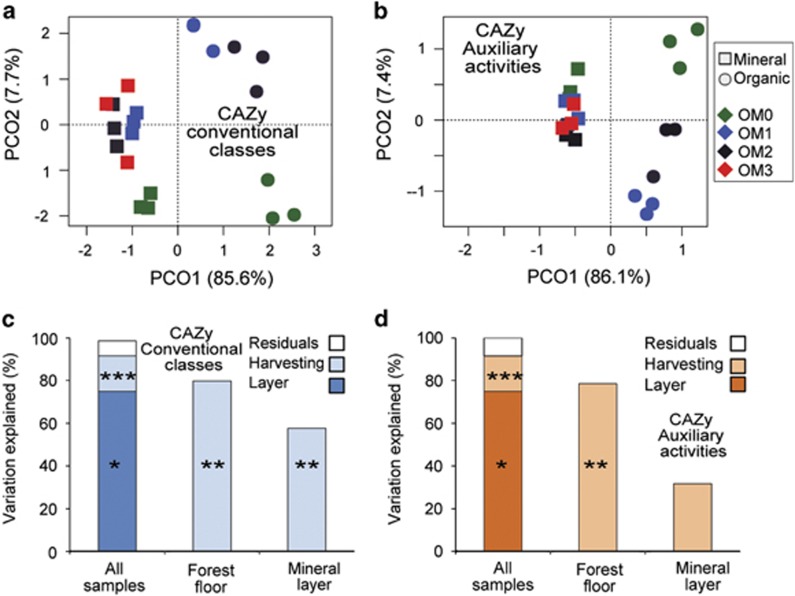
To evaluate the similarity of the metabolic potential of communities, we compared profiles based on relative abundances of CAZy gene families. Unconstrained ordinations revealed that gene family profiles clustered according to soil layer and, to a lesser extent, harvesting treatments (Figures 4a and b). Accordingly, profiles differed most significantly between soil layers (Figures 4c and d and Table 1). Within each layer, profiles generally differed significantly among harvesting treatments, with the greatest differences occurring in the organic layer. The patterns of profile similarity were congruent between the four conventional classes and AA, as the respective dissimilarity matrices were all highly correlated (average r=0.85) on the basis of Mantel tests (Figure 3f). The greatest correlation was among the GH, CE and AA classes.
Figure 4.
Variation of CAZy gene family profiles. Principal coordinates ordinations of gene family profiles of conventional CAZy classes (a) and the AA class (b). Percentages indicate the amount of that variability explained by the corresponding axis. Variation of gene family profiles for conventional CAZy classes (c) and the AA class (d) explained by soil layer and harvesting treatments, according to permutational multivariate analysis of variance (PERMANOVA) (*P<0.05, **P<0.01, ***P<0.001).
Table 1. Differences in gene family profiles of conventional and AA classes attributable to soil layer and harvesting treatments.
|
Conventional classes |
Auxiliary activities |
|||
|---|---|---|---|---|
| F or t | R2 | F or t | R2 | |
| All samples | ||||
| Layer | 21.982* | 0.76 | 22.51* | 0.75 |
| Harvesting treatment | 6.87*** | 0.17 | 5.54*** | 0.17 |
| Residuals | 0.07 | 0.08 | ||
| Mineral layer | ||||
| Harvesting treatment | 3.63** | 0.58 | 1.24 | NS |
| OM0 vs OM1 | 2.26* | |||
| OM0 vs OM2 | 2.86* | |||
| OM0 vs OM3 | 2.02* | |||
| OM1 vs OM2 | 2.23* | |||
| OM1 vs OM3 | 1.25 | |||
| OM2 vs OM3 | 0.91 | |||
| Organic layer | ||||
| Harvesting treatment | 11.85** | 0.80 | 10.98** | 0.79 |
| OM0 vs OM1 | 4.82* | 4.92** | ||
| OM0 vs OM2 | 2.22* | 2.21* | ||
| OM1 vs OM2 | 3.09* | 2.50* | ||
Abbreviations: AA, auxiliary activity; NS, not significant; OM, organic matter.
Permutational multivariate analysis of variance (PERMANOVA) based on Bray–Curtis distances. Multivariate dispersion was significantly different (P<0.05) when analyzing all samples together but nonsignificant when each soil layer was analyzed separately. Global test results yielded pseudo-F values, whereas pairwise comparisons yielded t-values (italicized). P-values are based on permutation for the general tests, and on Monte Carlo permutation for pairwise tests. *P<0.05; **P<0.01; ***P<0.001.
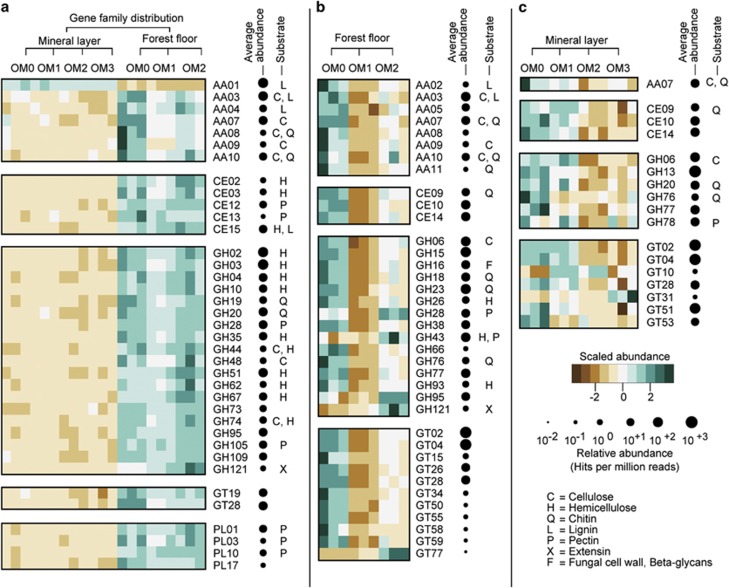
Trends in distributions of gene families were not obvious, and hence to discover those families most strongly associated with soil layers or harvesting treatments, a machine learning algorithm was used. Random forest analysis with Boruta feature selection identified predictive gene families that could best classify samples according to soil layer or treatment (Figure 5 and Supplementary Table S5). Classification error rates were low overall, and lower for layers than treatments (Table 2), consistent with the greater variability in gene family profiles explained by layer (Figure 4c). Many of these predictive families (41%) are associated with degradation of plant cell wall components, including lignin, cellulose, hemicellulose and pectin, whereas 10% are associated with fungal cell wall degradation (Figure 5). Those families predictive of layer were overwhelmingly enriched in the organic versus mineral layer, whereas the majority of those predictive of harvesting treatment were enriched in the reference. Interestingly, the OM1 harvesting treatment caused a greater reduction in the abundances of indicator CAZy families in the organic layer than did the OM2 treatment that removed more organic matter from the system.
Figure 5.
Distribution of gene families predictive of soil layers and harvesting treatments. Relative abundances of 37 CAZy gene families that were the strongest of 122 predictors of soil layers (a). Relative abundances of 44 CAZy gene families that are predictors of OM removal treatments in organic (b) and mineral (c) layers. Predictors were identified by random forest analysis with Boruta feature selection (average Z-scores of 1000 runs >4). Heatmaps show scaled relative abundances (normalized and centered) of gene families. Circle areas are proportional to the average relative abundance of each gene family in all samples, with the largest value being 909 hits per million reads for GH13. Substrate utilization is based on the CAZy database and literature as cited in Supplementary Table S5 that also includes the complete list of predictive families.
Table 2. Classification error for random forest analysis.
| Factor tested |
Classification error (%) |
|
|---|---|---|
| Conventional classes | Auxiliary activities | |
| Layer | 0 (0) | 0 (0) |
| Organic matter (OM) removal | ||
| Within mineral layer | 8.3 (45.0) | 58.3 (80.3) |
| Within organic layer | 0 (14.4) | 0 (12.2) |
Shown are error rates for reduced models that included only variables deemed as significant predictor by Boruta feature selection. Errors for classification using all variables are shown in parenthesis.
Discussion
Biodegradation potential differs greatly between soil horizons
Our results collectively indicate major differences between organic and mineral soil layers with respect to carbohydrate and lignin degradation potential. Such differences are not unexpected, but they reveal how specific gene families are distributed as part of the functional organization of the soil profile. The higher abundance of CAZy genes and greater richness of CAZy gene families in the organic versus mineral layer (Figure 1c), as well as the strong association of predictive gene families with the organic layer (Figure 4a), are all consistent with more biomass decomposition occurring in the organic versus mineral layer. This is further consistent with the greater impact of harvesting and varied levels of OM removal on biomass-degrading populations in the organic layer, as evidenced by decreases in abundances and richness of CAZy genes. Soil, especially forest soil, is described as a physically and biologically structured environment in which several key properties, such as organic carbon, nitrogen and microbial biomass, decrease with depth (Paul and Clark, 1996). Accordingly, we also observed very different chemical conditions in the organic versus mineral soil layers, with many nutrient concentrations >10-fold higher in the organic layer (Figure 1b and Supplementary Table S1). The similar abundance of AA genes in both layers is unexpected, because these genes also support biomass decomposition, particularly degradation of lignin and humic matter, that are prevalent in the organic layer of forest soils (Paul and Clark, 1996). However, more detailed analysis showed that the dominant AA family, AA1, a group that includes laccases and ferroxidases, was more abundant in the mineral layer than in the organic one. These results agree with studies on enzymatic activities in forest soil that have shown that in contrast to enzymatic activities that target labile carbon compounds, activities that target lignin such as phenol oxidase and peroxidase increase with depth (Brockett et al., 2012; Daradick, 2007) probably because of leaching of phenolic compounds from the organic layer and their preferential sorption to mineral surfaces (Kaiser et al., 2001).
Other studies have also shown the compartmentalization of microbial diversity in soil layers on the basis of culture-dependent studies (Axelrood et al., 2002), and more recently in molecular studies showing differences across forest soil profiles in terms of bacterial and fungal community composition (Hartmann et al., 2009; Baldrian et al., 2012), community gene expression (Baldrian et al., 2012) and carbohydrate utilization potential (Uroz et al., 2013). The latter study also showed differential distribution of CAZy families between organic and mineral layers of a spruce forest soil and identified 25 CAZy families as indicators of soil layer (Uroz et al., 2013). Notably, 21 of these indicators were among the families that we found predictive of layer (Supplementary Table S5). Thus, this study indicates that degradative functions of these families are similarly distributed among different forest types and that distributions of gene families with key functions may be more universal than distributions of microbial taxa.
Forest harvesting has long-term effects on biodegradation potential
This study supports the hypothesis that long-term shifts in forest soil microbial community composition because of harvesting also alter the capacity of communities for degradation of biomass. Specifically, harvesting appears to reduce the capacity for carbohydrate and lignin degradation, based on reductions in the relative abundance of total CAZy genes in the metagenome (Figure 1c). Distributions of gene families predictive of harvesting treatments (Figures 5b and c) indicate that harvesting specifically diminishes the genetic potential for hemicellulose, cellulose, lignin and pectin degradation. Differences in gene family profiles (Figure 4) further indicate that harvesting not only reduced degradative populations but also shifted their proportions relative to one another. Collectively, these impacts suggest substantial alteration of carbon cycling in the soil, particularly the organic layer, for many years following forest harvesting. Litter bag decomposition experiments have yielded mixed results (Prescott, 2005), with some showing reduced decomposition rates and enzymatic activities because of forest harvesting (Prescott et al., 2000; Waldrop et al., 2003). However, litter bag experiments have not shown such long-term effects as the present study that is the first to provide evidence that, for at least 12 years after harvesting a forest, the genetic potential for biomass decomposition was diminished in the soil community.
As we evaluated functional gene families, their richness can be used as a measure of functional redundancy within a community. It was previously hypothesized that the high microbial diversity in soils provides them with functional stability (Loreau, 2000), but that the level of redundancy depends strongly on the specificity of the function (Chaer et al., 2009). Thus, reductions in the functional redundancy of particular CAZy families that we observed may indicate reduced stability of the corresponding metabolic processes to further disturbances.
Differential effects of stem-only (OM1) versus whole-tree (OM2) harvesting in the organic layer were unexpected. In OM1, retention of crowns and branches minimized removal of OM, yet this treatment had a greater impact than OM2 on the genetic potential for biomass degradation. This impact of OM1 was evident as reduced relative abundances of CAZy genes, greater alteration of gene family profiles and reduced relative abundances of predictive gene families (Figures 1,3,4 and 5). One possibility is that the residues on the ground in OM1 delayed the establishment of grasses and shrubs between the growing planted trees, thereby limiting primary production and consequent organic inputs to the soil from living plants. This is consistent with recent reports (Koranda et al., 2011; Clemmensen et al., 2013) that have revealed the importance of root exudates to soil microbial metabolism.
The PL class was exceptional in that its relative abundance was increased by harvesting (Figure 3d). This increase was mainly because of increases in PL1, PL4 and PL11 that all catalyze pectin degradation. Within the PLs, we found changes in the dominant rhamnogalacturan lyase, the enzyme responsible for attacking the branched regions of pectin (Garron and Cygler, 2010). In all but the most extreme harvesting treatment, the dominant rhamnogalacturan lyases were predominantly of bacterial origin (PL11), but in OM3, the dominant rhamnogalacturan lyases were from a family dominated by fungal sequences (PL4). Although PLs are ubiquitous among all trophic groups, analysis of over 1300 sequenced genomes revealed that PLs are most abundant in phytopatogens and saprophytes (Lombard et al., 2010). Thus, harvesting may have selected for one or both of these trophic groups.
As expansion of specific CAZy families has been associated with ecological differentiation among fungal groups (Eastwood et al., 2011; Floudas et al., 2012) we tracked the distribution of such signature CAZy families. Five CAZy families associated with white rot fungi showed abundance reduction in the organic layer because of harvesting, including type II peroxidases (AA2), cellobiose dehydrogenase (AA3) and lytic polysaccharide monooxigenases (AA9, AA10, AA11). In contrast, GH28 whose expansion is associated with brown rot fungi and ectomycorhizae increased with harvesting in the organic layer. Nevertheless, the dominant AA group, which includes laccases and peroxidases, was not changed by harvesting.
This study suggests that soil microbial communities are more sensitive than above-ground biomass to harvesting and might be responsive indicators of disturbance and functionality of soil systems. Whereas a meta-analysis of 45 LTSP sites found no consistent effect of harvesting on tree biomass (Ponder et al., 2012), a meta-analysis of 139 reports of soil microbial responses to forest disturbance found that harvesting significantly affects microbial biomass, substrate-induced respiration and microbial community structure (Holden and Treseder, 2013). The present study is important in this context, because it demonstrates long-term effects of harvesting (>10 years) and shows that these effects extend to genes critical for carbon cycling. Only four previous studies have shown long-term effects of harvesting on soil communities, and these studies examined only biomass (Tan et al., 2008) or taxonomic composition of communities (Hassett and Zak, 2005; Chatterjee et al., 2009; Hartmann et al., 2012). It remains to be determined whether the effects of harvesting on soil communities are predictive or causative of reductions in reforestation rates or tree productivity. If so, a question of practical importance is whether proposed modifications to harvesting practices (Kohm and Franklin, 1997; Rosenvald and Lõhmus, 2008) might preserve intact soil communities in a way that facilitates their recovery in harvested areas.
Edaphic factors
Through selective pressure on the soil community, soil properties presumably drive the observed differences among metagenomes in biomass degradation genes. At 11 years after harvesting, total carbon, the C/N ratio and manganese remained depleted in the organic layer of harvested treatments versus the reference (Figure 1b). Soil carbon reflects the amount of organic matter, whereas C/N reflects the bioavailability and nutrient content of organic matter, both key selective factors for heterotrophic communities (Paul and Clark, 1996; Moorhead and Sinsabaugh, 2006). Manganese is an important cofactor for oxidative enzymes, especially ones involved in lignin degradation. In other studies, manganese concentration was positively correlated with plant litter and humic acid decomposition rates and negatively correlated with the stable fraction of litter (Steffen et al., 2002; Berg et al., 2013). The greater effect of harvesting on edaphic factors in the organic versus mineral layer is consistent with greater effects of harvesting on the CAZy genes in the organic versus mineral layer (Figures 1c, 4b and c). However, in comparing the harvesting treatments, the soil chemistry data correlated poorly with the various measures of CAZy gene relative abundances (not shown). Thus, there was no direct relationship between the degree of organic matter removal during harvesting and the impact of harvesting on the genetic potential for biomass decomposition.
Broader implications
Soil metagenomic studies have often been limited to discovery-based approaches because of high production costs, poor representation of organisms and environments in public databases and the complexity of many microbial communities. Here we took a hypothesis-based approach by using a curated database, statistical replication, and a robust experimental design. Our approach permitted accurate identification of proteins, while avoiding PCR amplification bias and primer limitations. As sequence read lengths increase, this approach will allow more accurate gene identification, as predicted in our validation experiments.
Long-term microbial ecology studies are rare, but here we show for the first time that tree harvesting causes long-term changes in functional profiles of the soil microbial communities. Although we did not perform metagenome analysis on samples from replicate experimental sites within the interior Douglas-fir biogeoclimatic zone, a previous study of community composition via high-throughput amplicon sequencing did so (Hartmann et al., 2012). That study demonstrated consistent effects of forest harvesting treatments on soil community composition at triplicate sites, including the site sampled for the current study. Thus, the observed effects of OM removal on biomass degradation capacity observed in this study are very likely representative of effects across this biogeoclimatic zone. An important outstanding question is whether the effects of harvesting observed in this study are representative of other ecozones.
Acknowledgments
This study was supported by grants from Genome Canada and Genome BC as well as a postdoctoral fellowship from the Tula foundation. We thank Charles Howes and Kishori Kunwar for bioinformatics support, Pedro Dimitriu for discussion of statistical analyses, Patrick Chain for metagenome assembly and Melanie Scofield, Jean Mather, Hilary Leung and Matt Zustovic for sampling.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62: 245–253. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Aust J Ecol 26: 32–46. [Google Scholar]
- Axelrood PE, Chow ML, Arnold CS, Lu K, McDermott JM, Davies J. (2002). Cultivation-dependent characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can J Microbiol 48: 643–654. [DOI] [PubMed] [Google Scholar]
- Baldrian P, Kolarik M, Stursova M, Kopecky J, Valaskova V, Vetrovsky T. (2012). Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6: 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg B, Erhagen B, Johansson M-B, Vesterdal L, Faituri M, Sanborn P et al. (2013). Manganese dynamics in decomposing needle and leaf litter — a synthesis. Can J For Res 43: 1127–1136. [Google Scholar]
- Berlemont R, Martiny AC. (2012). Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol 79: 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonan GB. (2008). Forests and climate change: forcings, feedbacks, and the climate benefits of forests. Science 320: 1444–1449. [DOI] [PubMed] [Google Scholar]
- Bouma J, Broll G, Crane TA, Dewitte O, Gardi C, Schulte RP et al. (2012). Soil information in support of policy making and awareness raising. Curr Opin Environ Sustain 4: 552–558. [Google Scholar]
- Breiman L. (2001). Random forests. Mach Learn 45: 5–32. [Google Scholar]
- Brockett BFT, Prescott CE, Grayston SJ. (2012). Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol Biochem 44: 9–20. [Google Scholar]
- Busse MD, Beattie SE, Powers RF, Sanchez FG, Tiarks AE. (2006). Microbial community responses in forest mineral soil to compaction, organic matter removal, and vegetation control. Can J For Res 36: 577–588. [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaer G, Fernandes M, Myrold D, Bottomley P. (2009). Comparative resistance and resilience of soil microbial communities and enzyme activities in adjacent native forest and agricultural soils. Microb Ecol 58: 414–424. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Ingram LJ, Vance GF, Stahl PD. (2009). Soil processes and microbial community structures in 45- and 135-year-old lodgepole pine stands. Can J For Res 39: 2263–2271. [Google Scholar]
- Clarke KR, Gorley RN. (2006), PRIMER v6: User Manual/Tutorial. PRIMER-E, Plymouth.
- Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H et al. (2013). Roots and associated fungi drive long-term carbon sequestration in Boreal Forest. Science 339: 1615–1618. [DOI] [PubMed] [Google Scholar]
- Daradick SP. (2007) Soil Microbial Enzyme Activity and Nutrient Availability in Response to Green Tree Retention Harvesting in Coastal British Columbia. The University of British Columbia: Vancouver, British Columbia: Canada. [Google Scholar]
- Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A et al. (2011). The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333: 762–765. [DOI] [PubMed] [Google Scholar]
- Egnell G. (2011). Is the productivity decline in Norway spruce following whole-tree harvesting in the final felling in boreal Sweden permanent or temporary? For Ecol Manag 261: 148–153. [Google Scholar]
- FAO. (2012) State of the World's Forests, 2012. Food and Agriculture Organization of the United Nations: Rome. [Google Scholar]
- Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B et al. (2012). The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336: 1715–1719. [DOI] [PubMed] [Google Scholar]
- Foley JA. (2005). Global consequences of land use. Science 309: 570–574. [DOI] [PubMed] [Google Scholar]
- Garron M-L, Cygler M. (2010). Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20: 1547–1573. [DOI] [PubMed] [Google Scholar]
- Hannam K, Bersh S, Curran M, Chapman W, Dubé S, Hope G et al. (2008). The effects of soil compaction and organic matter retention on long-term soil productivity in British Columbia. BC Ministry of Forests and Range, 1–30.
- Hartmann M, Howes CG, VanInsberghe D, Yu H, Bachar D, Christen R et al. (2012). Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J 6: 2199–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Lee S, Hallam SJ, Mohn WW. (2009). Bacterial, archaeal and eukaryal community structures throughout soil horizons of harvested and naturally disturbed forest stands. Environ Microbiol 11: 3045–3062. [DOI] [PubMed] [Google Scholar]
- Hassett JE, Zak DR. (2005). Aspen harvest intensity decreases microbial biomass, extracellular enzyme activity, and soil nitrogen cycling. Soil Sci Soc Am J 69: 227. [Google Scholar]
- Hatakka A. (2005). Biodegradation of lignin. In: Steinbüchel A, Steinbüchel A, Hofrichter M (eds) Biopolymers Online. Wiley-VCH Verlag GmbH & Co KGaA: Weinheim, Germany. Available from http://doi.wiley.com/10.1002/3527600035.bpol1005 (Accessed 10 September 2013). [Google Scholar]
- Holden SR, Treseder KK. (2013). A meta-analysis of soil microbial biomass responses to forest disturbances. Front Microbiol 4 doi:doi:10.3389/fmicb.2013.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope GD. (2006). Establishment of long-term soil productivity studies on acidic soils in the interior douglas-fir zone. LTSP Res Note 4: 1–4. [Google Scholar]
- Kaiser K, Guggenberger G, Zech W. (2001). Isotopic fractionation of dissolved organic carbon in shallow forest soils as affected by sorption. Eur J Soil Sci 52: 585–597. [Google Scholar]
- Kohm KA, Franklin JF. (1997). Creating a Forestry for the 21st Century: The Science of Ecosystem Management. Island Press: Washington, DC. [Google Scholar]
- Koranda M, Schnecker J, Kaiser C, Fuchslueger L, Kitzler B, Stange CF et al. (2011). Microbial processes and community composition in the rhizosphere of European beech – the influence of plant C exudates. Soil Biol Biochem 43: 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursa MB, Rudnicki WR. (2010). Feature selection with the Boruta package. J Stat Softw 36: 1–13. [Google Scholar]
- Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. (2013). Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J 432: 437–444. [DOI] [PubMed] [Google Scholar]
- Loreau M. (2000). Biodiversity and ecosystem functioning: recent theoretical advances. Oikos 91: 3–17. [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead DL, Sinsabaugh RL. (2006). A theoretical model of litter decay and microbial interaction. Ecol Monogr 76: 151–174. [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P et al. (2009). vegan: Community Ecology Package. R package version 1: 15–3. Available from http://vegan.r-forge.r-project.org/. [Google Scholar]
- Pan Y, Birdsey RA, Fang J, Houghton R, Kauppi PE, Kurz WA et al. (2011). A large and persistent carbon sink in the world's forests. Science 333: 988–993. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M. (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul EA, Clark FE. (1996) Soil Microbiology and Biochemistry, 2nd edn. Academic Press: San Diego. [Google Scholar]
- Ponder Jr F, Fleming RL, Berch S, Busse MD, Elioff JD, Hazlett PW et al. (2012). Effects of organic matter removal, soil compaction and vegetation control on 10th year biomass and foliar nutrition: LTSP continent-wide comparisons. For Ecol Manag 278: 35–54. [Google Scholar]
- Prescott CE. (2005). Do rates of litter decomposition tell us anything we really need to know? For Ecol Manag 220: 66–74. [Google Scholar]
- Prescott CE, Blevins LL, Staley CL. (2000). Effects of clear-cutting on decomposition rates of litter and forest floor in forests of British Columbia. Can J For Res 30: 1751–1757. [Google Scholar]
- R Core Team. (2012) R: A Language and Environment for Statistical Computing. Vienna: Austria. Available from http://www.R-project.org. [Google Scholar]
- Rosenvald R, Lõhmus A. (2008). For what, when, and where is green-tree retention better than clear-cutting? A review of the biodiversity aspects. For Ecol Manag 255: 1–15. [Google Scholar]
- Shallom D, Shoham Y. (2003). Microbial hemicellulases. Curr Opin Microbiol 6: 219–228. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Durbin R. (2010). Efficient construction of an assembly string graph using the FM-index. Bioinformatics 26: i367–i373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NR, Kishchuk BE, Mohn WW. (2007). Effects of wildfire and harvest disturbances on forest soil bacterial communities. Appl Environ Microbiol 74: 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KT, Hatakka A, Hofrichter M. (2002). Degradation of humic acids by the litter-decomposing basidiomycete Collybia dryophila. Appl Environ Microbiol 68: 3442–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Kelly SD, Kemner KM, Banfield JF. (2004). Enzymatic U(VI) reduction by Desulfosporosinus species. Radiochim Acta 92: 11–16. [Google Scholar]
- Tan X, Chang SX, Kabzems R. (2008). Soil compaction and forest floor removal reduced microbial biomass and enzyme activities in a boreal aspen forest soil. Biol Fert Soils 44: 471–479. [Google Scholar]
- Thiffault E, Hannam KD, Paré D, Titus BD, Hazlett PW, Maynard DG et al. (2011). Effects of forest biomass harvesting on soil productivity in boreal and temperate forests—a review. Environ Rev 19: 278–309. [Google Scholar]
- Tuck CO, Perez E, Horvath IT, Sheldon RA, Poliakoff M. (2012). Valorization of biomass: deriving more value from waste. Science 337: 695–699. [DOI] [PubMed] [Google Scholar]
- Uroz S, Ioannidis P, Lengelle J, Cébron A, Morin E, Buée M et al. (2013). Functional assays and metagenomic analyses reveals differences between the microbial communities inhabiting the soil horizons of a Norway spruce plantation. PloS One 8: e55929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrop MP, McColl JG, Powers RF. (2003). Effects of forest postharvest management practices on enzyme activities in decomposing litter. Soil Sci Soc Am J 67: 1250. [Google Scholar]
- Zhao Z, Liu H, Wang C, Xu J-R. (2013). Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.