Significance
The innate immune system protects the host against infections with a diverse set of microbes that include intracellular bacterial and protozoan pathogens residing within pathogen-containing vacuoles (PVs). Because PVs provide an intracellular niche permissive for microbial growth, their destruction is critical for host defense. In mammals, PV destruction is dependent on immunity-related GTPases and guanylate binding proteins (GBPs). Although it has been shown that GBPs translocate to and eliminate PVs, the mechanisms by which GBPs specifically bind to PVs were unknown. Here, we describe an immune pathway that results in the decoration of PVs with a small protein called ubiquitin. Ubiquitin-decorated PVs are subsequently recognized by GBPs, resulting in the elimination of PVs and their microbial inhabitants.
Keywords: interferon, Toxoplasma, Chlamydia, GBPs, immunity-related GTPase
Abstract
Many microbes create and maintain pathogen-containing vacuoles (PVs) as an intracellular niche permissive for microbial growth and survival. The destruction of PVs by IFNγ-inducible guanylate binding protein (GBP) and immunity-related GTPase (IRG) host proteins is central to a successful immune response directed against numerous PV-resident pathogens. However, the mechanism by which IRGs and GBPs cooperatively detect and destroy PVs is unclear. We find that host cell priming with IFNγ prompts IRG-dependent association of Toxoplasma- and Chlamydia-containing vacuoles with ubiquitin through regulated translocation of the E3 ubiquitin ligase tumor necrosis factor (TNF) receptor associated factor 6 (TRAF6). This initial ubiquitin labeling elicits p62-mediated escort and deposition of GBPs to PVs, thereby conferring cell-autonomous immunity. Hypervirulent strains of Toxoplasma gondii evade this process via specific rhoptry protein kinases that inhibit IRG function, resulting in blockage of downstream PV ubiquitination and GBP delivery. Our results define a ubiquitin-centered mechanism by which host cells deliver GBPs to PVs and explain how hypervirulent parasites evade GBP-mediated immunity.
Pathogen-containing vacuoles (PVs) provide a safe haven to many intracellular bacterial and protozoan pathogens (1). Within the vacuolar enclosure of PVs, these pathogens can accumulate nutrients required for microbial growth. Moreover, life within the vacuolar niche shields microbes from cytoplasmic immune sensors that, once activated, can trigger proinflammatory and cell-autonomous immune responses (1). Accordingly, many intracellular pathogens such as the bacterium Chlamydia trachomatis and the protozoan Toxoplasma gondii have successfully adapted to a vacuolar lifestyle.
For the host to successfully combat infections with PV-resident microbes, the innate immune system must target PVs and its inhabitants for destruction. Critical mediators of host-directed attacks on PVs are two families of IFNγ-inducible GTPases: immunity-related GTPases (IRGs) and guanylate binding proteins (GBPs) (2). Members of both GTPase families play roles in host-mediated lysis of PVs, a process resulting in the release of microbes into the host cell cytoplasm, subsequent killing of PV-expelled microbes, and host cell death (3–8). Additionally, GBPs help deliver cytosolic subunits of the antimicrobial NADPH oxidase NOX2 for assembly on phagosomal membranes, orchestrate the capture of PV-resident microbes inside degradative autophagolysosomes, and promote the activation of canonical and noncanonical inflammasome pathways (5, 8–12). As a critical first step underlying most if not all of these known GBP-controlled cell-autonomous immune responses, GBPs must locate to their intracellular microbial targets.
GBPs belong to the dynamin superfamily of large GTPases (13). Similar to other members of the dynamin superfamiliy, GBPs can assemble as oligomers in a nucleotide-dependent fashion (13). Binding of GTP results in dimer formation; subsequent GTP hydrolysis prompts conformational changes that enable GBPs to assemble as tetramers (14, 15). Mutations in the G domain that reduce nucleotide binding affinities and hydrolytic activity block GBP oligomerization, constrain the localization of GBPs to the cytoplasm, and prevent GBPs from binding to PV membranes (9, 15–18). These observations support a model in which GBP monomers are diffusely distributed in the cytoplasm and GBP oligomers associate with membranes. However, these observations fail to account for the specificity with which oligomeric GBPs agglomerate on PV membranes.
PVs formed by C. trachomatis and T. gondii recruit not only GBPs but also members of the IRG family of IFNγ-inducible GTPase (4, 19). The IRG protein family can be divided into two subgroups: IRGM and GKS proteins (20). Whereas GKS proteins feature the canonical glycine–lysine–serine (GKS) P-loop sequence, IRGM proteins have a substitution of a lysine for a methionine in their P-loop sequence (20). IRGM and GKS proteins also differ in their subcellular localization: IRGM proteins associate with endomembranes, whereas monomeric GDP-bound GKS proteins predominantly reside within the host cell cytoplasm (4, 17, 21, 22). Once GKS proteins transition into a GTP-bound active state, they can bind to PV membranes (21). IRGM proteins inhibit this activation step and thereby guard IRGM-decorated membranes against GKS protein targeting (17, 21). Because PV membranes surrounding either C. trachomatis or T. gondii are largely devoid of IRGM proteins, they are the preferred GKS binding substrate following a “missing-self” principle of immune targeting (17, 23). In IRGM-deficient cells, however, GKS proteins enter the active state prematurely, form protein aggregates, mislocalize, and thus fail to bind to PVs (17, 21). Although these previous observations help explain how IRGM proteins promote the delivery of GKS proteins to PVs, IRGM proteins also control the subcellular localization of GBPs through an uncharacterized mechanism (6, 17, 24–26).
Here, we report a previously unidentified host-directed ubiquitination pathway involved in innate immunity. We demonstrate that Chlamydia- and Toxoplasma-containing vacuoles become ubiquitin-decorated upon IFNγ priming of their host cells. IFNγ-dependent association of ubiquitin with PVs requires IFNγ-inducible IRG proteins and the E3 ligase tumor necrosis factor (TNF) receptor associated factor 6 (TRAF6). Experimental removal of the IFNγ-inducible ubiquitination pathway dramatically diminishes the p62-dependent delivery of GBPs to PVs and thereby renders host cells more susceptible to infections. Thus, our observations imply that ubiquitin serves as a host-induced pattern that marks intracellular structures as immune targets for members of the GBP family of host defense proteins.
Results
IFNγ Priming Triggers Ubiquitin Decoration of PVs and p62 Recruitment.
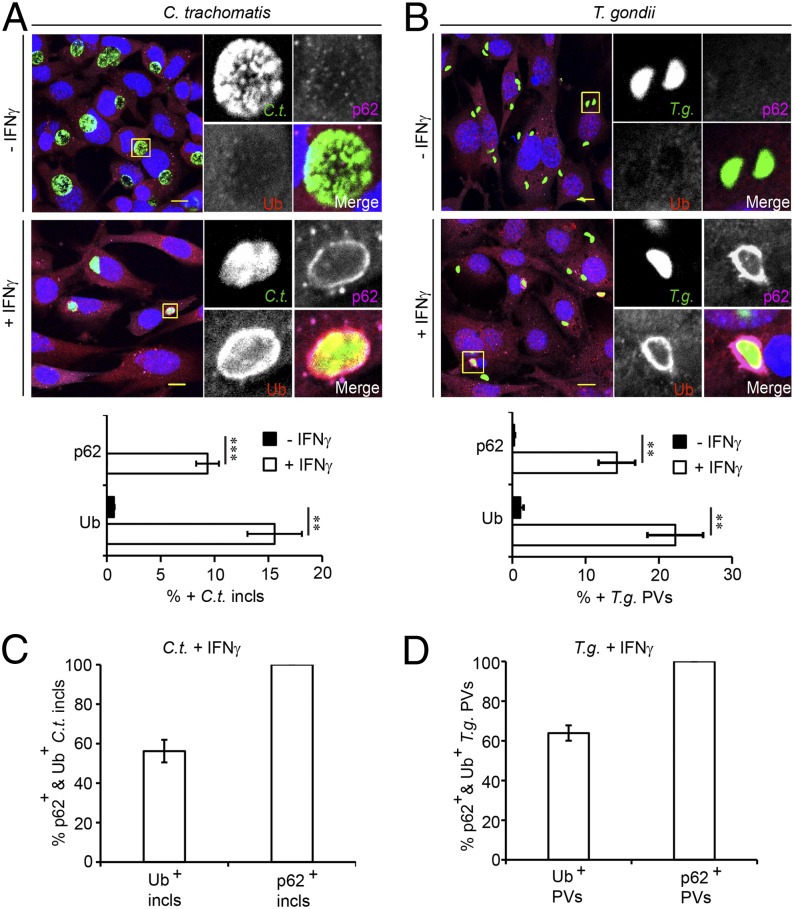
IFNγ priming induces GBP-mediated cell-autonomous immunity that results in the degradation of C. trachomatis inside autophagolysosomes (24, 27). Because antimicrobial autophagy (also called xenophagy) often depends on host-mediated ubiquitination of invading microbes (28), we asked whether IFNγ priming would lead to the accumulation of ubiquitin on C. trachomatis PVs (referred to henceforth as “inclusions” in agreement with accepted nomenclature). We observed only infrequent (<1%) association of ubiquitin with inclusions in unprimed mouse embryonic fibroblasts (MEFs) but a 10-fold increase in ubiquitin-positive inclusions in IFNγ-primed MEFs (Fig. 1A). Similarly, PVs containing the type II T. gondii strain Prugniaud A7 (Pru) became ubiquitin-labeled upon IFNγ priming in both MEFs (> 20% Ub-positive PVs) and primary bone marrow-derived macrophages (BMMs) (Fig. 1B and Fig. S1). These data suggest that IFNγ-induced PV ubiquitination is a general host response active against both bacterial and protozoan pathogens in varying cell types.
Fig. 1.
IFNγ priming triggers PV ubiquitination and p62 recruitment. (A) WT MEFs were infected with GFP+ C. trachomatis L2 and treated with IFNγ (100 U/mL) at 3 hpi or left untreated. At 20 hpi, cells were stained for ubiquitin (red), p62 (magenta), and DNA (blue). Colocalization of Ubiquitin (Ub) and p62 with inclusions (incls) was quantified as described in Experimental Procedures. (B) WT MEFs were left unprimed or primed overnight with IFNγ (200 U/mL) and infected with GFP+ T. gondii Pru. Cells were stained for ubiquitin, p62, and DNA at 1 hpi and assessed for colocalization with T. gondii PVs. (C and D) Randomly selected ubiquitin- or p62-positive PVs were assessed for costaining with the second marker p62 or ubiquitin, respectively. At least 100 PVs were counted for each condition. **P < 0.005 and ***P < 0.0005 (two-tailed unpaired t test). Data are representative of three independent experiments (mean ± SD; n = 3). (Scale bar, 10 μm.)
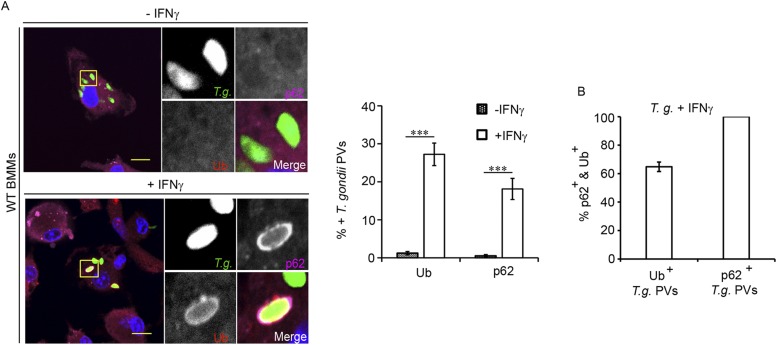
Fig. S1.
p62 colocalizes with ubiquitin-decorated T. gondii PVs in IFNγ-primed BMMs. (A) Unprimed and IFNγ-primed C57BL/6J-derived BMMs were infected with GFP+ T. gondii Pru. Colocalization of ubiquitin (red) and p62 (magenta) with T. gondii PVs was monitored at 1 hpi. ***P < 0.0005 (two-tailed unpaired t test). (Scale bar, 10 µm.) (B) Randomly selected ubiquitin- or p62-positive PVs were assessed for costaining with the second marker p62 or ubiquitin, respectively. At least 100 PVs were counted for each condition. Data are representative of three independent experiments (mean ± SD; n = 3).
The ubiquitin-binding protein p62/SQSMT-1 (sequestosome-1, also known as p62) promotes the xenophagic elimination of intracellular bacterial pathogens (29–32). We therefore tested the hypothesis that IFNγ priming would promote not only PV ubiquitination but also p62 recruitment. In support of our hypothesis, we found that p62 associated with inclusions as well as T. gondii PVs in IFNγ-primed but not in unprimed MEFs (Fig. 1 A and B). We also observed a dramatic increase in p62-decorated T. gondii PVs in BMMs (Fig. S1), thereby demonstrating that IFNγ-induced targeting of p62 to PVs is not cell type-dependent. Regardless of the cell type and pathogen used, we always recorded a lower percentage of p62-positive vacuoles than ubiquitin-positive vacuoles. Moreover, we found that all monitored p62-positive PVs stained positive for ubiquitin, whereas only ∼60% of ubiquitin-positive PVs were decorated with p62 (Fig. 1 C and D and Fig. S1). These observations suggest that PV ubiquitination is a prerequisite for p62 binding to PVs.
p62 Associates with PVs Through Its Ubiquitin-Binding Domain.
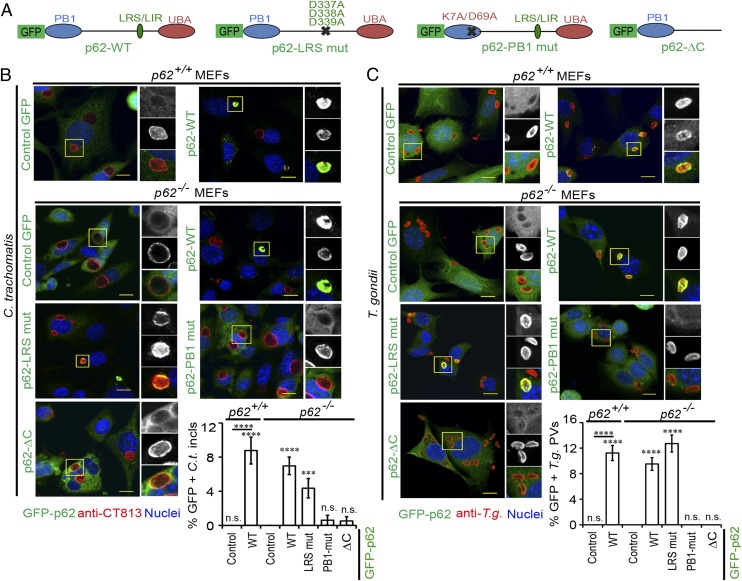
The p62 protein contains a ubiquitin-associated domain (UBA), which mediates ubiquitin binding (33). To determine whether p62 recruitment to PVs is mediated by ubiquitin binding, we asked whether UBA and/or other domains such as the LC3-recognition sequence (LRS) of p62 were required for PV targeting. To answer these questions, we ectopically expressed wild-type (WT) and mutant p62 variants fused to GFP in WT (p62+/+) and p62−/− cells and monitored their subcellular localization (Fig. 2 A–C). Previously characterized mutations in LRS (p62-LRS mut) known to disrupt LC3 binding (34) had no discernible effect on the association of p62 with inclusions or T. gondii PVs (Fig. 2 B and C). However, a deletion of the C terminus containing the UBA domain (p62-ΔC) eliminated sustained association of p62 with PVs (Fig. 2 B and C). In addition to the p62-ΔC mutant, the p62-PB1 mutant defective for protein oligomerization also failed to bind to PVs (Fig. 2 B and C). These results suggest that ubiquitin binding as well as PB1-mediated protein oligomerization are both critical for the stable association of p62 with PVs.
Fig. 2.
Functional UBA and PB1 domains are required for p62 localization to PVs. (A) Schematic representation of GFP–p62 fusion proteins: WT, p62 LRS mutant (D337A, D338A, and D339A), p62 PB1 mutant (K7A and D69A), and p62ΔC (1–265 amino acids). (B and C) WT p62+/+ and p62−/− MEFs were engineered to express the indicated p62 variants and activated with IFNγ. (B) Cells infected with C. trachomatis L2 were stained with Hoechst (blue) and antibody directed against the Chlamydial inclusion protein CT813 (red). GFP colocalizations with C. trachomatis are shown [***P < 0.0005 and ****P < 0.0001 (one-way ANOVA) relative to p62−/− MEFs expressing control GFP alone]. (C) Localization of GFP to T. gondii Me49 PVs was monitored at 1 hpi [****P < 0.0001 (one-way ANOVA) relative to p62−/− MEFs expressing GFP control alone]. Data are representative of three independent experiments (mean ± SD; n = 3). n.s., not significant. (Scale bar, 10 μm.)
The Delivery of p62 to PVs Is Independent of GBPs.
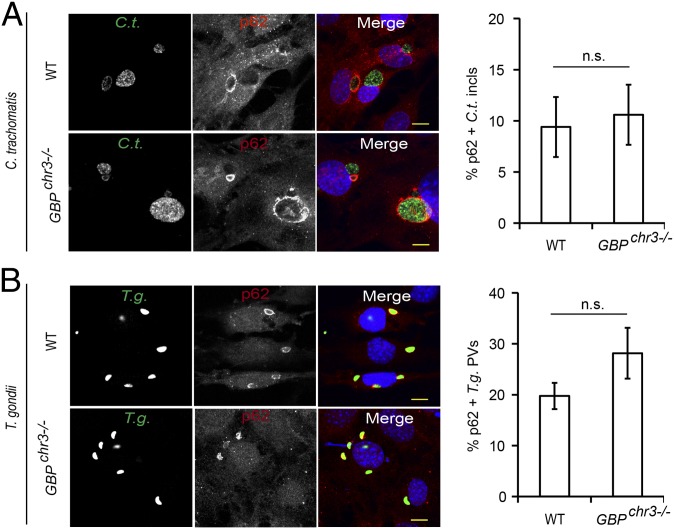
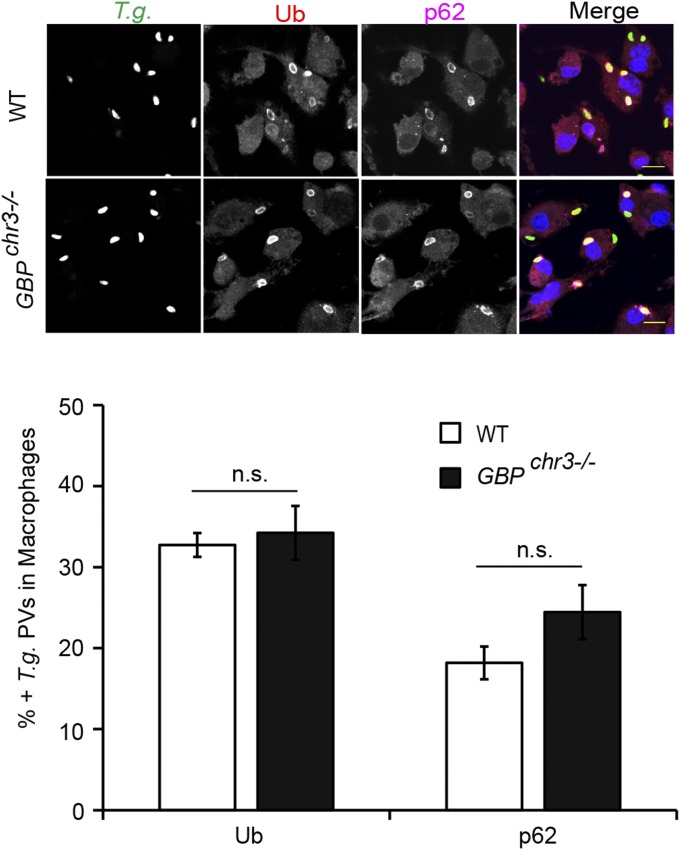
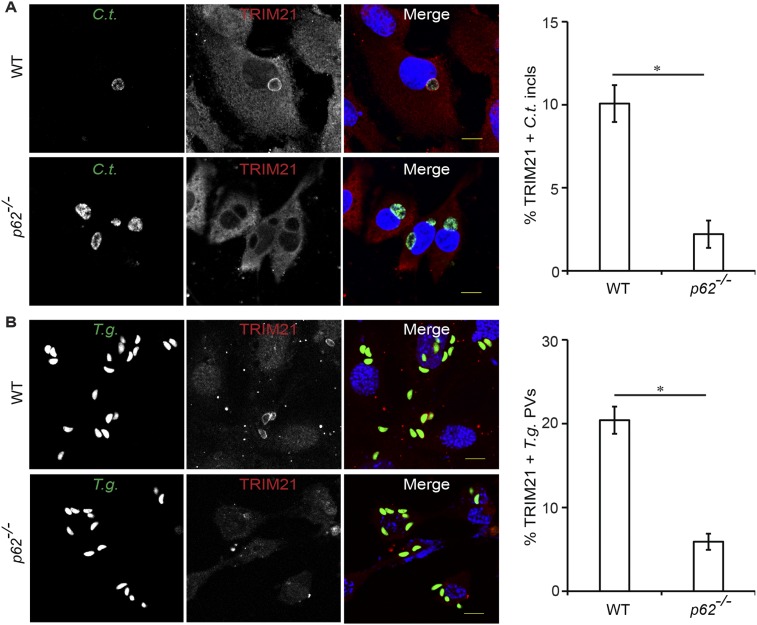
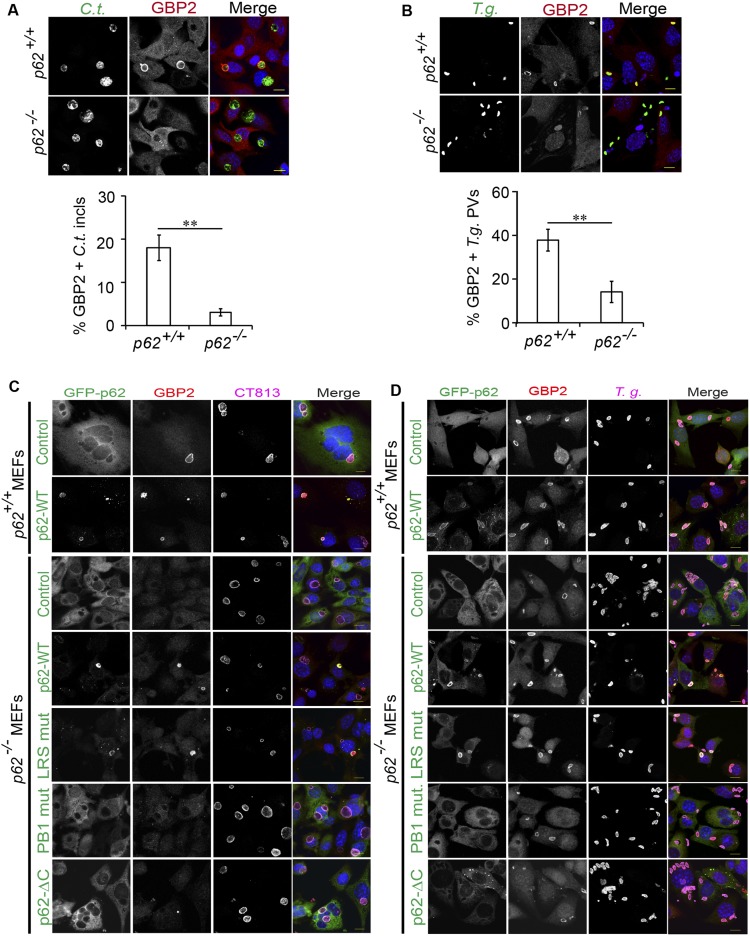
Because a previous report had identified p62 as a protein interaction partner of GBP1 (9), we considered the hypothesis that GBP1 could have an auxiliary function in the recruitment of p62 to PVs. To test our hypothesis, we asked whether the recruitment of p62 to PVs required the expression of GBP1 or other members of the GBP protein family. We found that the association of p62 with either inclusions or T. gondii PVs was unchanged in MEFs deficient in GBP1 and four additional GBP genes encoded on chromosome 3 (Fig. 3 A and B). Similarly, the absence of the GBPchr3 gene cluster also failed to alter the rate of p62 recruitment to T. gondii PVs in BMMs (Fig. S2), demonstrating that p62 binds to PVs independently of chromosome 3-encoded GBPs.
Fig. 3.
Binding of p62 to PVs is independent of GBPs. (A and B) IFNγ-activated WT and coisogenic GBPchr3−/− MEFs were infected with (A) GFP+ C. trachomatis or (B) GFP+ T. gondii Pru and stained with anti-p62 (red) and Hoechst (blue). Localization of endogenous p62 to inclusions at 20 hpi or to T. gondii PVs at 1 hpi was quantified. Data are representative of three independent experiments (mean ± SD; n = 3; n.s., not significant). (Scale bar, 10 μm.)
Fig. S2.
Ubiquitination and p62 recruitment to T. gondii PVs in macrophages are independent of GBPs. Macrophages derived from WT and GBPchr3−/− mice were primed overnight with IFNγ (200 U/mL) and infected with GFP+ T. gondii (Pru). Ubiquitination and localization of endogenous p62 to T. gondii PVs were monitored at 1 hpi. Representative confocal images and analyzed data from three independent experiments are shown (mean ± SD; n = 3).
p62-Dependent and p62-Independent Pathways Mediate Ubiquitin Deposition on PVs.
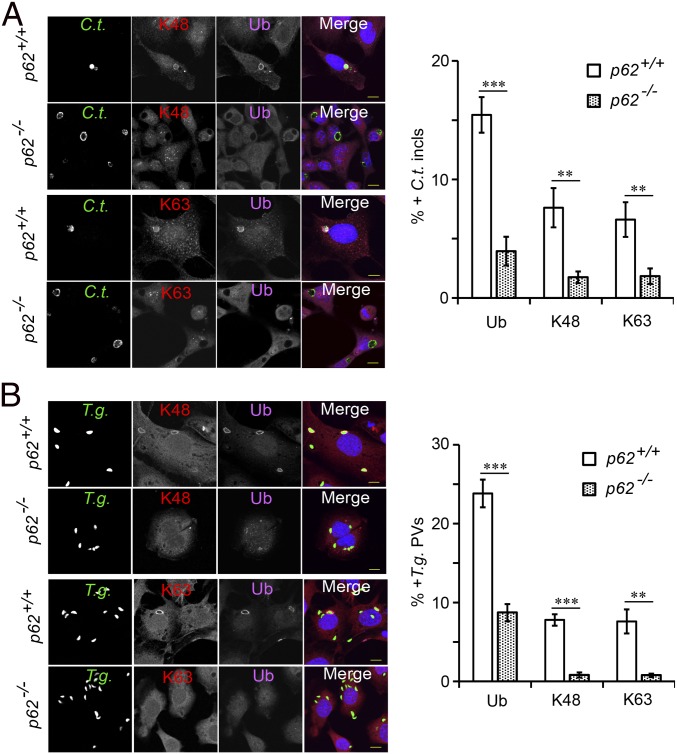
We next set out to define the function of PV-associated p62. Previous work had defined p62 as a multifunctional adaptor protein that can form distinct signal-organizing nodes modulating the cellular response to stimuli such as cytokines or amino acids (35–37). Central to its role as a signal rheostat is the ability of p62 to solicit ubiquitin E3 ligases to p62-orchestrated signaling hubs (38). We thus asked whether p62 itself could regulate levels of IFNγ-induced ubiquitin-positive PVs and thereby promote GBP recruitment to PVs. In support of this model, we observed an approximately threefold reduction in the percentage of ubiquitin-positive inclusions in p62−/− MEFs (Fig. 4A). Likewise, the association of ubiquitin with T. gondii PVs was diminished in p62−/− MEFs (Fig. 4B), collectively demonstrating that extensive ubiquitin association with PVs requires p62 expression. However, a proportion of PVs remained ubiquitin-associated in p62−/− cells, demonstrating the existence of both p62-dependent and p62-independent PV ubiquitin-labeling pathways. To further characterize the p62-dependent pathway, we stained IFNγ-primed cells using K63 linkage- and K48 linkage-specific antiubiquitin antibodies. We observed the accumulation of K48-linked and K63-linked poly-ubiquitin signals on PVs formed in p62+/+ cells. These signals were decreased by 4–8-fold in p62−/− cells (Fig. 4 A and B). These results show that p62 is largely responsible for the sustained decoration of inclusions and T. gondii PVs with K48-linked and K63-linked poly-ubiquitin chains.
Fig. 4.
p62 promotes PV polyubiquitination. (A) WT p62+/+ and p62−/− MEFs were infected with GFP+ C. trachomatis and treated with IFNγ at 3 hpi. Cells were stained with either anti–Ub-K48 (red) or anti–Ub-K63 (red), anti-ubiquitin (magenta), and Hoechst (blue). Colocalization of total ubiquitin and K48- and K63-linked ubiquitin with inclusions was monitored at 20 hpi. (B) WT p62+/+ and p62−/− MEFs were primed overnight with IFNγ, infected with GFP+ T. gondii Pru, and evaluated for colocalization at 1 hpi. **P < 0.005, ***P < 0.0005 (two-tailed unpaired t test). Data are representative of three independent experiments (mean ± SD; n = 3). (Scale bar, 10 μm.)
The p62-Interacting E3 Ubiquitin Ligase TRAF6 Promotes PV Ubiquitination.
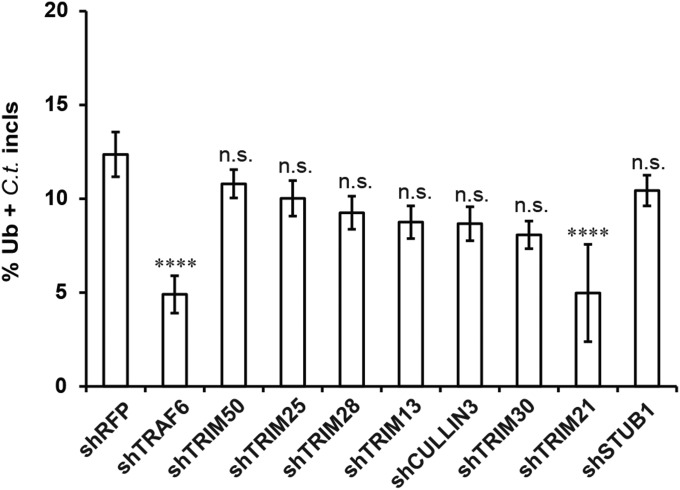
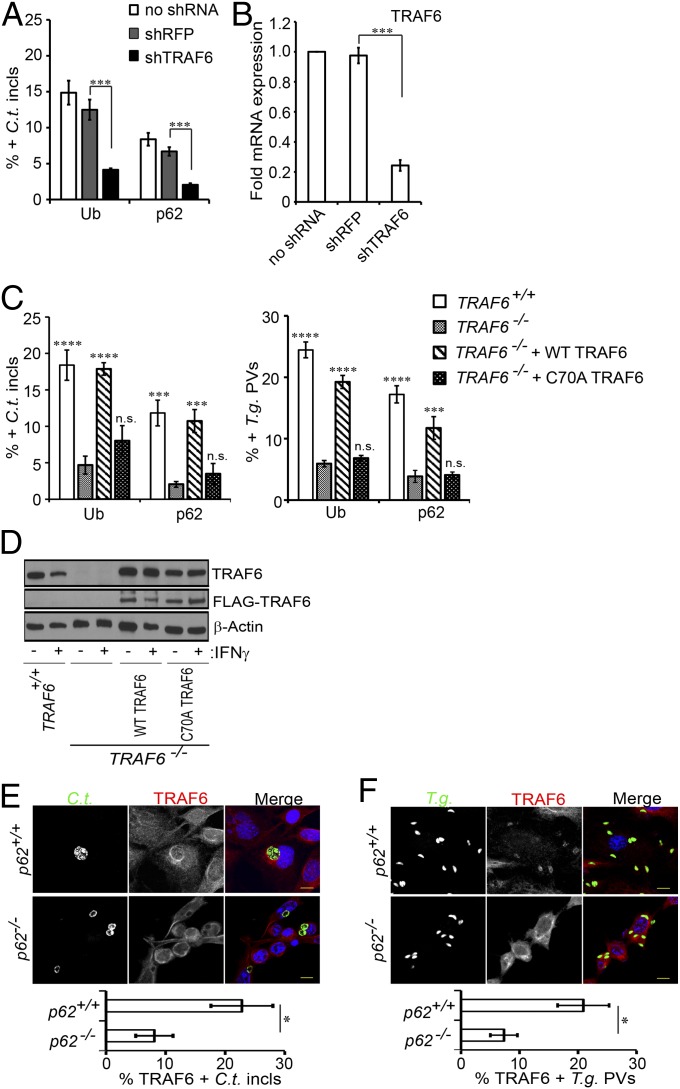
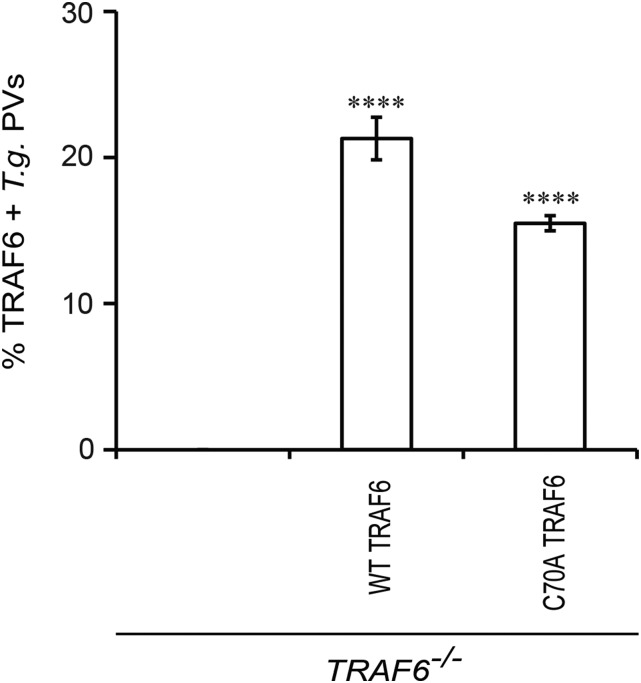
Because our data indicated that p62 facilitated PV ubiquitination, we hypothesized that p62 recruited E3 ubiquitin ligases to PVs, resulting in the deposition of ubiquitin at these vacuoles. To test our hypothesis, we first screened a set of known or predicted p62-associated E3 ligases for their role in IFNγ-dependent ubiquitin labeling of inclusions. We observed a reduction of 50% or more in ubiquitin-decorated inclusions when cells were treated with shRNAs against either TRIM21 or TRAF6 (Fig. S3 and Fig. 5 A and B). Taking advantage of established tools, we focused our follow-up studies on TRAF6. Using gene-deleted TRAF6−/− MEFs, we independently confirmed that TRAF6 promoted the association of ubiquitin with PVs (Fig. 5C). Complementation of TRAF6−/− MEFs with ectopically expressed WT TRAF6 protein but not a catalytically inactive (C70A) mutant (Fig. 5D) restored ubiquitin staining of PVs (Fig. 5C). Together, these data identify TRAF6 as an E3 ligase involved in the ubiquitination of inclusions and T. gondii PVs.
Fig. S3.
Screening of E3-ubiquitin ligases. WT MEFs were transduced with lentiviral vectors encoding shRNAs against indicated genes or an ORF encoding red fluorescent protein (as negative control). Cells were infected with C. trachomatis and treated with IFNγ. Localization of ubiquitin (Ub) to inclusions was monitored at 20 phi. ****P < 0.0001; n.s., not significant (one-way ANOVA). Data are representative of three independent experiments (mean ± SD; n = 3).
Fig. 5.
The E3 ligase TRAF6 binds to PVs and promotes PV ubiquitination. (A) MEFs were transduced with lentiviral vectors encoding shRNAs against TRAF6 or red fluorescent protein (as negative control), infected with C. trachomatis, and treated with IFNγ. Localization of ubiquitin (Ub) and p62 to inclusions was monitored at 20 phi. (B) TRAF6 mRNA expression was monitored by quantitative PCR in transduced and control cells. (C) PVs formed in IFNγ-primed WT TRAF6+/+, coisogenic TRAF6−/−, and TRAF6−/− MEFs complemented with WT FLAG-TRAF6 or FLAG-TRAF6 (C70A) mutant were scored for ubiquitin and p62 colocalization. ***P < 0.0005; ****P < 0.0001; n.s., not significant (one-way ANOVA) relative to TRAF6−/− MEFs. (D) Protein expression of TRAF6, FLAG-tagged TRAF6, and β-actin in cell lysates prepared from unprimed and overnight IFNγ-primed cells was detected by Western blotting. (E and F) IFNγ-activated WT p62+/+ and p62−/− MEFs were infected either with GFP+ C. trachomatis (E) or with GFP+ T. gondii Pru (F), and localization of endogenous TRAF6 to PVs was quantified. *P < 0.05 (two-tailed unpaired t test) relative to WT MEFs. (Scale bar, 10 μm.)
TRAF6 and p62 Recruitment to PVs Is Codependent.
Once we had identified TRAF6 as an E3 ligase partially responsible for decorating PVs with ubiquitin, we set out to test the hypothesis that p62 facilitates ubiquitination through the recruitment of TRAF6 to PVs. To test our hypothesis, we monitored the subcellular localization of TRAF6 in WT (p62+/+) and p62−/− MEFs. We found that TRAF6 localized to both inclusions and T. gondii PVs in IFNγ-primed cells (Fig. 5 E and F), further highlighting its role in innate immune responses directed against intracellular PVs. Autoubiquitination was not essential for TRAF6 translocation to PVs, as catalytically dead C70A mutant TRAF6 localized to T. gondii PVs (Fig. S4). Recruitment of not only TRAF6 but also Trim21 to PVs was reduced in p62−/− MEFs (Fig. 5 E and F and Fig. S5), supporting our initial hypothesis that p62 delivers E3 ligases to PVs. Because TRAF6 and Trim21 recruitment to PVs was reduced but not abolished in p62−/− cells, we conclude that both p62-dependent and p62-independent pathways execute the deployment of E3 ligases to PVs.
Fig. S4.
Catalytically inactive TRAF6 (C70A) colocalizes with PVs. Parental TRAF6−/− MEFs and TRAF6−/− MEFs ectopically expressing either WT TRAF6 or the enzymatically inactive C70A mutant were treated overnight with IFNγ (200 U/mL) and infected with the type II strain of T. gondii. At 1 hpi, cells were stained with anti-TRAF6 and assessed for TRAF6 localization to T. gondii PVs. ****P < 0.0001 (one-way ANOVA). Data are representative of three independent experiments (mean ± SD; n = 3).
Fig. S5.
p62 promotes TRIM21 recruitment to PVs. (A) WT p62+/+ and p62−/− MEFs were infected with GFP+ C. trachomatis and treated with IFNγ (100 U/mL) at 3 hpi. Cells were fixed at 20 hpi and stained with anti-TRIM21 (red) and Hoechst (blue). Localization of endogenous TRIM21 to inclusions was quantified. (B) IFNγ-primed (200 U/mL, overnight) p62+/+ and p62−/− MEFs were infected with GFP+ T. gondii Pru. Localization of endogenous TRIM21 to T. gondii PVs was monitored at 1 hpi. *P < 0.05 (two-tailed unpaired t test). Data are representative of two independent experiments (mean ± SD; n = 3). (Scale bar, 10 μm.)
TRAF6 catalyzes the formation of K63-linked polyubiquitin chains (35). Because the UBA domain of p62 preferentially binds to K63-linked polyubiquitin chains (39), TRAF6 should promote the tethering of p62 to PVs. In support of this model, we observed a reduction in the number of p62-postive PVs in TRAF6-deficient cells (Fig. 5 A and C), which was reversed in the presence of an ectopically expressed WT form of TRAF6 but not in the presence of a catalytically inactive (C70A) mutant (Fig. 5C). Together, these data indicated that p62 and TRAF6 docking to PVs was codependent. We conclude that codependent recruitment of p62 and TRAF6 establishes a positive feedback loop to assure the rapid accumulation of ubiquitin at PVs in IFNγ-primed cells.
TRAF6 and p62 Promote Cell-Autonomous Immunity and GBP Recruitment to PVs.
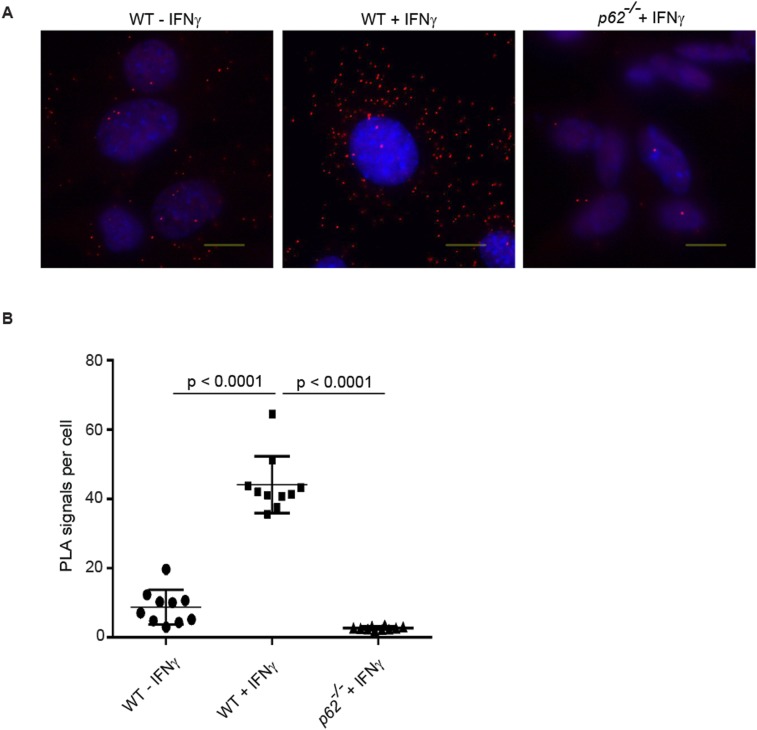
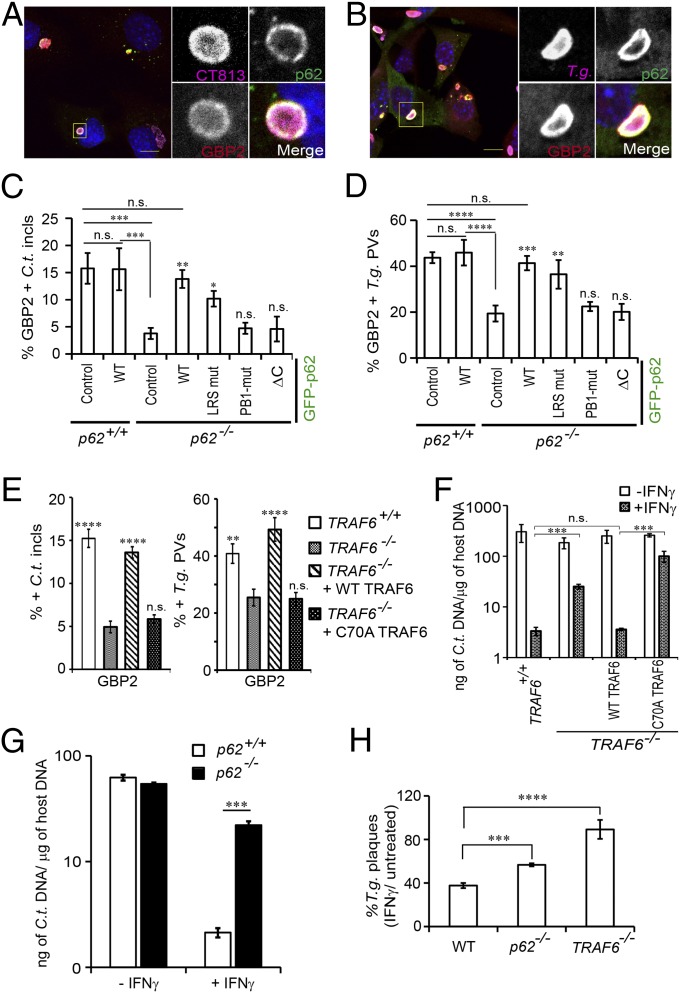
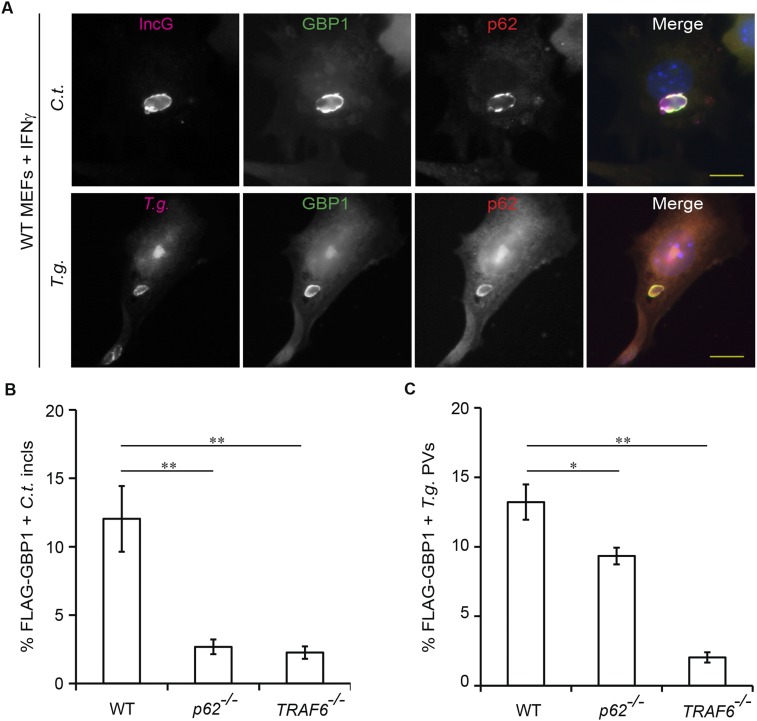
GBP1 and p62 coimmunoprecipitate in IFNγ-primed uninfected host cells (9). Using in situ proximity ligation assays (PLAs), we found that endogenous GBP2 resided in close proximity to p62 throughout the cytoplasm of uninfected cells (Fig. S6), suggesting that GBPs preform complexes with p62 before their translocation to PVs. We next asked whether this association was maintained as p62 translocates to PVs. In support of this model, we observed that p62 colocalized with GBP1 and GBP2 both on inclusion membranes and on T. gondii PVs (Fig. 6 A and B and Fig. S7A). These observations led us to hypothesize that p62 escorts GBP proteins to PVs. In support of our hypothesis, we found that FLAG-tagged GBP1 and endogenous GBP2 associated with inclusions less frequently in p62−/− MEFs relative to WT MEFs (Fig. 6C and Figs. S7B and S8A). In the absence of p62, the percentage of GBP1- and GBP2-positive T. gondii PVs was also significantly reduced (Fig. 6D and Figs. S7C and S8B), indicating a general role for p62 in directing GBPs to parasitophorous membranes. Recruitment of GBP2 was rescued in p62−/− MEFs when we ectopically expressed WT p62 or the PV-associated p62 LIR mutant but not when we expressed the p62-PB1 or p62-ΔC mutants defective for PV translocation (Fig. 6 C and D and Fig. S8 C and D). These data suggest that p62 escorts GBPs to PVs.
Fig. S6.
In situ PLA assessing colocalization of GBP2 with p62. (A) Representative images of PLA staining for GBP2–p62 complexes. (B) WT and p62−/− MEFs (as negative control) were treated overnight with 100 U/mL of IFNγ or left untreated. Cells were fixed and stained with rabbit anti-GBP2 antibody and mouse anti-p62 antibody and processed for PLA. Cell nuclei were counterstained with Hoechst (blue). Each red dot indicates localization of GBP2 and p62 in close proximity to another (<40 nm). A significant increase in PLA signal was observed in WT MEFs treated with IFNγ. ****P < 0.0001, one-way ANOVA.
Fig. 6.
P62 and TRAF6 elicit GBP targeting to PVs and cell-autonomous host defense. (A and B) GFP-p62 and GBP2 colocalize with (A) CT813-positive inclusions at 20 hpi and with (B) T. gondii (T.g.) PVs at 1 hpi in IFNγ-activated cells. (C and D) WT p62+/+ and coisogenic p62−/− MEFs engineered to express the indicated p62 variants fused to GFP were infected with either (C) C. trachomatis or (D) T. gondii strain Me49. The percentages of GBP2+ PVs for each experimental condition were quantified. *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001; n.s., not significant (one-way ANOVA) relative to p62−/− MEFs expressing control GFP. (E) PVs formed in IFNγ-primed WT TRAF6+/+, coisogenic TRAF6−/−, and TRAF6−/− MEFs complemented with WT FLAG-TRAF6 or FLAG-TRAF6 (C70A) mutant were scored for GBP2 colocalization. **P < 0.005; ****P < 0.0001; n.s., not significant (one-way ANOVA) relative to TRAF6−/− MEFs. (F and G). MEFs of the indicated genotypes were primed overnight with IFNγ or left unprimed and then infected with C. trachomatis at an MOI of 2. Bacterial burden was assessed by quantitative PCR at 24 hpi. ***P < 0.0005; n.s., not significant (two-tailed unpaired t test). (H) MEFs of the indicated genotypes were primed overnight with IFNγ or left unprimed and then infected with GFP+ T. gondii Pru. At 72 hpi, the number of GFP+ T. gondii plaques was quantified by microscopy. Data show the number of plaques in IFNγ-primed MEFs relative to the number of plaques formed in unprimed MEFs of the same genotype. ***P < 0.0005, ****P < 0.0001 (one-way ANOVA); mean ± SD; n = 3. (Scale bar, 10 μm.)
Fig. S7.
p62 and TRAF6 promote GBP1 recruitment to PVs. (A) Representative images of IFNγ-treated WT MEFs expressing GFP-GBP1 and infected with either C. trachomatis or T. gondii Me49. GFP-GBP1 and p62 colocalize with IncG-positive inclusions at 20 hpi and with T. gondii (T.g.) PVs at 1 hpi in IFNγ-treated cells. (B) WT, p62−/−, and TRAF6−/− MEFs expressing FLAG-GBP1 were infected with GFP+ C. trachomatis and treated with IFNγ (100 U/mL) at 3 hpi. Cells were fixed at 20 hpi, stained with anti-FLAG (red)/Hoechst (blue), and the number of FLAG-positive inclusions was quantified. (C) IFNγ-primed (200 U/mL, overnight) WT, p62−/−, and TRAF6−/− MEFs expressing FLAG-GBP1 were infected with GFP+ T. gondii Pru. Number of FLAG-positive T. gondii PVs was assessed at 1 hpi. *P < 0.05; **P < 0.005 (one-way ANOVA). Graphs represent the mean ± SD derived from three independent experiments. (Scale bar, 10 μm.)
Fig. S8.
p62 promotes GBP2 recruitment to PVs. (A) WT p62+/+ and p62−/− MEFs were infected with GFP+ C. trachomatis, left untreated, or treated with IFNγ (100 U/mL) at 3 hpi. Cells were fixed at 20 hpi, stained with anti-GBP2 (red)/Hoechst (blue), and localization of endogenous GBP2 to inclusions was quantified. (B) IFNγ-primed (200 U/mL, overnight) and unprimed p62+/+ and p62−/− MEFs were infected with GFP+ T. gondii Pru. Localization of endogenous GBP2 to T. gondii PVs was monitored at 1 hpi. (C and D) Representative images of IFNγ-treated p62+/+ and p62−/− MEFs expressing the indicated p62 variants and infected with either C. trachomatis (C) or T. gondii Me49 (D). (Scale bar, 10 μm.) Frequency of GBP2 colocalizations with PVs was quantified, and data are shown in Fig. 6 C and D. **P < 0.005 (two-tailed unpaired t test). Data are representative of three independent experiments (mean ± SD; n = 3). (Scale bar, 10 μm.)
Because TRAF6 promotes the deposition of p62 onto PVs, we expected that TRAF6 would also be required for the effective loading PVs with GBPs. As predicted, we found that GBP1 and GBP2 associated with PVs less frequently in TRAF6−/− MEFs than they did in WT (TRAF6+/+) cells (Fig. 6E and Fig. S7 B and C). Complementation of TRAF6−/− MEFs with ectopically expressed WT TRAF6 protein but not a catalytically inactive (C70A) mutant restored GBP2 recruitment to PVs (Fig. 6E).
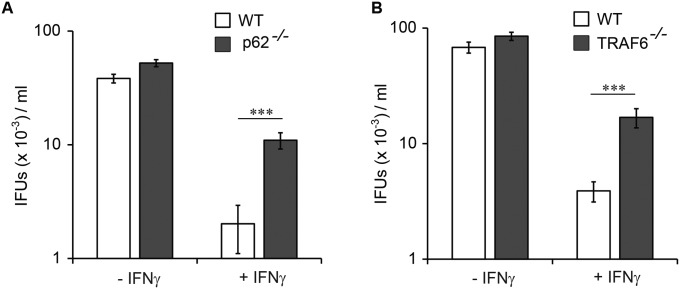
Because GBPs mediate IFNγ-induced cell-autonomous resistance to C. trachomatis infections (24, 27, 40), we next asked whether the diminished targeting of GBPs to PVs in p62−/− and TRAF6−/− cells would debilitate the cell-autonomous defense system of the host cell. We found that IFNγ-primed cells lacking either p62 or enzymatically active TRAF6 elicited a less effective cell-autonomous immune response to C. trachomatis (Fig. 6 F and G and Fig. S9) and to T. gondii (Fig. 6H) than WT cells. Taken together, these data demonstrate that TRAF6-mediated ubiquitin labeling of PVs is critical for the delivery of GBP1 and GBP2 to PVs and the execution of cell-autonomous immunity.
Fig. S9.
P62 and TRAF6 promote cell-autonomous host defense against C. trachomatis. (A) p62−/− MEFs and (B) TRAF6−/− MEFs were primed overnight with IFNγ or left unprimed and then infected with C. trachomatis at an MOI of 2. Infectious titers were quantified after 40 h of infection (as described in Experimental Procedures) and presented as IFUs per mL of inoculum. ***P < 0.005 (two-tailed unpaired t test). Graphs represent the mean ± SD derived from at least two independent experiments, assayed in triplicate.
TRAF6 and p62 Promote the Disruption of C. trachomatis Inclusions in IFNγ-Primed Cells.
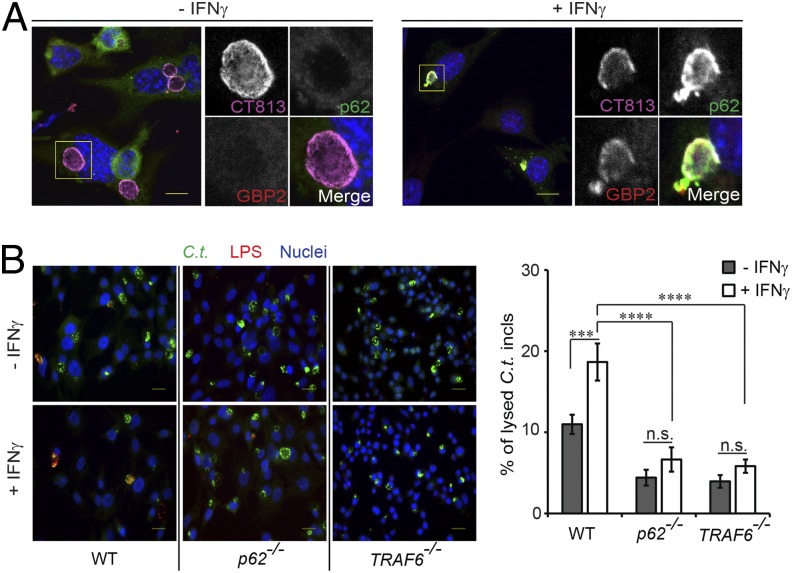
GBPs promote the destruction of T. gondii PVs and Salmonella-containing vacuoles in IFNγ-primed cells (5, 6). To assess whether IFNγ priming also results in the destruction of inclusions, we stained cells for the Chlamydial inclusion membrane protein CT813 (41). Whereas inclusions formed in unprimed cells generally displayed a continuous staining pattern of vacuolar CT813, inclusions formed in IFNγ-primed cells frequently appeared to be damaged based on the disrupted vacuolar staining of CT813 (Fig. 7A). To better quantitate our observations, we adapted an inclusion integrity assay based on differential permeabilization with digitonin. We found that treatment of C. trachomatis-infected cells with IFNγ resulted in a measurable increase in the number of lysed inclusions at 20 h postinfection (hpi) (Fig. 7B). The IFNγ-dependent increase in the number of damaged inclusions was dependent on the GBP recruitment factors p62 and TRAF6 (Fig. 7B). We further observed that already in the absence IFNγ treatment, the percentage of lysed inclusions was elevated in WT MEFs relative to p62−/− and TRAF6−/− MEFs at 20 hpi (Fig. 7B), possibly due to autocrine type I IFN signaling known to induce GKS expression (20). Together, these data indicate that p62 and TRAF6 promote the destruction of PVs in IFN-activated cells and suggest that the association of p62 with ruptured inclusions (Fig. 7A) precedes the process of inclusion lysis.
Fig. 7.
TRAF6 and p62 promote the disruption of C. trachomatis inclusions in IFNγ-primed cells. (A) Representative confocal images of intact inclusions (umprimed) and disrupted inclusions (IFNγ-primed) in WT MEFs expressing GFP-p62 at 20 hpi. Cells were stained for the inclusion membrane marker CT813 as well as for GBP2. (Scale bar, 10 μm.) (B) MEFs of the indicated genotypes were infected with C. trachomatis at an MOI of 2.5 and were treated with IFNγ (100 U/mL) at 3 hpi or left untreated. Ruptured inclusions were visualized through selective permeabilization of the plasma membrane with digitonin followed by staining of ruptured inclusions with anti-Chlamydia LPS. Representative epifluorescence images are shown. Quantified data are representative of three independent experiments. ***P < 0.0005; ****P < 0.0001; n.s., not significant (one-way ANOVA); mean ± SD; n = 3. (Scale bar, 20 μm.)
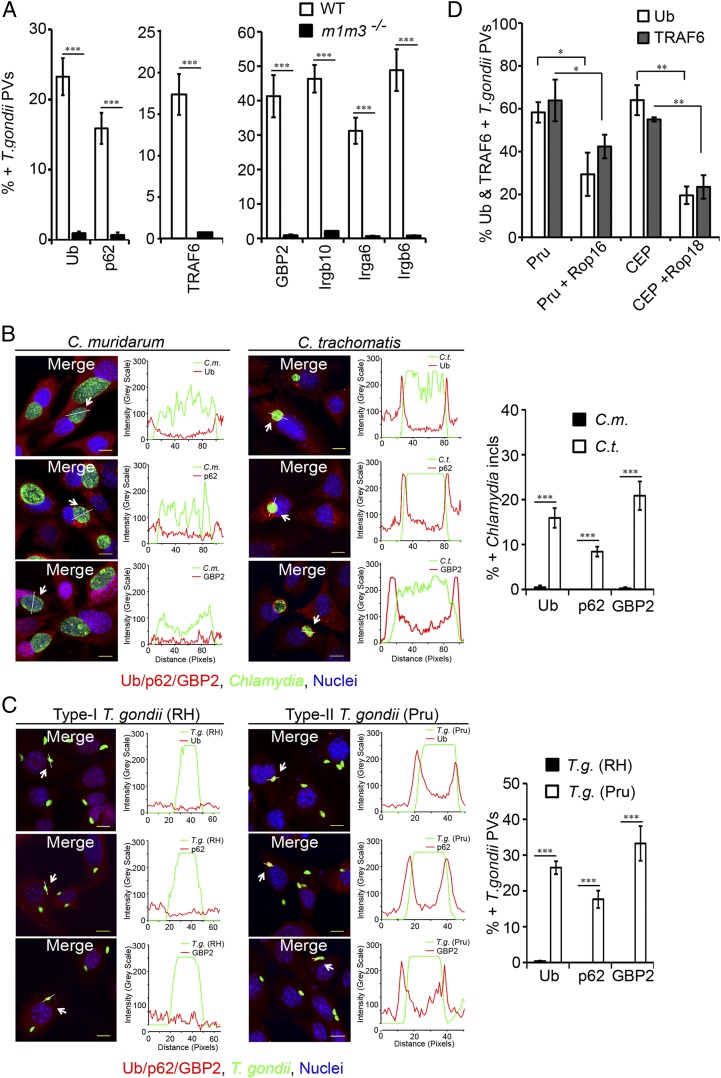
IFNγ-Inducible IRGs Control IFNγ-Inducible Decoration of PVs with Ubiquitin.
So far, our results revealed a previously unidentified IFNγ-inducible ubiquitination pathway operated in part by the E3 ligase TRAF6 and the adaptor protein p62. We next set out to identify the IFNγ-inducible components of the TRAF6-/p62-dependent ubiquitination pathway. As observed previously (17), we found that the expression of regulatory IRGM proteins is required for the efficient translocation of GBPs to PVs (Fig. 8A). We therefore asked whether IFNγ-inducible IRGM proteins were also required for PV ubiquitin staining. We found that cells lacking expression of the two IRGM proteins IRGM1 and IRGM3 (m1/m3−/−) failed to ubiquitinate T. gondii PVs (Fig. 8A). Similarly, targeting of p62 and TRAF6 to T. gondii PVs was severely compromised in m1/m3−/− cells (Fig. 8A). These data show that IFNγ-inducible association of ubiquitin with PVs is controlled by IRGM proteins and further argues that PV ubiquitination licenses PVs for GBP binding.
Fig. 8.
Hypervirulent strains of T. gondii and Chlamydia interfere with IRG-mediated PV ubiquitination. (A) Localization of ubiquitin (Ub), p62, GBP2, TRAF6, and the GKS proteins Irgb10, Irga6, and Irgb6 to type II T. gondii PVs in IFNγ-primed WT and m1m3−/− MEFs was assessed at 1 hpi. (B) WT MEFs were infected with either C. muridarum or C. trachomatis and treated with IFNγ at 3 hpi. At 20 hpi, inclusions were labeled with anti-Chlamydial LPS (green) and monitored for colocalizaiton with ubiquitin (red) or p62 (red) or GBP2 (red). ***P < 0.0005 relative to C. trachomatis infection (two-tailed unpaired t test). Line tracings of fluorescent intensities are shown. (C) IFNγ-primed WT MEFs were infected with either GFP+ type I strain T. gondii RH or GFP+ type II strain Pru and assessed for colocalization with ubiquitin (red) or p62 (red) or GBP2 (red) at 1 hpi. (D) IFNγ-primed WT MEFs were infected with the indicated transgenic T. gondii strains and assessed for the localization of ubiquitin and TRAF6 to PVs. Experimental data for D were generated in the E.-M.F. laboratory. *P < 0.05; **P < 0.005; ***P < 0.0005 (two-tailed unpaired t test); mean ± SD; n = 3. (Scale bar, 10 μm.)
Hypervirulent Strains of Chlamydia and T. gondii Block the Formation of Ubiquitin-Decorated PV Ubiquitination Through GKS Inhibition.
IRGM proteins promote loading of PVs with “effector IRGs” also known as GKS proteins (17, 21) (Fig. 8A). Because IFNγ-inducible GKS proteins directly associate with PV membranes, we hypothesized that one or multiple of the known 18 GKS proteins facilitate PV ubiquitination. To test this model, we exploited previous observations showing that hypervirulent T. gondii type I strains and the rodent-adapted pathogen Chlamydia muridarum can inhibit the translocation of GKS proteins to PVs (19, 42, 43). We observed that C. muridarum inclusions and PVs formed by the hypervirulent T. gondii type I strain RH were resistant to PV ubiquitination, p62, and GBP deposition to PVs (Fig. 8 B and C). These data show that C. muridarum and T. gondii RH interfere with the IFNγ-inducible cell-autonomous host response upstream of PV ubiquitin labeling and implicate GKS proteins as mediators of PV ubiquitination.
Although it is unknown how C. muridarum interferes with IFNγ-inducible cell-autonomous immunity (44), critical virulence factors that mediate immune evasion have been identified for hypervirulent T. gondii strains: Previous studies reported that the secreted rhoptry protein kinase 18 (ROP18) of type I strains can phosphorylate the nucleotide binding site of GKS proteins; this phosphorylation event prevents GKS protein oligomerization and binding to PV membranes (45–47). In addition to ROP18, the kinase ROP16 also blocks GKS proteins from docking to PVs (18). To assess the consequences of GKS protein inhibition, we monitored the effects of transgenic ROP18 or ROP16 expression on TRAF6 and ubiquitin PV staining. We observed a reduction in the percentages of TRAF6- and ubiquitin-positive PVs for the avirulent T. gondii type II strain Pru and type III strain CEP expressing either ROP16 (Pru + ROP16) or ROP18 (CEP + ROP18) as transgenes (Fig. 8D). Together, these data support a model in which PV-bound GKS proteins facilitate TRAF6 recruitment to PVs, thereby promoting PV ubiquitination and GBP deposition.
Discussion
GBPs are critical regulators of host resistance and inflammation (2). GBPs exert their antimicrobial and proinflammatory activities in part by binding to and disrupting PV membranes (5, 6). Additionally, GBPs can translocate to cytoplasmic bacterial pathogens, where they induce bacteriolysis (8, 12). The mechanisms by which GBPs recognize either PVs or cytoplasmic bacteria are currently unknown. Our study characterizes PV labeling with ubiquitin as an IRG-mediated immune function that triggers the deposition of GBPs on Chlamydia- and Toxoplasma-containing vacuoles. Thus, our work denotes polyubiquitin as a marker for the immunetargeting of intracellular pathogens by GBPs.
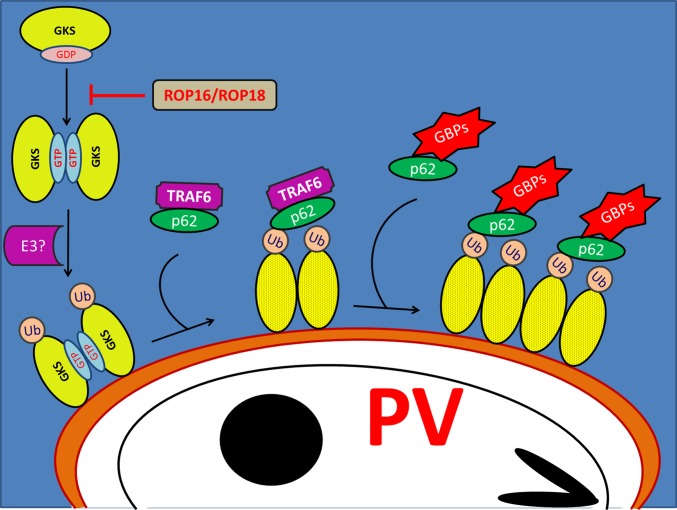
We previously showed that IRGs control GBP localization to PVs, albeit by an unknown mechanism (17). Here, we demonstrate that IRG proteins promote the deposition of ubiquitin on PVs and thereby mark PVs for GBP binding (Fig. S10). We identified TRAF6 as an E3 ligase that translocates to PVs in an IRG-dependent manner and promotes ubiquitin labeling of PVs, p62 binding, the recruitment of GBPs, and the lysis of inclusions. In addition to TRAF6, other IRG-dependent E3 ligases must exist, as we observed residual IRG-dependent PV ubiquitination in the absence of TRAF6. One such candidate E3 ligase to be involved in the ubiquitination of PVs is Trim21, yet this observation will need to be explored further. The identification of additional ubiquitinating enzymes in future studies will help build a more detailed understanding of the network of IRG-directed host responses and will likely extend the list of E3 ligases that mark invading pathogens with ubiquitin as an “eat me” or “kill me” signal.
Fig. S10.
GKS proteins control the recruitment of E3 ligases, p62, and GBPs to PVs. A subset of IRGs known as GKS proteins recognize C. trachomatis or T. gondii PVs through a missing-self principle and bind to PVs as GTP-bound dimers or multimers. PV-bound GKS proteins recruit TRAF6 and other E3 ligases to PVs, resulting in the decoration of PVs with ubiquitin. The ubiquitin-binding protein p62 enhances TRAF6 recruitment to PVs as part of a positive feedback mechanism. P62 is also found in cytoplasmic protein complexes together with GBPs and may escort GBPs to ubiquitin-decorated PVs. Hypervirulent T. gondii strains inject rhoptry protein kinases ROP16 and ROP18 into the host cell cytoplasm to block GKS protein activation and downstream effector mechanisms including ubiquitination and GBP targeting to PVs.
Our studies indicate that the GKS subset of IRG proteins is essential for ubiquitin-coating of PVs in mouse cells and that GKS-dependent ubiquitination drives GBP translocation to PVs (Fig. S10). GBP translocation to T. gondii PVs and C. trachomatis inclusions has also been observed in humans cells (48, 49), yet human cells do not encode GKS proteins (20). These combined observations pose the conundrum of explaining how human GBPs target PVs independently of GKS proteins. Possibly, GKS-independent ubiquitination of PVs could facilitate the translocation of GBPs to PVs in human cells. The coating of intracellular pathogens with ubiquitin is indeed a conserved defense mechanism found in host organisms as diverse as fruit flies and humans (28, 50). Specifically, the E3 ligases Parkin and LRSAM1 were found to be required for the ubiquitination of intracellular Mycobacterium tuberculosis and Salmonella enterica in mouse and human cells (50, 51). Both of these pathogens are subjects of GBP-mediated immune responses (5, 9, 10), yet the mechanisms by which the host can tag M. tuberculosis and S. enterica with GBPs are unknown. Based on the data presented here, it is tempting to speculate that Parkin- and LRSAM1-mediated ubiquitination could initiate GBP-orchestrated immune responses.
Our discovery of a previously unidentified ubiquitination pathway automatically prompts many questions, some of which we will discuss here briefly: One obvious question relates to the nature of the ubiquitination substrate. Potential substrates are GKS proteins, which become K63-polyubiquitinated when entering the active state (26). Accordingly, GKS proteins could fulfill a dual function in recruiting TRAF6 and serving as TRAF6 substrates. Autoubiquitinated TRAF6 itself could be among the K63-ubiquitinated proteins associated with PV. Our data suggest that the build-up of K63-linked polyubiquitin enlists p62 to PVs. However, p62 also recruits more TRAF6 protein, thereby creating an amplification loop that assures rapid ubiquitination of PVs. These observations suggest either that initial PV ubiquitination is p62-independent or that p62 transiently associates with unubiquitinated PVs to recruit the ubiquitin machinery.
In addition to K63-linked polyubiquitin, we also observed the deposition of K48-linked polyubiquitin on PVs. We found that the attachment of K48-linked polyubiquitin was largely p62-dependent, suggesting that PV-bound p62 recruits E3 ligases other than K63-specific TRAF6. We identified Trim21 as such a PV-resident E3 ligase that is known to interact with p62 and to form K48-linked polyubiquitin chains (52–54). Further defining the function of PV-associated E3 ligases such as Trim21 will doubtlessly reveal novel insights into host defense mechanisms directed at vacuolar pathogens.
A second set of questions explores the detailed molecular mechanisms by which ubiquitinated PVs recruit GBPs to their surrounding membranes. We propose that p62 preassembles with a subset of GBPs and then escorts these GBPs to PVs. In support of this model, GBP1 and GBP2 interact with p62 in cytoplasmic speckles (9) (Fig. S6). These speckles form independently of an infection and can be biochemically isolated (9), thus arguing that p62–GBP1/2 complexes are assembled before their translocation to PVs. Additional types of GBPs can be recruited through heterotypic protein–protein interactions between distinct members of the GBP protein family, as such interactions have been described previously (16, 18). Moreover, additional ubiquitin-binding proteins like NDP52 may aid in the delivery of GBPs to PVs.
A third area of inquiry would explore the existence of additional PV-associated patterns, which cooperatively or independently promote the recruitment of GBPs to PVs or cytoplasmic pathogens. One such pattern appears to be provided by lipidated ubiquitin-like ATG8 proteins that decorate inclusions and T. gondii PVs. Interference with ATG8 lipidation diminishes GBP and GKS protein recruitment to PVs, demonstrating the importance of these molecules in immunotargeting of PVs (24, 25, 42, 48, 55–57). Whether and how the ATG8 pathway intersects with the newly discovered IRG-driven ubiquitination pathway will need to be explored further in the future. Lastly, additional patterns associated with intracellular pathogens may directly or indirectly elicit GBP-driven host responses.
As expected for a central branch of the innate immune response, host-adapted pathogens have evolved mechanisms to interfere with the delivery of GBPs to PVs. We confirmed previous observations that hypervirulent strains of T. gondii block GBP accumulation on PV membranes (18, 25, 58). In light of our discovery of the causal link between the deposition of GKS proteins, PV ubiquitination, and p62-dependent GBP delivery, we were able to explain how hypervirulent strains of T. gondii can escape GBP-mediated immunity through the inhibition of GKS proteins. As future studies will likely discover additional microbial strategies to escape from GBP-rendered immunity, the GBP protein family emerges as a central player in the continuous arms race between the mammalian host and microbes.
Experimental Procedures
Host Cell Culture, Bacterial, and Protozoan Strains and Infections.
MEFs derived from WT, Irgm1/m3−/−, TRAF6−/−, p62−/−, and GBPchr3−/− mice were previously described (6, 59–61). Primary murine BMMs were isolated from the tibia and femurs of 2–4-mo-old mice as described before (10). MEFs and African green monkey kidney Vero cells were cultured in DMEM supplemented with 10% (vol/vol) heat-inactivated FBS. C. trachomatis LGV-L2, C. muridarum, and T. gondii tachyzoites of the type I strain RH, the type II strains Pru and ME49 strain, and the type III strain CEP were propagated in Vero cells, as described (19, 24). Transgenic T. gondii strains expressing Rop16 and Rop18 were reported previously (18). GFP-expressing T. gondii tachyzoites were a generous gift from John Boothroyd, Stanford University, Stanford, CA (62, 63). A previously described GFP expression vector (64) was introduced into LGV-L2 for visualizing C. trachomatis. Infections with C. trachomatis were performed at a nominal multiplicity of infection of 1–5 followed by treatment with 100 U/mL of IFNγ at 3 hpi or as indicated, essentially as described (17). For T. gondii infections, cells were incubated overnight with or without 200 U/mL of IFNγ and asynchronously (or synchronously in the E.-M.F. laboratory) infected with tachyzoites at a nominal multiplicity of infection of 5–10 for 1 h.
Immunocytochemistry.
Immunocytochemistry was performed essentially as described previously (17). Cells were washed thrice with PBS, pH 7.4, before fixation. Cells were fixed either with methanol or with 3% (vol/vol) formaldehyde and 0.025% (vol/vol) glutaraldehyde for 20 min at room temperature. In all experiments involving formaldehyde/glutaraldehyde fixation, fixed cells were permeabilized/blocked with 0.05% (wt/vol) saponin and 2% (wt/vol) BSA/PBS (SBP) for 30 min at room temperature. Cells were stained with the indicated primary antibodies, followed by Alexa Fluor-conjugated secondary antibodies (Molecular Probes/Invitrogen). Nucleic and bacterial DNA were stained with Hoechst 33258 according to the manufacturer’s protocol. Stained cells were washed with PBS, mounted on microscope slides with Mowiol (Sigma), and allowed to cure overnight. Cells were imaged using either a Zeiss LSM 510 inverted confocal microscope or a Zeiss Axioskop 2 upright epifluorescence microscope. Colocalization of proteins with PVs was quantified in at least three independent experiments. In each experiment, at least 10 randomly selected fields were imaged for each experimental condition and cell type. To determine the frequency with which ubiquitin, p62, TRAF6, GBP, and GKS proteins colocalize with PVs, at least 100 PVs were assessed for each experimental condition and cell type for every biological replicate. Differential interference contrast images were used to identify extracellular T. gondii tachyzoites. The fraction of ubiquitin-, p62-, TRAF6-, GBP-, or GKS-positive vacuoles was determined for each field by dividing the number of labeled vacuoles by the total number of vacuoles. To perform line tracings—that is, analyze the fluorescence signal intensity profiles of pixels along a selection from images—we used ImageJ software.
Antibodies and Immunoblotting.
The primary antibodies used for immunocytochemistry include anti-Irga6 mouse monoclonal antibody 10D7 (65) at 1:10, anti-Irgb10 rabbit polyclonal antiserum (19) at 1:1,000, anti-Irgb6 rabbit polyclonal antisera (26) at 1:1,000, anti-Gbp2 rabbit polyclonal (17) at 1:1,000, rabbit anti-p62/SQSTM1 (MBL International) at 1:500, mouse anti-Ubiquitin (FK2, ENZO) at 1:500, rabbit monoclonal anti–ubiquitin-K48 (Millipore) at 1:500, rabbit monoclonal anti-K63 (Millipore) at 1:100, anti-TRAF6 rabbit polyclonal (Abnova) at 1:200, anti-CT813 mouse monoclonal (41) at 1:25, mouse monoclonal anti-Chlamydial LPS (1681; Santa Cruz) at 1:50, anti-T. gondii mouse monoclonal antibody (Pierce) at 1:500, anti-T. gondii rabbit polyclonal antibody (Biogenex) at 1:500, and anti-TRAF6 (Abnova) at 1:200. To assess protein expression levels, samples from whole cell lysates were analyzed by SDS/PAGE and Western blotting. Blots were probed with primary antibodies specific for TRAF6 (Santa Cruz) at 1:200, anti-FLAG (Sigma) at 1:500, and anti–β-actin (Sigma) at 1:5,000. Binding of secondary HRP-labeled goat anti-rabbit or goat anti-mouse antibodies (Thermo Scientific) was analyzed using SuperSignal West Pico or West Femto Chemiluminescent Substrate (Thermo Scientific).
Retroviral Infections and Generation of Stable Cell Lines.
Stable cell lines were generated using a retroviral expression system. pMXs and packing vectors were transfected into HEK-293T cells to generate recombinant retroviruses. MEFs were infected with recombinant retroviruses and selected in medium containing 5 μg/mL puromycin. GFP-p62 expression vectors (34) and FLAG-tagged TRAF6 expression vectors (61) were previously reported. To interfere with gene expression, MEFs were transduced with lentivirus prepared in HEK-293T cells and subsequently selected for shRNA expression with puromycin (5 μg/mL) for at least 48 h. shRNA constructs are listed in Table S1.
Table S1.
List of shRNAs used for screening E3 ligase function
| Target genes | shRNA sequences | Sequence ID |
| TRAF6 | CCGGCCCAGGCTGTTCATAATGTTACTCGAGTAACATTATGAACAGCCTGGGTTTTTG | NM_009424.2–1227s1c1 |
| TRAF6 | CCGGGCCTGCATCATCAAATCCATACTCGAGTATGGATTTGATGATGCAGGCTTTTTG | NM_009424.2–680s1c1 |
| TRIM50 | CCGGCGGATTATTAATGAGTCCGATCTCGAGATCGGACTCATTAATAATCCGTTTTTG | NM_178240.1–487s1c1 |
| TRIM25 | CCGGCCTGGAGTACAAACTGAGGAACTCGAGTTCCTCAGTTTGTACTCCAGGTTTTTG | NM_009546.1–597s1c1 |
| TRIM28 | CCGGCCACCAGTCTTCAAGGTCTTTCTCGAGAAAGACCTTGAAGACTGGTGGTTTTTG | NM_011588.1–2065s1c1 |
| TRIM13 | CCGGGCAGGTCAATTACTCCCTAAACTCGAGTTTAGGGAGTAATTGACCTGCTTTTTG | NM_023233.1–331s1c1 |
| CULLIN3 | CCGGGTGCGAGAAGATGTACTAAATCTCGAGATTTAGTACATCTTCTCGCACTTTTTG | NM_016716.4–384s21c1 |
| TRIM30 | CCGGGAGAACCAAATACAGATCAATCTCGAGATTGATCTGTATTTGGTTCTCTTTTTG | NM_009099.2–742s21c1 |
| TRIM21 | CCGGTTGTCTCCTTCTACAACATAACTCGAGTTATGTTGTAGAAGGAGACAATTTTTG | NM_009277.3–1421s21c1 |
| TRIM21 | CCGGTGACAATGAGGAGAGGTTTAGCTCGAGCTAAACCTCTCCTCATTGTCATTTTTG | NM_009277.3–1131s21c1 |
| TRIM21 | CCGGGAACCTGGACACGTTAGATATCTCGAGATATCTAACGTGTCCAGGTTCTTTTTG | NM_009277.3–945s21c1 |
| STUB1 | CCGGGAGAGTTATGATGAGGCCATTCTCGAGAATGGCCTCATCATAACTCTCTTTTT | NM_019719.2–833s1c1 |
In Situ PLA and Quantification.
To perform PLA, we used the Duolink in Situ Detection Kit (Sigma). Briefly, we fixed and permeabilized cells following a standard immunofluorescence protocol for mouse anti-p62 and rabbit anti-GBP2 antibodies. Subsequent incubation steps, PLA probe dilutions and incubation times, rolling circle amplification times, and polymerase concentrations were used according to the manufacturer’s instructions. For dual recognition of GBP2 and p62, the experiments were performed using anti-rabbit PLUS and anti-mouse MINUS secondary probes. Fluorescence images were acquired on a Carl-Zeiss Axio Observer.Z1 microscope using a 63× oil objective for high-magnification images. For all experiments, quantifications were performed from at least 10 images. High-resolution images from single scans were analyzed in Fiji (ImageJ, NIH) to calculate the number of PLA punctae. Images were first smoothed, and a threshold was selected manually to discriminate PLA punctae from background fluorescence. Once selected, this threshold was applied uniformly to all images in the sample set. The built-in macro “Analyze Particles” was used to count and characterize all objects within images. Objects larger than 200 pixels were rejected, effectively removing cell nuclei from the analysis. The remaining objects were counted as PLA punctae.
Quantitative PCR.
Total nucleic acid was prepared from trypsinized cell pellets using the QIAamp DNA Mini Kit from Qiagen. Samples were then subjected to singleplex quantitative PCR on an ABI 7000 Sequence Detection System to assess the amount of 16S Chlamydia and GAPDH host DNA in the sample. Chlamydia 16S DNA was detected through use of the following primer sequences, as described (19): 16S forward primer 5′-GGA GGC TGC AGT CGA GAA TCT-3′, reverse primer 5′-TTA CAA CCC TAG AGC CTT CAT CAC A-3′, and dual-labeled probe 5′-[6-FAM]-TCG TCA GAC TTC CGT CCA TTG CGA-[TAMRA]-3′. Mouse GAPDH DNA was detected using the Rodent GAPDH Control Reagent Kit from Applied Biosystems. Standard curves were generated in parallel from known amounts of C. trachomatis and murine DNA, and these curves were used to calculate the amount (pg) of Chlamydia DNA per unit mass (µg) of mouse DNA in the samples.
Quantification of Intracellular Ruptured C. trachomatis Inclusions.
For quantification of ruptured C. trachomatis inclusions, we adapted and modified a published protocol for Digitonin-mediated permeabilization of plasma membranes (8). MEFs were infected with GFP+ C. trachomatis at a multiplicity of infection (MOI) of 2 followed by treatment with 100 U/mL of IFNγ at 3 hpi or left untreated. At 20 hpi, cells were washed with KHM buffer (110 mM potassium acetate, 20 mM Hepes, and 2 mM MgCl2, pH 7.3), followed by incubation for 3 min in KHM buffer with 12.5 μg/mL digitonin (Sigma). Cells were immediately washed three times with KHM buffer and then stained with mouse monoclonal anti-Chlamydial LPS in KHM buffer with 2% BSA for 20 min. Next, cells were washed with PBS, subsequently fixed with 4% PFA for 12 min, followed by staining with goat anti-mouse Alexa Fluor 568-conjugated secondary antibody for 30 min. Images were acquired on a Carl-Zeiss Axio Observer.Z1 microscope, and the percentage of LPS-stained inclusions was quantified.
Quantification of C. trachomatis IFUs.
MEFs were primed with IFNγ (100 U/mL) overnight or left unprimed and subsequently infected with GFP+ C. trachomatis in 24-well plates. At 40 hpi, infected monolayers were washed before cells lysis with water and physical dislodgement into 0.5 mL SPG buffer. SPG suspensions were transferred to sterile reaction tubes and sonicated. Samples were stored at –80 °C until further processing. For quantification, thawed samples were serially diluted and used to reinfect fresh, confluent monolayers of Vero cells in triplicate. The next day, samples were fixed with MeOH, and cell nuclei were stained with Hoechst. Inclusions were visualized under the microscopes. Per data point (original well infected) inclusions were counted in 10 randomly selected fields, and the number of inclusion forming units (IFUs) was calculated based on inclusion count and dilution factor.
Quantification of T. gondii Replication.
WT, p62−/−, and TRAF6−/− MEFs were primed overnight with IFNγ or left unprimed in 24-well plates. Cells were infected with 200 tachyzoites per well of the strain GFP+ T. gondii Pru. At 72 hpi, images of at least 20 fields per condition were acquired on a Carl-Zeiss Axio Observer.Z1 microscope using a 10× objective. The number of GFP+ T. gondii Pru plaques was counted by a person other than the experimenter in a blinded fashion. The number of plaques in IFNγ-primed MEFs was normalized over the number of plaques in corresponding unprimed MEFs.
Statistical Analysis.
Prism 6.0a software (GraphPad Software) and Excel were used for statistical data analysis. To assess statistical significance of differences between two sets of independent data points, unpaired two-tailed t test was used. To analyze the differences among group means, one-way ANOVA was used. Values of P < 0.05 were considered significant. Results are represented as means ± SD.
Acknowledgments
We thank Drs. John Boothroyd, Bryant Darnay, Kirk Jensen, Ashok Kumar, Tak Mak, Michael Reese, Jeroen Saeij, Greg Taylor, Masahiro Yamamoto, and Guangming Zhong for sharing mice, cell lines, expression constructs, antibodies, and T. gondii strains. We thank David Tobin for his careful and critical reading of our manuscript. This research was supported by American Heart Association Predoctoral Award 12PRE10440003 (to D.M.P.-M.), a National Science Foundation predoctoral award (to R.F.), a Medical Research Council (MRC) studentship and Boehringer Ingelheim Fonds PhD Fellowship (to C.F.), a Wellcome Trust Career Development Award and an MRC Grant MC_UP_1202/12 (to E.-M.F.), and National Institute Health Grant R01AI103197 (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515966112/-/DCSupplemental.
References
- 1.Kumar Y, Valdivia RH. Leading a sheltered life: Intracellular pathogens and maintenance of vacuolar compartments. Cell Host Microbe. 2009;5(6):593–601. doi: 10.1016/j.chom.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12(4):432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ling YM, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203(9):2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martens S, et al. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 2005;1(3):e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meunier E, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509(7500):366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto M, et al. A cluster of interferon-γ-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37(2):302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Zhao YO, Khaminets A, Hunn JP, Howard JC. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog. 2009;5(2):e1000288. doi: 10.1371/journal.ppat.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meunier E, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16(5):476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim BH, et al. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332(6030):717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 10.Pilla DM, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci USA. 2014;111(16):6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shenoy AR, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336(6080):481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 12.Man SM, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16(5):467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403(6769):567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440(7080):101–104. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 15.Vöpel T, et al. Mechanism of GTPase-activity-induced self-assembly of human guanylate binding protein 1. J Mol Biol. 2010;400(1):63–70. doi: 10.1016/j.jmb.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 16.Britzen-Laurent N, et al. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One. 2010;5(12):e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haldar AK, et al. IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog. 2013;9(6):e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virreira Winter S, et al. Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS One. 2011;6(9):e24434. doi: 10.1371/journal.pone.0024434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coers J, et al. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J Immunol. 2008;180(9):6237–6245. doi: 10.4049/jimmunol.180.9.6237. [DOI] [PubMed] [Google Scholar]
- 20.Bekpen C, et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: Loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6(11):R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunn JP, et al. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 2008;27(19):2495–2509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor GA, et al. The inducibly expressed GTPase localizes to the endoplasmic reticulum, independently of GTP binding. J Biol Chem. 1997;272(16):10639–10645. doi: 10.1074/jbc.272.16.10639. [DOI] [PubMed] [Google Scholar]
- 23.Coers J. Self and non-self discrimination of intracellular membranes by the innate immune system. PLoS Pathog. 2013;9(9):e1003538. doi: 10.1371/journal.ppat.1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haldar AK, Piro AS, Pilla DM, Yamamoto M, Coers J. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PLoS One. 2014;9(1):e86684. doi: 10.1371/journal.pone.0086684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selleck EM, et al. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog. 2013;9(4):e1003320. doi: 10.1371/journal.ppat.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traver MK, et al. Immunity-related gtpase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J Biol Chem. 2011;286(35):30471–30480. doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Zeer MA, Al-Younes HM, Lauster D, Abu Lubad M, Meyer TF. Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins. Autophagy. 2013;9(1):50–62. doi: 10.4161/auto.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle KB, Randow F. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol. 2013;16(3):339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Dupont N, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6(2):137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Yoshikawa Y, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11(10):1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 31.Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183(9):5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 32.Mostowy S, et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem. 2011;286(30):26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271(34):20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 34.Itakura E, Mizushima N. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192(1):17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 36.Linares JF, et al. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51(3):283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19(7):1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53(2):167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Seibenhener ML, et al. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24(18):8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464(7289):721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, et al. The hypothetical protein CT813 is localized in the Chlamydia trachomatis inclusion membrane and is immunogenic in women urogenitally infected with C. trachomatis. Infect Immun. 2006;74(8):4826–4840. doi: 10.1128/IAI.00081-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khaminets A, et al. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell Microbiol. 2010;12(7):939–961. doi: 10.1111/j.1462-5822.2010.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y, et al. Virulent Toxoplasma gondii evade immunity-related GTPase-mediated parasite vacuole disruption within primed macrophages. J Immunol. 2009;182(6):3775–3781. doi: 10.4049/jimmunol.0804190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coers J, Starnbach MN, Howard JC. Modeling infectious disease in mice: Co-adaptation and the role of host-specific IFNgamma responses. PLoS Pathog. 2009;5(5):e1000333. doi: 10.1371/journal.ppat.1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fentress SJ, et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe. 2010;8(6):484–495. doi: 10.1016/j.chom.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinfeldt T, et al. Phosphorylation of mouse immunity-related GTPase (IRG) resistance proteins is an evasion strategy for virulent Toxoplasma gondii. PLoS Biol. 2010;8(12):e1000576. doi: 10.1371/journal.pbio.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hermanns T, Muller UB, Konen-Waisman S, Howard JC, Steinfeldt T. The Toxoplasma gondii rhoptry protein ROP18 is an Irga6-specific kinase and regulated by the dense granule protein GRA7. Cell Microbiol. 2015 doi: 10.1111/cmi.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohshima J, et al. Role of mouse and human autophagy proteins in IFN-γ-induced cell-autonomous responses against Toxoplasma gondii. J Immunol. 2014;192(7):3328–3335. doi: 10.4049/jimmunol.1302822. [DOI] [PubMed] [Google Scholar]
- 49.Tietzel I, El-Haibi C, Carabeo RA. Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One. 2009;4(8):e6499. doi: 10.1371/journal.pone.0006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manzanillo PS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501(7468):512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huett A, et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12(6):778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JY, Ozato K. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-kappaB activity. J Immunol. 2009;182(4):2131–2140. doi: 10.4049/jimmunol.0802755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Connor C, et al. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5alpha. J Virol. 2010;84(12):5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, et al. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14(2):172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Zeer MA, Al-Younes HM, Braun PR, Zerrahn J, Meyer TF. IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS One. 2009;4(2):e4588. doi: 10.1371/journal.pone.0004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi J, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014;40(6):924–935. doi: 10.1016/j.immuni.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Z, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4(5):458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Degrandi D, et al. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J Immunol. 2007;179(11):7729–7740. doi: 10.4049/jimmunol.179.11.7729. [DOI] [PubMed] [Google Scholar]
- 59.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 60.Coers J, et al. Compensatory T cell responses in IRG-deficient mice prevent sustained Chlamydia trachomatis infections. PLoS Pathog. 2011;7(6):e1001346. doi: 10.1371/journal.ppat.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamothe B, et al. The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL. J Biol Chem. 2008;283(36):24871–24880. doi: 10.1074/jbc.M802749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim SK, Karasov A, Boothroyd JC. Bradyzoite-specific surface antigen SRS9 plays a role in maintaining Toxoplasma gondii persistence in the brain and in host control of parasite replication in the intestine. Infect Immun. 2007;75(4):1626–1634. doi: 10.1128/IAI.01862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pernas L, et al. Toxoplasma effector MAF1 mediates recruitment of host mitochondria and impacts the host response. PLoS Biol. 2014;12(4):e1001845. doi: 10.1371/journal.pbio.1001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, et al. Development of a transformation system for Chlamydia trachomatis: Restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7(9):e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martens S, et al. Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes: Retargeting to plasma membrane induced by phagocytosis. J Immunol. 2004;173(4):2594–2606. doi: 10.4049/jimmunol.173.4.2594. [DOI] [PubMed] [Google Scholar]