Abstract
AIM: To assess the incidence of MLH1 (the human MutL homologue) and MSH2 (the human MutS homologue) protein expression in Turkish patients with sporadic colorectal cancers and to compare their survival and clinicopathological features.
METHODS: We validated the tissue microarray technology in 77 colorectal carcinomas by analyzing the immunohistochemical expression of proteins involved in two main pathways of colorectal carcinogenesis: p53 protein for loss of heterozygosity tumors; MLH1 and MSH2 proteins for microsatellite instability (MSI).
RESULTS: Our analysis showed that 29 (39.2%) had loss of MLH1 expression, 5 (6.8%) had loss of MSH2 expression and 2 cases had loss of expression of both proteins. We found that 60% of MSH2-negative tumors were located in the right side of the colon; all MSH2-negative cases were women. In addition, the loss of MSH2 expression was correlated with low p53 expression. Neither MLH1 nor MSH2 expressions were associated with prognosis, although there seemed a tendency of longer survival (71.7 ± 8.65 mo vs 47.08 ± 5.26 mo) for the patients with MLH1-negative versus MLH1-positive carcinomas. There were not significant differences in overall and recurrence-free survival among MLH1/MSH2-positive and -negative cases.
CONCLUSION: Our data supports that Turkish patients with MLH1- and MSH2-defective tumors have some distinct features from each other. Although prognostic importance remains controversial, immunohistochemical analysis of mismatch repair genes may be used as a routine histopathological examination of sporadic colorectal carcinomas.
Keywords: Colorectal carcinoma, MLH1, MSH2, Immunohistochemistry, Prognosis
INTRODUCTION
In humans, mismatch repair (MMR) system is mediated by at least six genes, including human MutL homologue (hMLH1), human MutS homologue (hMSH2), hMSH3, hMSH6, hPMS1, and hPMS2[1]. MMR deficiency leads to the accumulation of base-base mismatches and short insertion/deletion mispairs, generated as a consequence of DNA replication errors and homologous recombinations. Most cell deficient in hMLH1 and hMSH2 genes often display a high level of genomic instability, characterized by changes in repeated numbers of simple repetitive sequences, microsatellite instability (MSI)[2,3]. The detection of MSI status is based on molecular analysis. According to this, three tumor phenotypes have been defined: Microsatellite stable (MSS), low-frequency MSI (MSI-L) and high-frequency MSI (MSI-H). A germline mutation in one of MMR genes, accompanied by somatic inactivation of the other allele, may result in high level of microsatellite instability (MSI-H)[4-6].
In patients with hereditery nonpolyposis colon carcinoma (HNPCC), MSI was detected in 80% of the tumor samples[7]. It has been reported that MMR genes are involved in the 10%-15% of sporadic colorectal carcinomas[3,8]. In sporadic MSI-H colorectal carcinomas, the mechanism of development of mutator phenotype is inactivation of MLH1 by promoter hypermethylation. Sporadic MSI-H tumors tend to be poorly differentiated and/or mucinous subtype[9-11]. They are usually diploid; p53 mutations, loss of heterozygosity at 18q, APC mutations and K-ras mutations are found less frequently than in MSS tumors[12,13]. In literature, there are controversial results about the prognostic importance of MSI status in sporadic colorectal carcinoma.
Establishing the presence of MSI requires polymerase chain reaction-based technology, examining DNA sequences of intact and tumor tissue. This is an expensive and time-consuming procedure that is not readily available in all pathology laboratories. Immunohistochemically identifying MSI tumors is a less costly alternative procedure. There are some studies which have shown a high correlation of MLH1 and MSH2 immunohistochemical patterns of expression with the DNA analyses. MSI-H correlates with loss of immunohistochemical staining of either MLH1 or MSH-2 in the tumor nuclei[14-17]. The sensitivity of the immunohistochemical technique for detecting MSI-H tumors is about 80% to 100%[15-17].
To our knowledge, this is the first published study in Turkey concerning investigation of MSI status in sporadic colorectal carcinomas. Therefore, it is important to find out prognostic importance of MSI status in patients with colorectal carcinoma in Turkish population to perform their appropriate treatment and follow-up. In this study, we determined (1) the frequency of MLH1- and MSH2-deficient colorectal carcinomas in Turkish patients, (2) the relationship between MLH1/MSH2 expression and clinicopathological features, and (3) the predictive and prognostic relevance of loss of MLH1 and/or MSH2 expression in recurrence-free and overall survival.
MATERIALS AND METHODS
Tissue specimens
A total of 77 colorectal carcinoma specimens were obtained from the archives of the Department of Pathology of Cerrahpasa Medical College, Istanbul University. Patients with familial adenomatosis polyposis or inflammatory bowel disease were excluded. Tumors were staged according to the TNM staging system[18]. Tumor type and grade of differentiation were determined by criteria of the World Health Organization[19]. Peritumoral Crohn’s-like reaction, pattern of growth and lymphocytic infiltration were also evaluated according to literature[20,21]. Tissue microarray (TMA) was applied to study normal colorectal mucosa and colorectal carcinomas. The patients had received neither chemotherapy nor radiation therapy before tumor resection. Their pathological specimens and slides were revised. After revision, two slides and corresponding two paraffin blocks were chosen, one for normal colorectal mucosa and one for colorectal carcinomas tissues for each case. Corresponding areas for normal mucosa and carcinomas were marked on chosen paraffin blocks. In TMA, 2-mm cores were taken from these selected areas, two for normal mucosa and two for cancer areas, and then they were embedded into microarray block. Each block containing 60 cores also contained other tissues (for example, sausage blocks included tissues from breast, thyroid, prostate, skin, gastric mucosa, lymph node, etc) for negative and positive controls.
Immunohistochemistry
Formalin-fixed paraffin-embedded TMA blocks were cut into 3-μm thick sections and mounted on polarized glass slides. After mounting, they were kept in an oven at 56°C overnight. Sections were deparaffinized in xylene and rehydrated. They were incubated in a microwave containing 10 g/L EDTA solution 3 times for 5 min each, and then kept in room temperature for 20 min and placed in 10 mL/L hydrogen peroxide for 10 min. After being washed with distilled water and phosphate-buffered saline (PBS), sections were incubated overnight at 4°C with primary antibodies of MSH2 (Ab-1, Clone 2MSH01, NeoMarkers; 1:25) and MLH1 (Clone 14, Zymed Lab; 1:50) with pre-antibody blocking solution (Immunovision-Sitogen). Primary antibody was replaced with PBS for a negative control. After being washed with PBS, the sections were incubated with primary antibody enhancer for 30 min and then with HRP polymer for 30 min. After being washed thrice with PBS for 10 min each, the sections were stained with a streptavidin-peroxidase detection system.
Immunostaining for p53 (p53 AB-5 clone DO-7, Neomarkers; 1:100) was almost the same as above-mentioned, except using citrate buffer solution instead of 10 g/L EDTA solution in microwave.
Immunohistochemical evaluation
Stained tissues in tissue microarray slides (TMS) were scored under the light microscope and the extent and intensity of staining with MLH1, MSH2 and p53 antibodies were evaluated independently by two pathologists (SE and EU) without knowledge of clinicopathological data. Tumors showing loss of nuclear MLH1 or MSH2 expressions were classified as MLH1- or MSH2-negative, respectively. Nuclear immunostaining of normal epithelial cells, lymphocytes and stromal cells served as internal positive controls. For p53, tumors showing a proportion of stained nuclei of > 10% were classified as p53-positive.
Statistical analysis
Spearman rank, Kendall correlation test, Cox regression and Kaplan-Meier test were used, when appropriate. P < 0.05 was considered statistically significant.
RESULTS
There were positive staining of normal control mucosa in the lower third of the epithelium and nuclear staining of lymphocytes from a control germinal centre (Figure 1A and B). Three MLH1/MSH2-stained cases were excluded from the study because the quality of immunostaining was unsatisfactory. Of the remaining 74 colorectal adenocarcinomas, 42 (56.7%) cases demonstrated normal expression of both MLH1 and MSH2 gene products (MLH1+/MSH2+) (Figure 2A and B). Loss of MLH1 or MSH2 expression was detected in 32 (43.2%) of all cases examined. Complete loss of MLH1 expression (Figure 3A) and normal immunoreactivity for MSH2 were observed in 27 (36.48%) cases of adenocarcinoma, while 5 (6.8%) cases displayed complete loss of MSH2 expression (Figure 3B) and normal immunoreactivity for MLH1. Two (2.07%) adenocarcinoma cases showed lack of both MLH1 and MSH2 expressions.
Figure 1.
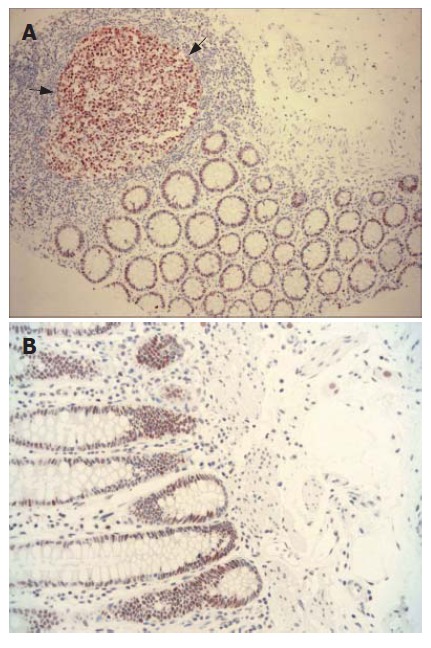
MLH1 and MSH2 expression. A: Nuclear MLH1 expression detected in germinal centre of lymphoid follicule (dark arrows) and in epithelia of normal colonic mucosa (× 100); B: Crypt epithelia showing normal positive nuclear staining with MSH2 (× 200).
Figure 2.
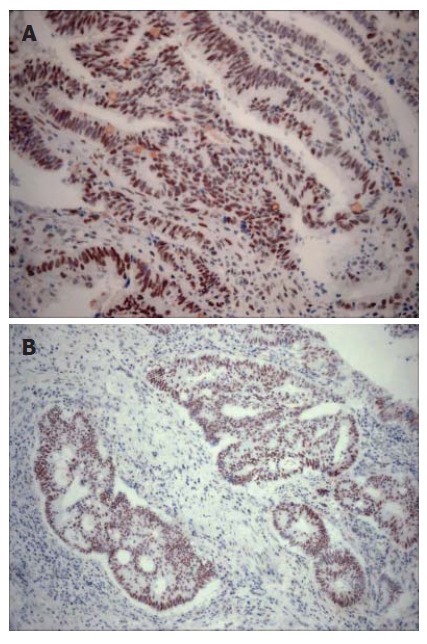
MLH1 and MSH2 expression. A: Extensive nuclear staining with MLH1 in adenocarcinoma of colon (× 200); B: Tumor cells showing strong positive nuclear staining with MSH2 (× 100).
Figure 3.
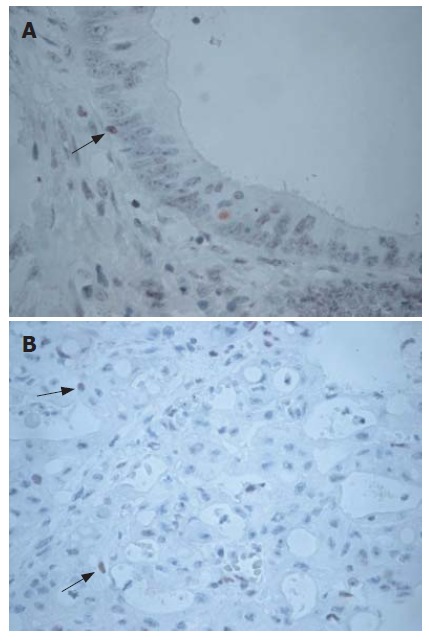
Loss of MLH1 and MSH2 expression in colorectal cancer. A: Loss of staining with MLH1 in cancer cells, although lymphocytes (arrow) show positive staining (× 400); B: Adenocarcinoma with complete loss of MSH2 expression. Nuclear staining of lymphocytes (arrows) in the stroma served as internal positive control (× 200).
Immunohistochemical pattern of MLH1/MSH2 expressions was found to be related to some clinical and pathological variables (Tables 1 and 2). MSH2-negative carcinomas occurred only in women (P = 0.049). In addition, MSH2-negative carcinomas developed more frequently in patients ≥ 50 years than did MSH2-positive (P ≥ 0.05). Majority of MSH2-negative tumors (60%) were located in the right colon (P > 0.05). However, no any preferential location for MLH1-negative cases was observed. There was no significant correlation between MLH1/MSH2 expression and tumor size and tumor type, while a significant relation was detected between tumor invasion and MSH2 expression (Table 2).
Table 1.
Clinicopathological parameters and MLH1 and MSH2 expressions n (%)
| MLH1 (+) | MLH1 (-) | P | MSH2 (+) | MSH2 (-) | P | |
| Gender | 0.943 | 0.049 | ||||
| Male | 26 (57.8) | 17 (58.6) | 38 (55.1) | 0 (0) | ||
| Female | 19 (42.2) | 12 (4.4) | 31 (44.9) | 5 (100) | ||
| Age (yr) | 0.602 | 0.579 | ||||
| < 50 | 15 (33.3) | 8 (27.6) | 22 (31.9) | 1 (20) | ||
| ≥ 50 | 30 (66.7) | 21 (72.4) | 47 (68.1) | 4 (80) | ||
| Localization | 0.630 | 0.244 | ||||
| Right colon | 10 (22.2) | 9 (31) | 16 (23.2) | 3 (60) | ||
| Left colon | 11 (24.4) | 9 (31) | 20 (29) | 0 (0) | ||
| Rectum | 16 (35.6) | 7 (24.1) | 22 (31.9) | 1 (20) | ||
| Colon (NOS) | 8 (17.8) | 4 (13.8) | 11 (15.9) | 1 (20) | ||
| Tumor size | 0.757 | 0.969 | ||||
| < 5 cm | 17 (37.8) | 12 (41.4) | 27 (39.1) | 2 (40) | ||
| > 5 cm | 28 (62.2) | 17 (58.6) | 42 (60.9) | 3 (60) | ||
| Macroscopic type | 0.479 | 0.144 | ||||
| Ulcerofungating | 22 (48.8) | 17 (57.1) | 36 (52.1) | 3 (60) | ||
| Ulceroinfiltrative | 22 (48.8) | 12 (42.9) | 33 (47.9) | 1 (20) | ||
| Polypoid | 1 (2.4) | 0 (0) | 0 (0) | 1 (20) | ||
| Tumor type | 0.206 | 0.219 | ||||
| Adenocarcinoma | 35 (77.7) | 27 (85.1) | 58 (84) | 4 (80) | ||
| Mucinous AdenoCarcinoma | 8 (17.9) | 2 (14.9) | 10 (14) | 0 (0) | ||
| Adeno+neuroendocrine | 1 (2.2) | 0 (0) | 0 (0) | 1 (20) | ||
| Undifferentiated carcinoma | 1 (2.2) | 0 (0) | 1 (2) | 0 (0) | ||
| Grade | 0.805 | 0.743 | ||||
| 1 | 4 (10.5) | 4 (14.8) | 7 (11.7) | 1 (20) | ||
| 2 | 32 (84.2) | 21 (77.8) | 49 (81.7) | 4 (80) | ||
| 3 | 2 (5.3) | 2 (7.4) | 4 (6.7) | 0 (0) | ||
| Tumor necrosis | 0.451 | 0.660 | ||||
| - | 5 (11.1) | 5 (17.2) | 9 (13) | 1 (20) | ||
| + | 40 (88.9) | 24 (82.8) | 60 (87) | 4 (80) | ||
| Stromal desmoplasia | 0.258 | 0.556 | ||||
| Mild | 12 (26.7) | 5 (17.2) | 15 (21.7) | 2 (40) | ||
| Moderate | 28 (62.2) | 23 (79.3) | 48 (69.6) | 3 (60) | ||
| Severe | 5 (11.1) | 1 (3.4) | 6 (8.7) | 0 (0) | ||
| Stromal inflammatory reaction | 0.512 | 0.352 | ||||
| Mild | 9 (20) | 9 (31) | 18 (26.1) | 0 (80) | ||
| Moderate | 31 (68.9) | 18 (62.1) | 45 (65.2) | 4 (80) | ||
| Intense | 5 (11.1) | 2 (6.9) | 6 (8.7) | 1 (20) |
MLH: MutL homologue; MSH: MutS homologue; NOS: Not otherwise specified.
Table 2.
MLH1/MSH2 expression and other clinicopathological parameters
| MLH1 (+) | MLH1 (-) | P | MSH2 (+) | MSH2 (-) | P | |
| PT Crohn1 | 0.571 | 0.492 | ||||
| - | 42 (93.3) | 26 (89.7) | 63 (91.3) | 3 (60) | ||
| + | 3 (6.7) | 3 (10.3) | 6 (8.7) | 2 (40) | ||
| PT Lymph2 | 0.953 | 0.172 | ||||
| - | 37 (82.2) | 24 (82.8) | 58 (84.1) | 3 (60) | ||
| + | 8 (17.8) | 5 (17.2) | 11 (15.9) | 2 (40) | ||
| Tumor border | 0.747 | 0.184 | ||||
| Expansive | 7 (15.6) | 3 (10.3) | 8 (11.6) | 2 (40) | ||
| Infiltrating | 17 (37.8) | 13 (44.8) | 29 (42) | 1 (20) | ||
| Both | 21 (46.7) | 13 (44.8) | 32 (46.4) | 2 (40) | ||
| Invasion level | 0.865 | 0.002 | ||||
| Submucosa | 1 (2.2) | 0 (0) | 0 (0) | 1 (20) | ||
| Muscularis | 7 (15.6) | 4 (13.8) | 10 (14.5) | 1 (20) | ||
| Subserosa | 33 (73.3) | 22 (75.9) | 52 (75.4) | 3 (60) | ||
| Serosa | 4 (8.9) | 3 (10.3) | 7 (10.1) | 0 (0) | ||
| PNI3 | 0.633 | 0.394 | ||||
| - | 27 (60) | 19 (65.5) | 42 (60.9) | 4 (80) | ||
| + | 18 (40) | 10 (34.5) | 27 (39.1) | 1 (20) | ||
| LVI4 | 0.522 | |||||
| - | 7 (15.6) | 3 (10.3) | 9 (13) | 4 (80 | 0.660 | |
| + | 38 (84.4) | 26 (89.7) | 60 (87) | 1 (20) | ||
| BVI5 | 0.114 | 0.927 | ||||
| - | 38 (84.4) | 20 (69) | 54 (78.3) | 4 (80) | ||
| + | 7 (15.6) | 9 (31) | 15 (21.7) | 1 (20) | ||
| LN metastasis | 0.146 | 0.816 | ||||
| - | 25 (58.1) | 21 (75) | 43 (65.2) | 3 (60) | ||
| + | 18 (41.9) | 7 (25) | 23 (34.8) | 2 (20) | ||
| Survival | 0.035 | 0.562 | ||||
| Disease-free | 19 (45.2) | 13 (50) | 31 (48.4) | 2 (40) | ||
| Recurrence/metastasis | 2 (4.8) | 6 (23.1) | 7 (10.9) | 1 (20) | ||
| Extend | 21 (50) | 7 (26.9) | 26 (40.6) | 2 (40) | ||
| Stage (AJCC- 2002)6 | 0.119 | 0.520 | ||||
| 1 | 5 (11.6) | 3 (10.7) | 6 (9.1) | 2 (40) | ||
| 2 | 18 (41.8) | 17 (60.7) | 34 (51.5) | 1 (20) | ||
| 3 | 16 (37.2) | 5 (17.8) | 20 (30.3) | 1 (20) | ||
| 4 | 4 (9.4) | 3 (10.7) | 6 (9.1) | 1 (20) | ||
| p53 expression | 0.227 | < 0.05 | ||||
| ≤ 10% | 27 (60) | 16 (57.1) | 0 (0) | 5 (100) | ||
| > 10 % | 18 (40) | 12 (42.9) | 74 (100) | 0 (0) |
Peritumoral Crohn-like inflammation;
Peritumoral lymphocytic inflammation;
Perineural invasion;
Lymph vessel invasion;
Blood vessel invasion;
American Joint Committee of Cancer.
All MSH2-negative tumors had no or low p53 expression, while MSH2-positive cases had p53 expression ≥ 10% (P < 0.05). MLH1/MSH2 expressions were not significantly associated with other histopathological variables, such as perineural invasion, lymphatic/blood vessel invasion, peritumoral Crohn’s-like lymphoid reaction. On the other hand, 80% of cases with loss of MSH2 expression had no perineural, lymphatic and blood vessel invasion (Table 2).
Our study included 32 (41.6%) females and 45 (58.4%) males, with average age of 56 (range, 23-77) years. The mean follow-up in surviving patients was 34 (range, 3-97) mo. The patients with MSH2-negative/MLH-positive carcinomas more frequently died of disease (average 12.5 ± 1.06 mo post-operatively) than the patients with MLH1-positive/MSH2-positive carcinomas (47.08 ± 5.26 mo post-operatively) (Table 3). However, overall and disease-free survival analysis did not show significant differences among the four groups of patients
Table 3.
Relation of MLH1/MSH2 expression and survival time (mean ± SD, mo)
| n | Survival time | P | |
| MLH1 (+)/MSH2 (+) | 40 | 47.08 ± 5.26 | 0.065 |
| MLH1 (+)/MSH2 (-) | 3 | 12.50 ± 1.06 | |
| MLH1 (-)/MSH2 (+) | 24 | 71.71 ± 8.65 | |
| MLH1 (-)/MSH2 (-) | 2 | 51.00 ± 16.26 |
In COX regression analysis, only lymph node metastasis and stage were found as independent prognostic factors in all clinicopathological features. But loss of MLH1 and MSH2 expressions did not show prognostic significance (Table 4).
Table 4.
Results of COX regression analysis
| P | |
| Loss of MLH1 expression | 0.127 |
| Loss of MSH2 expression | 0.325 |
| p53 over-expression | 0.417 |
| Stromal reaction | 0.124 |
| Stromal inflammatory reaction | 0.407 |
| Level of invasion | 0.572 |
| Perineural invasion | 0.267 |
| Lymphatic invasion | 0.085 |
| Blood vessel invasion | 0.769 |
| Peritumoral Crohn’s-like inflammation | 0.247 |
| Peritumoral lymphocytic inflammation | 0.952 |
| Stage | 0.000 |
| Lymph node metastasis | 0.000 |
DISCUSSION
One of the two different pathogenetic pathways in colorectal carcinogenesis is mutator pathway which is characterized by explicit microsatellite instability (MSI). Mutator pathway covers DNA mismatch repair genes like MLH1 and MSH2. Identification of MSI status of large bowel adenocarcinomas is clinically important, since patients with MSI carcinomas demonstrated several distinct features compared to microsatellite stability (MSS) carcinomas[22-25]. It has also been suggested that MSI carcinomas might be particularly sensitive to 5-fluorouracil-based adjuvant chemotherapy[26,27]. Besides, patients with MSI tumors are considered to be at risk of developing metachronous colorectal cancers and need long-term colonoscopic surveillance[28].
As mentioned in previous studies, immunohisto-chemical analysis of MLH1 and MSH2 protein expression represents a rapid, easier and less costly alternative method for detection of colorectal tumors of the mutator phenotype[3,15,16,29-32]. This analysis could be performed in the histopathology laboratories as routine immunohistochemical staining, while genetic analysis of MSI status is time-consuming and expensive and requires specialized equipment[3,28]. This difference is important for developing countries like Turkey. Lindor et al[11] showed that absence of expression of MLH1 or MSH2 had a 100% specificity and 92.3% sensitivity for predicting a tumor with MSI-H phenotype. So, immunohistochemical analysis of MLH1/MSH2 proteins can be used as a prescreening method for mutation analysis of mismatch repair genes[33-35]. The inactivation of MLH1 and MSH2 genes is resulted in loss of expression of these proteins by immunohistochemistry.
We report here our first results of MMR (mismatch repair) analysis in Turkish sporadic colorectal carcinomas. To our knowledge, this TMA-based study about MSI status was completely performed for the first time in Turkey. We found loss of MLH1 expression in 29 (39.2%) and loss of MSH2 expression in 5 (6.8%) of cases. In addition, loss of either MLH1 or MSH2 expression was seen in 32 (43.2%) of cases, while loss of expression of both MLH1 and MSH2 was detected in 2 (3.1%) of cases. A previous study demonstrated that 351 (87.3%) cases expressed either one or both of MLH1 and MSH2; MLH1 and MSH2 were not expressed in 35 (8.7%) and 19 (4.7%) cases, respectively; and 3 cases showed neither MLH1 nor MSH2 expression[33].
Lanza et al[28] found that 106 (80.3%) MSI-H carcinomas showed complete loss of MLH1 expression, 14 (10.6%) displayed complete loss of MSH2 expression, 12 (9.1%) MSI-H carcinomas demonstrated normal expression of both MLH1 and MSH2, but no MSI-H tumors showed lack of both MLH1 and MSH2 expression. In contrast, nuclear immunoreactivity for MLH1 and MSH2 proteins was observed in all MSS and MSI-L tumors analyzed[28]. The common finding of all these studies is that MLH1 extinction is more frequent than MSH2 extinction. This supports the hypothesis that involvement of the MLH1 gene is prevalent in the development of sporadic large bowel MSI cancers.
In our study, unlike previous studies, MLH1-negative cases had no gender, age or tumor localization predominance. On the other hand, loss of MSH2 seemed to be related to some parameters more than loss of MLH1; for example, all MSH2-negative cases were women, 60% of them were located in the right colon and no serosal involvement was detected in MSH2-negative cases. Other interesting but statistically insignificant findings were both MLH1-negative/MSH-nagative cases had peritumoral Crohn’s-like lymphocytic infiltration and prominent intratumoral neutrophilic infiltration.
Although there was no statistical significance, 80% of MSH2-negaative cases did not show perineural invasion, lymphatic invasion and blood vessel invasion. Bernardo et al[36] found that only vascular invasion was significantly correlated with MSH2 expression. Similarly, Wright et al[14] found that in MLH1- and MSH2-negative carcinomas, extramural vascular, lymphatic and perineural invasion were all significantly less than the others.
In our study, 72.4% of MLH1-negative and 80% of MSH2-negative cases were ≥ 50 years. Tumor size was ≥ 5 cm in 58.6% of MLH1-negative cases and 60% of MSH2-negative cases. But there was no significant correlation between these parameters. Lanza et al[28] showed that MLH1- and MSH2-negative carcinomas were located in the proximal colon, more often of > 7 cm in diameter, poorly-differentiated, and had expanding pattern of growth and intense peritumoral Crohn’s-like lymphoid reaction.
Several studies demonstrated that MSI tumors were correlated with clinicopathological features, such as right colon location, mucinous type, expansive borders, peritumoral Crohn’s-like lymphoid reaction and peritumoral lymphoid response[10,29,31,37]. A Japanese study of a series of colorectal carcinomas did not find any correlation between MSI status and any clinicopathological features except for tumor location in the proximal colon[38].
The p53 gene is mutated in 70% of colorectal carcinomas[39]. Over-expression of p53 has been used as an indicator of p53 mutational status in many studies[33]. In the present study, there was no significant difference between p53 expression and MLH1 expression, whereas a significant correlation between low p53 expression and loss of MSH2 expression was detected (P < 0.05). Park et al[33] demonstrated that there was a significant difference in p53 expression between the MLH1-positive group and MLH1-deficient group, indicating a correlation of loss of MLH1 or MSH2 expression with low p53 expression. We found that all MSH2-negative tumors showed low p53 immunostaining. This finding supports that different carcinogenic pathways, different molecular changes and different genes can be affected. Thus, an inverse correlation between genetic alterations of p53 and the mismatch repair system may simply reflect different carcinogenic pathways.
It has been reported that survival in patients with colorectal cancer with MMR gene defect is better than without one[24,27]. In our study, loss of MLH1/MSH2 expression had no significant correlation with the survival. It has been clearly elucidated that tumor stage and lymph node metastasis are the independent prognostic factors for colorectal carcinomas. Hameed et al[40] found similar results in their study. Chapusot et al[41] found that three independent factors were significantly associated with the loss of expression of MLH1 and MSH2: proximal location, the presence of Crohn’s-like lymphoid reaction and poor differentiation.
Interestingly, in our study overall survival of the cases with loss of only MLH1 or both MLH1 and MSH2 expression was longer than those with both MLH1- and MSH2-positive or with only MSH2-negative expression. Although this finding is not statistically significant (P = 0.065), we think that this analysis should be repeated and combined with the result of genetic analysis which will be planned for near future. This finding showed some similarities with other studies. Lanza et al[28] found that cases with MLH1-negative carcinomas more often died of disease, but the survival of the cases was not statistically significant. However, Gafa et al[37] found that cases with both MLH1- and MSH2-negative carcinoma showed a better clinical outcome and survival.
In conclusion, our findings suggest that the assessment of MSI status using immunohistochemistry is important in genetic and biologic characterization of colorectal carcinomas. Turkish patients with colorectal cancers show some similarities with other populations in terms of histopathological features and MSI status. Although prognostic importance remains controversial, immunohistochemical analysis of MMR genes may be used as routine histopathological examination of colorectal cancer tissues. However, genetic analysis should be combined with these results.
COMMENTS
Background
Human colorectal cancer (CRC) is one of the leading cancers in most countries. Understanding pathogenetic pathways in colorectal carcinogenesis is important for diagnosis and treatment of these patients. DNA mismatch repair (MMR) genes like MLH1 and MSH2 identify of MSI status of large bowel adenocarcinomas as microsatellite stable (MSS) or MSI. MMR deficiency leads to the accumulation of base-base mismatches and short insertion/deletion mispairs, generated as a consequence of DNA replication errors and homologous recombinations.
Research frontiers
Simply, microsatellite instability (MSI) tumors are seen more frequently in hereditary poliposis colon cancers. Last studies showed that MSI was found also in sporadic CRC. So, searching MSI status of CRC is clinically important, since patients with carcinomas demonstrated several distinct features compared to MSS carcinomas. In this point immunohistochemistry is useful method since it is easy, cheap and reliable method to detect these patients.
Innovations and breakthroughs
This analysis was performed for the first time in Turkey. Loss of MLH1 and/or MSH2 expression is important finding to predict their different morphology and behaviour in CRC cases.
Applications
Searching MSI status of CRC should be performed more commonly. Immunohistochemical analysis of MSI status of CRC cases may be used as screening method in developing countries.
Peer review
This is the first study of MMR deficiency and its prognostic importance in sporadic CRCs in Turkish population, which makes it valuable. The study assessed the incidence of MLH1 and MSH2 expression losses in Turkish sporadic CRCs and the data was compared with survival and clinicopathological features of the patients.
Footnotes
Supported by the Research Foundation of Istanbul University, No T-493/25062004
S- Editor Zhu LH L- Editor Kumar M E- Editor Liu Y
References
- 1.Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ. 2001;8:1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- 2.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 3.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, Halling KC, McDonnell SK, Schaid DJ, Walsh Vockley C, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69:780–790. doi: 10.1086/323658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haydon AM, Jass JR. Emerging pathways in colorectal-cancer development. Lancet Oncol. 2002;3:83–88. doi: 10.1016/s1470-2045(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 6.Redston M. Carcinogenesis in the GI tract: from morphology to genetics and back again. Mod Pathol. 2001;14:236–245. doi: 10.1038/modpathol.3880292. [DOI] [PubMed] [Google Scholar]
- 7.Vasen HF, Wijnen JT, Menko FH, Kleibeuker JH, Taal BG, Griffioen G, Nagengast FM, Meijers-Heijboer EH, Bertario L, Varesco L, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–1027. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 8.Liu B, Nicolaides NC, Markowitz S, Willson JK, Parsons RE, Jen J, Papadopolous N, Peltomäki P, de la Chapelle A, Hamilton SR. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 9.Hatch SB, Lightfoot HM, Garwacki CP, Moore DT, Calvo BF, Woosley JT, Sciarrotta J, Funkhouser WK, Farber RA. Microsatellite instability testing in colorectal carcinoma: choice of markers affects sensitivity of detection of mismatch repair-deficient tumors. Clin Cancer Res. 2005;11:2180–2187. doi: 10.1158/1078-0432.CCR-04-0234. [DOI] [PubMed] [Google Scholar]
- 10.Kim H, Jen J, Vogelstein B, Hamilton SR. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- 11.Young J, Simms LA, Biden KG, Wynter C, Whitehall V, Karamatic R, George J, Goldblatt J, Walpole I, Robin SA, et al. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:2107–2116. doi: 10.1016/S0002-9440(10)63062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, et al. Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology. 1996;111:307–317. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 13.Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, Robertson MA, Nichols MF, Gruenthal KM, Lynch BJ, et al. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158:1517–1524. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright CL, Stewart ID. Histopathology and mismatch repair status of 458 consecutive colorectal carcinomas. Am J Surg Pathol. 2003;27:1393–1406. doi: 10.1097/00000478-200311000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Cawkwell L, Gray S, Murgatroyd H, Sutherland F, Haine L, Longfellow M, O'Loughlin S, Cross D, Kronborg O, Fenger C, et al. Choice of management strategy for colorectal cancer based on a diagnostic immunohistochemical test for defective mismatch repair. Gut. 1999;45:409–415. doi: 10.1136/gut.45.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 17.Plevová P, Krepelová A, Papezová M, Sedláková E, Curík R, Foretová L, Navrátilová M, Novotný J, Zapletalová J, Palas J, et al. Immunohistochemical detection of the hMLH1 and hMSH2 proteins in hereditary non-polyposis colon cancer and sporadic colon cancer. Neoplasma. 2004;51:275–284. [PubMed] [Google Scholar]
- 18.Greene FL, Page DL, Fleming ID, Fritz AG, Balch CM, Haller DG, Morrow M. AJCC cancer staging handbook TNM classification of malignant tumors. 6th ed. New York: Springer-Verlag; 2002. pp. 127–138. [Google Scholar]
- 19.Hamilton SR, Aaltonen LA. Tumors of the Digestive system, Pathology and Genetics, World Health Organization Classification of Tumors. Lyon: IARC Press; 2000. pp. 103–119. [Google Scholar]
- 20.Jass JR, Atkin WS, Cuzick J, Bussey HJ, Morson BC, Northover JM, Todd IP. The grading of rectal cancer: historical perspectives and a multivariate analysis of 447 cases. Histopathology. 1986;10:437–459. doi: 10.1111/j.1365-2559.1986.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 21.Graham DM, Appelman HD. Crohn's-like lymphoid reaction and colorectal carcinoma: a potential histologic prognosticator. Mod Pathol. 1990;3:332–335. [PubMed] [Google Scholar]
- 22.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 23.Halling KC, French AJ, McDonnell SK, Burgart LJ, Schaid DJ, Peterson BJ, Moon-Tasson L, Mahoney MR, Sargent DJ, O'Connell MJ, et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999;91:1295–1303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 24.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 25.Wright CM, Dent OF, Barker M, Newland RC, Chapuis PH, Bokey EL, Young JP, Leggett BA, Jass JR, Macdonald GA. Prognostic significance of extensive microsatellite instability in sporadic clinicopathological stage C colorectal cancer. Br J Surg. 2000;87:1197–1202. doi: 10.1046/j.1365-2168.2000.01508.x. [DOI] [PubMed] [Google Scholar]
- 26.Elsaleh H, Joseph D, Grieu F, Zeps N, Spry N, Iacopetta B. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 27.Hemminki A, Mecklin JP, Järvinen H, Aaltonen LA, Joensuu H. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–928. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- 28.Lanza G, Gafà R, Maestri I, Santini A, Matteuzzi M, Cavazzini L. Immunohistochemical pattern of MLH1/MSH2 expression is related to clinical and pathological features in colorectal adenocarcinomas with microsatellite instability. Mod Pathol. 2002;15:741–749. doi: 10.1097/01.MP.0000018979.68686.B2. [DOI] [PubMed] [Google Scholar]
- 29.Kruschewski M, Noske A, Haier J, Runkel N, Anagnostopoulos Y, Buhr HJ. Is reduced expression of mismatch repair genes MLH1 and MSH2 in patients with sporadic colorectal cancer related to their prognosis? Clin Exp Metastasis. 2002;19:71–77. doi: 10.1023/a:1013853224644. [DOI] [PubMed] [Google Scholar]
- 30.Stone JG, Robertson D, Houlston RS. Immunohistochemistry for MSH2 and MHL1: a method for identifying mismatch repair deficient colorectal cancer. J Clin Pathol. 2001;54:484–487. doi: 10.1136/jcp.54.6.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jover R, Payá A, Alenda C, Poveda MJ, Peiró G, Aranda FI, Pérez-Mateo M. Defective mismatch-repair colorectal cancer: clinicopathologic characteristics and usefulness of immunohistochemical analysis for diagnosis. Am J Clin Pathol. 2004;122:389–394. doi: 10.1309/V9PG-K2Y2-60VF-VULR. [DOI] [PubMed] [Google Scholar]
- 32.Thibodeau SN, French AJ, Roche PC, Cunningham JM, Tester DJ, Lindor NM, Moslein G, Baker SM, Liskay RM, Burgart LJ, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res. 1996;56:4836–4840. [PubMed] [Google Scholar]
- 33.Park IJ, Kim HC, Kim JS, Yu ES, Yu CS, Kim JC. Correlation between hMLH1/hMSH2 and p53 protein expression in sporadic colorectal cancer. Hepatogastroenterology. 2005;52:450–454. [PubMed] [Google Scholar]
- 34.Lamberti C, Kruse R, Ruelfs C, Caspari R, Wang Y, Jungck M, Mathiak M, Malayeri HR, Friedl W, Sauerbruch T, et al. Microsatellite instability-a useful diagnostic tool to select patients at high risk for hereditary non-polyposis colorectal cancer: a study in different groups of patients with colorectal cancer. Gut. 1999;44:839–843. doi: 10.1136/gut.44.6.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendriks Y, Franken P, Dierssen JW, De Leeuw W, Wijnen J, Dreef E, Tops C, Breuning M, Bröcker-Vriends A, Vasen H, et al. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol. 2003;162:469–477. doi: 10.1016/S0002-9440(10)63841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernardo CG, González JJ, Sanz L, Barbón E, Noval JG, Fresno MF, Aza J. Mismatch repair protein MSH2 expression and prognosis of colorectal cancer patients. Int J Biol Markers. 2004;19:190–195. [PubMed] [Google Scholar]
- 37.Gafà R, Maestri I, Matteuzzi M, Santini A, Ferretti S, Cavazzini L, Lanza G. Sporadic colorectal adenocarcinomas with high-frequency microsatellite instability. Cancer. 2000;89:2025–2037. [PubMed] [Google Scholar]
- 38.Senba S, Konishi F, Okamoto T, Kashiwagi H, Kanazawa K, Miyaki M, Konishi M, Tsukamoto T. Clinicopathologic and genetic features of nonfamilial colorectal carcinomas with DNA replication errors. Cancer. 1998;82:279–285. [PubMed] [Google Scholar]
- 39.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 40.Hameed F, Goldberg PA, Hall P, Algar U, van Wijk R, Ramesar R. Immunohistochemistry detects mismatch repair gene defects in colorectal cancer. Colorectal Dis. 2006;8:411–417. doi: 10.1111/j.1463-1318.2006.00956.x. [DOI] [PubMed] [Google Scholar]
- 41.Chapusot C, Martin L, Mungra N, Rageot D, Bouvier AM, Bonithon Kopp C, Ponnelle T, Faivre J, Piard F. Sporadic colorectal cancers with defective mismatch repair display a number of specific morphological characteristics: relationship between the expression of hMLH1 and hMSH2 proteins and clinicopathological features of 273 adenocarcinomas. Histopathology. 2003;43:40–47. doi: 10.1046/j.1365-2559.2003.01641.x. [DOI] [PubMed] [Google Scholar]


