Abstract
AIM: To establish stock of clinical Helicobacter pylori (H. pylori) isolates, to perform cagA and vacA typing of these isolates, to evaluate the relationship between genotypes of cagA and vacA and upper gastrointestinal diseases and to assess the association of vacA genotypes with presence of the pathogenicity marker-cagA.
METHODS: Clinical H.pylori strains were isolated from the antrum of 259 patients in Clumbia agar. The isolated H. pylori strains were identified by histology, and16SrRNA PCR. CagA genotypes were detected by colony hybridization, the probe was derived from the cloned plasmid PcagA, and digested by EcoRI-HindIII and the isolated PcagA DNA fragment was radioactively labelled by the random priming method. vacA genes types (s,m)and subtypes (s1a, s1b, s2) were typed by PCR. Vacuolating toxin was detected with neutral red absorb test. The results were treated statistically by χ2 test, t test, and rank sum test.
RESULTS: A total of 192 clinical H.pylori strains were isolated and the stock of Helicobacter pylori was established. The total positive rate of cagA was 87% in all gastric diseases, and 95% in gastric cancer group. There was a difference between gastric cancer group and the other groups (P < 0.05) except duodenal ulcer group. The expression of type s1 of vacA was more than type s2 (P < 0.05), and, the expression of type m1 was equal to type m2. In gastric cancer group, there was a difference between s1a and s1b (P < 0.05), and s1a was more than s1b. Vacuolating toxins were more in Xi’an area isolates.
CONCLUSION: The cagA+ vacA type s1 clinical isolates are more in Xi’an area, but this can not serve as an index to predict gastric cancer.
INTRODUCTION
Helicobacter pylori (H. pylori) is an important human pathogen that causes chronic gastritis and is associated with development of peptic ulcer diseases, and gastric malignancies[1]. Epidemiological studies have shown that H. pylori is a class 1 carcinogen for gastric adenocarcinoma, which is one of the most common cancers worldwide, and the odds ratio for developing gastric cancer is 3.8 to 8.7 in H.pylori infected subjects[2-6]. The pathophysiological mechanism by which H.pylori leads to gastric cancer has not yet been defined, although many hypotheses have been put forward.
The genetic variability among H.pylori strains is relatively high[7]. Approximately 50%-60% of H. pylori strains contain cytotoxin-associated (cagA) gene and consequently produce 128 ku CagA protein[8,9]. The presence of cagA is associated with gastric cancer, gastric mucosal atrophy, and duodenal ulcer. CagA is a part of a large genomic entity, designating the pathogenicity (cag) island[10,11], which contains multiple genes that are related to the virulence and pathogenicity of H. pylori strains. Therefore, the presence of cagA can be considered as a marker for this genomic pathogenicity island and is associated with more virulent H.pylori strains.
Another important virulence factor, produced by approximately 50% of H. pylori strains, is a cytotoxin that induces formation of vacuolates in mammalian cells in vitro and leads to cell death[12]. The toxin is encoded by vacA gene, which is virtually present in all H.pylori strains[13]. The existence of different allelic variants in two parts of this gene has been described[14-17]. The N-terminal signal (s) region occurs as either a s1a, s1b or s2 allele. The middle (m) region is present as a m1 or m2 allele. The mosaic structure of vacA gene accounts for differences in cytotoxin production between strains[18-20].
The aim of this study was to detect and type the virulence-associated cagA and vacA genes of clinical H.pylori isolates in Xi’an area of China. Finally, the clinical relevance of vacA and cagA genotyping was investigated with gastric cancer and precancerous conditions.
MATERIALS AND METHODS
Bacterial isolation and culture
A total of 192 H.pylori isolates were obtained from patients undergoing upper gastrointestinal examination. The diagnosis obtained by endoscopy and histology was recorded for all the patients from whom the strains were isolated. Following primary isolation, H.pylori strains were grown on Columbia agar with 50 mL/L frozen-melting sheep blood, 100 mL/L fetal bovine serum, and Skirrow’s antibiotic supplement in a microaerophilic atmosphere for 5 days at 37 °C, then frozen at -70 °C. Most of the strains were frozen four to six passages after primary isolation. Subsequent analyses were performed on strains derived from the frozen stocks. The isolated H.pylori strains were identified by histology, and16SrRNA PCR.
Plasmid DNA preparation
A signal bacteria clone was incubated at 37 °C overnight, harvested, and suspended in 0.5 mol/L EDTA-1M Tris-HCl (pH 8.0), 10 mol/L NaOH and 100 g/L SDS was added, mixed, then 5 mol/L KAc and ice HAc were added. The mixture was centrifuged at 12000 r/min for 10 min. The supernatant was extracted with phenol-chloroform followed by with chloroform. After extraction, the DNA solution was mixed with absolute ethanol and 3 mol/L NaAc at -20 °C for 20 min, washed with 700 mL/L ethanol, and dried. The DNA pellet was suspended in TE (10 mmol/L Tris-HCl, 1 mmol/L pH8.0 EDTA), and stored at -20 °C.
DNA probes
The probe was derived from the cloned plasmid PcagA, and digested by EcoRI-HindIII. The enzyme digestion segment of PcagA was retrieved by DNA purification reagent kit (Baotaike Biotechnology Company). The isolated PcagA DNA fragment was radioactively labelled by the random priming method.
Colony hybridization
SDS of 100 g/L was added in fresh H.pylori liquid for 10 min, 2×SSC (1×SSC is 0.15 mol/L NaCl plus 0.015 mol/L sodium citrate) for 5 min, then H.pylori degeneration liquids were dotted on NC membranes which had been treated by 2×SSC, heated at 80 °C for 2 h. The membranes were put into pre-hybridization liquid (1×Denhardt, 1 g/L SDS, 5×SSC, 500 g/L deionised fomamide) at 42 °C for 3 h, then α-32P labeled probes were added at 43.5 °C overnight. The membrane filters were subsequently washed once in 2×SSC-5 g/L SDS and three times in 0.1×SSC-1 g/L SDS, then exposed to X-ray film and their radioactivity was self-developing at -20 °C for 48 h. The film was developed and then fixed.
H.pylori chromosomal DNA extracts
Fresh H. pylori strains were harvested, suspended in 0.1 mol/ L NaCl-10 mmol/L Tris-HCl-1 mmol/L EDTA (pH 8.0), and incubated at 37 °C for 15 min. NH4Ac was added and the mixture was placed on ice for 5 min and extracted once with chloroform, mixed with isopropanol and placed on ice for 10 min. After extraction, the DNA solution was mixed with absolute ethanol at -20 °C for 1 h, washed with 700 mL/L ethanol, and dried. The DNA pellet was suspended in distilled water and stored at -20 °C.
VacA genotyping
The vacA was typed by PCR. Table 1 shows the primer sequence of vacA. And Table 2 shows the system of PCR. The reaction condition was at 94 °C for 1 min, at 52 °C for 1 min, and at 72 °C for 1 min for 35 cycles and extension at 72 °C for 6 min. The PCR product was analyzed by 20 g/L agarose gel. The reference H.pylori strains were 60190 (s1a/m1), 84183 (s1b/m1) and 86313 (s2/m2).
Table 1.
Oligonucleotide primers used for vacA typing
| Gene and region amplified | Genotype identified | Primer designation | Primer sequence | Size of PCR product(bp) |
| Mid-region | m1/m2 | VAG-F | 5’CAATCTGTCCAATCAAGCGAG3’ | 567/642 |
| VAG-R | 5’GCGTCAAAATAATTCCAAGG3’ | |||
| Signal sequence | s1/s2* | VA1-F | 5’ATGGAAATACAACAAACACAC3’ | 259/286* |
| VA1-R | 5’CTGCTTGAATGCGCCAAAC3’ | |||
| s1a | SS1-F# | 5’GTCAGCATCACACCGCAAC3’ | 190 | |
| s1b | SS3-F# | 5’AGCGCCATACCGCAAGAC3’ | 187 |
vacA types s1 and s2 were differentiated on the size of the PCR product. # Used with reverse primer VA1-R.
Table 2.
The system of PCR
| Constituents | Volume/per tube (μL) | Final concentration |
| Water | 36 | |
| 10×PCR buffer | 5 | 1× |
| 4dNTP, 2.5mmol/L /each | 4 | 0.2mmol/L each |
| primer 1.25 μmol/L | 1 | 0.5 μmol/L |
| primer 2.25 μmol/L | 1 | 0.5 μmol/L |
| MgCl2, 50 mmol/L | 1.5 | 1.5 mmol/L |
| Taq DNA polymerase, 5MU/L | 0.5 | 0.1 MU/L |
| Template DNA | 1 |
Cytotoxicity test on hela cells
Hela cells were cultured in plastic flasks in Dulbecco’s modified Eagle’s medium (DMEM) containing 25 mmol/L HEPES buffer (N -2 - hydroxyethylpiperazine- N’ -2 - ethanesulfonic acid, pH7.2) and 100 g/L fetal bovine serum. Cells were maintained at 37 °C in a 50 mL/L CO2 atmosphere. After cultured for 24 h, the cells were suspended with trypsin-EDTA and seeded in 96-well titration plates to make the density of cells of 104 per well. The supernatant prepared by water extracts from different strains was diluted two-fold from 1:2 to 1:32 and incubated with Hela cells for 12 h, then added 0.5 g/L neutral red- normal saline for 5 min, washed 3 times with 2 g/L BAS-normal saline and added HCl-ethanol. The absorbance was detected at 540 nm. When the detected value was more than 3 times of the negative control, it was defined as a positive result.
Statistical analysis
χ2 test, t test, and rank sum test were used for statistical analysis.
RESULTS
Presence of cagA gene
The presence of cagA gene was investigated in all clinical isolates by colony hybridization. EcoRI-HindIII-digested chromosomal DNA (Figure 1) was probed with an α 32P-labelled DNA fragement internal to cagA. Typical examples of the results obtained with colony hybridization are reported in Figure 2. Of the 192 clinical isolates, 165 strains (86%) had a positive hybridization dot, while 27 strains (14%) did not show hybridization dot. In the 192 strains, the positive rate of gastric cancer group was 95%, chronic superficial gastritis (CSG) 77%, chronic atrophic gastritis (CAG) 86%, gastric ulcer (GC) 69%, and duodenal ulcer (DU) 95%. There were differences between CAG and GU (P < 0.05), and between GU and DU (P < 0.05), but there were no statistical differences between CSG and CAG (P > 0.05), and between CSG and GU (P > 0.05).
Figure 1.
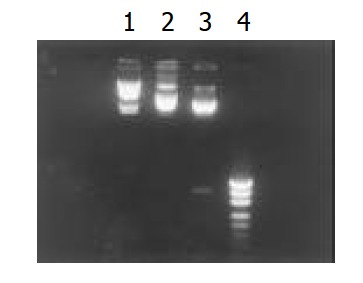
Restriction endonuclease EcoRI, HindIII digests of PcagA. 1. λDNA HindIIImarker, 2. PcagA, 3. Fragement di-gested by EcoRI-HindIII, 4. PUC18/MSPI marker.
Figure 2.
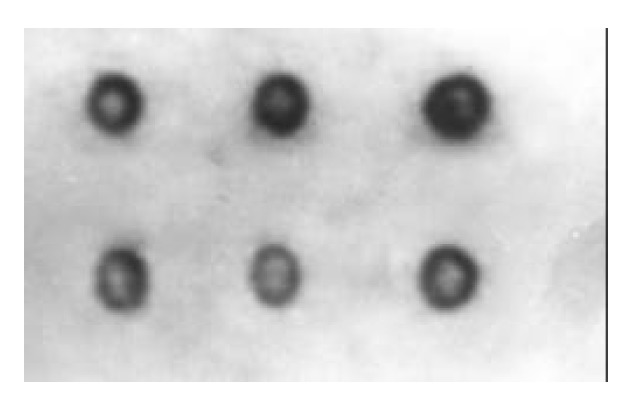
Colony hybridization detection of cagA. The positive hybridization dot.
Presence of signal sequence typing of vacuolating toxin gene
The signal sequence gene was typed by PCR in all H. pylori clinical isolates. The products of s1 and s2 were 259 bp and 286 bp, respectively. In the 192 strains, 174 strains (90.6%) were s1 type, and 18 strains (9.4%) were s2. The subtypes of s1 were typed by two pairs of primers (SS1-F/VA1-R and SS3- F/VA1-R) (Table 3 and Figure 3). The product of s1a and s1b was 190 bp and 187 bp, respectively. In the 174 s1 type strains, 111 strains (63.8%) were s1a, and 63 strains (36.2%) were s1b (Figure 4, Figure 5).
Table 3.
H.pylori signal sequence typing and gastric diseases
| s type | GC | CSG | CAG | GU | DU | Total(%) |
| s1a | 43a | 14 | 37 | 5 | 12 | 111 (57.8) |
| s1b | 10 | 13 | 25 | 8 | 7 | 63 (32.8) |
| s2 | 3 | 4 | 7 | 3 | 1 | 18 (9.4) |
| Total | 56 | 31 | 69 | 16 | 20 | 192 (100.0) |
P < 0.05 vs GU, CSG, CAG.
Figure 3.
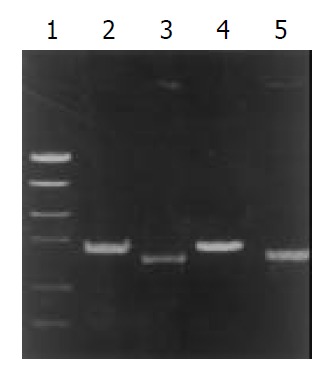
PCR typing of vacA signal sequence. 1.PCR marker, 2. Type s2, 3. Type s1, 4. Standard strain 86313, 5. Standard strain 60 190.
Figure 4.
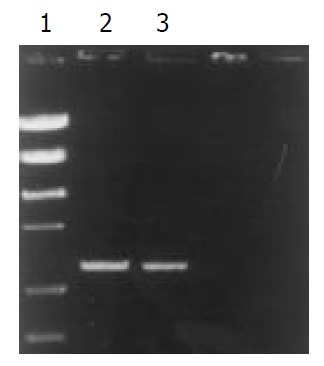
PCR typing of vacA s1a. 1. PCR marker, 2. Clinical isolates, 3. Standard strain 84 183.
Figure 5.
PCR typing of vacA mid-region. 1. PCR marker, 2. Clinical isolates, 3. Standard strain 84 183.
Presence of middle region typing of vacuolating toxin gene
The middle region gene was also typed by PCR in all H. pylori clinical isolates. The products of m1 and m2 were 567 bp and 642 bp, respectively. In the 192 strains, 99 strains (51.6%) were m1 type, and 93 strains(48.4%) were m2, (Table 4 and Figure 6).
Table 4.
H.pylori signal sequence typing and gastric diseases
| m type | GC | CSG | CAG | GU | DU | Total(%) |
| m1 | 31 | 13 | 36 | 7 | 12 | 99 (51.6) |
| m2 | 25 | 18 | 33 | 9 | 8 | 93 (48.4) |
| Total | 56 | 31 | 69 | 16 | 20 | 192 (100.0) |
No difference at all.
Figure 6.
PCR typing of vacA s1a. 1. PCR marker, 2.Type m2, 3. Type m1, 4. Standard strain 86 313, 5. Standard strain 60 190.
Presence of middle region typing and signal sequence of vacuolating toxin gene
In the 192 strains, 65 strains (33.9%) were sla/m1, 34 strains (17.7%) s1b/m1, 46 strains (24.0%) s1a/m2, 29 strains (15.1%) s1b/m2, and 18 strains (9.4%) s2/m2.
Relationship between vacA typing and detecting of vacuolating cytotoxin activity in vitro
The maximum diluting times of positive result were the value of vacuolating toxin. Based on the maxium diluting unit, the strains were divided into three types: high toxin ( ≥ 8), low toxin (1-8) and none toxin (less than 1). In the 192 strains, high toxin was 90 strains (46.9%), low toxin 62 (32.3%) and none toxin 40 (20.8%, Table 5).
Table 5.
Relationship between vacA typing and grade of vacu-olating cytotoxin activity
| vacA type |
Grade of vacuolating cytotoxin activity |
||
| None | Low | High | |
| s1a | 5 | 38 | 68 |
| s1b | 17 | 24 | 22 |
| s2 | 18 | 0 | 0 |
| m1 | 10 | 30 | 59 |
| m2 | 30 | 32 | 31 |
P < 0.05, each group vs s1a, s1b, s2.
Relationship between cagA and vacA subtype
In the 165 cagA gene positive strains, 165 strains (100%) were vacA s1 type; 97 strains (58.8%) vacA m1 type, and 78 strains (46.2%, Table 6) vacA m2 type.
Table 6.
Relationship between vacA typing, cagA gene and gastric diseases for 192 H. pylori isolates
| vacA type |
cagA+ |
Total(%) | ||||
| GC | CSG | CAG | GU | DU | ||
| s1a | 43 | 14 | 37 | 4 | 12 | 110 (66.7) |
| s1b | 10 | 10 | 22 | 6 | 7 | 55 (33.3) |
| s2 | 0 | 0 | 0 | 0 | 0 | 0 (0.0) |
| m1 | 31 | 12 | 36 | 6 | 12 | 97 (58.8) |
| m2 | 22 | 12 | 23 | 4 | 7 | 78 (46.2) |
DISCUSSION
H. pylori is the major causative agent of chronic superficial gastritis and plays a central role in the etiology of peptic ulcer disease. Evidence suggests that H. pylori infection pre-exists in gastric carcinoma and precancerous lesions, and is a risk for development of gastric carcinoma. Cittelly showed that lower frequencies of cytotoxic genotypes such as cagA and vacA s1m1 were observed in patients with NAG (non atrophic gastritis), when compared to patients with GC (gastric cancer) or PU (peptic ulcer). He suggested that vacA and cagA could be used as markers for increased virulence. In 1994, H. pylori was designated as a class I carcinogen by the World Health Organization[21,22]. H. pylori infection is high among Chinese people (the infected rate is 40%-70%), and also high in gastric cancer. The mortality of gastric cancer ranks the first among malignancies. The results of epidemiologic surveys on the relationship between H. pylori and gastric cancer show that infection of H.pylori in Lanzhou, an area with a high occurrence of gastric cancer, is higher than that in Guangzhou, an area with a low occurrence of gastric cancer[23,24].
Recent researches suggested that the positive cytotoxin-associated gene A of H.pylori was about 60%. This toxin can induce severe inflammation of gastric mucosa and is related with peptic ulcer and gastric cancer[25]. We detected the cagA gene in 192 clinical strains by colony hybridization, with a total positive rate of 86%. In gastric cancer and duodenal ulcer, H.pylori infection rate (91%) was the highest in all gastric diseases. In gastric ulcer group it was the lowest, only 69%. Our results suggest that positive cagA of H.pylori isolated strains is related with gastric diseases in Xi’an area, Because the expression of cagA positive strains in gastric cancer group was higher than that in chronic gastritis group and the difference was apparent (P < 0.05). CagA can increase the serious consequences of carcinogenesis from chronic inflammation. The detectable rate of CagA antibody in human group in high occurrence region of gastric cancer was obviously higher than that in low occurrence area. Some researches have also proved that cagA positive strains are easy to cause inflammation of gastric mucosa and can stimulate hyperplasia of gastric mucosal cells. Many studies have shown that there is a relationship between cagA positive antibody and peptic ulcer and gastric cancer. One research suggested that development of more prominent gastritis and severe atrophy in cagA (+) patients was an indicator for the importance of cagA rather than H. pylori load. Our research was consistent with theirs[26,27].
All strains have the gene encoding toxin vacA, but its structure varies, especially in the mid-region which may be type m1 or m2 and the region encoding the signal sequence (type s1a, s1b or s2). The final structure is a mosaic, and all combinations of signal sequence and mid-region types are found except s2/m1. A strain’s vacA structure determines its in vitro cytotoxin activity, with type m1 vacA being more active than type m2, type s1a being more active than type s1b, and type s2 vacA not producing detectable activity. Our results showed that the main genotype was s1a in gastric cancer group, and there was an apparent difference between s1a and s1b (P < 0.05), and the s2 genotype was seldom found. In the meantime, there was no apparent difference between m1 and m2 (P > 0.05). The expression of s1a type was different among gastric cancer, gastric ulcer, and chronic gastritis. In gastritis group, there was no difference between s1a type and s1b type. Andreson’s research showed that the presence of the cagA gene was correlated with that of vacA signal sequence type s1a. However, no clear differences were found in the distribution of cagA and vacA genotypes among patients with peptic ulcer or chronic gastritis in Estonia. The reason why the different regional distribution can cause different genotype of H.pylori is not clear[28].
Our results showed that the infected H.pylori in Xi’an area was mainly toxin strains. In the meantime, the expression of s1a and s1b in the toxin strains was obviously different (P < 0.05), and there was no significant difference between m1 and m2 (P > 0.05). This suggests that s2 type can not produce detectable vacuolating toxin, and s1 type can produce the toxin. The middle-region is not obviously related to toxin production. The toxin produced by s2 type may not effectively pass through cellular membrane[29].
In cagA positive strains, the main vacA subtype was s1a, about 67%, and s1b was only 33%, and there was an apparent difference between these two subtypes (P < 0.05). But there was no difference between m1 and m2 (P > 0.05). We found that s1 type was related with gastric cancer[30].
Inactivation of multiple antioncogenes and H.pylori infection may be involved in the development and progress of gastric carcinoma, and H.pylori infection may be associated with the inactivation of some oncogenes[31]. The genotype expression of H.pylori in gastric cancer was mainly cagA+/s1a strains in Xi’an area, but this can not serve as an index to predict gastric cancer. The main genotype of vacA in chronic gastritis, gastric ulcer and duodenal ulcer is s1, but there is no difference between s1a, s1b, m1 and m2.
Footnotes
Edited by Ma JY
References
- 1.YaoYL , Zhang WD. The relationship between Helicobacter pylori and gastric cancer. Shijie Huaren Xiaohua Zazhi. 2001;9:1045–1049. [Google Scholar]
- 2.Zhuang XQ, Lin SR. Research of Helicobacter pylori infection in precancerous gastric lesions. World J Gastroenterol. 2000;6:428–429. doi: 10.3748/wjg.v6.i3.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandenplas Y. Helicobacter pylori infection. World J Gastroenterol. 2000;6:20–31. doi: 10.3748/wjg.v6.i1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, Yuan Y, Gao H, Dong M, Wang L, Gong YH. Apoptosis, proliferation and p53 gene expression of H. pylori associated gastric epithelial lesions. World J Gastroenterol. 2001;7:779–782. doi: 10.3748/wjg.v7.i6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao H, Wang JY, Shen XZ, Liu JJ. Effect of Helicobacter pylori infection on gastric epithelial cell proliferation. World J Gastroenterol. 2000;6:442–444. doi: 10.3748/wjg.v6.i3.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhuang XQ, Lin SR. Study on the relationship between Helicobactr pylori and gastric cancer. Shijie Huaren XiaoHuaZazhi. 2000;8:206–207. [Google Scholar]
- 7.Figueiredo C, Van Doorn LJ, Nogueira C, Soares JM, Pinho C, Figueira P, Quint WG, Carneiro F. Helicobacter pylori genotypes are associated with clinical outcome in Portuguese patients and show a high prevalence of infections with multiple strains. Scand J Gastroenterol. 2001;36:128–135. doi: 10.1080/003655201750065861. [DOI] [PubMed] [Google Scholar]
- 8.Li XJ, Yan XJ, Liu ZG, Su CZ. Expression, purification and clini-cal research of Helicobacter pylori cytotoxin-associated gene A. Shijie Huaren Xiaohua Zazhi. 2002;10:271–274. [Google Scholar]
- 9.Abasiyanik MF, Sander E, Salih BA. Helicobacter pylori anti-CagA antibodies: prevalence in symptomatic and asymptomatic subjects in Turkey. Can J Gastroenterol. 2002;16:527–532. doi: 10.1155/2002/589087. [DOI] [PubMed] [Google Scholar]
- 10.Kidd M, Lastovica AJ, Atherton JC, Louw JA. Conservation of the cag pathogenicity island is associated with vacA alleles and gastroduodenal disease in South African Helicobacter pylori isolates. Gut. 2001;49:11–17. doi: 10.1136/gut.49.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu WJ, Li NS, Wang SL, Liu J, Tu ZX, Xu GM. Difference be-tween cagA and cagM in cag I of Helicobacter pylori isolated from Chinese patients. Shijie Huaren Xiaohua Zazhi. 2001;9:405–409. [Google Scholar]
- 12.Ye GA, Zhang WD, Liu LM, Shi L, Xu ZM, Chen Y, Zhou DY. vacA gene of Helicobacter pylori. Shijie Huaren Xiaohua Zazhi. 2001;9:593–595. [Google Scholar]
- 13.van Doorn LJ. Detection of Helicobacter pylori virulence-associated genes. Expert Rev Mol Diagn. 2001;1:290–298. doi: 10.1586/14737159.1.3.290. [DOI] [PubMed] [Google Scholar]
- 14.Lin CW, Wu SC, Lee SC, Cheng KS. Genetic analysis and clinical evaluation of vacuolating cytotoxin gene A and cytotoxin-associated gene A in Taiwanese Helicobacter pylori isolates from peptic ulcer patients. Scand J Infect Dis. 2000;32:51–57. doi: 10.1080/00365540050164227. [DOI] [PubMed] [Google Scholar]
- 15.Fallone CA, Barkun AN, Göttke MU, Best LM, Loo VG, Veldhuyzen van Zanten S, Nguyen T, Lowe A, Fainsilber T, Kouri K, et al. Association of Helicobacter pylori genotype with gastroesophageal reflux disease and other upper gastrointestinal diseases. Am J Gastroenterol. 2000;95:659–669. doi: 10.1111/j.1572-0241.2000.01970.x. [DOI] [PubMed] [Google Scholar]
- 16.Shiesh SC, Sheu BS, Yang HB, Tsao HJ, Lin XZ. Serologic response to lower-molecular-weight proteins of H. pylori is related to clinical outcome of H. pylori infection in Taiwan. Dig Dis Sci. 2000;45:781–788. doi: 10.1023/a:1005460130305. [DOI] [PubMed] [Google Scholar]
- 17.Yao YL, Xu B, Song YG, Zhang WD. Overexpression of cyclin E in Mongolian gerbil with Helicobacter pylori-induced gastric precancerosis. World J Gastroenterol. 2002;8:60–63. doi: 10.3748/wjg.v8.i1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji KY, Hu FL. The development of Helicobacter pylori and cytokines. Shijie Huaren Xiaohua Zazhi. 2002;10:503–508. [Google Scholar]
- 19.Hou P, Tu ZX, Xu GM, Gong YF, Ji XH, Li ZS. Helicobacter pylori vacA genotypes and cagA status and their relationship to associated diseases. World J Gastroenterol. 2000;6:605–607. doi: 10.3748/wjg.v6.i4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.She FF, Su DH, Lin JY, Zhou LY. Virulence and potential pathogenicity of coccoid Helicobacter pylori induced by antibiotics. World J Gastroenterol. 2001;7:254–258. doi: 10.3748/wjg.v7.i2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue FB, Xu YY, Wan Y, Pan BR, Ren J, Fan DM. Association of H. pylori infection with gastric carcinoma: a Meta analysis. World J Gastroenterol. 2001;7:801–804. doi: 10.3748/wjg.v7.i6.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cittelly DM, Huertas MG, Martínez JD, Oliveros R, Posso H, Bravo MM, Orozco O. [Helicobacter pylori genotypes in non atrophic gastritis are different of the found in peptic ulcer, premalignant lesions and gastric cancer in Colombia] Rev Med Chil. 2002;130:143–151. [PubMed] [Google Scholar]
- 23.Wang GT. The relationship between CagA/VacA strains and chronic gastric diseases. Shijie Huaren XiaoHuaZazhi. 2001;9:1335–1338. [Google Scholar]
- 24.Chen JP, Shen DM, Yang ZB. CagA Hp broth culture filtrates induced malignant transformation on human gastric epithelial cells. Shijie Huaren Xiaohua Zazhi. 2001;9:617–621. [Google Scholar]
- 25.Wang CD, Li JS, Chen LY. CagA and VacA do not predict Helicobacter pylori-associated gastroduodenal diseases. Shijie Huaren Xiaohua Zazhi. 2002;10:533–535. [Google Scholar]
- 26.Cai L, Yu SZ, Zhang ZF. Helicobacter pylori infection and risk of gastric cancer in Changle County,Fujian Province,China. World J Gastroenterol. 2000;6:374–376. doi: 10.3748/wjg.v6.i3.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou JC, Xu CP, Zhang JZ. The research development of cagA/ CagA of Helicobacter pylori mocular biology. Shijie Huaren XiaoHuaZazhi. 2001;9:560–562. [Google Scholar]
- 28.Demirtürk L, Ozel AM, Yazgan Y, Solmazgül E, Yildirim S, Gültepe M, Gürbüz AK. CagA status in dyspeptic patients with and without peptic ulcer disease in Turkey: association with histopathologic findings. Helicobacter. 2001;6:163–168. doi: 10.1046/j.1523-5378.2001.00024.x. [DOI] [PubMed] [Google Scholar]
- 29.Andreson H, Lõivukene K, Sillakivi T, Maaroos HI, Ustav M, Peetsalu A, Mikelsaar M. Association of cagA and vacA genotypes of Helicobacter pylori with gastric diseases in Estonia. J Clin Microbiol. 2002;40:298–300. doi: 10.1128/JCM.40.1.298-300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SM, Park J, Kim JG, Yoo BC. Relevance of vacA genotypes of Helicobacter pylori to cagA status and its clinical outcome. Korean J Intern Med. 2001;16:8–13. doi: 10.3904/kjim.2001.16.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang XD, Fang DC, Li W, Du QX, Liu WW. A study on relation-ship between infection of Helicobacter pylori and inactivation of antioncogenes in cancer and pre-cancerous lesion. Shijie Huaren Xiaohua Zazhi. 2001;9:984–987. [Google Scholar]


