Significance
Actin filaments are the fundamental building blocks for cellular motility in muscle cells. However, the regulation of actin filaments in sarcomeres is poorly understood. Lmod is a class of potent tandem-G-actin–binding nucleators in muscle cells. Lmod mutations, deletion, or instability lead to lethal nemaline myopathy. Here we report the crystal structure of actin–Lmod2 complex and complementary functional studies. Our data collectively unraveled the mechanisms of Lmod’s essential functions as an actin nucleator and filament length regulator, thus providing a solid foundation for understanding the molecular basis of disease-causing mutations and malfunctions of Lmod that often are life threatening. Most importantly, our results suggest a previously unrecognized level of regulation in mammalian signal transduction mediated by certain tandem-G-actin–binding nucleators.
Keywords: actin nucleation, nemaline myopathy, pointed-end elongation
Abstract
Leiomodin (Lmod) is a class of potent tandem-G-actin–binding nucleators in muscle cells. Lmod mutations, deletion, or instability are linked to lethal nemaline myopathy. However, the lack of high-resolution structures of Lmod nucleators in action severely hampered our understanding of their essential cellular functions. Here we report the crystal structure of the actin–Lmod2162–495 nucleus. The structure contains two actin subunits connected by one Lmod2162–495 molecule in a non–filament-like conformation. Complementary functional studies suggest that the binding of Lmod2 stimulates ATP hydrolysis and accelerates actin nucleation and polymerization. The high level of conservation among Lmod proteins in sequence and functions suggests that the mechanistic insights of human Lmod2 uncovered here may aid in a molecular understanding of other Lmod proteins. Furthermore, our structural and mechanistic studies unraveled a previously unrecognized level of regulation in mammalian signal transduction mediated by certain tandem-G-actin–binding nucleators.
In response to environmental or cellular signals, eukaryotic cells use actin nucleators to convert globular actin monomers (G-actin) into actin oligomers (actin nuclei), which then quickly lead to actin filaments (F-actin). Actin-related protein 2/3 (Arp2/3), formins, and tandem-G-actin–binding proteins are the three classes of known actin nucleators in nonmuscle cells (1–7). Arp2/3-mediated actin nucleation produces branched actin networks, whereas formins and tandem-G-actin–binding nucleators result in long, unbranched actin filaments (1–7). In muscle cells, the specific mechanisms for actin nucleation and maintenance in sarcomeres were poorly understood (8). Recent studies have uncovered actin nucleation activities of the nebulin–N-WASP complex (9) and of formin proteins FHOD3 (10–12), mDia2, DAAM, FMNL1, and FMNL2 (13, 14) in sarcomeres. In particular, leiomodin (Lmod) has been identified as a class of potent tandem-G-actin–binding nucleators in muscle cells (15, 16); Lmod1 is found in smooth muscle of many human tissues, and Lmod2 and Lmod3 are found in cardiac and skeletal muscle (17). Lmod2 knockdown severely compromises sarcomere organization and assembly in muscle cells (15), whereas mutations, deletions (18), or instability (19) in Lmod3 underlies severe, often lethal, human nemaline myopathy.
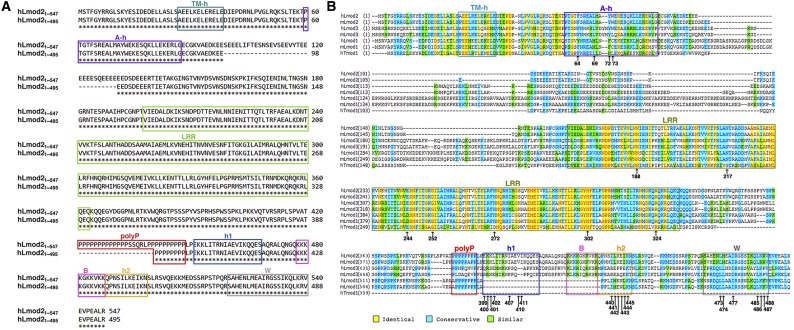
Full-length human Lmod2 is predicted to have 547 residues with two regions of low sequence complexity, an acidic region between residues 97–138 and a polyproline (polyP) region between residues 421–448 (Fig. S1A). Probably because low-complexity regions tend to be intrinsically disordered, previous studies of human Lmod2 used a protein construct that deleted residues 99–130 in the acidic region and residues 421–440 in the polyP region, resulting in Lmod21–495 (15, 16). Another study on chicken Lmod2 removed 12 residues in the polyP region (20). In all cases, Lmod2 remained fully functional (15, 16, 20). Therefore, in the present study we focused on the human Lmod21–495 construct as previously used (Fig. S1A) (15, 16).
Fig. S1.
Lmod sequences. (A) Sequence alignment of human Lmod21–547 and Lmod21–495. (B) Sequence alignment of human Tmod1 (hTmod1), human Lmod (hLmod), and mouse Lmod (mLmod) isoforms 1–3. The human Lmod21–495 sequence is at the top, and the residues that were tested by mutagenesis in this study are indicated by arrows.
Human Lmod21–495 has three actin-binding sites (15). The first ∼340 residues are about 45% identical to the pointed-end capping protein tropomodulin 1 (Tmod1) (21) and contain a tropomyosin-binding helix (TM-h) and two actin-binding sites [an actin-binding helix (A-h) and a leucine-rich repeat (LRR) domain] (Fig. 1A and Figs. S1 and S2A). The C-terminal ∼150-residue extension of Lmod2 includes two predicted short helices (h1 and h2), a basic segment (B) harboring the nuclear localization sequence (16), and a Wiskott–Aldrich syndrome protein-homology 2 (W) domain (Fig. 1A and Figs. S1 and S2A). Thus, Lmod2 has the capacity to bind three actin subunits and one tropomyosin (15). Unexpectedly, tropomyosin promoted Lmod2-mediated actin nucleation only weakly (15). In sharp contrast, tropomyosin substantially enhanced the binding of Lmod2 to the pointed end of preformed actin filament for controlled elongation in cardiac muscle (16, 20). In the absence of high-resolution structures of the actin–Lmod complex, however, rationalization of these seemingly contradictory findings is difficult.
Fig. 1.
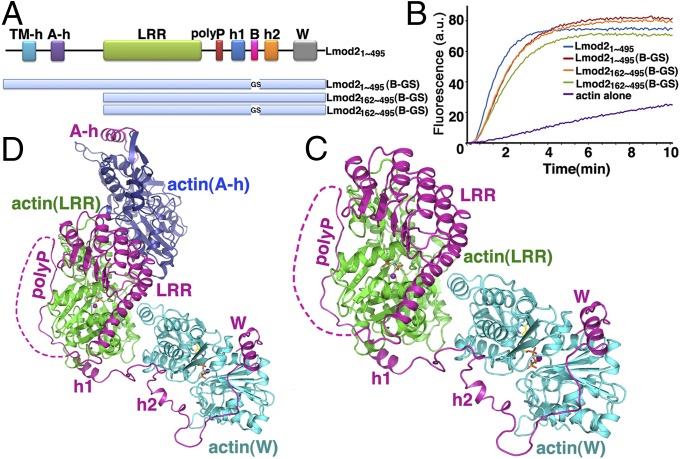
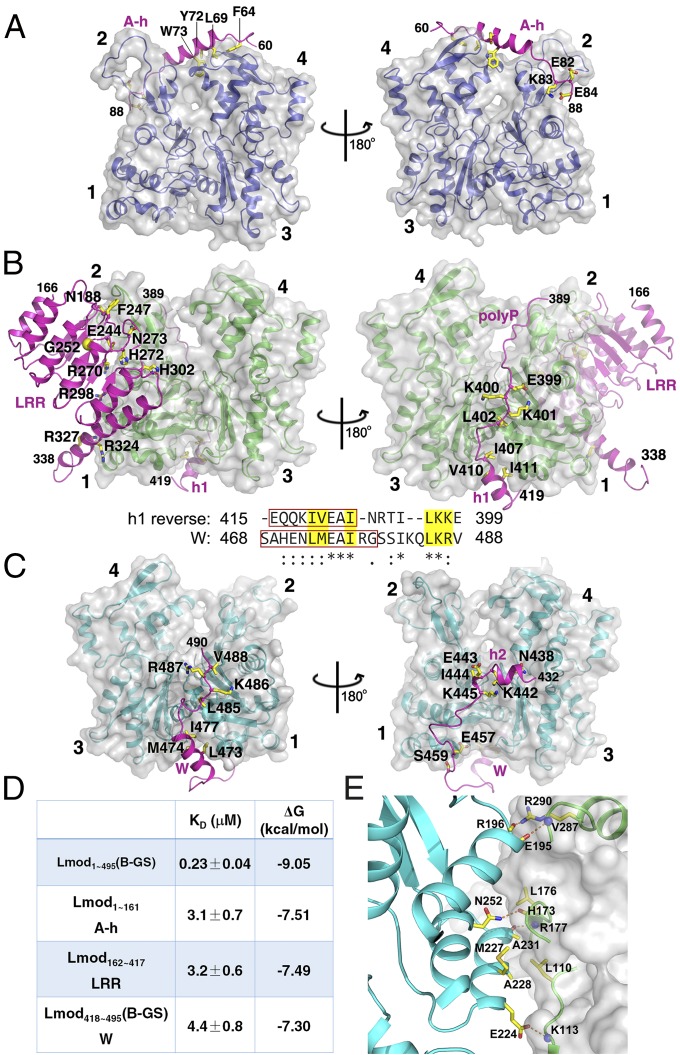
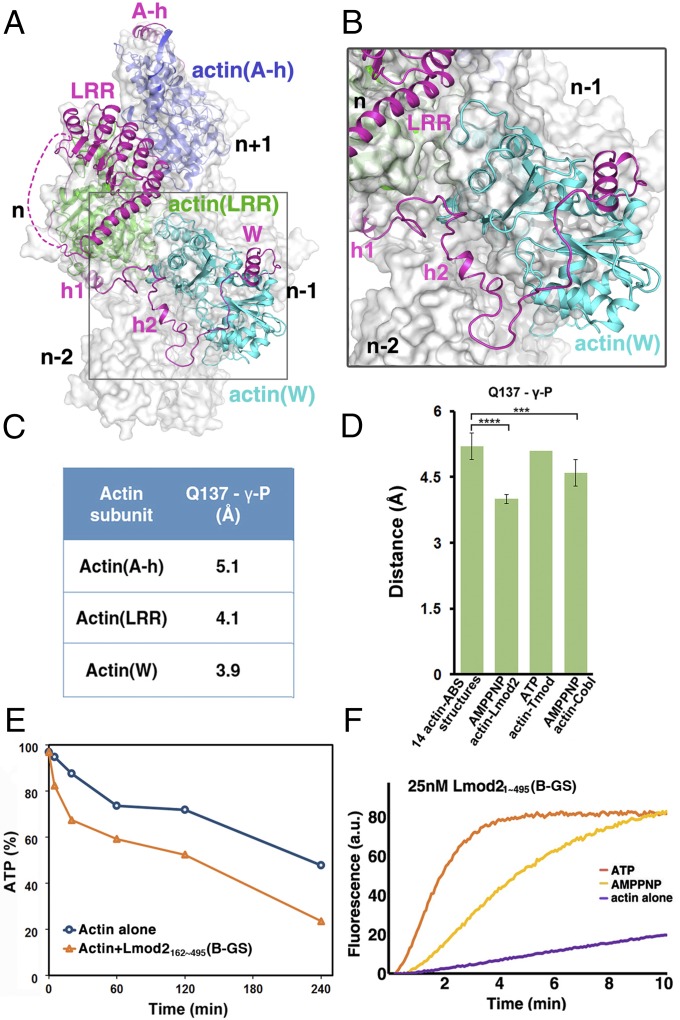
Structure of actin–Lmod2. (A) Domain organization of human Lmod21–495 and constructs used in this study. (B) Pyrene-based activity assay of Lmod21–495 and its various constructs. a.u., arbitrary units. (C) The crystal structure of actin–Lmod2162–495(B-GS). All residues are visualized except an internal flexible region (residues 339–388) between LRR and polyP, the extreme four N-terminal residues (162–165), and five C-terminal residues (491–495). AMPPNP is shown as ball-and-stick models, and the Mg2+ ions are shown as purple spheres. (D) The modeled structure of actin–Lmod21–495 in which the actin(A-h)–A-h complex structure was borrowed from the Tmod1 structure (PDB ID code: 4PKG) and combined with our crystal structure of actin–Lmod2162–495(B-GS). See also Movies S1–S3.
Fig. S2.
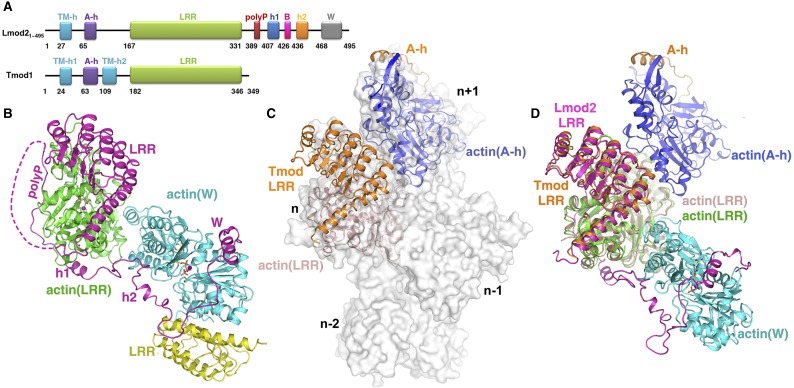
Structure of actin–Lmod2162–495(B-GS). (A) Comparison of domain organization of human Lmod21–495 and Tmod1. (B) The structure of actin–Lmod2162–495(B-GS) as observed in the asymmetric unit. It contains two actin subunits (in green and cyan) bound with one Lmod2162–495(B-GS) (in magenta) and one extra LRR domain from degradation (in yellow). (C) The modeled actin–Tmod1 structure is superimposed on subunits n+1 and n of the pointed end of the actin filament. (D) Superposition of our actin–Lmod2162–495(B-GS) structure with the modeled actin–Tmod1 structure at the actin(LRR)–LRR. Lmod2 is shown in magenta, and the interacting actin(LRR) and actin(W) are in green and cyan, respectively. Tmod1 is in orange, and its interacting actin(A-h) and actin(LRR) are in blue and pink, respectively.
Historically study of the crystallographic structure of the complexes of actin nucleators with oligomeric actin or of F-actin–binding proteins with F-actin was difficult because actin dimers and trimers are kinetically unstable, and actin tetramers rapidly polymerize into F-actin that is refractory to crystallization (22). Indeed, although Arp2/3 has been subjected to intensive structural studies (23–25), the crystal structure of the actin–Arp2/3 complex has eluded investigation so far. Before our study (26), the only available crystal structure of this kind was the yeast formin Bni1p FH2 domain that binds to two crystallographically related tetramethylrhodamine-modified actin (TMR–actin) subunits in a pseudo short-pitch fashion (27). However, the large size of TMR likely interferes with its interaction with actin and with actin function, thus limiting the use of TMR–actin in investigations of crystal structure.
To enable crystallographic studies of biological actin complexes, our group recently has developed a double-mutant strategy in which actin-binding proteins and two types of nonpolymerizable actin mutants are combined to form stable complexes amenable to crystallization (26). This strategy made possible the rapid determination of the first two crystal structures of oligomeric actin with tandem-G-actin–binding nucleator complexes: a mammalian nucleator Cordon-bleu (Cobl) (26) and a bacterial effector Vibrio parahaemolyticus protein L (VopL) (28). Importantly, the observed non–filament-like conformation in actin–Cobl and the filament-like conformation in actin–VopL together suggest that both types of conformation are fully accessible to an actin complex obtained via the double-mutant strategy; thus the observed structure most likely reflects its native functional state.
Here we report the crystal structure of the actin–Lmod2 nucleus and complementary functional studies. Our data not only unraveled the atomic mechanisms of Lmod’s essential functions in muscle cells but also suggested a previously unrecognized level of regulation in mammalian signal transduction mediated by certain tandem-G-actin–binding nucleators.
Results
Overall Structure of the Actin–Lmod2 Complex.
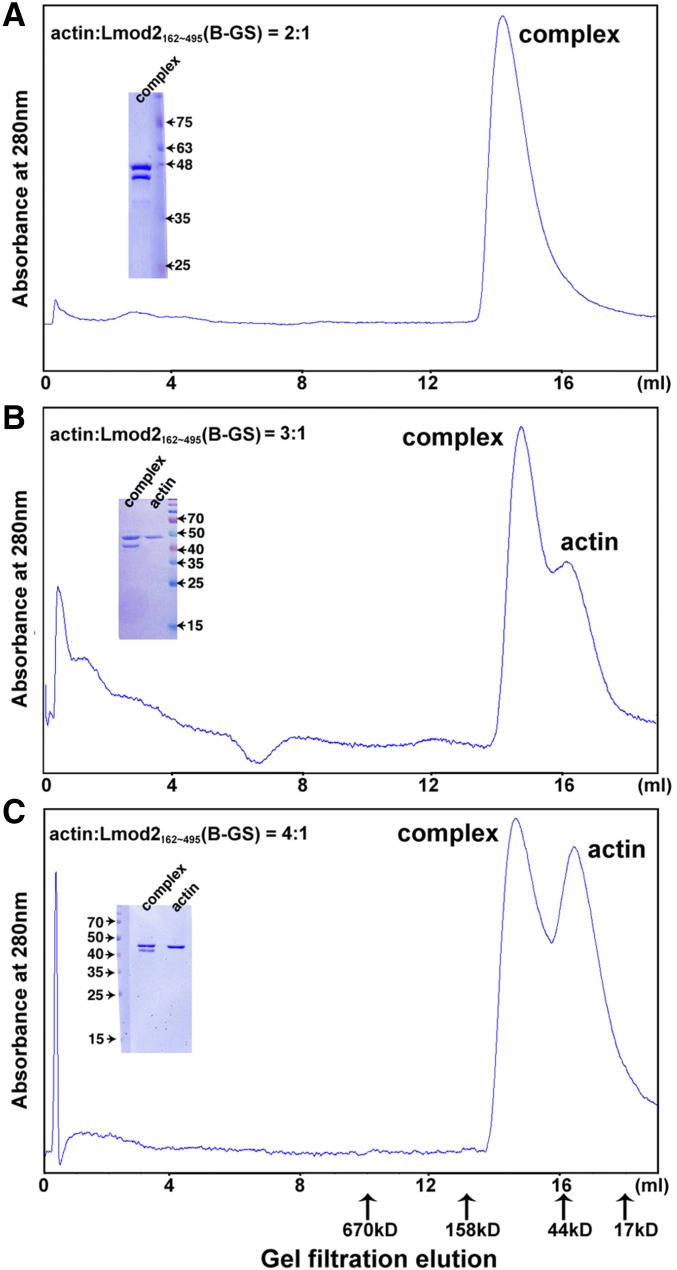
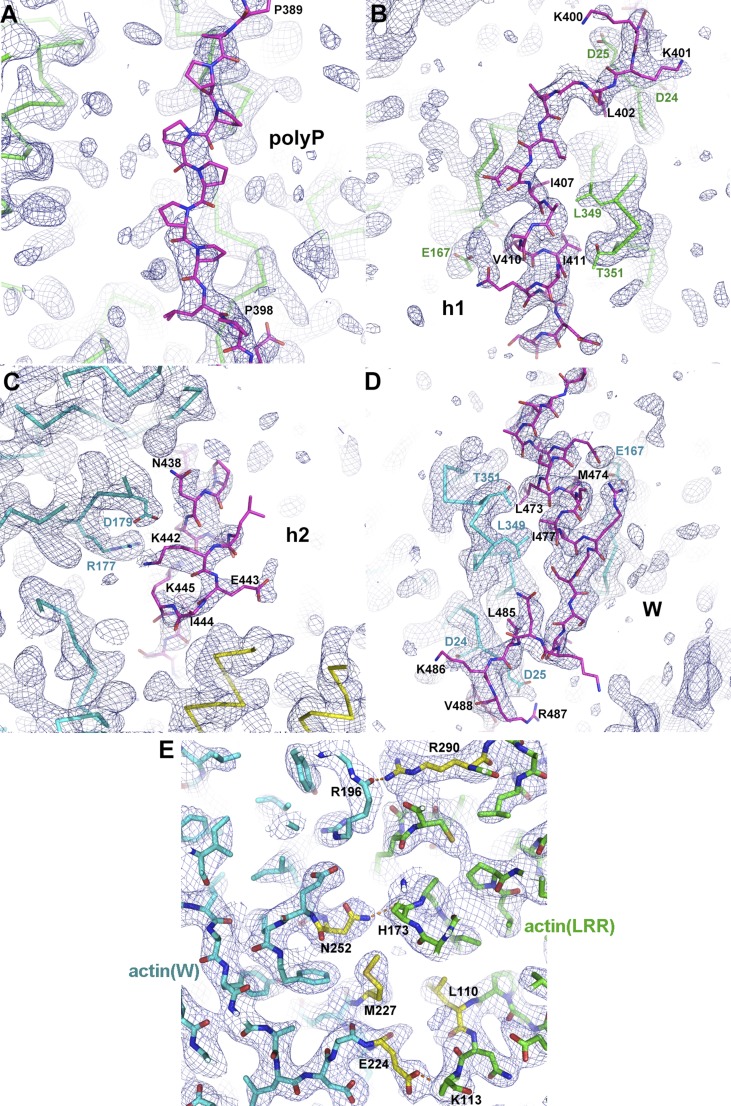
In our attempt to obtain crystals of the actin–Lmod2 complex, we found that the multiple lysine residues in the B segment led to severe protein degradation. Therefore we replaced the region 406KKKKGKKVKK415 with the sequence 406GSGSGGSVGS415, resulting in the Lmod21–495(B-GS) construct (Fig. 1A). This construct remained highly active in actin nucleation (Fig. 1B), as did Lmod2162–495 and Lmod2162–495(B-GS), the minimal nucleating fragments that comprise the second and third actin-binding sites (Fig. 1 A and B). In agreement with the earlier report that Lmod2162–495 interacts with two actin subunits (15), the mixture of nonpolymerizable actin mutants and Lmod2162–495(B-GS) at 2:1 molar ratio was eluted from a Superdex S200 size-exclusion column as a single peak (Fig. S3A), suggesting a well-defined 2:1 actin–Lmod2162–495(B-GS) complex in solution. In contrast, the mixture of actin and Lmod2162–495(B-GS) at higher molar ratios (3:1 or 4:1) generated two elution peaks, one corresponding to the 2:1 actin–Lmod2162–495(B-GS) complex and the other comprising excessive actin monomers (Fig. S3 B and C). Using the 2:1 actin–Lmod2162–495(B-GS) complex peak eluted from size-exclusion chromatography in the presence of AMPPNP, a nonhydrolysable ATP analog, we have determined the crystal structure of the actin–Lmod2162–495(B-GS) complex to a 2.98-Å resolution (Table S1).
Fig. S3.
Size-exclusion chromatography profiles of actin–Lmod2162–495(B-GS) at molar ratios of 2:1 (A), 3:1 (B), and 4:1 (C). The peaks of the 2:1 actin–Lmod2162–495(B-GS) complex and actin monomers are labeled, and corresponding Coomassie-stained SDS/PAGE gels are shown.
Table S1.
Statistics of data collection and structural refinement
| Data collection statistics | |
| Ligand | AMPPNP |
| Space group | P1 |
| Unit cell | |
| a, b, c, Å | 63.35, 65.65, 81.92 |
| α, β, γ, o | 101.29, 90.94, 107.97 |
| Resolution, Å* | 51.10–2.98 (3.14–2.98) |
| Number of unique reflections* | 25,184 (3,658) |
| Multiplicity* | 3.6 (3.6) |
| Completeness, %* | 97.4 (97.2) |
| Rmerge, %* | 13.1 (51.4) |
| I/σ, I* | 10.5 (2.1) |
| Structural refinement statistics | |
| Resolution, Å | 2.98 |
| Number of atoms | 9,323 |
| Rwork/Rfree, %† | 24.82/25.72 |
| Rmsd bond length, Ň | 0.010 |
| Rmsd bond angle, o ‡ | 1.289 |
| Mean B-value, Å2 § | 50.34 |
| Ramachandran plot, % favored/disallowed | 98.0/0.3 |
Values in parentheses are for the highest resolution shell.
The Rfree was calculated by using 5% data that were omitted from structural refinement.
The RMSD of bond lengths and angles from ideal geometry for the final model.
The mean temperature factor for all of the atoms of the polypeptide chains in the asymmetric unit.
Consistent with the 2:1 stoichiometry of actin:Lmod2162–495(B-GS) that was used for crystallization, the structure contains one Lmod2162–495(B-GS) molecule that interacts with two actin subunits: actin(LRR), which associates with Lmod2 in the region of LRR–polyp-h1, and actin(W), which binds to Lmod2 in the region of h2-W (Fig. 1C and Fig. S2B). Additionally, an extra LRR domain in the structure (likely resulting from partial degradation of Lmod2162–495(B-GS) during crystallization as confirmed by SDS/PAGE) binds to the actin(W) subunit (Fig. S2B).
Recently, the crystal structures of actin with human Tmod1 A-h and Tmod1 LRR were determined separately (29). Structural modeling suggested that the Tmod1 A-h and LRR respectively bind to the first (subunit n+1) and second (subunit n) actin protomer at the pointed end of the filament (Fig. S2C) (29). Given that (i) the first ∼340 residues of Lmod21–495 share a high (∼45%) sequence identity with human Tmod1; (ii) in the absence of the W domain, Lmod2 behaved just as did Tmod1 (16, 20); and (iii) similar to Tmod1, Lmod2 is capable of binding to the pointed end of a preformed actin filament (20), we expect that Lmod21–340 binds to subunits n+1 and n in a conformation similar to that of human Tmod1. Indeed, the actin(LRR)–LRR complex in the modeled actin–Tmod1 structure (Fig. S2C) (29) was readily superimposable to that in our actin–Lmod2162–495(B-GS) structure (Fig. 1C), with rmsds of 1.2 Å and 0.9 Å for actin(LRR) and LRR, respectively (Fig. S2D). Because the Lmod2 construct in our study lacks the N-terminal 161 residues (including the A-h helix), this superposition allowed us to borrow the actin(A-h)–A-h complex structure from the modeled actin–Tmod1 structure (Fig. S2C), which combined with our actin–Lmod2162–495(B-GS) structure (Fig. 1C), provides a plausible structural model of Lmod21–495 bound with three actin subunits (Fig. 1D).
The Actin–Lmod2 Interfaces.
The A-h region from Tmod1 contains residues 58–99 in which residues 58–84 are highly similar to the corresponding region (residues 60–86) of Lmod2 (Fig. S1B). We therefore modeled the actin(A-h)–A-h complex structure for Lmod2 by keeping only residues 58–84 (Tmod1 numbering) and in silico replacing Tmod1 residues with the corresponding Lmod2 residues if they differed. The modeled actin(A-h)–A-h complex structure readily accommodated the introduced Lmod2 residues, including bulky substitutions such as H72Y and L73W (Lmod2 numbering) (Fig. 2A).
Fig. 2.
Actin–Lmod2 and actin–actin interfaces. (A) Detailed interactions of actin(A-h) with the Lmod2 A-h region (residues 60–86) in the modeled actin(A-h)–Lmod2 A-h structure. (B) Detailed interactions between actin(LRR) and Lmod2 LRR-polyP-h1 observed in the crystal structure of actin–Lmod2162–495(B-GS). Also shown is the alignment of h1 in reverse order (h1 reverse) and the W domain. The helix in each region is highlighted in a red box. Asterisks indicate identical residues and colons indicate similar residues. Residues mediating important actin–Lmod2 interactions are highlighted in yellow background. (C) Detailed interactions between actin(W) and Lmod2 h2-W observed in the crystal structure of actin–Lmod2162–495(B-GS). The four subdomains of actin are labeled 1–4. (D) The dissociation constants and binding energies of Lmod21–495(B-GS) and its individual actin-binding sites with actin. (E) Detailed interactions between actin(LRR) (shown in green ribbons and as a gray surface) and actin(W) (in cyan) observed in the crystal structure of actin–Lmod2162–495(B-GS). Inter-actin hydrogen bonds are shown as dashed orange lines.
In the actin–Lmod2162–495(B-GS) crystal structure, actin(LRR) interacts with Lmod2 in the region of LRR, polyP, and h1 (Fig. 2B and Fig. S4 A and B). The Lmod2 LRR domain interacts with actin at subdomains 1 and 2. An LRR motif contains a module of the β-strand–loop–α-helix in which the loop within the motif is the “ascending” loop and the loop connecting two neighboring LRR motifs is the “descending” loop (30). The ascending loops mediate most of the interactions with actin (Fig. 2B). Interestingly, the reverse sequence of the h1 helix and its neighboring regions (residues 399–415) (h1 reverse) is quite similar to the helix and LKRV motif (comprised of residues L485, K486, R487, and V488) of the W domain (Fig. 2B). It binds to the cleft between subdomains 1 and 3 in an opposite direction from the W domain. This binding mode also is seen for other actin-binding proteins including the α-helix at residues 121–139 of gelsolin (29). Located between the LRR domain and the h1 helix is the shortened polyP region in which a stretch of proline residues extends along the surface of the actin(LRR) domain (Fig. 2B and Fig. S4A).
Fig. S4.
Electron density maps of representative regions in the actin–Lmod2162–495(B-GS) structure. (A) polyP. (B) h1. (C) h2. (D) The W domain of Lmod2. (E) The actin(LRR)–actin(W) interface. The composite omit 2Fo-Fc electron density maps are contoured at 1.0σ. Actin(LRR), actin(W), and Lmod2162–495(B-GS) are shown as green, cyan, and magenta, respectively. In E some of the interacting residues are shown in yellow, and inter-actin hydrogen bonds are drawn as orange dashed lines.
Lmod2 binds to actin(W) through the h2–W region in the actin–Lmod2162–495(B-GS) structure (Fig. 2C and Fig. S4 C and D). The W domain interacts with actin via the hydrophobic residues L473, M474, and I477 on the α-helix (residues 468–479) and via the residues L485, K486, R487, and V488 that form the LKRV motif (Fig. 2C and Fig. S4D). In addition, the small α-helix, h2, at residues 436–441, associates with actin through mostly ionic interactions (Fig. 2C and Fig. S4C).
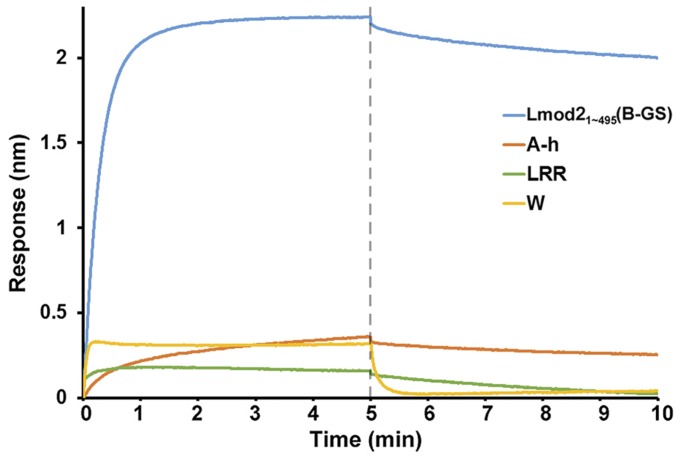
Each of the three actin-binding sites has a similar dissociation constant with actin monomers (at a Kd of 3–4 μM), corresponding to a binding energy (ΔG) of −7.30 to −7.51 kcal/mol (Fig. 2D and Fig. S5). In contrast, the Kd of the full-length Lmod21–495(B-GS) is about 10 times lower, with a ΔG of −9.05 kcal/mol, which is significantly smaller than the sum of binding energies by all three actin-binding sites (at ΔG of −22.30 kcal/mol). One possible reason for this difference is the entropic cost of ordering Lmod2 and multiple actin subunits into a compact complex; another is that not all three actin-binding sites may contribute equally to the formation of the actin–Lmod2 complex.
Fig. S5.
Association and dissociation data of actin with Lmod21–495(B-GS) or its individual actin-binding sites. A range of different actin concentrations were measured; the data shown here used 2.3 μM actin.
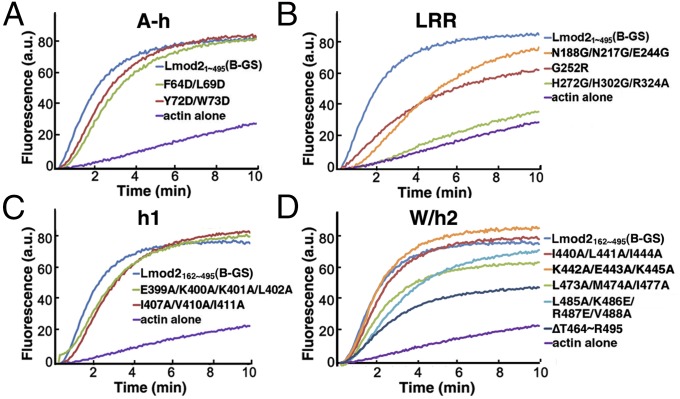
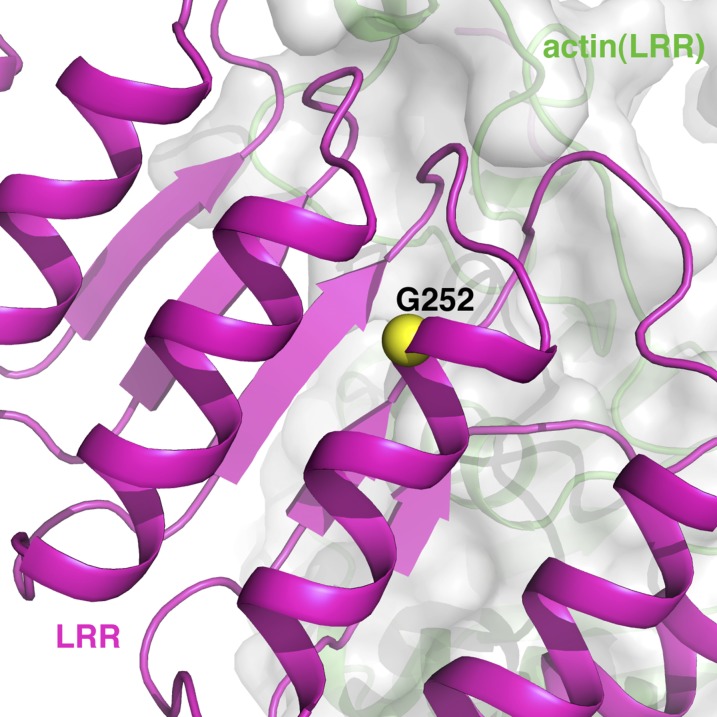
The key residues of Lmod2 that are involved in interacting with actin subunits (Fig. 2) are highly conserved overall among human and mouse Lmod isoforms (Fig. S1B). They were subjected to site-directed mutagenesis followed by pyrene-based activity assays (Fig. 3). The mutations on the h2 helix had the smallest impacts on actin polymerization activity (Fig. 3D), agreeing with the weak interactions of this region with actin(W) (Fig. 2C). The mutations on the A-h and h1 regions resulted in small decreases in actin polymerization activity (Fig. 3 A and C), consistent with the dispensable nature of A-h in Lmod2-mediated actin nucleation (15). The largest reductions in actin polymerization activity were observed for mutations on the LRR domain (Fig. 3B) and the W domain (Fig. 3D). For instance, the triple mutation H272G/H302G/R324A located at the second half of the LRR domain (Fig. S1B) almost completely diminished Lmod2 activity (Fig. 3B). Also tested was G252R, a single mutation on the LRR domain that, when present in Lmod3, caused lethal nemaline myopathy in multiple patients (18). G252 is completely buried at the N terminus of helix 250–262 within the LRR domain (Fig. 2B and Fig. S6). The small-to-large mutation G252R introduced a charged residue and may substantially destabilize the local structure of this region. The drastic decrease in actin polymerization activity by this single mutation (Fig. 3B) helped explain its observed lethality in patients. Similarly, deletion of the 32 C-terminal residues belonging to the W domain (ΔT464–R495) caused significant loss in actin nucleation activity (Fig. 3D), as is consistent with its important roles in Lmod2-mediated nucleation (15). Within the W domain, mutations in the helix region (L473A/M474A/I477A) and in the LKRV motif (L485A/K486E/R487E/V488A) resulted in substantial reduction in pyrene-based activities (Fig. 3D).
Fig. 3.
Pyrene-based activity of 25 nM Lmod2 and various mutants. Shown are mutations in the A-h region (A), LRR domain (B), h1 (C), and W/h2 (D).
Fig. S6.
The location of G252 in the actin–Lmod2162–495(B-GS) structure. Its buried location and the small-to-large/charged mutation of G252R likely destabilize the local structure to abolish the nucleation activity of Lmod2.
A previously unidentified Actin–Actin Interface.
In the actin–Lmod2162–495(B-GS) crystal structure (Fig. 1C), actin(LRR) contributes three loop regions, residues 110–113, 173–177, and 287–290, to interact with actin(W) in the regions of a short α-helix (residues 222–233), N252 on helix 252–262, and the C terminus of helix 184–196. Both hydrophobic interactions [involving residues L110 and L176 on actin(LRR) and residues M227, A228, and A231 on actin(W)] and hydrophilic interactions (i.e., five intermolecular hydrogen bonds) were observed at the interface (Fig. 2E and Fig. S4E). This interface between actin(LRR) and actin(W) led to the burial of accessible surface areas of 497 Å2 for actin(LRR) and 560 Å2 for actin(W).
Using the model of F-actin from X-ray fiber diffraction [Protein Data Bank (PDB) ID code: 2ZWH] (31), we estimated that each actin subunit buries ∼1,010 Å2 or ∼455 Å2 when an actin subunit is added to form a longitudinal or short-pitch dimer, respectively. However, neither of these actin dimers is sufficiently stable in solution. Therefore, although the interactions between actin(LRR) and actin(W) in the actin–Lmod2162–495(B-GS) structure are specific, Lmod2 must have served to hold the actin(LRR)–actin(W) dimer together to allow the complex to elute as a well-defined single peak in size-exclusion chromatography (Fig. S3).
Actin(LRR) and Actin(W) Are Poised for ATP Hydrolysis.
Similar to the modeled actin–Tmod1 structure (Fig. S2C) (29), actin(A-h) and actin(LRR) of the modeled actin–Lmod21–495 structure are expected to locate at subunit n+1 and n, respectively, at the pointed end of actin filament. However, this location leads to a partial overlap of actin(W) with actin subunit n−1 (Fig. 4 A and B), suggesting a non–filament-like conformation of the experimental actin–Lmod2162–495(B-GS) structure and the modeled actin–Lmod21–495 nucleus.
Fig. 4.
The actin–Lmod2 nucleus is in a non–filament-like conformation in which Lmod2-bound actin subunits are poised for efficient ATP hydrolysis. (A) The modeled structure of actin–Lmod21–495 superimposed on actin filament. Actin subunits in the filament are labeled as n−2 to n+1 from the barbed end to the pointed end. (B) Enlarged view of the boxed region in A to highlight the steric clash between actin(W) and subunit n−1 of actin filament. (C) The distances of Q137–γ-P in the actin–Lmod21–495 structural model. Actin(A-h) is from the modeled actin(A-h)–Lmod2 A-h structure, and actin(LRR) and actin(W) are from the experimental actin–Lmod2162–495(B-GS) crystal structure. (D) Statistic comparison of known structures of actin with actin-binding proteins (****P < 0.001; ***P < 0.005; two-tailed Student’s t test). Structures used include 14 actin-ABS structures, actin(LRR) and actin(W) in AMPPNP–actin–Lmod2 as determined in this study, ATP–actin–Tmod (PDB ID code: 4PKG), and AMPPNP–actin–Cobl (PDB ID code: 4JHD) (Table S2). The data are presented as mean ± SD. (E) ATP hydrolysis of nonpolymerizable actin mutants in the absence (blue circles) and presence (orange triangles) of Lmod2162–495(B-GS). (F) Pyrene-based actin polymerization activity of 25 nM Lmod21–495(B-GS) in the presence of ATP or AMPPNP.
The Lmod2162–495(B-GS) construct used here is shorter than the predicted full-length human Lmod21–547 by 20 residues within the polyP region (Fig. S1A), and it is reasonable to ask whether this 20-residue deletion might be the cause of the non–filament-like conformation of the actin–Lmod2 structure. The non–filament-like conformation of the actin–Lmod2 nucleus arises from the relative organization of actin(LRR) and actin(W) that differs from F-actin (Fig. 4A). The 20-residue deletion is in the disordered region (indicated by a dashed line in Fig. 4A) immediately proceeding the structured polyP region, and the entire polyP region is located between the LRR domain and the h1 helix, both of which bind to the same actin(LRR) subunit. Thus, the disordered polyP region is locked on an opposite face of F-actin away from the actin(W) subunit and is highly unlikely to influence the relative location of actin(W) in the actin–Lmod2 nucleus.
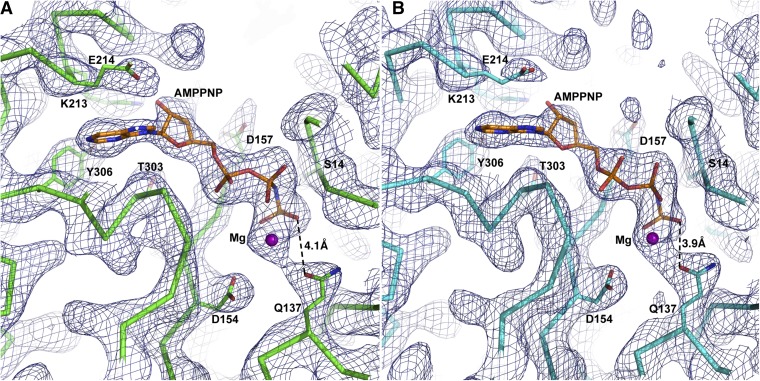
The next question is how the non–filament-like conformation of actin–Lmod2 nucleus complexes serves as the seed for productive actin polymerization. The actin–Cobl nucleus also has a non–filament-like conformation in which ATP hydrolysis is required to discharge the bound W domain, thus releasing the steric clash and allowing rapid growth at the barbed end (26). We first measured the distances between Q137 and the γ-phosphate group (γ-P) of AMPPNP (ATP) in different structures since previous studies suggested that shorter distances correspond to more effective ATP hydrolysis (31–34). Compared with the 14 ATP–actin structures bound with individual actin-binding domains (Q137–γ-P distances averaged at 5.2 ± 0.3 Å), actin(LRR) and actin(W) in the actin–Lmod2162–495(B-GS) structure have significantly shorter Q137–γ-P distances (at an averaged distance of 4.0 ± 0.1 Å; P < 0.001 in two-tailed Student’s t test) (Fig. 4 C and D, Fig. S7, and Table S2) and thus presumably are poised for efficient ATP hydrolysis. The more accessible position of Q137 in the actin(LRR) and actin(W) domains of the actin–Lmod2162–495(B-GS) crystal structure is remarkably similar to the position of Q137 in the other two known complex structures of actin with tandem-G-actin–binding nucleators, AMPPNP–actin–Cobl–2W (PDB ID code: 4JHD) (Fig. 4D and Table S2) (26) and ATP–actin–VopL (PDB ID code: 4M63) (Table S2) (28). Thus the roles of these actin nucleators are likely twofold: (i) to hold multiple G-actin monomers together into an actin nucleus and (ii) to prime the actin nucleus for efficient ATP hydrolysis that eventually may allow the release and recycling of actin nucleators. In marked contrast, actin(A-h) borrowed from the actin–Tmod1 A-h structure (PDB ID code: 4PKG) (29) has a Q137–γ-P distance similar to the 14 ATP–actin structures bound with individual actin-binding domains (Fig. 4 C and D). Whether actin(A-h) assumes a shorter Q137–γ-P distance in the native actin–Lmod21–495 complex awaits elucidation in future studies.
Fig. S7.
ATP-binding sites in actin(LRR) (A) and actin(W) (B) in the actin–Lmod2162–495(B-GS) structure. The composite omit 2Fo-Fc electron density maps are contoured at 1.0σ. Actin(LRR) and actin(W) are shown as green and cyan, respectively. The distances between Q137 and γ-P of AMPPNP are shown as dashed lines with the distance values labeled.
Table S2.
Known structures of actin with actin-binding domains
| PDB ID code | Ligand | Ion | Q137–γ-P distance, Å |
| Structures of actin with individual actin-binding domains (n = 14) | |||
| 3UE5 | ATP | Ca | 4.9 |
| 3MMV | ATP | 5.5 | |
| 3MN5 | ATP | Ca | 4.1 |
| 3MN6 | ATP | 5.3 | |
| 3MN7 | ATP | 5.3 | |
| 3MN9 | ATP | 5.3 | |
| 2D1K | ATP | Ca | 5.5 |
| 2FF6 | ATP | Ca | 5.1 |
| 2A3Z | ATP | Ca | 5.4 |
| 2A40 | ATP | Ca | 5.3 |
| 5.3 | |||
| 2A41 | ATP | Ca | 5.3 |
| 1T44 | ATP | Ca | 5.1 |
| 4PKG | ATP | Ca | 5.1 |
| 4PKI | ATP | Ca | 5.3 |
| Mean | 5.2 | ||
| SD | 0.3 | ||
| AMPPNP–actin–Lmod2162–495 (B-GS) structure | |||
| 4RWT | |||
| Actin(LRR) | AMPPNP | Mg | 4.1 |
| Actin(W) | AMPPNP | Mg | 3.9 |
| Mean | 4.0 | ||
| SD | 0.1 | ||
| AMPPNP–actin–Cobl-2W structure | |||
| 4JHD | |||
| Chain A | AMPPNP | Mg | 5.0 |
| Chain B | AMPPNP | Mg | 4.5 |
| Chain D | AMPPNP | Mg | 4.2 |
| Chain E | AMPPNP | Mg | 4.6 |
| Mean | 4.6 | ||
| SD | 0.3 | ||
| ATP-actin-VopL structure | |||
| 4M63 | |||
| Chain C | ATP | Ca | 4.4 |
| Chain D | ATP | Ca | 4.6 |
| Chain E | ATP | Ca | 3.6 |
| Mean | 4.2 | ||
| SD | 0.5 | ||
To confirm that Lmod2 indeed positions actin(LRR) and actin(W) for efficient ATP hydrolysis, we monitored the rate of ATP hydrolysis in reactions that contain actin in the presence or absence of Lmod2162–495(B-GS). For this set of experiments, we used nonpolymerizable actin mutants to ensure the formation of actin–Lmod2 nuclei while preventing actin polymerization. In doing so, ATP hydrolysis was limited to two main sources: spontaneous hydrolysis in the test condition or upon Lmod2 stimulation. As expected, spontaneous ATP hydrolysis that increased with time was observed in the test condition (in F-buffer) for both samples (Fig. 4E). However, in addition to the spontaneous ATP hydrolysis, the actin–Lmod2162–495(B-GS) sample exhibited significantly accelerated ATP hydrolysis within the first 20 min. The maximal difference between these two samples was reached at ∼20 min and was maintained for the rest of the monitored period (20–240 min) (Fig. 4E). Lmod2-mediated pyrene-based actin polymerization activity took ∼20 min to reach equilibrium (Fig. 3); therefore, the observed accelerated ATP hydrolysis in the actin–Lmod2162–495(B-GS) sample is most likely the result of Lmod2 stimulation. In contrast, when ATP hydrolysis was blocked, e.g., by the use of nonhydrolysable AMPPNP, Lmod2-mediated actin polymerization activity was drastically impaired (Fig. 4F).
Discussion
The Structure of the Actin–Lmod2162–495(B-GS) Nucleus Is Incompatible with Tropomyosin Binding.
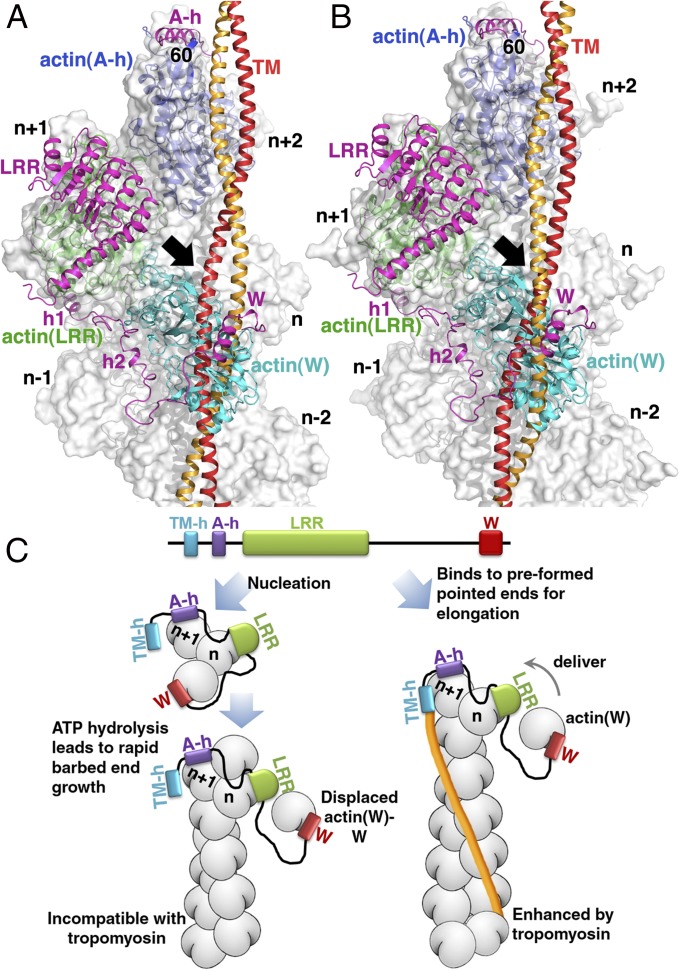
Of note, the observed actin–Lmod2162–495 nucleus structure is not compatible with the binding of tropomyosin, because superimposing the A- and M-states of tropomyosin (35, 36) onto the modeled actin–Lmod21–495 structure results in steric clash with actin(W) (Fig. 5 A and B). This incompatibility explains tropomyosin’s very weak promotion of nucleation at low concentrations but strong inhibition at high concentrations (15). This steric clash, together with the partial overlap of actin(W) with subunit n−1 of actin filament (Fig. 4 A and B), suggests that Lmod2 most likely adopts a distinct conformation when binding to the pointed end of preformed, tropomyosin-decorated actin filaments in maintaining their lengths (16, 20) (see below).
Fig. 5.
Mechanisms of Lmod2-mediated de novo nucleation and pointed-end elongation. (A and B) The actin(W) subunit of the actin–Lmod21–495 nucleus structural model is incompatible with tropomyosin in the A or M states on actin filament. The actin–Lmod21–495 structural model is superimposed onto the cryo-EM structures of actin-tropomyosin in the A (apo) state (PDB ID code: 3J8A) (A) and myosin-bound M (rigor)-state (PDB ID code: 4A7F) (B). The resulting steric clash of actin(W) with tropomyosin is highlighted by an arrow. The N terminus of Lmod2 A-h (residue 60) is shown to indicate the close proximity of Lmod2 TM-h (N-terminal to A-h) with tropomyosin. (C) A model for Lmod2-mediated de novo nucleation (Left) and pointed-end elongation (Right). A partial dissociation of Lmod2 from the nucleus likely releases the steric clash, thus allowing rapid growth at the barbed end. The less compact conformation of Lmod2 also allows controlled pointed-end elongation.
Model of Lmod2 Functions in de Novo Nucleation and Pointed-End Elongation.
Our data support a hypothetical model for Lmod2-mediated de novo nucleation/polymerization (15) and controlled pointed-end elongation (Fig. 5C) (20). The experimental actin–Lmod2162–495(B-GS) structure (Fig. 1C) and the modeled actin–Lmod21–495 structure (Fig. 1D) represent a de novo nucleus in which actin(LRR) and actin(W) have significantly closer Q137–γ-P distances poised for efficient ATP hydrolysis (Fig. 4 C and D). Hydrolysis of ATP into ADP and inorganic phosphate in actin(LRR) and actin(W) and the subsequent release of inorganic phosphate may change the conformation of actin subdomain 2 (37, 38), result in a partial dissociation of Lmod2 from the nucleus, thereby releasing the steric clash to allow fast growth at the barbed end (Fig. 5C, Left). The less compact structure of Lmod2 with a displaced W domain also may be the conformation that binds to preformed actin filaments, where the W domain controls elongation at the pointed end by providing a constant supply of actin monomers (Fig. 5C, Right). This delivery function of the W domain is analogous to that proposed for the tandem-W domains in the bacterial effector VopL (28). The Lmod2-mediated pointed-end elongation of the preformed filament is enhanced by tropomyosin via the TM-h site located at the N terminus of Lmod2 (Fig. 5C, Right) (20), as suggested by their spatial proximity on actin filament (Fig. 5 A and B). This model explains the observation that the W domain is crucial for the unique “non-capping” and pointed-end elongation functions of Lmod2, whereas in its absence, Lmod21–342 (16) and Lmod21–462 (20) behave in the same way as Tmod1. The high level of conservation among Lmod proteins in sequence and functions suggests that the mechanistic insights of Lmod2 learned from this study may provide a mechanistic understanding of other Lmod proteins. For instance, the structure of actin–Lmod2162–495(B-GS) and the functional study of Lmod2 G252R mutant explain the lethal consequence of Lmod3 G252R in patients.
The Non–Filament-Like Conformation of Actin and Tandem-G-Actin–Binding Nucleator Complexes in Mammals.
It is interesting that the actin nucleus assembled by the C-terminal domain of the bacterial effector VopL (28) is in a filament-like conformation that presumably allows rapid formation of actin filament beneficial to pathogen invasion. However, in sharp contrast, the only two structurally characterized mammalian tandem-G-actin–binding nucleators, Lmod2 in this study and Cobl in our previous study (26), both adopt a non–filament-like conformation in which the actin subunits bound by tandem-G-actin–binding sites are poised for efficient ATP hydrolysis (Fig. 4). The non–filament-like conformation of these actin nuclei understandably blocks rapid actin polymerization at the barbed end. However, subsequent ATP hydrolysis in nucleator-bound actin subunits by the better positioned Q137 and the release of inorganic phosphate likely would destabilize the actin- nucleator complexes, leading to partial or complete removal of the nucleator and allowing rapid growth at the barbed end. Therefore, it is intriguing to speculate that the non–filament-like conformation of the actin nucleus and the better positioned Q137 in nucleator-bound actin subunits together may constitute an elegant sensing mechanism for at least some of the mammalian tandem-G-actin–binding nucleators in which productive actin polymerization would be possible only in the presence of persistent signals, thus offering an important level of regulation in actin-mediated signal transduction pathways.
Materials and Methods
Proteins.
Nonpolymerizable Drosophila 5C actin mutants were purified as previously described (26). Wild-type actin was purified from rabbit skeletal muscle, and pyrene-labeled actin was purchased from Cytoskeleton, Inc. The cDNA encoding human Lmod21–495 (UniProt ID Q6P5Q4.2) (Fig. S1A) was codon-optimized and synthesized using a two-step gene-assembly method (39) to obtain the same construct as reported earlier (15, 16).
Crystallization, Data Collection, and Structural Refinement.
The actin–Lmod2162–495(B-GS) complex at a stoichiometry of 2:1 was purified by size-exclusion chromatography in F-buffer (10 mM Hepes, 100 mM KCl, 1 mM MgCl2, 0.2 mM AMPPNP, 1 mM DTT, pH 7.6) and concentrated to 10 mg/mL for crystallization screening. Plate-like crystals were obtained using the vapor-diffusion hanging-drop method. X-ray diffraction data were collected at 100 K at the 21ID-F beamline at Advanced Photon Source. Data processing and structural refinement were accomplished as previously described (26).
Pyrene-Based Actin Polymerization Assay.
The pyrene-actin polymerization assay was performed using wild-type actin as previously described (26). A range of protein concentrations was used for each Lmod2 construct.
Binding Affinity of Actin with Lmod2 or Its Fragments.
The binding affinity of actin with Lmod21–495(B-GS) and its fragments was measured using Octet RED96 (Pall ForteBio Corp.). Nonpolymerizable actin 5C mutant (actin II) was used in the assays to allow interaction with Lmod2 but prevent actin polymerization. The association and dissociation reactions were conducted in 20 mM Tris⋅HCl, 50 mM NaCl, pH 8.0 at 25 °C. The dissociation constants were calculated as Kd = Koff/Kon from which the binding energy (ΔG) was derived using ΔG = RT ln(KD/cθ), where R is the ideal gas constant, T is the temperature (298 K), and the standard reference concentration cθ = 1 M.
ATP Hydrolysis in Actin Stimulated by Lmod2162–495(B-GS).
This assay was carried out essentially as described earlier (40). Both the actin with Lmod2162–495(B-GS) and actin alone samples were exchanged into F-buffer and placed on ice. To determine the rate of ATP hydrolysis, samples were incubated on ice, and 50-μL samples were taken at various time points and loaded on a Mono Q 4.6/100 PE column (GE Healthcare). ATP and ADP standards were at 25 μM.
SI Materials and Methods
Proteins.
Nonpolymerizable Drosophila 5C actin mutants were purified as previously described (26). Wild-type actin was purified from rabbit skeletal muscle, and pyrene-labeled actin was purchased from Cytoskeleton, Inc. The cDNA encoding human Lmod21–495 (UniProt ID Q6P5Q4.2) (Fig. S1A) was codon-optimized and synthesized using a two-step gene-assembly method (39) to obtain the construct as reported earlier (15, 16). The Lmod21–495(B-GS) mutant was obtained by replacing the B segment 406KKKKGKKVKK415 with 406GSGSGGSVGS415 using site-directed mutagenesis with the QuikChange kit (Agilent Technologies). Four constructs, Lmod21–495, Lmod21–495(B-GS), Lmod2162–495, and Lmod2162–495(B-GS), were cloned into the pET-45b vector (Clontech Laboratories) with a short N-terminal his6-tag. Various mutations of Lmod21–495(B-GS) or Lmod2162–495(B-GS) were introduced by site-directed mutagenesis and verified by DNA sequencing. Fragments of Lmod21–495(B-GS), including residues 1–161, 162–417, and 418–495, were cloned into the pGEX-6p1 vector (Clontech Laboratories) with an N-terminal GST-tag for binding affinity assays. All the protein constructs were transformed into Escherichia coli Rosetta (DE3)-competent cells, and overexpression was achieved by induction with 0.5 mM isopropyl β-d-1-thiogalactopyranoside for 5 h. Cells were lysed by sonication in the lysis buffer containing 50 mM Tris⋅HCl and 300 mM NaCl, pH 8.0. The lysate was clarified by centrifugation to remove insoluble cell debris. The clarified supernatant was incubated with Ni-NTA metal-chelating Sepharose (Clontech Laboratories) for his6-tagged proteins or Glutathione Agarose (GenScript USA Inc.) for GST-tagged proteins and was washed with 50 mM Tris⋅HCl and 300 mM NaCl, pH 8.0 to remove unbound proteins. Target proteins were eluted and further purified by ion-exchange columns. A final step of size-exclusion chromatography by Superdex 200 10/300 GL was used to remove any aggregated or degraded proteins.
Crystallization, Data Collection, and Structural Refinement.
Lmod2162–495(B-GS) was mixed with actin 5C mutants in a molar ratio of 1:2, and the complex was purified by size-exclusion chromatography using Superdex 200 10/300 GL in F- buffer (10 mM Hepes, 100 mM KCl, 1 mM MgCl2, 0.2 mM AMPPNP, 1 mM DTT, pH 7.6). This purified actin–Lmod2162–495(B-GS) complex was concentrated to 10 mg/mL to set up crystallization screening. Plate-like crystals of the actin–Lmod2162–495(B-GS) complex were obtained using the vapor-diffusion hanging-drop method in a solution of 17–19% PEG3350, 0.25 M magnesium acetate, and 3% (vol/vol) 2,2,2-trifluoroethanol. X-ray diffraction data were collected at 100 K at Advanced Photon Source (the 21ID-F beamline). The data were processed using MOSFLM (41), scaled by SCALA, and truncated to structure factor amplitude by TRUNCATE in CCP4 (42). The atomic coordinates of an actin monomer (PDB ID code: 3TU5) were used as the search model for molecular replacement by PHENIX (43). Each asymmetric unit contains one actin–Lmod2162–495(B-GS) complex, with actin(W) also bound by an extra LRR domain derived from degradation. The model then was subjected to iterative cycles of positional and B-factor refinement using REFMAC5 (44) in CCP4 (42) and manual model building in O (45) guided by composite omit 2Fo−Fc maps. In addition, the normal mode-based X-ray refinement method (46) was used to provide better descriptions of B factors, resulting in enhanced densities for flexible regions.
Pyrene-Based Actin Polymerization Assay.
The pyrene-actin polymerization assay was performed using wild-type actin as previously described (26). Briefly, freshly thawed pyrene-labeled G-actin (Cytoskeleton, Inc.) was incubated on ice for 1 h in G buffer [2 mM Tris⋅HCl (pH 8.0), 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT] to depolymerize actin oligomers. The mixture of labeled and unlabeled G-actin was centrifuged at 21,000 × g for 30 min at 4 °C to remove residual actin filaments. Actin polymerization was initiated by adding 4 μL of an actin mixture containing 45 μM unlabeled G-actin and 5 μM pyrene-labeled G-actin (final concentration, 2 μM actin) into 96-μL reaction mixtures containing test proteins in F buffer (including 0.2 mM ATP or another nucleotide). The reaction of actin polymerization was followed by monitoring the change in fluorescence intensity over time with an excitation wavelength of 355 nm and an emission wavelength of 410 nm in a FLUOstar Omega fluorescence spectrometer (BMG Labtech GMBH). A range of protein concentrations was used for each Lmod2 construct.
Binding Affinity of Actin with Lmod2 or Its Fragments.
The binding affinity of actin with Lmod21–495(B-GS) and its fragments was measured using bioLayer interferometry (BLI) technology on an Octet RED96 system (Pall ForteBio Corp.). The nonpolymerizable actin 5C mutant (actin II) was used in the assays to allow the interaction with Lmod2 but prevent actin polymerization. For association, the Lmod21–495(B-GS) protein or its individual actin-binding domain [Lmod21–161, Lmod2162–417, or Lmod2418–495(B-GS)] was loaded onto biosensors and incubated in the binding buffer (20 mM Tris⋅HCl, 50 mM NaCl, pH 8.0) containing actin II in at least three different concentrations at 25 °C. The binding generally reached equilibrium within 5 min. For dissociation, the biosensors then were incubated in the binding buffer in the absence of actin and were monitored for 5 min. Because of the interference of nucleotides with BLI signals, no nucleotide was included in the buffer. The dissociation constants were calculated as Kd = Koff/Kon in which Kon and Koff were measured from the slope of the association and dissociation curves, respectively. The binding energy (ΔG) was calculated from the Kd using the following equation: ΔG = RT ln(KD/cθ), where R is the ideal gas constant, T is the temperature (298 K), and the standard reference concentration cθ = 1 M.
ATP Hydrolysis in Actin Stimulated by Lmod2162–495(B-GS).
This assay was carried out essentially as described earlier (40). Briefly, a 50% slurry of Dowex AG 1-X8 was used to remove free external nucleotides from nonpolymerizable actin mutants by the addition of 10% in volume. The suspension was mixed for 1 min on ice and centrifuged for 20 s at 10,000 × g. The Dowex treatment was performed a total of three times. The final supernatant was centrifuged for 30 min at 300,000 × g. The actin mutant concentration was adjusted to 4 mg/mL. Both the actin samples with Lmod2162–495(B-GS) and actin alone were exchanged into F-buffer and placed on ice. To determine the rate of ATP hydrolysis, samples were incubated on ice, and 50-μL samples were taken at various time points. Fifty microliters of 10% perchloric acid was added. After 5 min on ice, 20 μL of 4 M potassium acetate in 10 M potassium hydroxide was added, and the sample was centrifuged for 10 min at 21,000 × g followed by 10 min at 300,000 × g. The supernatant was diluted to 200 μL with water and loaded on a Mono Q 4.6/100 PE column preequilibrated with 5 mM triethylammonium bicarbonate (pH 8.5) (Sigma-Aldrich) using an AKTA-FPLC system (GE Healthcare). The nucleotides were eluted with a 10-mL gradient of 5 mM to 0.5 M triethylammonium bicarbonate (pH 8.5), followed by an additional 5 mL of 0.5 M triethylammonium bicarbonate (pH 8.5). ATP and ADP standards were at 25 μM.
Supplementary Material
Acknowledgments
Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and Michigan Technology Tri-Corridor Grant 085P1000817. J.M. was supported by National Institutes of Health Grant R01-GM067801 and Welch Foundation Grant Q-1512. Q.W. was supported by National Institutes of Health Grant R01-AI067839 and Welch Foundation Grant Q-1826.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank database www.pdb.org (ID code 4RWT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512464112/-/DCSupplemental.
References
- 1.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 2.Chhabra ES, Higgs HN. The many faces of actin: Matching assembly factors with cellular structures. Nat Cell Biol. 2007;9(10):1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 3.Chesarone MA, Goode BL. Actin nucleation and elongation factors: Mechanisms and interplay. Curr Opin Cell Biol. 2009;21(1):28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qualmann B, Kessels MM. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19(6):276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez R. Structural insights into de novo actin polymerization. Curr Opin Struct Biol. 2010;20(2):217–225. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126(Pt 1):1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ono S. Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton (Hoboken) 2010;67(11):677–692. doi: 10.1002/cm.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takano K, et al. Nebulin and N-WASP cooperate to cause IGF-1-induced sarcomeric actin filament formation. Science. 2010;330(6010):1536–1540. doi: 10.1126/science.1197767. [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi K, et al. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J Biol Chem. 2009;284(43):29873–29881. doi: 10.1074/jbc.M109.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kan-o M, et al. Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One. 2012;7(4):e34765. doi: 10.1371/journal.pone.0034765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iskratsch T, et al. Formin follows function: A muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J Cell Biol. 2010;191(6):1159–1172. doi: 10.1083/jcb.201005060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosado M, et al. Critical roles for multiple formins during cardiac myofibril development and repair. Mol Biol Cell. 2014;25(6):811–827. doi: 10.1091/mbc.E13-08-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molnár I, et al. DAAM is required for thin filament formation and Sarcomerogenesis during muscle development in Drosophila. PLoS Genet. 2014;10(2):e1004166. doi: 10.1371/journal.pgen.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chereau D, et al. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320(5873):239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skwarek-Maruszewska A, et al. Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres. Mol Biol Cell. 2010;21(19):3352–3361. doi: 10.1091/mbc.E10-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: Larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73(2):127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 18.Yuen M, et al. Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J Clin Invest. 2014;124(11):4693–4708. doi: 10.1172/JCI75199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garg A, et al. KLHL40 deficiency destabilizes thin filament proteins and promotes nemaline myopathy. J Clin Invest. 2014;124(8):3529–3539. doi: 10.1172/JCI74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukada T, et al. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J Cell Sci. 2010;123(Pt 18):3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashiro S, Gokhin DS, Kimura S, Nowak RB, Fowler VM. Tropomodulins: Pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton (Hoboken) 2012;69(6):337–370. doi: 10.1002/cm.21031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81(2):667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci USA. 2004;101(44):15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26(3):449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294(5547):1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, et al. Structural basis of actin filament nucleation by tandem W domains. Cell Reports. 2013;3(6):1910–1920. doi: 10.1016/j.celrep.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433(7025):488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 28.Zahm JA, et al. The bacterial effector VopL organizes actin into filament-like structures. Cell. 2013;155(2):423–434. doi: 10.1016/j.cell.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao JN, Madasu Y, Dominguez R. Mechanism of actin filament pointed-end capping by tropomodulin. Science. 2014;345(6195):463–467. doi: 10.1126/science.1256159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol Life Sci. 2008;65(15):2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457(7228):441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 32.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467(7316):724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 33.Vorobiev S, et al. The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci USA. 2003;100(10):5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders MG, Voth GA. Water molecules in the nucleotide binding cleft of actin: Effects on subunit conformation and implications for ATP hydrolysis. J Mol Biol. 2011;413(1):279–291. doi: 10.1016/j.jmb.2011.07.068. [DOI] [PubMed] [Google Scholar]
- 35.von der Ecken J, et al. Structure of the F-actin-tropomyosin complex. Nature. 2015;519(7541):114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behrmann E, et al. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150(2):327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strzelecka-Gołaszewska H, Moraczewska J, Khaitlina SY, Mossakowska M. Localization of the tightly bound divalent-cation-dependent and nucleotide-dependent conformation changes in G-actin using limited proteolytic digestion. Eur J Biochem. 1993;211(3):731–742. doi: 10.1111/j.1432-1033.1993.tb17603.x. [DOI] [PubMed] [Google Scholar]
- 38.Orlova A, Egelman EH. A conformational change in the actin subunit can change the flexibility of the actin filament. J Mol Biol. 1993;232(2):334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 39.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164(1):49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 40.Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J Biol Chem. 2006;281(42):31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 41.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 46.Poon BK, et al. Normal mode refinement of anisotropic thermal parameters for a supramolecular complex at 3.42-A crystallographic resolution. Proc Natl Acad Sci USA. 2007;104(19):7869–7874. doi: 10.1073/pnas.0701204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.