Significance
During cell division, multisubunit kinetochores partition chromosomes while maintaining a grip on dynamic microtubules under tension. Previous work in Caenorhabditis elegans showed that the central kinetochore component, Mis12/MIND (Mtw1, Nsl1, Nnf1, Dsn1) complex, increases microtubule binding of outer kinetochore complexes, but the mechanism for this enhancement remains unknown. Here, we identify new contacts between MIND and the outer kinetochore Ndc80 (Ndc80, Nuf2, Spc24, Spc25) complex that are essential for interaction in vitro and for cell viability. Using single-molecule microscopy, we demonstrate that a single MIND complex enhances the microtubule binding of a single Ndc80 complex. Our results suggest a molecular mechanism for enhancing kinetochore–microtubule attachment by a central kinetochore component.
Keywords: kinetochore, Ndc80 complex, MIND/Mis12 complex, mitosis, microtubules
Abstract
Multiple protein subcomplexes of the kinetochore cooperate as a cohesive molecular unit that forms load-bearing microtubule attachments that drive mitotic chromosome movements. There is intriguing evidence suggesting that central kinetochore components influence kinetochore–microtubule attachment, but the mechanism remains unclear. Here, we find that the conserved Mis12/MIND (Mtw1, Nsl1, Nnf1, Dsn1) and Ndc80 (Ndc80, Nuf2, Spc24, Spc25) complexes are connected by an extensive network of contacts, each essential for viability in cells, and collectively able to withstand substantial tensile load. Using a single-molecule approach, we demonstrate that an individual MIND complex enhances the microtubule-binding affinity of a single Ndc80 complex by fourfold. MIND itself does not bind microtubules. Instead, MIND binds Ndc80 complex far from the microtubule-binding domain and confers increased microtubule interaction of the complex. In addition, MIND activation is redundant with the effects of a mutation in Ndc80 that might alter its ability to adopt a folded conformation. Together, our results suggest a previously unidentified mechanism for regulating microtubule binding of an outer kinetochore component by a central kinetochore complex.
During mitosis, kinetochores coordinate the movement of replicated chromosomes into two daughter cells and ensure that the genome is equally segregated upon division. Kinetochores maintain a grip on dynamic microtubules that are constantly growing and shortening, and they also ensure that each chromatid is properly attached to microtubules emanating from only one pole. These attachments must be strong enough to withstand the mechanical tension associated with bipolar chromosome alignment, and yet they must be quickly released in response to signals that detect improper attachments (1, 2). Each kinetochore is a macromolecular structure composed of 40 different types of proteins assembled into repeating subcomplexes that span from the centromeric DNA to the microtubule (2). There is an intrinsic hierarchy, with few DNA-binding elements expanding out to multiple microtubule attachment complexes (3, 4). How subcomplexes are held together and function as a cohesive molecular unit is unclear. Assembling kinetochore proteins in vitro allows us to map their interconnectivity and directly probe how each component contributes to microtubule attachment strength. By systematically rebuilding a kinetochore in vitro, we aim to gain a clear understanding of force transmission throughout the kinetochore and to discern the precise role of each component in kinetochore function.
Previous work has characterized the individual and combined activities of the outer microtubule-binding kinetochore complexes (1, 2). How central kinetochore complexes contribute to establishing and maintaining microtubule attachment is less clear. Work from Caenorhabditis elegans identified a conserved core microtubule-binding “KMN” network composed of Knl1/Spc105, Mis12/MIND (Mtw1, Nsl1, Nnf1, Dsn1) complex, and Ndc80 (Ndc80, Nuf2, Spc24, Spc25) complex (5). In C. elegans, the 4-protein Ndc80 complex and Knl1 bind directly to microtubules, but MIND does not. Instead, MIND serves as a structural linker that connects DNA-binding components with the microtubule-binding complexes (6, 7). The KMN complex binds microtubules with a higher affinity than Ndc80 complex or Knl1 alone, demonstrating that MIND can facilitate the synergistic binding of outer kinetochore complexes. This finding highlighted the importance of cooperation between kinetochore complexes and suggested that central complexes can enhance kinetochore–microtubule attachment indirectly by acting through the microtubule-binding components. However, the mechanisms underlying this enhancement remain unknown.
Two hypotheses can explain how a central kinetochore complex could increase the microtubule binding activity of an outer kinetochore component. One possibility is that it oligomerizes the outer kinetochore component to increase avidity. Another possibility is that the central kinetochore component induces structural changes in the outer component that enhance its binding to microtubules. Although MIND has been shown to enhance the microtubule binding of Knl1 in cosedimentation assays (5), this method cannot distinguish which mechanism is responsible. Whether MIND can similarly affect the Ndc80 complex (Ndc80c) by either mechanism has not been tested.
Within the kinetochore, outer components are present in higher copy numbers relative to central components, suggesting that oligomerization may be a mechanism to enhance microtubule binding. Additionally, substantial evidence suggests that the outer kinetochore Ndc80 complex undergoes conformational changes throughout mitosis (8–10). The Ndc80 complex hinges about a flexible “loop” region and exists in both a folded conformation and an elongated state (8, 11). In vivo evidence supports the physiological significance of these conformational changes (10, 12), underscoring the importance of different conformations of the Ndc80 complex. However, it is unknown whether these structural changes correlate with changes in microtubule affinity. The Ndc80 complex also interacts with a second kinetochore receptor, CENP-T/Cnn1 (13–16). It is unclear how central kinetochore components influence the activity of the Ndc80 complex throughout mitosis.
Here, we have reconstituted the MIND–Ndc80 (MN) co-complex using recombinant yeast components and have used cross-linking analysis to identify a network of interactions between the two complexes that is more extensive than previously recognized. Using single-molecule fluorescence microscopy, we found that MIND enhances the microtubule-binding activity of the Ndc80 complex. This enhancement does not require oligomerization of the Ndc80 complex. Instead, a single MIND complex binds a single Ndc80 complex far from its microtubule-binding domain and confers increased microtubule interaction of the co-complex. In addition, MIND activation is redundant with the effects of a mutation in Ndc80 that hinders its ability to adopt a folded conformation, suggesting that MIND might promote an unfolded conformation of Ndc80 complex with higher affinity for microtubules. Finally, we used optical tweezers to show that the MIND–Ndc80 linkage can support the high levels of tension generated by the components of the kinetochore–microtubule interface, establishing the MIND complex as a key component of the force transmission pathway within the kinetochore.
Results
The MIND and Ndc80 Complexes Are Connected by an Extensive Interaction Network.
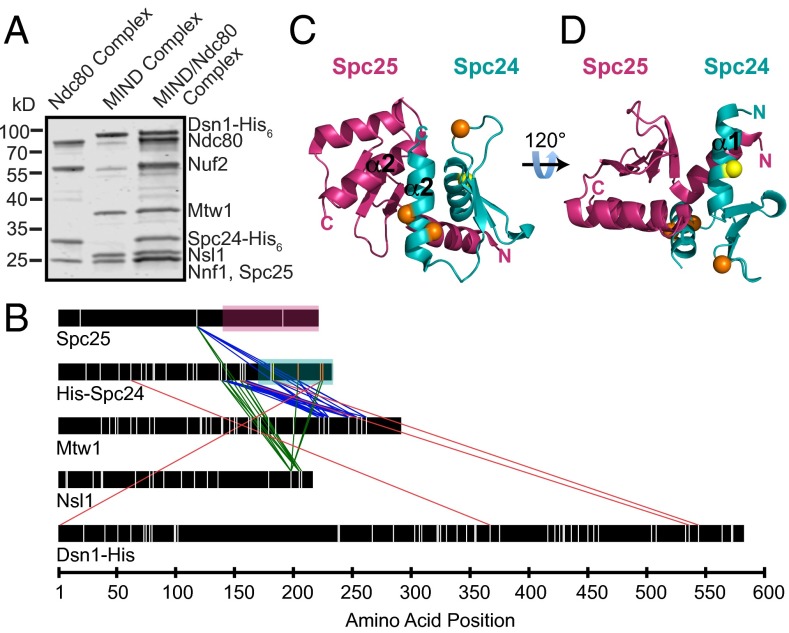
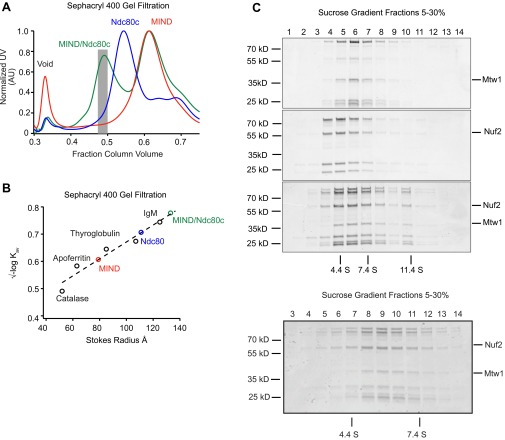
To study the interaction between the MIND and Ndc80 complexes in vitro, we assembled a stable MIND–Ndc80 (MN) co-complex with a 1:1 stoichiometry (17, 18) (Fig. 1A and Fig. S1). By pairing gel filtration and velocity sedimentation experiments, we found that MN exhibits a frictional ratio of 2.7 (19), consistent with its extremely elongated conformation as seen previously by negative-stain electron microscopy (17, 18, 20) (Fig. S1).
Fig. 1.
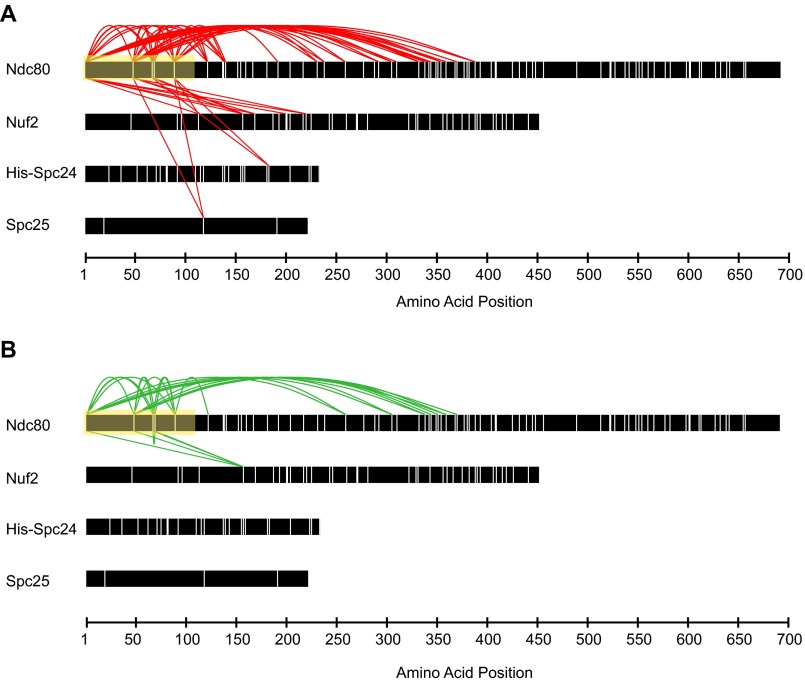
Cross-linking analysis identifies previously unidentified regions of interaction between the MIND and Ndc80 complexes. (A) Coomassie-stained gel showing Ndc80 complex (left lane), MIND complex (middle lane), and MIND/Ndc80 co-complex (right lane). (B) Cross-links between Ndc80 and MIND complexes are shown as colored lines. Lysine residues within each protein are marked as vertical white lines with four exceptions: cross-linked Spc24 lysines highlighted in yellow and orange in C and D are color-coded to match. Regions of Spc25 and Spc24 corresponding to the crystal structure in C and D are highlighted with magenta or teal boxes, respectively. For clarity, only cross-links between Ndc80 complex and MIND are shown; all others are omitted. (C and D) Spc24/Spc25 globular domain crystal structure depicting cross-linked lysines. Spc25 amino acids133–221 are shown in magenta, and Spc24 amino acids 155–213 are shown in teal. Visible N and C termini are marked. (C) The predicted binding pocket for the Ndc80 complex formed by the α2-helices (labeled) of Spc24 and Spc25. Nsl1 cross-links to two lysines (orange) in the Spc24 α2-helix, suggesting it may bind within the hydrophobic pocket. Nsl1 also cross-links to a third lysine (orange) in the disordered Spc24 loop. (D) A 120° rotation of C depicting the lysine residue (yellow) within the α1-helix of Spc24 that cross-links to both Mtw1 and Dsn1.
Fig. S1.
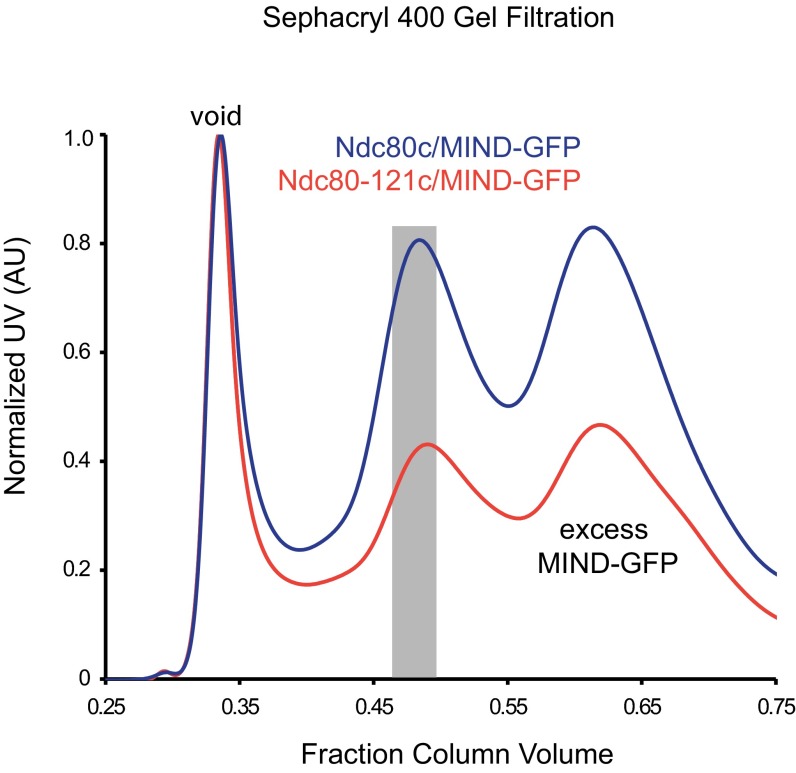
MIND/Ndc80c co-complex associates with 1:1 stoichiometry. (A) Representative Sephacryl 400 gel filtration elution profiles for Ndc80c (green), MIND (blue), and MIND/Ndc80c co-complex (red). Note that, for the MIND/Ndc80c experiment, complexes were mixed in a 2.5:1 molar ratio (MIND:Ndc80c); therefore, excess MIND can be seen eluting as a separate peak. Gray bar indicates fractions pooled and used for subsequent MIND/Ndc80c experiments. (B) Stokes radius vs. √–logKav plotted for indicated standard proteins (catalase, 52 Å; apoferritin, 63 Å; thyroglobulin, 85 Å; fibrinogen, 107 Å; IgM, 130 Å) and fit with a linear regression. Stokes radii for Ndc80c, MIND, and MIND/Ndc80c were calculated using the equation derived from the linear fit: Ndc80c = 111 Å, MIND = 79 Å, MIND/Ndc80c = 134 Å. (C) Velocity sedimentation analysis of Ndc80c, MIND, and MIND/Ndc80c run on a 5–30% sucrose gradient split into 16 sequential fractions; n = 2. Representative elution fractions for each complex across gradient are shown in Coomassie-stained gel; fraction 1 corresponds to the top of the gradient. Some stoichiometric MIND/Ndc80c oligomers are visible in later fractions. BSA (4.4 S), aldolase (7.4 S), and catalase (11.4 S) were run as standards (positions indicated by vertical lines), and a linear fit was determined to find the S values for experimental complexes: Ndc80c = 4.24 S, MIND = 5.42 S, MIND/Ndc80c = 5.89 S. Using the S value and stokes radius, the molecular mass of the MIND/Ndc80c complex was determined to be 331.8 kDa, within 0.04% of the expected molecular mass of a stoichiometric MIND/Ndc80c complex (333 kDa). The frictional ratio of MIND/Ndc80c was also calculated using the S value and stokes radius. MIND/Ndc80c exhibits a frictional ratio of 2.7, indicating that it is highly elongated. (Below) A sucrose gradient from a repeat experiment split into 18 sequential fractions to more clearly visualize the MIND/Ndc80c peak centered at 5.89 S.
We generated a comprehensive map of interactions between the two complexes by treating MN with the cross-linking agent disuccinimidyl suberate and identifying the cross-linked peptides by mass spectrometry (10) (Fig. 1B and Tables S1–S3). The C-terminal regions of Mtw1, Nsl1, and Dsn1 interface with two highly conserved amphipathic helices of Spc24 (Fig. 1 B and C). Specifically, Nsl1 seems to bind a hydrophobic pocket between the α2-helices of Spc24 and Spc25, previously recognized as a putative interaction site (21) (orange circles, Fig. 1C), but for which no binding partner had been identified. Mtw1 binds the α1-helix on the opposite side of Spc24, in the same region shown to interact with another Ndc80c receptor, CENP-T/Cnn1 (yellow circle, Fig. 1D) (22). Because Spc25 contains only three lysines (Fig. 1B), our lysine-specific cross-linker provided limited information about MIND–Spc25 interactions. Previous work identified the C-terminal region of Dsn1 as being important for interaction with Spc24 and Spc25 and suggested that it also shares the binding site with Cnn1 (20, 22). Our cross-linking results support this interaction although we detected far fewer cross-links between Dsn1 and Spc24 than between Nsl1 or Mtw1 and Spc24. Small regions of Nsl1, Mtw1, and Dsn1 also cross-linked to disordered segments of Spc24 (138–154) and Spc25 (128–132) not depicted in the crystal structure (Fig. 1C) (21). Our cross-linking analysis demonstrates that, whereas MIND and Cnn1 share an overlapping binding site within the Ndc80 complex, MIND also forms a second distinct connection to Ndc80c.
Table S1.
Intercomplex cross-links between the Ndc80 tail domain and Ndc80, Nuf2, Spc24, and Spc25 in the presence of MIND
| Protein | Amino acid position | Protein | Amino acid position |
| Ndc80 | 1 | Ndc80 | 48 |
| Ndc80 | 1 | Ndc80 | 67 |
| Ndc80 | 1 | Ndc80 | 69 |
| Ndc80 | 1 | Ndc80 | 89 |
| Ndc80 | 1 | Ndc80 | 122 |
| Ndc80 | 1 | Ndc80 | 140 |
| Ndc80 | 1 | Ndc80 | 292 |
| Ndc80 | 1 | Ndc80 | 305 |
| Ndc80 | 1 | Ndc80 | 310 |
| Ndc80 | 1 | Ndc80 | 332 |
| Ndc80 | 1 | Ndc80 | 338 |
| Ndc80 | 1 | Ndc80 | 342 |
| Ndc80 | 1 | Ndc80 | 344 |
| Ndc80 | 1 | Ndc80 | 354 |
| Ndc80 | 1 | Ndc80 | 359 |
| Ndc80 | 1 | Ndc80 | 370 |
| Ndc80 | 1 | Ndc80 | 388 |
| Ndc80 | 1 | Nuf2 | 113 |
| Ndc80 | 1 | Nuf2 | 157 |
| Ndc80 | 1 | Nuf2 | 169 |
| Ndc80 | 1 | Nuf2 | 200 |
| Ndc80 | 1 | Nuf2 | 220 |
| Ndc80 | 48 | Ndc80 | 67 |
| Ndc80 | 48 | Ndc80 | 69 |
| Ndc80 | 48 | Ndc80 | 89 |
| Ndc80 | 48 | Ndc80 | 122 |
| Ndc80 | 48 | Ndc80 | 138 |
| Ndc80 | 48 | Ndc80 | 140 |
| Ndc80 | 48 | Ndc80 | 231 |
| Ndc80 | 48 | Ndc80 | 238 |
| Ndc80 | 48 | Ndc80 | 259 |
| Ndc80 | 48 | Ndc80 | 292 |
| Ndc80 | 48 | Ndc80 | 305 |
| Ndc80 | 48 | Ndc80 | 332 |
| Ndc80 | 48 | Ndc80 | 338 |
| Ndc80 | 48 | Ndc80 | 354 |
| Ndc80 | 48 | Ndc80 | 388 |
| Ndc80 | 48 | Nuf2 | 157 |
| Ndc80 | 48 | Nuf2 | 169 |
| Ndc80 | 48 | Nuf2 | 220 |
| Ndc80 | 48 | Spc25 | 118 |
| Ndc80 | 67 | Spc24 | 183 |
| Ndc80 | 67 | Ndc80 | 69 |
| Ndc80 | 67 | Ndc80 | 89 |
| Ndc80 | 67 | Nuf2 | 157 |
| Ndc80 | 69 | Ndc80 | 89 |
| Ndc80 | 69 | Ndc80 | 122 |
| Ndc80 | 69 | Nuf2 | 157 |
| Ndc80 | 89 | Spc24 | 183 |
| Ndc80 | 89 | Ndc80 | 122 |
| Ndc80 | 89 | Ndc80 | 138 |
| Ndc80 | 89 | Ndc80 | 140 |
| Ndc80 | 89 | Ndc80 | 192 |
| Ndc80 | 89 | Ndc80 | 231 |
| Ndc80 | 89 | Ndc80 | 238 |
| Ndc80 | 89 | Ndc80 | 259 |
| Ndc80 | 89 | Nuf2 | 113 |
| Ndc80 | 89 | Spc25 | 118 |
The Ndc80 tail domain (amino acids 1–113) cross-links to each of the four proteins in the Ndc80 complex (diagram shown in Fig. S8).
Table S3.
Cross-links between the Ndc80 complex and MIND
| Protein | Amino acid position | Protein | Amino acid position |
| Dsn1 | 2 | Spc24 | 225 |
| Dsn1 | 367 | Spc24 | 62 |
| Dsn1 | 536 | Spc24 | 155 |
| Dsn1 | 544 | Spc24 | 183 |
| Mtw1 | 222 | Spc24 | 155 |
| Mtw1 | 222 | Spc24 | 159 |
| Mtw1 | 226 | Spc24 | 143 |
| Mtw1 | 226 | Spc24 | 155 |
| Mtw1 | 226 | Spc24 | 159 |
| Mtw1 | 226 | Spc24 | 183 |
| Mtw1 | 226 | Spc24 | 204 |
| Mtw1 | 226 | Spc25 | 118 |
| Mtw1 | 230 | Spc24 | 155 |
| Mtw1 | 230 | Spc24 | 159 |
| Mtw1 | 230 | Spc24 | 183 |
| Mtw1 | 230 | Spc24 | 204 |
| Mtw1 | 247 | Spc24 | 139 |
| Mtw1 | 247 | Spc24 | 143 |
| Mtw1 | 247 | Spc24 | 155 |
| Mtw1 | 247 | Spc24 | 159 |
| Mtw1 | 247 | Spc25 | 118 |
| Mtw1 | 254 | Spc24 | 143 |
| Mtw1 | 254 | Spc24 | 155 |
| Mtw1 | 254 | Spc24 | 159 |
| Mtw1 | 254 | Spc24 | 183 |
| Mtw1 | 254 | Ndc80 | 577 |
| Mtw1 | 254 | Spc25 | 118 |
| Mtw1 | 258 | Ndc80 | 409 |
| Mtw1 | 262 | Spc24 | 143 |
| Mtw1 | 262 | Spc24 | 204 |
| Mtw1 | 262 | Spc25 | 118 |
| Nnf1 | 1 | Nuf2 | 187 |
| Nnf1 | 26 | Ndc80 | 137 |
| Nsl1 | 198 | Spc24 | 139 |
| Nsl1 | 198 | Spc24 | 143 |
| Nsl1 | 198 | Spc24 | 204 |
| Nsl1 | 198 | Spc24 | 223 |
| Nsl1 | 198 | Spc24 | 225 |
| Nsl1 | 198 | Spc25 | 118 |
| Nsl1 | 205 | Spc24 | 139 |
| Nsl1 | 205 | Spc24 | 143 |
| Nsl1 | 205 | Spc24 | 155 |
| Nsl1 | 205 | Spc24 | 159 |
| Nsl1 | 207 | Spc24 | 155 |
Proteins in the MIND complex (Dsn1, Mtw1, Nnf1, and Nsl1) cross-link to proteins in the Ndc80 complex (diagram shown in Fig. 1).
Table S2.
Intercomplex cross-links between the Ndc80 tail domain and Ndc80, Nuf2, Spc24, and Spc25 in the absence of MIND
| Protein | Amino acid position | Protein | Amino acid position |
| Ndc80 | 1 | Ndc80 | 48 |
| Ndc80 | 1 | Ndc80 | 67 |
| Ndc80 | 1 | Ndc80 | 69 |
| Ndc80 | 1 | Ndc80 | 89 |
| Ndc80 | 1 | Ndc80 | 259 |
| Ndc80 | 1 | Ndc80 | 305 |
| Ndc80 | 1 | Ndc80 | 338 |
| Ndc80 | 1 | Ndc80 | 359 |
| Ndc80 | 1 | Ndc80 | 370 |
| Ndc80 | 1 | Nuf2 | 157 |
| Ndc80 | 48 | Ndc80 | 67 |
| Ndc80 | 48 | Ndc80 | 69 |
| Ndc80 | 48 | Ndc80 | 89 |
| Ndc80 | 48 | Ndc80 | 259 |
| Ndc80 | 48 | Ndc80 | 305 |
| Ndc80 | 48 | Ndc80 | 338 |
| Ndc80 | 48 | Ndc80 | 344 |
| Ndc80 | 48 | Ndc80 | 351 |
| Ndc80 | 48 | Ndc80 | 359 |
| Ndc80 | 48 | Nuf2 | 157 |
| Ndc80 | 67 | Ndc80 | 69 |
| Ndc80 | 67 | Ndc80 | 89 |
| Ndc80 | 67 | Nuf2 | 157 |
| Ndc80 | 69 | Ndc80 | 89 |
| Ndc80 | 69 | Nuf2 | 157 |
| Ndc80 | 89 | Ndc80 | 122 |
With the addition of MIND to the Ndc80 complex, the number of cross-links of the Ndc80 tail domain is decreased.
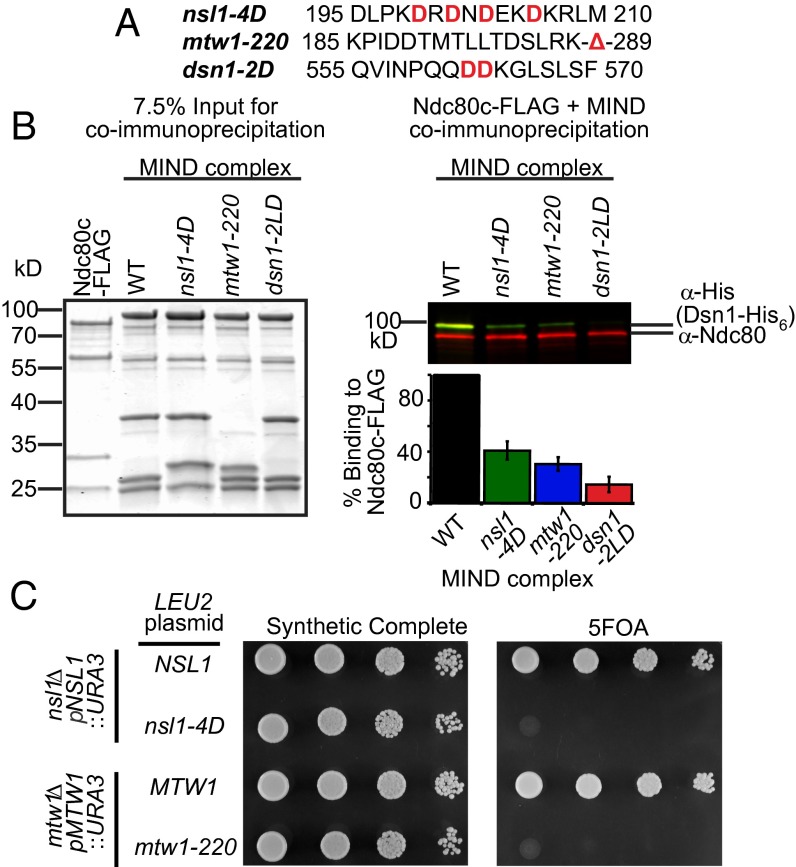
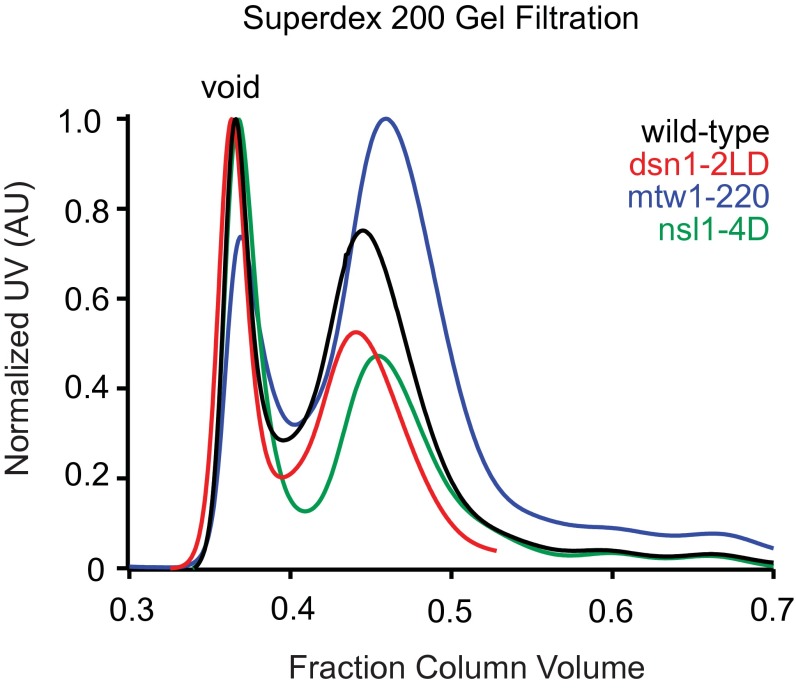
Using our cross-links as a guide, we generated three sets of mutations within Dsn1, Nsl1, and Mtw1 to determine whether these regions are required to form the MN co-complex (Fig. 2A). Lysines K198, K205, and K207 of Nsl1 lie within a predicted amphipathic helix and displayed multiple cross-links to the Spc24 pocket. Therefore, we mutated hydrophobic residues V199, Y201, V203, and V206 to aspartic acid to disrupt the hydrophobic side of the putative Nsl1 amphipathic helix (nsl1-4D). Second, we truncated the 62 C-terminal residues of Mtw1 (mtw1-220), from which multiple cross-links to Spc24 were identified. Third, we analyzed the effects of the L562D/L563D mutation in Dsn1 (dsn1-2LD) that was previously suggested to disrupt the interaction between the MIND and Ndc80 complexes in vivo (22). None of these mutations interfered with the assembly of the MIND complex, as indicated by the normal migration of all three mutant complexes in size-exclusion chromatography experiments (Fig. S2).
Fig. 2.
The Mtw1 C terminus and a putative alpha-helix in Nsl1 are required for in vitro binding to the Ndc80 complex and are essential in vivo. (A) MIND complex protein sequences, with mutated amino acids shown in red. (B) Immunoprecipitation assays with FLAG–Ndc80 complex immobilized on anti-FLAG beads and the indicated MIND–His6 complexes (WT, nsl1-4D, mtw1-220, and dsn1-2LD) added in solution. (Left) Coomassie-stained gel shows 7.5% input of protein complexes used in immunoprecipitation. (Right) Western blot of immunoprecipitation assay. Copurifying MIND complex was visualized by anti-His (staining Dsn1-His6, green); Ndc80 complex was detected with anti-Ndc80 (red). (Below Right) Quantification of immunoprecipitation experiments. WT MIND binding was normalized to 100% for each experiment, and MIND mutant binding is shown as a percentage of WT binding (n = 2 and error bars denote SEM.) (C) Plasmid shuffle assay of cells spotted in 10-fold dilutions on synthetic complete (SC) media (Left) or SC media supplemented with 5-FOA (Right). (Top spots) nsl1Δ cells containing WT NSL1 on a URA3 plasmid and either WT or nsl1-4D on a LEU2 plasmid. WT NSL1 fully supports growth on 5-FOA media whereas nsl1-4D is lethal. (Bottom spots) mtw1Δ cells with WT MTW1 on a URA3 plasmid and either WT or mtw1-220 on a LEU2 plasmid. mtw1-220 cannot support growth on 5-FOA whereas MTW1 is viable.
Fig. S2.
Gel filtration profiles of MIND complex mutants. Gel filtration elution profiles of MIND–His complex mutants run on a Superdex 200 column. Each complex elutes as a distinct peak (wild-type, black; dsn1-2LD, red; mtw1-220, blue; nsl1-4D, green).
The ability of mutant MIND complexes to bind the Ndc80 complex was quantified in vitro by immunoprecipitation (Fig. 2A). Relative to WT MIND, all three mutant versions of the MIND complex were impaired in coimmunoprecipitation with the Ndc80 complex (Fig. 2B, Right). dsn1-2LD was previously shown to cause lethality (22), and we tested whether nsl1-4D or mtw1-220 was also detrimental to cell growth. We deleted the endogenous copy of NSL1 or MTW1 and asked whether a mutated allele (nsl1-4D or mtw1-220) could support growth. Cells containing only nsl1-4D or mtw1-220 alleles failed to grow whereas those also containing WT copies of NSL1 or MTW1 grew normally (Fig. 2C). Thus, the Mtw1 C terminus and an amphipathic helix in Nsl1 are essential for the formation of the MN co-complex. Together, these results reveal an extensive protein interaction network, centered on a conserved binding pocket on the Spc24-Spc25 heterodimer, that connects the MIND and Ndc80 complexes.
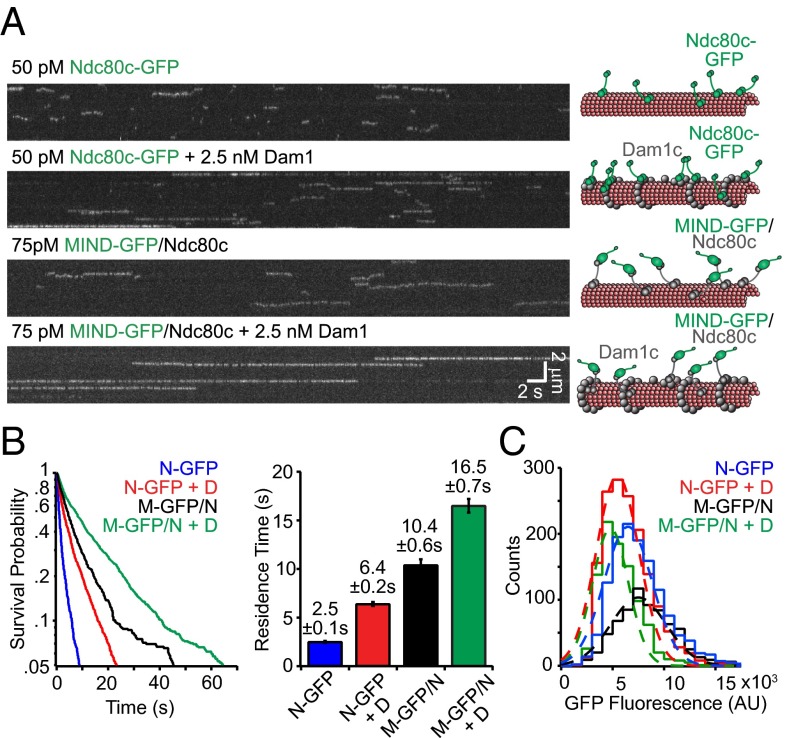
MIND Activates Microtubule Binding by Ndc80c via a Mechanism Distinct from Dam1c Activation.
We next used the MIND-GFP/Ndc80 co-complex to determine whether MIND influences the microtubule-binding properties of the Ndc80 complex. The KMN network binds synergistically to microtubules, and MIND can directly affect the activity of Knl1 in vitro (5). It is unknown whether MIND can similarly influence the behavior of the Ndc80 complex. In nematodes, the MIND and Ndc80 complexes do not directly interact without Knl1, but they do form a stable co-complex in many other organisms, including yeast and humans (17, 18, 20). We therefore assessed how the MIND complex influences Ndc80 complex microtubule binding at the single-molecule level using total internal reflection fluorescence (TIRF) microscopy. As shown previously, Ndc80c-GFP alone has a relatively weak affinity for microtubules (23) and exhibited a mean residence time of 2.5 ± 0.1 s on the microtubule lattice (Fig. 3 A and B). Previous work found that addition of the 10-member outer kinetochore Dam1 complex increased the residence time of Ndc80c 2.6-fold, to 6.4 ± 0.2 s (Fig. 3 A and B) (24). Surprisingly, we found that MIND also dramatically affected the microtubule binding of Ndc80c because the residence time of MIND-GFP/Ndc80c complexes was 10.4 ± 0.6 s, fourfold longer than that of Ndc80c-GFP (Fig. 3B). By contrast, MIND-GFP alone did not interact with microtubules (5), even when added at high concentrations (Fig. S3), indicating that MIND activates the microtubule-binding activity of the Ndc80 complex.
Fig. 3.
MIND and Dam1c individually and additively enhance the binding of Ndc80 complex to microtubules. (A) Representative TIRF kymographs of Ndc80c-GFP, Ndc80c-GFP plus Dam1c, MIND-GFP/Ndc80c, and MIND-GFP/Ndc80c plus Dam1c. Concentrations are noted and scale bars are indicated in white. Diagrams on right denote each GFP tagged complex (green) and untagged complex (gray) binding to microtubules (red) in kymograph on left. (B, Left) Survival probability vs. time is plotted for each complex, quantified from individual binding events. Ndc80c-GFP (N-GFP, n = 1,278), Ndc80c-GFP + Dam1c (N-GFP + D, n = 1,525), MIND-GFP/Ndc80c (M-GFP/N, n = 706), and MIND-GFP/Ndc80c + Dam1c (M-GFP/N + D, n = 924). (Right) Average residence time for each complex derived from distribution on left; error bars denote SD. (C) GFP fluorescence distribution for each GFP-tagged complex shown as histograms. Gaussian fits are shown with dotted lines and used to quantify the mean GFP fluorescence for each complex. The number of events is the same as in B. The mean fluorescence values for each complex are as follows: N-GFP = 7,000 ± 2,300 arbitrary units (AU), N-GFP + D = 6,000 ± 2,100 AU, M-GFP/N = 8,000 ± 2,700 AU, M-GFP/N + D = 5,300 ± 1,800 AU.
Fig. S3.
MIND-GFP does not bind to microtubules and does not affect Dam1c binding to microtubules. Representative TIRF kymographs of MIND-GFP (A), Dam1-GFP (B), or MIND-GFP plus Dam1c (C). Binding to region of interest (ROI) with microtubules or coverslip (random ROI, no microtubules) is shown. (A) MIND-GFP shows no binding preference between microtubules (Top) and coverslip (Bottom). (B) Dam1-GFP specifically binds to microtubules (Top) and shows little nonspecific binding to the coverslip (Bottom). (C) MIND-GFP shows no difference between microtubule (Top) and coverslip (Bottom) in the presence of excess Dam1.
The effects of MIND and Dam1c on the ability of Ndc80c to bind microtubules are additive. The average residence time for MIND-GFP/Ndc80c increases 1.5-fold in the presence of Dam1 complex to 16.5 ± 0.7 s (Figs. 3 A and B). This combinatorial effect suggests that Dam1c and MIND influence Ndc80c via independent mechanisms.
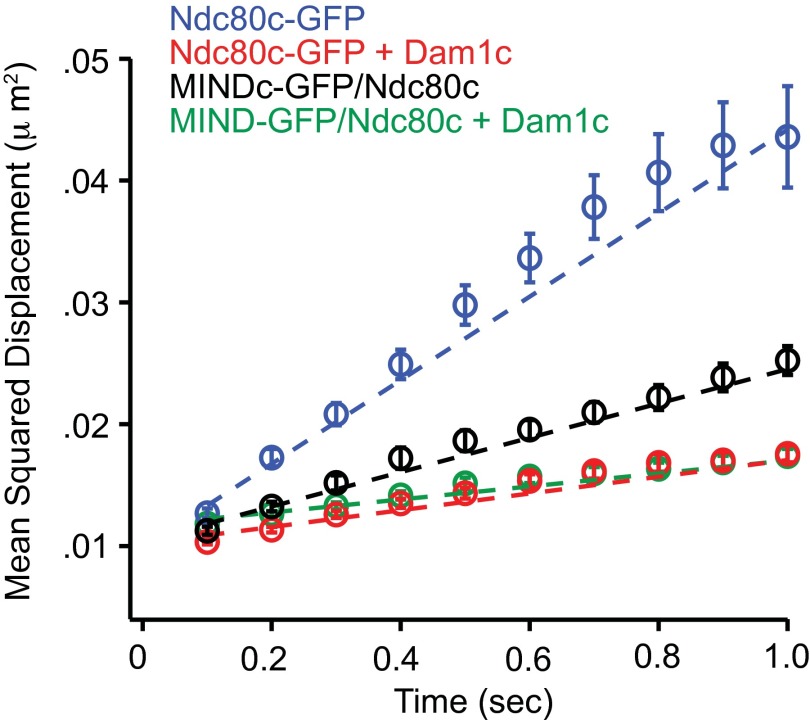
The observation that MIND and Dam1c influence Ndc80c independently was further supported by their disparate effects on the motility of Ndc80c along microtubules. MIND showed a milder effect on the diffusion of Ndc80c than does the Dam1 complex (Fig. S4). We also found that MIND and Dam1c differ in their ability to enhance the tracking of Ndc80c with disassembling microtubule tips. Dynamic microtubule extensions were assembled off of GMPCPP-stabilized microtubule seeds, and their disassembly was initiated by the removal of free tubulin from solution. Ndc80c-GFP alone tracks poorly with disassembling microtubule ends but can track robustly when artificially oligomerized by antibodies, or in the presence of Dam1c (23, 24) (Fig. S5). By contrast, MIND did not enhance the ability of Ndc80c to track disassembling microtubule ends (Fig. S5). Therefore, Dam1c and MIND have different effects on the microtubule binding, diffusion, and tip tracking of Ndc80c. These observations indicate that MIND and Dam1c do not simply influence the behavior of Ndc80c by differing degrees, but do so by distinct mechanisms.
Fig. S4.
MIND and Dam1c have different effects on the diffusion of Ndc80c. Mean squared displacement of indicated complexes plotted vs. time. Dotted lines show weighted linear fits used to calculate average diffusion constants, with error bars denoting SD. Diffusion constants for each complex are as follows: (blue) Ndc80c-GFP = 0.0343 ± 0.0012 μm2⋅s−1, (red) Ndc80c-GFP + Dam1c = 0.0069 ± 0.0002 μm2⋅s−1, (black) MIND-GFP/Ndc80c = 0.0141 ± 0.0004 μm2⋅s−1, (green) MIND-GFP/Ndc80c + Dam1c = 0.0053 ± 0.0002 μm2⋅s−1.
Fig. S5.
Unlike Dam1c, MIND does not enhance the ability of the Ndc80 complex to track with depolymerizing microtubule tips. Representative two-color TIRF kymograph of MIND-GFP/Ndc80c (Top) or Ndc80c-GFP plus Dam1c (Bottom) with depolymerizing microtubules (red). Ndc80c-GFP efficiently tracks with depolymerizing tips in the presence of Dam1c, with a mean tracking distance of 780 ± 120 nm (n = 16) whereas MIND-GFP/Ndc80c failed to track with depolymerizing microtubule tips. Error bars denote SEM. Scale bars are shown in white.
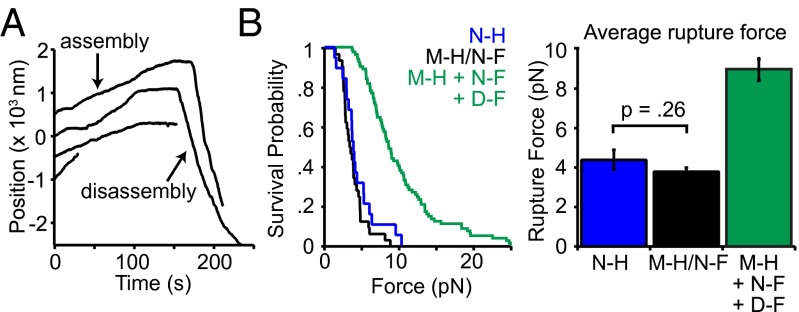
The MIND–Ndc80c Interface Can Bear Substantial Levels of Mechanical Load.
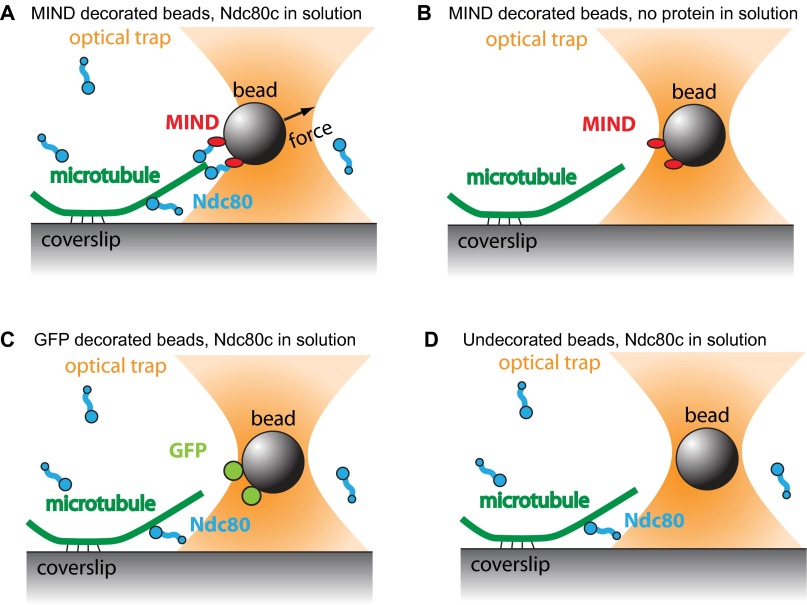
In vivo, kinetochores transmit tension from the microtubule interface to the centromeric DNA. Beyond the Ndc80 and Dam1 complexes (24), it is not known which kinetochore components participate in the force transmission pathway. We therefore asked whether MIND could directly support mechanical load transmitted through Ndc80c. Indeed, when bound to polystyrene beads via a His tag on MIND, MIND-His/Ndc80c-FLAG was able to couple beads to both assembling and disassembling microtubule tips against an applied load of ∼2.5 pN (Fig. 4A). Beads failed to couple to microtubules against force when decorated with the MIND–His complex in the absence of Ndc80c-FLAG (n = 30). Likewise, no coupling was observed when beads lacking MIND–His were incubated with Ndc80c-FLAG alone (GFP-His–coated beads, n = 79 and uncoated beads, n = 60) (Fig. S6). These controls rule out direct microtubule binding by MIND and/or nonspecific adsorption of Ndc80-Flag to the beads. Therefore, the applied load must be transmitted through the MIND–Ndc80c interface. To probe the strength of the MIND–Ndc80 linkage, we used a rupture force assay (25). MIND-His/Ndc80c-FLAG–coated beads were coupled to assembling microtubule tips and briefly subjected to a test force of ∼1 pN. Then, the load was increased at a constant rate (0.25 pN⋅s−1) until the bead detached from the microtubule. On average, MIND-His/Ndc80c-FLAG–mediated attachments ruptured at 3.8 ± 0.2 pN, comparable with the strength afforded by coupling Ndc80 complex directly to the bead, 4.5 ± 0.2 pN (not significantly different, P = 0.26) (Fig. 4B). When Dam1c-FLAG was added to the assay, the average rupture force of MIND-His/Ndc80c-FLAG beads increased to 9.0 ± 0.6 pN (Fig. 4B). Altogether, these results indicate that the linkage between MIND and Ndc80c can support substantial levels of tension, suggesting that MIND complex is a key participant in force transmission through the kinetochore.
Fig. 4.
The MIND/Ndc80c linkage can withstand substantial load. (A) Representative traces of bead position versus time for 20 nM MIND-His/Ndc80c-FLAG beads tracking with assembling and disassembling microtubule tips under 1.7–2.5 pN of force applied in the direction of microtubule assembly. Traces are arbitrarily offset on the y axis for visual clarity. (B, Left) Survival probability vs. force for 20 nM Ndc80c-His (N-H, n = 20), 20 nM MIND-His/Ndc80c-FLAG (M-H/N-F, n = 32), and 20 nM MIND-His beads with 40 nM Ndc80c-FLAG and 2 nM Dam1c-FLAG (M-H + N-F + D-F, n = 50). (Right) Average rupture force derived from the distributions on the Left. Ndc80c-His and MIND-His/Ndc80c-FLAG were not significantly different, P = 0.26, two-tailed Student’s t test. Error bars denote SEM.
Fig. S6.
The Ndc80 complex and MIND form a specific load-bearing linker in vitro. (A) MIND-His decorated polystyrene beads form a specific load-bearing linkage to Ndc80c-Flag and can withstand an applied load of ∼2.5 pN (n = 32). (B) MIND-His decorated beads cannot form load-bearing attachments to microtubules without Ndc80c-Flag in solution (n = 30). (C) Beads decorated with GFP-His are unable to couple to free Ndc80c-Flag in solution to create force-bearing attachments to microtubules (n = 79). (D) Undecorated beads do not interact with Ndc80c-Flag in solution to form microtubule attachments (n = 60).
A Single MIND Activates a Single Ndc80 Complex.
The Dam1 and Ndc80 complexes form a unit on microtubules that increases the number of microtubule-binding contacts. Unlike Dam1c, MIND does not form additional direct contacts with the microtubule but instead contributes to microtubule binding indirectly through Ndc80c. How might MIND activate microtubule binding by Ndc80c? One possible explanation is that MIND promotes oligomerization of Ndc80c on microtubules, thereby increasing avidity. MIND could drive oligomerization of Ndc80c by binding two or more Ndc80 complexes and/or by binding other MIND complexes.
First, we tested for oligomerization directly by performing a dual-label TIRF experiment to compare the residence time of GFP-tagged Ndc80 complexes alone to those that colocalized with MIND-SNAP (Fig. S7 A–C). By measuring fluorescence intensity, we found that Ndc80c-GFP remained monomeric when alone or bound to MIND-SNAP, but its residence time was significantly increased when in complex with MIND (Fig. S7 B and C). Second, we added excess Ndc80c in our TIRF assay in an attempt to both drive the association of MIND with multiple Ndc80c complexes and maximize the occupancy of Ndc80-binding sites within the MIND complex. Supplementing up to a 267-fold molar excess of Ndc80c did not affect the residence time of MN on microtubules (Fig. S7D). Third, under assay conditions where MIND enhanced microtubule binding by Ndc80c, MIND oligomerization was rare; the average fluorescence intensity of GFP-tagged MIND within MN co-complexes bound to microtubules was similar to that of monomeric GFP-tagged Ndc80 complexes (Fig. 3C). Altogether, these data demonstrate that, in our assay, MIND does not enhance Ndc80c microtubule binding by oligomerization. Instead, a single MIND complex directly enhances microtubule attachment by a single Ndc80 complex.
Fig. S7.
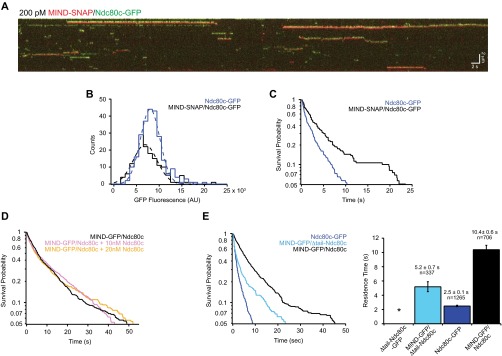
MIND does not enhance Ndc80 complex via oligomerization and does not require the Ndc80 N-terminal tail. (A) Representative two-color TIRF kymograph of individual MIND-SNAP/Ndc80c-GFP complexes binding to microtubules. MIND-SNAP is shown as red and Ndc80c-GFP shown as green. Note: Kymographs for each channel were overlaid with a slight vertical offset to visualize dually fluorescent complexes. Scale bars are shown in white. (B) GFP fluorescence distribution of individual events. Dotted lines show Gaussian fits used to determine mean GFP fluorescence for each complex. MIND-SNAP/Ndc80c-GFP, black histogram, n = 160 events, mean = 7,400 ± 2,600 AU; Ndc80-GFP, blue histogram, n = 248 events, mean = 8,500 ± 2,100 AU. Note that Ndc80c-GFP only events were measured from the same kymographs as MIND-SNAP/Ndc80c-GFP events. (C) Survival probability vs. time for Ndc80c-GFP and MIND-SNAP/Ndc80c-GFP. Ndc80c-GFP, blue line, n = 248 events, average residence time = 2.5 ± 0.2 s; MIND-SNAP/Ndc80c-GFP, black line, n = 160 events, average residence time = 6.2 ± 0.8 s. Error bars denote SD. As in B, the Ndc80c-GFP only events and MIND-SNAP/Ndc80c-GFP events were measured from the same kymographs. (D) Survival probability vs. time for 75 pM MIND-GFP/Ndc80c alone (black line), or with the addition of excess Ndc80c [10 nM (pink line) or 20 nM (gold line)] in solution. The mean residence times were as follows: MIND-GFP/Ndc80c, 12.1 ± 0.7 s, n = 604 events; MIND-GFP/Ndc80c plus 10 nM Ndc80c, 12.1± 0.9 s, n = 464 events; MIND-GFP/Ndc80c plus 20 nM Ndc80c, 11.9 ± 0.7 s, n = 610 events. (E, Left) Survival probability vs. time quantified from individual binding events for each complex noted in the legend. Note: Traces for Ndc80c-GFP and MIND-GFP/Ndc80c are repeated here (from Fig. 3B) for comparison. (Right) Average residence time with error bars denoting SD. For Δtail-Ndc80c-GFP, the * indicates no binding.
We hypothesized that MIND induces a conformational change in the Ndc80 complex that favors microtubule coupling by activation of the microtubule-binding domains. The Ndc80 complex binds microtubules primarily through the Ndc80 calponin homology (CH) domains and the Ndc80 N-terminal tail (Fig. 5A) (26–29). We first tested whether MIND activation requires the N-terminal tail of Ndc80. In our cross-linking analysis, the disordered Ndc80 N-terminal tail interacted with portions of Ndc80, Nuf2, Spc24, and Spc25 (Fig. S8A) (10). The presence of the MIND complex reduced cross-linking of the tail, potentially restricting its position in the complex (Fig. S8B and Tables S1–S3). As previously demonstrated (28, 29), tail-less Ndc80 complex (Δtail-Ndc80c-GFP) bound poorly to microtubules. Due to its extremely short interactions with microtubules at the single-molecule level, we were unable to accurately measure its residence time in the TIRF assay. However, MIND increased the residence time of Δtail-Ndc80c, yielding an average of 5.2 ± 0.7 s, indicating that the tail domain is not required for MIND-mediated enhancement (Fig. S7E).
Fig. 5.
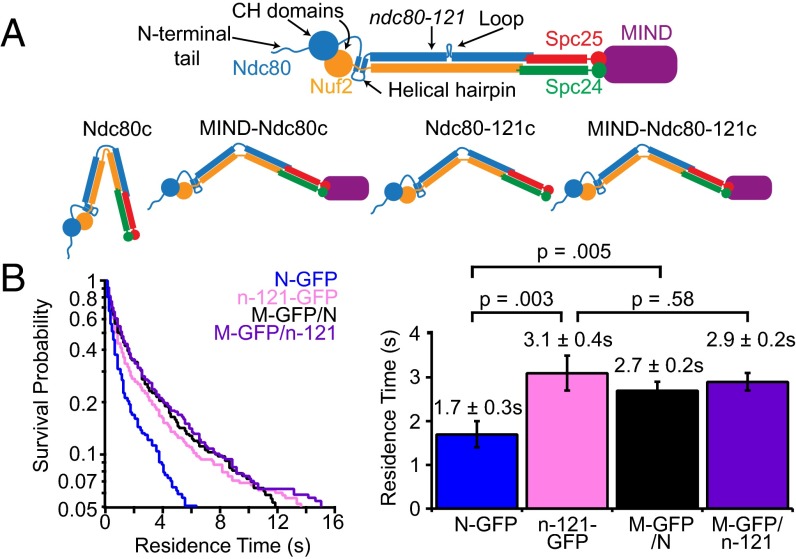
MIND activates Ndc80 complex binding via the same mechanism as ndc80-121. (A, Top) Diagram of Ndc80 complex denoting Ndc80 microtubule-binding domains (CH domain, N-terminal tail) and position of ndc80-121 mutation (Y465C/I469Q) near the loop domain. Modified from ref. 10 with permission from the Genetics Society of America. (Bottom) Diagram depicting proposed conformational changes of MIND-Ndc80c vs. MIND-Ndc80-121c. (B, Left) Survival probability vs. time quantified from individual binding events for Ndc80c-GFP (N-GFP, n = 355), ndc80-121c-GFP (n-121-GFP, n = 481), MIND-GFP/Ndc80c (M-GFP/N, n = 359), and MIND-GFP/ndc80-121 (M-GFP/n-121, n = 426) at the restrictive temperature (37 °C). (Right) Average residence times derived from distributions on Left; error bars denote SD. Ndc80c-GFP is statistically different from Ndc80-121c-GFP, P = 0.003 and MIND-GFP/Ndc80c, P = 0.005 (two-tailed Student’s t test). Ndc80-121c-GFP and MIND-GFP/Ndc80-121c are not statistically different (two-tailed Student’s t test, P = 0.58).
Fig. S8.
The disordered N-terminal Ndc80 tail cross-links extensively within the Ndc80 complex. Comparison of Ndc80 tail domain (amino acids 1–113, highlighted in yellow) cross-linking to proteins in Ndc80 complex alone (A) or when bound to MIND complex (B). The positions of lysine residues within each protein are marked as vertical white lines. For simplicity, cross-links not involving the tail domain are omitted from diagrams.
An alternative possibility is that MIND influences larger scale conformational changes in the Ndc80 complex. We recently identified a temperature-sensitive mutant of Ndc80 (ndc80-121) (10) that harbors two mutations near the loop domain, far from the microtubule and MIND-binding domains (Fig. 5A). Based on previous genetic analysis, it was hypothesized that this mutant adopts a conformation at 37 °C that enhances its microtubule binding. Consistent with this view, the Ndc80-121–GFP complex bound at the restrictive temperature (37 °C) to microtubules 1.5-fold longer than the WT complex (Fig. 5B). We then asked whether MIND can further enhance the ability of the Ndc80-121 complex to bind microtubules by measuring the residence time of a MIND-GFP/Ndc80-121 co-complex via TIRF. MIND-GFP/Ndc80-121c assembled as a stoichiometric complex and exhibited a similar gel filtration profile as MIND-GFP/Ndc80c, indicating that the ndc80-121 mutations did not affect interaction with MIND (Fig. S9). At 37 °C, MIND-GFP/Ndc80c also exhibited an average residence time 1.5-fold longer than Ndc80c-GFP alone, similar to the behavior of Ndc80-121–GFP (Fig. 5B). By contrast, MIND did not further enhance the binding of Ndc80-121c; MIND-GFP/Ndc80-121c residence time was indistinguishable from Ndc80-121 complex alone (not significantly different, P = 0.58) (Fig. 5B). These results suggest that both MIND and the ndc80-121 mutations alter the behavior of the Ndc80 complex by the same mechanism, which may involve promoting conformational activation of the Ndc80 complex.
Fig. S9.
Gel filtration profiles of MINDc-GFP/Ndc80c and MINDc-GFP/Ndc80-121c. Representative Sephacryl 400 gel filtration elution profiles for MINDc-GFP/Ndc80c co-complex (blue) and MINDc-GFP/Ndc80-121c (red). The gray-shaded bar indicates peaks that samples were taken from for experiments. Note that MIND-GFP/Ndc80c and MIND-GFP/Ndc80-121c elute at the same volume, indicating that the mutations in Ndc80-121c do not influence formation of higher order structures.
Discussion
MIND was previously identified as part of the core microtubule-binding KMN network, yet how MIND facilitates microtubule attachment has remained unclear. By reconstituting the yeast MIND/Ndc80 co-complex and using cross-linking analysis, we have identified an intricate set of interactions involving five of the eight proteins within the two complexes. In addition to the previously identified Spc24–Spc25 interface shared by both MIND and Cnn1 (15, 22), we found a unique connection between Nsl1 and a hydrophobic Spc24/Spc25 cleft. This identification of a second unique interface suggests that the Ndc80 complex may differentially interact with MIND and Cnn1, raising the possibility that each receptor might distinctly regulate Ndc80c function. Furthermore, Nsl1 has been identified as a link between human Mis12 and Ndc80 complexes (20), and our identification of its important contribution to the yeast MIND/Ndc80 interface establishes the structural conservation of this connection.
Using single-molecule techniques, we show that the MIND complex promotes the binding of Ndc80c to microtubules. This effect is additive, with the enhancement conferred by the Dam1 complex, providing further evidence that kinetochore components act cooperatively to form robust microtubule attachments. Finally, we demonstrate that the MIND/Ndc80c interface can withstand substantial load, implicating MIND as an integral component of the force transmission pathway of the kinetochore.
How does MIND enhance microtubule attachments? MIND binds Ndc80c far from its microtubule-binding domain (17, 18, 20). Consistent with this placement, we show here that MIND does not directly bind microtubules. Furthermore, MIND does not seem to enhance binding by organizing the Ndc80 N-terminal tail domain nor is oligomerization of Ndc80c required. Instead, we demonstrate that an individual MIND complex enhances the microtubule binding of a single Ndc80 complex. We show that MIND binding to the Spc24/Spc25 terminus of the Ndc80 complex confers increased affinity of the microtubule-binding domain.
The Ndc80 complex hinges about a flexible loop region, and in vitro and in vivo evidence supports the existence of both a folded conformation and an elongated state (8, 11). Here, we show that a temperature-sensitive NDC80 mutant (ndc80-121) that has been previously suggested to affect the stability of a folded complex at 37 °C exhibits increased microtubule affinity at 37 °C compared with the WT complex. Because addition of MIND does not further increase the affinity of the Ndc80-121 complex for microtubules, MIND and Ndc80-121c increase affinity by a redundant mechanism. We propose that both favor formation of an open, high-affinity conformation of the Ndc80 complex.
The physiological relevance of different conformational states of the Ndc80 complex has remained unclear. Deleting the loop domain of Ndc80 causes lethality, suggesting that flexibility of the complex is necessary in vivo (12). Additional in vivo evidence suggests that a folded Ndc80 complex is important during early mitosis whereas an elongated conformation exists at metaphase (8–10). We propose that the Ndc80 complex adopts different conformations to modulate the strength of its microtubule attachment. A folded Ndc80 complex that interacts less stably with microtubules might be favorable in prometaphase when erroneous attachments must be corrected by Ipl1/Aurora B kinase (30, 31). The Ndc80 complex adopts an elongated conformation during metaphase, which could promote strong microtubule interaction when kinetochore tension is highest. Folding of the complex at the Ndc80 loop domain positions Spc24 and Spc25 near the microtubule-binding domains of Ndc80, potentially obstructing full contact with microtubules. Indeed, it was previously demonstrated using C. elegans components that the Nuf2/Ndc80 dimer bound more tightly to microtubules than the four-member complex, suggesting intracomplex inhibition (5). This autoinhibitory conformation is reminiscent of kinesins and myosin V, which fold into inhibited states that prevent interaction between motor domains and the cytoskeleton in the absence of cargo (32–35). We propose that MIND binding to Spc24/Spc25 relieves this autoinhibition via steric hindrance or allosteric activation, by interfering with the intracomplex interactions that stabilize the folded state of Ndc80c, and thus promotes its binding to microtubules.
Autoinhibition of the Ndc80 complex could help during S-phase, when premature microtubule binding might interfere with kinetochore assembly. MIND-dependent relief of this autoinhibition could ensure that Ndc80 is activated only after it is successfully incorporated into the kinetochore. It could also explain why Ndc80 is detected only at the kinetochore whereas other microtubule binding components of the kinetochore are also detected all along the spindle microtubules (36). Cnn1 provides a distinct Ndc80 receptor during anaphase (13, 15, 16). It will therefore be interesting to learn whether and how Cnn1 affects Ndc80’s microtubule affinity.
Altogether, our results highlight the previously unidentified regulation of the microtubule-binding activity of an outer kinetochore component by a central kinetochore complex. We propose that modulating the conformation of microtubule couplers is a way to regulate the strength of microtubule attachments throughout mitosis.
Materials and Methods
Protein Expression and Purification.
His6-tagged MIND complex was expressed from a polycistronic pRSF vector in BL21 cells. The His6-tagged Ndc80 complex was expressed and purified as described (23, 37), and the FLAG–Ndc80 complex was expressed similarly. MIND/Ndc80 co-complex was prepared by running nickel-purified MIND-His6 and His6-Ndc80 complexes over a Sepharose 400 size exclusion column (GE Healthcare). See SI Materials and Methods for additional details.
Plasmid Shuffle Assay.
NSL1 or MTW1 was deleted in a diploid strain and transformed with a WT copy of NSL1 or MTW1 on a URA3 plasmid. Haploids were transformed with a WT or mutated gene (nsl1-4D or mtw1-220) on a LEU2 plasmid. Colonies were grown in SD-Leu media, plated in 10-fold dilutions on synthetic complete and 1 mg⋅mL−1 5-FOA plates, and growth was assessed after 48 h. See SI Materials and Methods for additional details.
Cross-Linking Analysis.
The MIND/Ndc80 co-complex was cross-linked for 2 min at 25 °C with disuccinimidyl suberate (0.3 mM final; Pierce). Reactions were quenched, and buffer was exchanged using protein desalting spin columns (Pierce). Cross-linked proteins were reduced with 10 mM DTT, alkylated with 15 mM iodoacetamide, and digested with trypsin (at a substrate-to-enzyme ratio of 60:1). Samples were acidified with 5 M HCl, and 0.75 µg of protein was loaded onto a fused-silica capillary tip column (75-µm i.d.) packed with 40 cm of Reprosil-Pur C18-AQ (3-µm bead diameter; Dr. Maisch). Cross-linked peptides were identified using the Kojak cross-link identification software (www.kojak-ms.org) (38) (SI Materials and Methods).
Immunoprecipitation.
Anti-FLAG M2 affinity gel was incubated with 600 nM FLAG–Ndc80 complex and then washed with Tris-buffered saline plus 0.1% Nonidet P-40. Then, 1 μm Dsn1-His6 tagged MIND complex was added, incubated for 1 h at 4 °C, and washed. Protein was eluted with 0.1 mg⋅mL−1 3X FLAG peptide and analyzed by Western blot using anti-His (Genscript) and anti-Ndc80 (a gift from Arshad Desai, Ludwig Cancer Research Center, University of California, San Diego). Total MIND complex binding was measured using Dsn1-His6 intensity normalized over Ndc80 intensity for each reaction (SI Materials and Methods).
TIRF Microscopy.
Custom instrumentation and flow cells were prepared as previously described (23, 39). All protein complexes were purified via gel filtration as described under Protein Expression and Purification. Protein mixtures were diluted into BRB80 assay buffer [80 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.9, 1 mM MgCl2, 1 mM EGTA] with 8 mg⋅mL−1 BSA (BB80), 1 mg⋅mL−1 κ-casein, and oxygen scavengers (200 μg⋅mL−1 glucose oxidase, 35 μg⋅mL−1 catalase, 25 mM glucose, 5 mM DTT) and then introduced into a flow cell with coverslip-anchored, taxol-stabilized, Alexa-568–labeled microtubules. GFP and Alexa Fluor-568 channels were imaged simultaneously using a cooled camera (iXon 887-BI; Andor). For experiments at 37 °C, an objective heater controller (Bioptechs Inc.) was used, and flow cells were heated to 37 °C before protein addition and imaging. Microtubule disassembly was induced by buffer exchange, as in ref. 24. Single-particle tracking and analysis were done using custom Labview (National Instruments) and Igor Pro (Wavemetrics) software as previously described (23, 39). See SI Materials and Methods for additional details.
Optical Bead Motility Assay.
Anti-His5 polystyrene beads (11 pM) were incubated with 40 nM His6-tagged Ndc80 or MIND complex as described (40–42), such that each bead was decorated with ∼1,800 protein complexes. Protein-coated beads were introduced into the flow chambers in BB80 with 1 mM GTP, 1.4 mg⋅mL−1 tubulin, 200 μg⋅mL−1 glucose oxidase, 35 μg⋅mL−1 catalase, 25 mM glucose, and 1 mM DTT. For MIND/Ndc80c reactions, His6-tagged MIND and FLAG-tagged Ndc80 complex were preassembled into a co-complex as described under Protein Expression and Purification before preparing beads. For assays containing MIND, Ndc80, and Dam1 complexes, 20 nM His6-MIND beads were prepared as described previously in this section, and then 40 nM free FLAG–Ndc80 complex and 2 nM free Dam1–FLAG complex were added. Assays were performed at 26 °C using custom instrumentation to capture and manipulate beads and analyzed as described (40). See SI Materials and Methods for additional details.
SI Materials and Methods
Protein Expression and Purification.
Dsn1-His6–tagged MIND complex was expressed from a polycistronic pRSF vector in BL21 cells and induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 14 h at 20 °C. Cells were lysed with a French press, and MIND-His was purified via a Ni-charged IMAC resin column (Bio-Rad) in 50 mM NaPO4, 200 mM NaCl, pH 7.0, supplemented with protease inhibitors (Roche), 5 mM imidazole, and 1 mM PMSF, and then washed and eluted with 300 mM imidazole. The MIND complex was purified using a Superdex 200 column (GE Healthcare), and concentration was measured by bicinchoninic acid (BCA) (Sigma). For TIRF assays, Mtw1-GFP–tagged MIND complex was purified via a His6-tag on Nnf1 as described previously in this section. Mtw1-SNAP–tagged Nnf1-His6–MIND complex was expressed and purified as described previously in this section. The MIND–SNAP complex was labeled with SNAP-Surface 549 dye (New England Biolabs) at a 2:1 dye:protein molar ratio overnight at 4 °C. His6-tagged Ndc80 and His6-tagged Ndc80-121 complex were expressed and purified as described (10, 37). FLAG-Spc24 Ndc80 complex was expressed similarly to His6-tagged complex, lysed using a French press in 50 mM Hepes, 200 mM NaCl, pH 7.6, and bound to anti-FLAG M2 affinity gel (A2220; Sigma). The affinity gel was washed, and protein was eluted with 0.1 mg⋅mL−1 3X FLAG Peptide (F4799; Sigma). The Ndc80–FLAG complex was purified using a Superdex 200 size exclusion column (GE Healthcare). The MIND/Ndc80 co-complex was prepared by combining nickel-purified MIND–His6 and Ndc80–His6 complexes in a 2.5:1 molar ratio and incubating for 15 min at room temperature. MIND-GFP/Ndc80-FLAG co-complex was prepared similarly. All resulting MIND/Ndc80 co-complexes were subsequently purified with a Sepharose 400 size exclusion column (GE Healthcare) in 50 mM NaPO4, 100 mM NaCl, pH 7.0.
Velocity Sedimentation.
Sucrose gradients [5–30% (wt/vol)] were generated by layering 250 μL of 5%, 13.8%, 21.6%, and 30% sucrose in 50 mM NaPO4, 100 mM NaCl, pH 7.0, and allowing a continuous gradient to form for 3 h at 4 °C. Then 1–2 μM purified MIND-His6 and Ndc80-His6 complex were mixed at 1:1 molar ratio for 15 min at room temperature before loading onto gradient. Reactions were ultracentrifuged at 166,000 × g at 4 °C for 5 h, and 16–18 sequential fractions were collected and analyzed by SDS/PAGE, staining with Coomassie Blue. Sedimentation coefficients were determined by comparing to elution of known standard proteins (BSA, catalase, and aldolase).
Plasmid Shuffle Assay.
Endogenous NSL1 or MTW1 was deleted in a diploid strain with a KanMX cassette and then transformed with a WT copy of NSL1 or MTW1 under its endogenous promoter on a URA3 plasmid. Cells were sporulated, and haploids were selected. Haploids containing both KanMX and URA3 markers were transformed with a WT or mutated gene (nsl1-4D or mtw1-220) under its endogenous promoters on a LEU2 plasmid. Positive colonies were selected on SD-Leu for 3 d at 30 °C. Individual clones were grown to log phase in SD-Leu media and then plated in 10-fold dilutions on synthetic complete and 1 mg⋅mL−1 5-FOA plates. Growth was assessed after 48 h at 30 °C.
Cross-Linking of Recombinant MIND/Ndc80 Complex and Mass Spectrometry Analysis.
The MIND/Ndc80 co-complex (26 µg in 133 µL of either 100 mM NaCl or 300 mM NaCl, 50 mM NaPO4 buffer, pH 7) was cross-linked for 2 min at room temperature with disuccinimidyl suberate (0.3 mM final; Pierce). The reaction mix was quenched with 10 µL of 500 mM NH4HCO3, and the buffer was exchanged to HB500 (40 mM Hepes, 500 mM NaCl, pH 7.5) using protein desalting spin columns (Pierce) according to the manufacturer’s protocol. Cross-linked proteins were subsequently reduced with 10 mM DTT for 30 min at 37 °C, alkylated with 15 mM iodoacetamide for 30 min at room temperature, and digested with trypsin (at a substrate to enzyme ratio of 60:1) overnight at room temperature with shaking. Samples were acidified with 5 M HCl and stored at −80 °C.
Samples (0.75 µg) were loaded onto a fused-silica capillary tip column (75-µm i.d.) packed with 40 cm of Reprosil-Pur C18-AQ (3-µm bead diameter; Dr. Maisch). Peptides were eluted from the column at 250 nL⋅min−1 using a gradient of 2–35% acetonitrile (in 0.1% formic acid) over 120 min, followed by 35–60% acetonitrile over 10 min. Mass spectrometry was performed on a Q-Exactive (Thermo Scientific), operated using data-dependent acquisition where a maximum of six MS/MS spectra were acquired per MS spectrum (scan range of m/z 400–1,600). At m/z 200, the resolution for MS and MS/MS was 70,000 and 35,000, respectively.
Cross-linked peptides were identified using the Kojak cross-link identification software (www.kojak-ms.org). The results of Kojak were exported directly to Percolator (43) to produce a statistically validated set of cross-linked peptide identifications at a false discovery rate threshold of 5%.
Immunoprecipitation.
Anti-FLAG M2 affinity gel (Sigma) was prepared according to the manufacturers instructions and then incubated with purified 600 nM Ndc80c–FLAG complex for 1 h at 4 °C. Beads were subsequently washed 3 × 5 min with Tris-buffered saline (TBS) plus 0.1% Nonidet P-40, and 1 μm Dsn1-His6–tagged purified MIND complex was added. Reactions were incubated for 1 h at 4 °C and washed 3 × 5 min with TBS plus 0.1% Nonidet P-40. Protein was eluted with 50 μL of TBS plus 0.1 mg⋅mL−1 3X FLAG peptide (Sigma) at room temperature for 30 min with gentle agitation. Eluate was removed and boiled with sample buffer and then run on SDS/PAGE and subjected to Western blot analysis using anti-His (Genscript) and anti-Ndc80 (a gift from Arshad Desai, Ludwig Cancer Research Center, University of California, San Diego). Total MIND complex binding was measured using Dsn1-His6 band intensity normalized over Ndc80 band intensity for each reaction. Binding of mutant MIND complexes to the Ndc80 complex was calculated as a percentage of WT MIND complex binding.
TIRF Microscopy.
Custom instrumentation and flow cells were prepared as previously described (23, 39). For assays, coverslips were rinsed twice with ddH20 and then incubated with “rigor” kinesin (44) in standard BB80 assay buffer (80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.9, 1 mM MgCl2, 1 mM EGTA, 8 mg⋅mL−1 BSA) for 5 min. Alexa Fluor-568–labeled taxol-stabilized bovine microtubules were adhered to coverslips and rinsed with BB80. All protein complexes were purified via gel filtration before TIRF microscopy. Dilutions of the protein complex of interest in BB80 supplemented with 1 mg⋅mL−1 κ-casein and oxygen scavengers (200 μg⋅mL−1 glucose oxidase, 35 μg⋅mL− catalase, 25 mM glucose, 5 mM DTT) were flowed in and immediately imaged. GFP and Alexa Fluor-568 channels were imaged simultaneously using a cooled camera (iXon 887-BI; Andor) at 10 frames⋅s−1 for 200 s. Controls assaying MIND-GFP alone, Dam1c alone, and Dam1c-GFP plus MIND maintained the same TIRF microscopy assay conditions as the experimental assays. Dam1c-GFP and MIND were combined immediately before imaging.
For imaging at 37 °C, an objective heater controller (Bioptechs Inc.) was used. Flow cells were preheated to 37 °C for at least 5 min before flowing in protein and imaging as described previously in this section. Type 37 immersion oil was used to compensate for the change in temperature. The laser power of the GFP channel was increased 2.7-fold to maintain the same GFP brightness as 25 °C reactions.
For disassembling microtubule assays, solutions were prepared as in ref. 24. Briefly, flow cells were washed and prepared with rigor kinesin as described previously in this section and then Alexa Fluor-568–labeled GMPCPP microtubule seeds were bound to coverslips and washed with BB80. Microtubule growth buffer (BB80 with 1 mM GTP and oxygen scavengers) containing ∼2 mg⋅mL−1 Alexa Fluor-647–labeled tubulin was introduced to the chamber and incubated with seeds for 10 min until extensions were present. The protein of interest was then introduced in BB80 with 1 mg⋅mL−1 κ-casein and oxygen scavengers, and imaging began immediately as the microtubules depolymerized upon buffer exchange.
TIRF Microscopy Analysis.
Single-particle tracking and analysis were done using custom Labview (National Instruments) and Igor Pro (Wavemetrics) software as previously described (23, 39). Briefly, kymographs were generated from 2,000-frame movies, and individual events were traced recording tracking position, total binding time, and GFP fluorescence. Binding events lasting less than 0.2 seconds or exhibiting a fluorescence signal less than 20% over background noise were excluded from analysis. Bootstrapping analysis was used to determine mean residence time and the error of the mean. The mean squared displacement of each complex was plotted against time, and a one-dimensional diffusion constant was calculated using a weighted linear fit. Histograms of GFP fluorescence intensity were generated and fit with a Gaussian to determine average fluorescence of single particles.
Microtubule tip-tracking analysis was performed by overlaying kymographs of GFP and 647 channels and then measuring the distance of GFP-particle tracking using Photoshop (Adobe).
Optical Trapping Bead Motility Assay.
Streptavidin-coated 0.44-μm polystyrene beads were coated with biotinylated anti-His5 (Qiagen) and 11-pM beads were incubated with 40 nM His6-tagged Ndc80 or MIND complex as described (40–42), such that each bead was decorated with ∼1,800 protein complexes. Flow cells were prepared using double-sided tape and plasma-cleaned coverslips and incubated with 30 μL of 1 mg⋅mL−1 biotinylated BSA (Vector Laboratories) for 10 min at 50 °C, followed by BRB80 (80 mM PIPES, pH 6.9, 1 mM MgCl2, 1 mM EGTA) wash, 30 μL of 1 mg⋅mL−1 avidin DN (Vector Laboratories) for 2 min at room temperature, and another BRB80 wash. GMPCPP biotinylated tubulin seeds in BRB80 were bound for 2 min and washed with 37 °C growth buffer (BB80 plus 1 mM GTP). Protein-coated beads were introduced into the flow chambers in growth buffer with 1.4 mg⋅mL−1 tubulin, 200 μg⋅mL−1 glucose oxidase, 35 μg⋅mL−1 catalase, 25 mM glucose, 1 mM DTT. For MIND/Ndc80c assays, His6-tagged MIND and FLAG-tagged Ndc80 complex were preassembled into a co-complex and subjected to size exclusion gel filtration as described under Protein Expression and Purification. For assays with MIND, Ndc80, and Dam1 complexes, 20 nM His6-tagged MIND beads were prepared as described previously in this section, and then 40 nM free FLAG-tagged Ndc80 complex and 2 nM free Dam1 complex were added. For controls (i.e., MIND decorated beads with no protein in solution and GFP decorated beads and undecorated beads, both with Ndc80c in solution), all beads and soluble proteins were maintained at the concentrations used in the experimental reactions. For each condition, a bead was manipulated to make contact with a microtubule. If the bead–microtubule contact could not withstand any amount of force, then it was considered to be a nonbinder.
Optical trap assays were performed at 26 °C using custom instrumentation to capture and manipulate beads as described (40).
Constant force assays using His6-Ndc80c beads and MIND-His/Ndc80c-FLAG beads were performed as described (40). Then, 2.5 pN of force was applied in the direction of microtubule growth through rounds of assembly and disassembly. Bead position vs. time was analyzed using custom Igor Pro software, and attachment lifetime was calculated. Rupture force assays were performed as described (24, 25, 45). Once beads were bound to microtubule tips, a test force of 1 pN was applied, and only beads that tracked with ∼100 nm of tip growth were subjected to ramping force of 0.25 pN⋅s−1 until detachment. Records of force vs. time were collected, and maximum rupture force was determined using custom Igor Pro software.
Acknowledgments
We thank Andrew Franck, Andrew Powers, Krishna Sarangapani, and Austin Kim for technical assistance and advice. We also thank the members of the T.N.D. laboratory, C.L.A. laboratory, and Seattle Mitosis Club for helpful discussions. This work was supported by National Institute of General Medical Sciences Grants F32 GM099223 (to E.M.K), T32 GM008268 (to N.T.U.), T32 GM007270 (to E.A.S), P41 GM103533 (to M.J.M), R01 GM040506 (to T.N.D.), and R01 GM079373 (to C.L.A.), and National Center for Research Resources Grant S10 RR26406 (to C.L.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513882112/-/DCSupplemental.
References
- 1.Cheeseman IM. The kinetochore. Cold Spring Harb Perspect Biol. 2014;6(7):a015826. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 2013;194(4):817–846. doi: 10.1534/genetics.112.145276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8(6):581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston K, et al. Vertebrate kinetochore protein architecture: Protein copy number. J Cell Biol. 2010;189(6):937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127(5):983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 6.Obuse C, et al. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol. 2004;6(11):1135–1141. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- 7.Screpanti E, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21(5):391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19(8):694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravamudhan P, Felzer-Kim I, Gurunathan K, Joglekar AP. Assembling the protein architecture of the budding yeast kinetochore-microtubule attachment using FRET. Curr Biol. 2014;24(13):1437–1446. doi: 10.1016/j.cub.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tien JF, et al. Kinetochore biorientation in Saccharomyces cerevisiae requires a tightly folded conformation of the Ndc80 complex. Genetics. 2014;198(4):1483–1493. doi: 10.1534/genetics.114.167775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HW, et al. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383(4):894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maure JF, et al. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21(3):207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schleiffer A, et al. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol. 2012;14(6):604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- 14.Rago F, Gascoigne KE, Cheeseman IM. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol. 2015;25(5):671–677. doi: 10.1016/j.cub.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishino T, et al. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 2013;32(3):424–436. doi: 10.1038/emboj.2012.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bock LJ, et al. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat Cell Biol. 2012;14(6):614–624. doi: 10.1038/ncb2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornung P, et al. Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J Mol Biol. 2011;405(2):548–559. doi: 10.1016/j.jmb.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maskell DP, Hu XW, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol. 2010;190(5):823–834. doi: 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrovic A, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190(5):835–852. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei RR, et al. Structure of a central component of the yeast kinetochore: The Spc24p/Spc25p globular domain. Structure. 2006;14(6):1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Malvezzi F, et al. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 2013;32(3):409–423. doi: 10.1038/emboj.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers AF, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136(5):865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189(4):713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akiyoshi B, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468(7323):576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133(3):427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467(7317):805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol. 2013;200(1):21–30. doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14(1):54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 30.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15(23):3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108(3):317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 32.Verhey KJ, Hammond JW. Traffic control: Regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10(11):765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 33.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1(5):293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 34.Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1(5):288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 35.Donovan KW, Bretscher A. Head-to-tail regulation is critical for the in vivo function of myosin V. J Cell Biol. 2015;209(3):359–365. doi: 10.1083/jcb.201411010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106(2):195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 37.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102(15):5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoopmann MR, et al. Kojak: Efficient analysis of chemically cross-linked protein complexes. J Proteome Res. 2015;14(5):2190–2198. doi: 10.1021/pr501321h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gestaut DR, Cooper J, Asbury CL, Davis TN, Wordeman L. Reconstitution and functional analysis of kinetochore subcomplexes. Methods Cell Biol. 2010;95:641–656. doi: 10.1016/S0091-679X(10)95032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franck AD, Powers AF, Gestaut DR, Davis TN, Asbury CL. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. 2010;51(2):242–250. doi: 10.1016/j.ymeth.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umbreit NT, Davis TN. Mitosis puts sisters in a strained relationship: Force generation at the kinetochore. Exp Cell Res. 2012;318(12):1361–1366. doi: 10.1016/j.yexcr.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umbreit NT, et al. Kinetochores require oligomerization of Dam1 complex to maintain microtubule attachments against tension and promote biorientation. Nat Commun. 2014;5:4951. doi: 10.1038/ncomms5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 44.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402(6763):778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 45.Sarangapani KK, Akiyoshi B, Duggan NM, Biggins S, Asbury CL. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc Natl Acad Sci USA. 2013;110(18):7282–7287. doi: 10.1073/pnas.1220700110. [DOI] [PMC free article] [PubMed] [Google Scholar]