Abstract
AIM: To refine the loss of heterozygosity on chromosome 5p15 and to identify the new tumor suppressor gene (s) in colorectal tumorigenesis.
METHODS: Sixteen polymorphic microsatellite markers were analyzed on chromosome 5 and another 6 markers were applied on chromosome 5p15 in 83 cases of colorectal and normal DNA by PCR. PCR products were electrophoresed on an ABI 377 DNA sequencer. Genescan 3.1 and Genotype 2.1 software were used for LOH scanning and analysis.
RESULTS: We observed 2 distinct regions of frequent allelic deletions on Chromosome 5, at D5S416 on 5p15 and D5S428-D5S410 on 5q. Another 6 polymorphric microsatellite markers were applied to 5p15 and the minimal region of frequent loss of heterozygosity was established on 5p15 spanning the D5S416 locus.
CONCLUSION: Through our detailed deletion mapping studies, we have found a critical and precise location of 5p deletions, 5p15.2-5p15.3, which must contain one or more unknown tumor suppressor gene (s) of colorectal cancer.
INTRODUCTION
Colorectal cancer is one of the most common malignant tumors threatening people’s health[1,2]. Its occurrence has been rising over the past 3 decades[3]. Now, colorectal cancer is the second cause of death in Western countries, and the fourth in China[4]. It is clear that improvement in its prognosis will not be achieved without a better understanding of its etiology and tumor biology. In recent years, the genetic basis of human tumors has been increasingly elucidated. A growing number of studies have shown that the molecular events controlling tumorigenesis involve abnormal cell growth promoted by activation of proto-oncogenes and/or inactivation of tumor suppressor genes[5,6]. Identification of novel tumor suppressor genes has been facilitated by loss of heterozygosity (LOH) studies that have guided the localization of minimally deleted regions on chromosomes[6,7]. In colorectal cancers, frequent allelic loss has been identified on chromosomes 5q (30%), 8p (40%), 17p (75%-80%), 18q (80%), and 22q (20%-30%)[8,9].
Previous allelotype analyses of chromosome 5 in colorectal cancer mainly focused on the long arm (5q), where a high frequency of allelic deletions of 5q21-22 was reported, implicating the APC/MCC genes[10-13]. However, alterations of the short arm of chromosome 5 have not been studied extensively in colorectal cancers. In this study, 16 polymorphic microsatellite markers were applied to chromosome 5. As a result of the high frequence of LOH on the loci of D5S416, another 6 microsatellite markers from chromosomal region 5p15, spanning the D5S416 were precisely localized.
MATERIALS AND METHODS
Materials
This study was based on 83 patients with colorectal cancer including 40 men and 43 women, who were treated at the Surgical Department in Shanghai First People’s Hospital, China, between 1998 and 1999. The patients’ age ranged from 31 to 84 years with a median of 66. All the patients were confirmed by pathology, and staged by Duke’s criteria. Eight, 21, 40, 14 cases were of Duke’s stages A, B, C and D, respectively. 23 cases were well differentiated, 39 moderately differentiated, 6 poorly differentiated adenocarcinoma, and 15 cases were mucinous adenocarcinoma. HNPCC patients were ruled out by Amsterdam criteria[14,15]. Each patient gave his were or her informed consent for the use of his or her tissues in this study.
Methods
DNA extraction The cancerous and adjacent normal tissues were freshly frozen within 30 min after being removed. These tissues were then cut into cubes of approximately 2 mm3 and immediately frozen in liquid nitrogen. DNA was extracted using standard methods with proteinase K digested and phenol/ chloroform purified.
Microsatellite markers and PCR Initially, 83 cases of colorectal cancer were analyzed by PCR using 16 microsatellite markers which were mapped to chromosome 5. DNA samples were analyzed as normal/tumor pairs using primers for the following microsatellite loci(location/heterozygosity): D5S1981-(5P15.3/0.73), D5S406-(5P15.3/0.78), D5S630-(5P15.3/0.89), D5S416(5P15.2/0.77), D5S419-(5P13.3/0.80), D5S418-(5P13.1/0.80), D5S407-(5q11.2/0.86), D5S647-(5q12.2/0.82), D5S424-(5q13.2/0.76), D5S428-(5q14.1/0.76), D5S2027-(5q21.1/0.78), D5S471-(5q23.1/0.75), D5S2115-(5q31.1/0.76), D5S410-(5q33.2/0.79), D5S400-(5q35.1/0.81), D5S408-(5q35.3/0.73). As a result of the high frequency of LOH in D5S416, 6 additional microsatellite markers from chromosomal region 5p15 were employed to span the D5S416 locus. DNA samples were also analyzed as normal/tumor pairs for the following microsatellite loci (location/heteozygosity): D5S1987-(5p15.3/0.87), D5S1991-(5p15.3/0.68), D5S1954- (5p15.3/0.69), D5S1963-(5p15.2/0.69), D5S2114-(5P15.2/0.87), D5S486-(5p15.1/0.74). Polymorphic microsatellite markers were analyzed by PCR (GeneAmp PCR System 9700, PE Applied Biosystems Fostercity CA, USA). PCR conditions were as follows: 5 µl total volume with approximately 1.4 ng of DNA as a template with 10 × standard buffer, 0.3 µl Mg2+, 0.8 µl deoxynucleotide triphosphates, 0.3 unit of Hot-start taq polymerase and 0.06 ml of each oligonucleotide primer, with the forward primer fluorescence labeled with HEX, FAM or NED. The “touch-down” was applied. Cycling conditions consisted of 3 stages: an initial denaturation at 96 °C for 12 min in stage I; 14 cycles each at 94 °C for 20 s, at 63-56 °C for 1 min (decrease 0.5 °C per cycle) and at 72 °C for 1 min in stage II, 35 cycles each at 94 °C for 20 s, at 56 °C for 1 min and at 72 °C for 1 min in stage III.
LOH analysis A portion of each PCR product (0.5 µl) was combined with 0.1 µl of Genescan 500 size standard (PE Applied Biosystems Fostercity CA, USA) and 0.9 ul of formamide loading buffer. After denaturation at 96 °C for 5 min, the products were electrophoresed on a 5% polyacrylamide gel on an ABI 377 DNA sequencer (PE Applied Biosystems Fostercity CA, USA) for 3 hours. Genotype 2.1 software displayed individual gel lanes as electrophoretograms with a given size, height, and area for each detected fluorescent peak. Stringent criteria were used to score the samples. Alleles were defined as the two highest peaks within the expected size range. A ratio of T1:T2/N1:N2 of less than 0.67 or greater than 1.50 was scored as a loss of heterozygosity (Figure 1). Most amplifications of normal DNA produced two PCR products indicating heterozygosity. A single fragment amplified from normal DNA (homozygote) and those PCR reactions in which fragments were not clearly amplified, were scored as not informative. The LOH frequency of a locus was equal to the ratio between allelic loss and informative cases.
Figure 1.
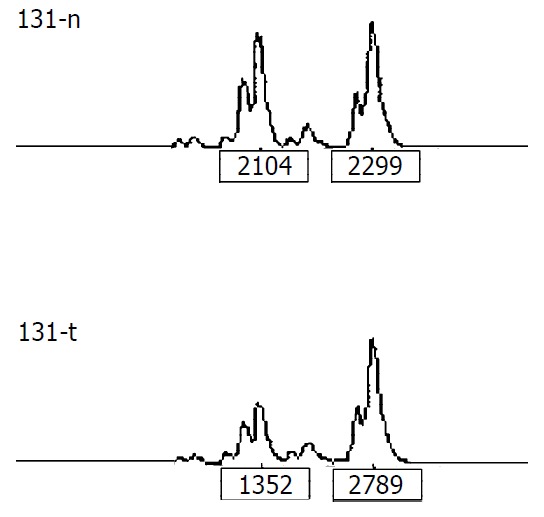
Representative LOH in tumor. “n” is normal DNA, "t" is tumor DNA. The peak heights are indicated below the corresponding alleles. Allele ratio = (t1:t2)/(n1:n2) = (1352:2789)/ (2104:2299) = 0.53. Allele ratio of less than 0.67 or greater than 1.50 was scored as LOH.
RESULTS
LOH analysis of colorectal cancers
Eighty-three colorectal cancers were analyzed for LOH at the 16 marker loci spanning chromosome 5 with the following most likely order: pter-D5S1981-D5S406-D5S630-D5S416-D5S419-D5S418-D5S407-D5S647-D5S424-D5S641-D5S428-D5S2027-D5S471-D5S2115-D5S410-D5S400-D5S408-qter[16] (Table 1 and Figure 2). We observed 2 distinct regions of frequent allelic deletions on chromosomes at D5S416 on 5p15 and D5S428-D5S410 on 5q.
Table 1.
The ratios of LOH of all loci on chromosome 5
| Loci | Map location | Number of informative cases (%) | LOH No (%) |
| D5S1981 | p15.3 | 24 (28.92) | 3 (12.50) |
| D5S406 | p15.3 | 26 (31.33) | 5 (19.23) |
| D5S630 | p15.3 | 65 (78.31) | 8 (12.31) |
| D5S416 | p15.2 | 54 (65.06) | 26 (48.15) |
| D5S419 | p13.3 | 63 (75.90) | 7 (11.11) |
| D5S418 | p13.1 | 66 (79.52) | 15 (22.73) |
| D5S407 | q11.2 | 51 (61.45) | 10 (19.61) |
| D5S647 | q12.2 | 74 (89.16) | 16 (21.62) |
| D5S424 | q13.2 | 36 (43.37) | 8 (22.22) |
| D5S428 | q14.1 | 61 (73.49) | 23 (37.70) |
| D5S2027 | q21.1 | 46 (55.42) | 15 (32.61) |
| D5S471 | q23.1 | 62 (74.70) | 24 (38.71) |
| D5S2115 | q31.1 | 59 (71.08) | 19 (32.20) |
| D5S410 | q33.2 | 18 (21.69) | 8 (44.44) |
| D5S400 | q35.1 | 28 (33.73) | 4 (14.39) |
| D5S408 | q35.3 | 61 (73.49) | 6 (9.84) |
Figure 2.
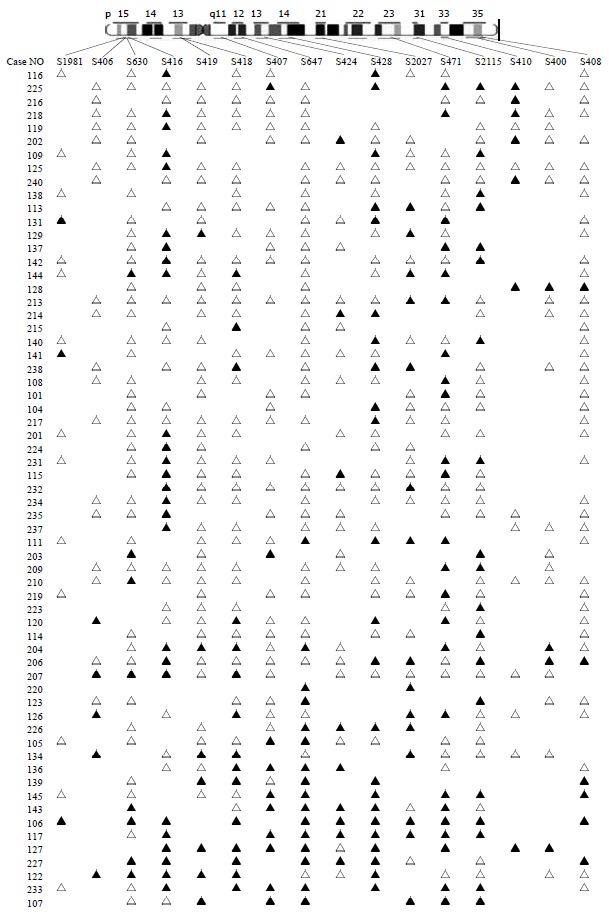
LOH map of chromosome 5 in colorectal cancer. ▲ LOH; △ retaining heterozygosity; neither ▲ or △ is non-informative. We can clearly figure out the two regions of the high rates of LOH on chromosome 5, one is D5S416 on 5p15, the other is D5S428-D5S410 on 5 q.
LOH mapping of 5p15 region
The chromosomal region spanning D5S416 locus on chromosome 5p was further investigated using a saturation mapping strategy with another 6 microstellite markers that are localized and closely spaced within this region (Figure 3). Eighty-three paired normal and tumor DNA samples were analyzed for LOH at these loci which further refined the resolution of the deletion map to a genomic segment of approximately 1 centimorgan (cM) and established the minimal region of frequent chromosomal loss at D5S416 locus. Based on the databases described previously, the most likely order of these markers was pter-D5S630-D5S1987-D5S1991-D5S1954-D5S1963-D5S416-D5S2114-D5S486[16] (Table 2). Overall, preferential deletions of D5S416 were observed in 26/54 colorectal cancers and deletions of the 2 markers, D5S1954 and D5S1963 (approximately 1 cM centromeric to D5S416), were seen in 33.3% (16/48) and 21.1% (12/57), respectively. Thus, the majority of observed interstitial deletions were localized within a 1 cM genomic segment encompassing the D5S416 locus.
Figure 3.
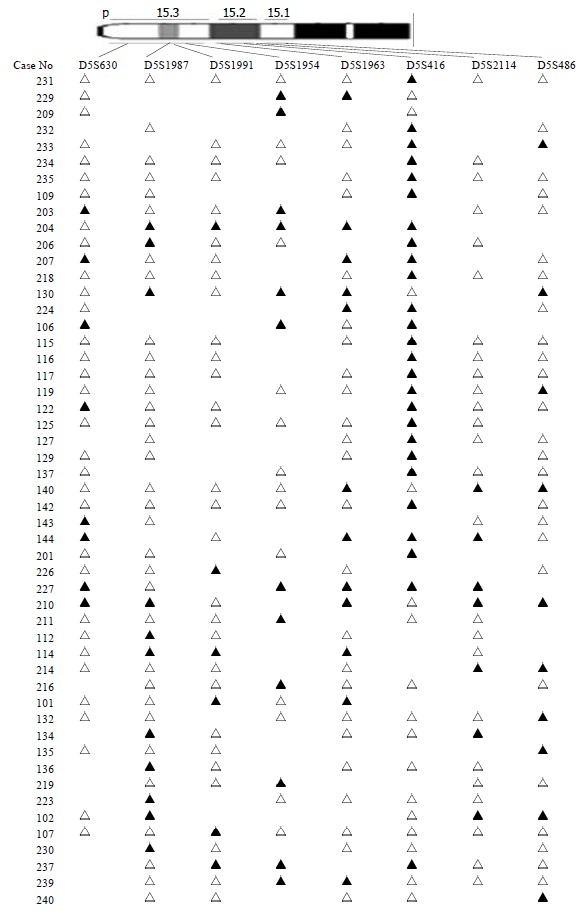
LOH map of 5p15 in colorectal cancer. ▲ LOH; △ retaining heterozygosity; neither ▲ or △ is non-informative.
Table 2.
The ratios of LOH of the loci on 5p15 region
| Loci | Map location | Space of two loci (cM) | Number of Informative cases (%) | LOH No (%) |
| D5S630 | p15.3 | 2.2 | 65 (78.31) | 8 (12.31) |
| D5S1987 | p15.3 | 4.7 | 67 (80.31) | 11 (16.42) |
| D5S1991 | p15.3 | 0.6 | 50 (60.24) | 6 (12.00) |
| D5S1954 | p15.3 | 0.1 | 48 (57.83) | 16 (33.33) |
| D5S1963 | p15.2 | 0.8 | 57 (68.67) | 12 (21.05) |
| D5S416 | p15.2 | 1.0 | 54 (65.06) | 26 (48.15) |
| D5S2114 | p15.2 | 2.2 | 53 (63.86) | 7 (13.21) |
| D5S486 | p15.1 | 60 (72.29) | 10 (16.67) |
DISCUSSION
Inactivation of tumor suppressor genes appears to be one of the genetic mechanisms involved in the development of colorectal cancer[17-19]. This process includes mutation of one allele, followed by a deletion of the remaining one (LOH) or homozygous deletion of both alleles[20]. Allelic deletions detected as LOH have been proved useful for mapping regions of DNA that contain tumor suppressor genes. LOH at specific chromosomal regions strongly suggests the existence of tumor suppressor genes at the relevant segment[21,22]. We performed deletion mapping analyses of chromosome 5 markers in 83 sporadic colorectal cancers, using 16 microsatellite markers. Analysis of the deletion map (Figure 2), together with the frequency of LOH obtained for each locus (Table 1) allowed us to identify two regions on chromosome 5 displaying high rates of LOH.
The 5q14-q22 region (D5S428-D5S410) has been frequently deleted in colorectal cancer. Ashton-Rickardt is the first author to describe the APC/MCC region as being frequently deleted in sporadic colorectal cancer[10]. Since then, several reports have indicated a high frequency of LOH at this region[11-13]. Our study also identified it, which proved our ways right.
The other region is 5p15 (D5S416). Further mapping of 5p15 region defined a minimal region of frequent deletion spanning the D5S416 locus (Figure 3). Based on this map, the majority of allelic deletions were localized within 1 cM chromosomal segment encompassing the 3 loci (D5S1954, D5S1963 and D5S416). Of these, D5S416 was the most frequently deleted locus (48.15%), while the other 2 markers, D5S1954 and D5S1963, demonstrated allelic losses of 33.33% and 21.05%, respectively. Thus, the high LOH frequency and the patterns of allelic losses of chromosome 5p15.2-5p15.3 suggest that this region may be preferentially deleted in colorectal cancer and could potentially harbor important tumor suppressor genes in colorectal development and progression. A database search has identified four candidate genes, PDCD6[23-27], TERT[28-29], T RIP13[30,31] and POLS[32-34] with suggestive tumor suppressor gene functions. The programmed cell death gene, PDCD6, is a member of the family of intracellular Ca2+-binding proteins and a part of the apoptotic machinery controlled by T-cell receptor (TCR) Fas, and glucocorticoid signals[23-27]. TERT encodes a reverse transcriptase required for the replication of chromosome termini and plays a role in telomere elongation. Although telomerase expression is a hallmark of cancer[35], the mice lacking the RNA component of telomerase (mTERC) exhibit progressive telomere shortening and chromosomal instability associated with epithelial tumors[28]. TRIP13, thyroid receptor interacting protein 13, is a transcription factor that regulates expression of a variety of specific target genes such as the human papilloma virus type16 E1(HPV16E1) which is important in cervical carcinoma[30,31]. POLS gene, a topoisomerase-related functional protein-4-1, encodes polymerase (DNA directed) signal within the deleted region. The yeast homolog TRF4 plays a critical role in chromosome segregation by coordinating between DNA replication and sister chromatid cohesion[32,33]. The data strongly support the importance of the POLS gene, which is critical for TRF4’s essential and repair functions. Cell cycle analysis has revealed that TRF4/POLS is associated with chromosomes in G1, S and G2 phases[34]. The presence of these genes at the 5p15.2-5p15.3 interval suggests that they may target tumor suppressor genes of these deletions in colorectal cancer. It will be helpful for us to find the tumor suppressor genes through studying their functions.
In summary, through our detailed deletion mapping studies, we have found a critical and precise location of 5p deletions, 5p15.2-5p15.3, which must contain one or more unknown tumor suppressor gene(s) of colorectal cancer.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30080016
Edited by Ma JY and Wang XL
References
- 1.Zhang ZS, Zhang YL. Progress in research of colorectal cancer in China. Shijie Huaren Xiaohua Zazhi. 2001;9:489–494. [Google Scholar]
- 2.Zhang YL, Zhang ZS, Wu BP, Zhou DY. Early diagnosis for colorectal cancer in China. World J Gastroenterol. 2002;8:21–25. doi: 10.3748/wjg.v8.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker N. [Epidemiology of colorectal cancer] Radiologe. 2003;43:98–104. doi: 10.1007/s00117-003-0868-9. [DOI] [PubMed] [Google Scholar]
- 4.Yu JP, Dong WG. Current situation about early diagnosis of can-cer of large intestine. Shijie Huaren Xiaohua Zazhi. 1999;7:553–554. [Google Scholar]
- 5.Lasko D, Cavenee W, Nordenskjöld M. Loss of constitutional heterozygosity in human cancer. Annu Rev Genet. 1991;25:281–314. doi: 10.1146/annurev.ge.25.120191.001433. [DOI] [PubMed] [Google Scholar]
- 6.Cliby W, Ritland S, Hartmann L, Dodson M, Halling KC, Keeney G, Podratz KC, Jenkins RB. Human epithelial ovarian cancer allelotype. Cancer Res. 1993;53:2393–2398. [PubMed] [Google Scholar]
- 7.Peng Z, Ling Y, Bai S. [Loss of heterozygosity on chromosome 3 in sporadic colorectal carcinoma] Zhonghua Yixue Zazhi. 2001;81:336–339. [PubMed] [Google Scholar]
- 8.Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R. Allelotype of colorectal carcinomas. Science. 1989;244:207–211. doi: 10.1126/science.2565047. [DOI] [PubMed] [Google Scholar]
- 9.Weber TK, Conroy J, Keitz B, Rodriguez-Bigas M, Petrelli NJ, Stoler DL, Anderson GR, Shows TB, Nowak NJ. Genome-wide allelotyping indicates increased loss of heterozygosity on 9p and 14q in early age of onset colorectal cancer. Cytogenet Cell Genet. 1999;86:142–147. doi: 10.1159/000015368. [DOI] [PubMed] [Google Scholar]
- 10.Ashton-Rickardt PG, Dunlop MG, Nakamura Y, Morris RG, Purdie CA, Steel CM, Evans HJ, Bird CC, Wyllie AH. High frequency of APC loss in sporadic colorectal carcinoma due to breaks clustered in 5q21-22. Oncogene. 1989;4:1169–1174. [PubMed] [Google Scholar]
- 11.Okamoto M, Sato C, Kohno Y, Mori T, Iwama T, Tonomura A, Miki Y, Utsunomiya J, Nakamura Y, White R. Molecular nature of chromosome 5q loss in colorectal tumors and desmoids from patients with familial adenomatous polyposis. Hum Genet. 1990;85:595–599. doi: 10.1007/BF00193581. [DOI] [PubMed] [Google Scholar]
- 12.Iacopetta B, DiGrandi S, Dix B, Haig C, Soong R, House A. Loss of heterozygosity of tumour suppressor gene loci in human colorectal carcinoma. Eur J Cancer. 1994;30A:664–670. doi: 10.1016/0959-8049(94)90541-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Zhou C, Ling Y, Qiu G, Bai S, Liu W, He L, Peng Z. [Allelic analysis on chromosome 5 in sporadic colorectal cancer patients] Zhonghua Zhongliu Zazhi. 2002;24:458–460. [PubMed] [Google Scholar]
- 14.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 15.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 16. Available from: http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?org=hum&chr=5. [Google Scholar]
- 17.Kataoka M, Okabayashi T, Johira H, Nakatani S, Nakashima A, Takeda A, Nishizaki M, Orita K, Tanaka N. Aberration of p53 and DCC in gastric and colorectal cancer. Oncol Rep. 2000;7:99–103. [PubMed] [Google Scholar]
- 18.Zhou CZ, Peng ZH, Zhang F, Qiu GQ, He L. Loss of heterozygosity on long arm of chromosome 22 in sporadic colorectal carcinoma. World J Gastroenterol. 2002;8:668–673. doi: 10.3748/wjg.v8.i4.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komarova NL, Lengauer C, Vogelstein B, Nowak MA. Dynamics of genetic instability in sporadic and familial colorectal cancer. Cancer Biol Ther. 2002;1:685–692. doi: 10.4161/cbt.321. [DOI] [PubMed] [Google Scholar]
- 20.Dong JT. Chromosomal deletions and tumor suppressor genes in prostate cancer. Cancer Metastasis Rev. 2001;20:173–193. doi: 10.1023/a:1015575125780. [DOI] [PubMed] [Google Scholar]
- 21.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 22.Kinzler KW, Nilbert MC, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hamilton SR, Hedge P, Markham A. Identification of a gene located at chromosome 5q21 that is mutated in colorectal cancers. Science. 1991;251:1366–1370. doi: 10.1126/science.1848370. [DOI] [PubMed] [Google Scholar]
- 23.Vito P, Lacanà E, D'Adamio L. Interfering with apoptosis: Ca(2+)-binding protein ALG-2 and Alzheimer's disease gene ALG-3. Science. 1996;271:521–525. doi: 10.1126/science.271.5248.521. [DOI] [PubMed] [Google Scholar]
- 24.Krebs J, Klemenz R. The ALG-2/AIP-complex, a modulator at the interface between cell proliferation and cell death A hypothesis. Biochim Biophys Acta. 2000;1498:153–161. doi: 10.1016/s0167-4889(00)00091-4. [DOI] [PubMed] [Google Scholar]
- 25.Jung YS, Kim KS, Kim KD, Lim JS, Kim JW, Kim E. Apoptosis-linked gene 2 binds to the death domain of Fas and dissociates from Fas during Fas-mediated apoptosis in Jurkat cells. Biochem Biophys Res Commun. 2001;288:420–426. doi: 10.1006/bbrc.2001.5769. [DOI] [PubMed] [Google Scholar]
- 26.Krebs J, Saremaslani P, Caduff R. ALG-2: a Ca2+ -binding modulator protein involved in cell proliferation and in cell death. Biochim Biophys Acta. 2002;1600:68–73. doi: 10.1016/s1570-9639(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 27.Kitaura Y, Satoh H, Takahashi H, Shibata H, Maki M. Both ALG-2 and peflin, penta-EF-hand (PEF) proteins, are stabilized by dimerization through their fifth EF-hand regions. Arch Biochem Biophys. 2002;399:12–18. doi: 10.1006/abbi.2001.2736. [DOI] [PubMed] [Google Scholar]
- 28.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 29.Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 2001;269:1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- 30.Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 31.Yasugi T, Vidal M, Sakai H, Howley PM, Benson JD. Two classes of human papillomavirus type 16 E1 mutants suggest pleiotropic conformational constraints affecting E1 multimerization, E2 interaction, and interaction with cellular proteins. J Virol. 1997;71:5942–5951. doi: 10.1128/jvi.71.8.5942-5951.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Castaño IB, De Las Peñas A, Adams C, Christman MF. Pol kappa: A DNA polymerase required for sister chromatid cohesion. Science. 2000;289:774–779. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- 33.Carson DR, Christman MF. Evidence that replication fork components catalyze establishment of cohesion between sister chromatids. Proc Natl Acad Sci U S A. 2001;98:8270–8275. doi: 10.1073/pnas.131022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Castaño IB, Adams C, Vu C, Fitzhugh D, Christman MF. Structure/function analysis of the Saccharomyces cerevisiae Trf4/Pol sigma DNA polymerase. Genetics. 2002;160:381–391. doi: 10.1093/genetics/160.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granger MP, Wright WE, Shay JW. Telomerase in cancer and aging. Crit Rev Oncol Hematol. 2002;41:29–40. doi: 10.1016/s1040-8428(01)00188-3. [DOI] [PubMed] [Google Scholar]


