Significance
Gene targets of de novo mutation in autistic children have a lighter load of rare disruptive variation than typical human genes. This finding suggests such mutations are under negative selection and autism genes are highly vulnerable to mutation. Disruptive variants in these genes have biased transmission: They are more frequently transmitted to affected children, and more often from mothers than from fathers. Targets of mutation in lower intelligence quotient (IQ) affected children have a lower load of disruptive mutations than targets of mutation in higher IQ affected children. Biased transmission is seen more frequently to affected children of lower IQ. These observations are consistent with a correlation between severity of mutations and phenotype, and based on them, we list candidate autism genes ordered by likelihood.
Keywords: autism spectrum disorder, gene vulnerability, disruptive mutations, biased transmission, autism genes
Abstract
We previously computed that genes with de novo (DN) likely gene-disruptive (LGD) mutations in children with autism spectrum disorders (ASD) have high vulnerability: disruptive mutations in many of these genes, the vulnerable autism genes, will have a high likelihood of resulting in ASD. Because individuals with ASD have lower fecundity, such mutations in autism genes would be under strong negative selection pressure. An immediate prediction is that these genes will have a lower LGD load than typical genes in the human gene pool. We confirm this hypothesis in an explicit test by measuring the load of disruptive mutations in whole-exome sequence databases from two cohorts. We use information about mutational load to show that lower and higher intelligence quotients (IQ) affected individuals can be distinguished by the mutational load in their respective gene targets, as well as to help prioritize gene targets by their likelihood of being autism genes. Moreover, we demonstrate that transmission of rare disruptions in genes with a lower LGD load occurs more often to affected offspring; we show transmission originates most often from the mother, and transmission of such variants is seen more often in offspring with lower IQ. A surprising proportion of transmission of these rare events comes from genes expressed in the embryonic brain that show sharply reduced expression shortly after birth.
The past decade has seen remarkable progress in understanding genetic causation of autism spectrum disorders (ASD), confirmatory of predictions made by a “unified” genetic theory of autism proposed in 2007 (1). This theory proposes that much of ASD is caused by new mutation, sometimes directly contributing to the disorder through germ-line mutation, or transmitted by parents, especially females, who carry a variant of recent vintage without experiencing severe consequences. The theory was based largely on three sets of observations: (i) low ASD incidence in females compared with males (2), (ii) apparently dominant transmission to male children in multiplex families (1), and (iii) greater incidence of de novo (DN) copy number mutation in children with ASD than in their siblings in simplex families (3, 4). Since then, evidence for causal DN mutation has accumulated (5–8). These damaging mutations generally affect only one allele, suggesting that gene targets are dosage-sensitive, prone to dominant negative mutation, or some combination of these factors.
A widely held genetic model for autism is that combinations of common variation are the major driving force. As we argue, there is little evidence for this belief. Damaging DN mutation contributes to at least 30% of ASD in simplex families (9). Among such damaging mutations are those mutations that are likely gene-disruptive (LGD) in that they create nonsense, splice-site, or small frame-shift variants (5). The estimates of contribution from DN mutations derive from the statistically robust increased difference in disruptive mutation frequency in affected vs. unaffected siblings, which we call the “ascertainment differential.” An ascertainment differential is not seen for mutations that are not disruptive, such as synonymous variants.
From recurrence and overlap analysis of DN LGD targets, we estimate ∼500 causative ASD target genes in the affected individuals with lower intelligence quotients (IQ), and these targets are enriched in certain functional classes (10). Because there are so many autism targets, the penetrance of any given disruptive mutation in a specific target cannot be individually observed at present. However, from the size of the causative target set, DN mutation rate, and ASD incidence rates, we can directly compute what we call “vulnerability” in these genes: the likelihood that a disruptive mutation in the gene results in ASD. We define an “autism gene” as one that, when mutated, may contribute to ASD diagnosis. We computed that roughly half of the time in males, a DN LGD mutation within an autism gene will produce severe ASD (10). Because people with ASD have lower fecundity than the general population, a disruptive mutation in an autism gene will be under strong purifying selection and quickly eliminated from the population (11). A clear prediction is that autism genes will have a smaller load of disruptive mutations than “typical” genes, as we first observed for fragile X mental retardation protein (FMRP)-associated genes (5, 12).
Indeed, recent reports indicate that the targets of disruptive DN mutation in affected children do have a lighter load of disruptive mutation in the human population (13, 14). The methods used for measuring the load used missense mutations as well as LGD mutations, in fairly complex formulations termed the “residual variation intolerance score” (RVIS) or gene constraint. The former coins the term “tolerance” to distinguish genes with high loads (high tolerance) and low loads (low tolerance), which we adopt here, although our tolerance score differs. They apply their particular tolerance score on targets from a subset of the Simons Simplex Collection (SSC), a collection of simplex autism families (5, 7, 8, 15). More recently, additional evidence has been shown for transmission of mutations in genes with low tolerance using the RVIS from mothers (16).
We describe here an independent study using a simpler measure of tolerance based on ratios of rare disruptive mutations to length in a given gene, using larger populations, and avoiding missense mutations in the tolerance score altogether, because the significance of missense mutations is very difficult to call. Using the LGD score, we obtain strong statistical evidence for low disruptive loads in autism genes, especially in the autism genes affecting children of lower IQ, and for preferential transmission of disruptive mutations in rarely disrupted genes from mothers to children with severe ASD. We also find a strong signal for biased transmission in the functional categories of genes previously associated with ASD (10). We use the tolerance score to reorder the likelihood of candidate autism genes among the known targets. We compare gene rankings based on tolerance for LGD mutations with the RVIS, a tolerance score derived largely from missense mutations.
Results
Sources of Human Sequence Variation from Whole-Exome Sequence Databases.
For our purposes, we consider only what we call ultrarare (UR) variation: variants found at very low frequency in genes that do not have a large load of other LGD variants. Common LGD variants might arise due to errors in the annotation of the transcriptome as coding variants that retain some protein function, or within genes that are not under strong purifying selection.
In this analysis, we used two distinct whole-exome sequence (WES) databases. The first set is derived from nearly 5,000 parents in a collection of families with only one child on the autism spectrum, the SSC (15). Obviously, some of these parents may be carriers of variation contributing to ASD, so statistics extracted from these families may overestimate the rare variation seen in candidate autism genes. The advantage of using the SSC is that the WES data were obtained with similar capture and sequencing platforms and were subject to a uniform analysis pipeline, and the coverage is known for every nucleotide position in every gene in every person within the targeted regions. Moreover, because we have data from families, we can make adjustments due to transmission, which aids our understanding of candidate target genes and transmission. Our second source is the exome variant sequence (EVS) database, incorporating about 6,000 people (evs.gs.washington.edu/EVS/). The EVS database is the database used in RVIS ranking (13).
We examined the parameters of variation between the two databases, and found them comparable (Table 1). We define UR as a variant observed only once in either population. The total number of UR synonymous variants is similar, as are the ratios of missense, synonymous, nonsense, and LGD mutations, as well as the ratios of transition and transversion UR variants. This observation is what we expect from two comparably sized statistical samples representing outbred gene pools. Given these findings, we combined the two datasets to increase statistical power. We also include the ratio of UR LGD mutations to UR synonymous mutations among DN variants in the children from the SSC (Table 1). Relative to the proportion of LGD variants to synonymous variants in parents, the proportion of LGD mutations to synonymous mutations among all DN variants is increased in all children. Of course, this increase is most notable in affected children, because alleles in parents show the effects of cumulative purifying selection over many generations. The excess of LGD mutations in DN mutations in children relative to parents reflects selective pressure yet to come, and thus, roughly speaking, the proportion that will be deleterious or, in some respect, affect fecundity. The majority of LGD mutations (and the subset of nonsense) will be harmful, but this scenario is not so for missense mutations. We estimate that one in five individuals is burdened with one DN mutation that reduces fecundity.
Table 1.
Global statistics for UR variants in this study
| Dataset | Individuals | syn | LGD | mis | non | LGD/syn | mis/syn | non/syn | Ti/Tv |
| UR variants in SSC parents | 4,942 | 211,780 | 28,056 | 394,923 | 10,355 | 0.132 | 1.865 | 0.049 | 2.52 |
| UR variants in EVS | ∼6,000 | 216,146 | 25,890 | 410,372 | 11,600 | 0.120 | 1.899 | 0.054 | 2.72 |
| DN variants in unaffected siblings | 1,875 | 486 | 176 | 1,131 | 59 | 0.362 | 2.327 | 0.121 | 2.60 |
| DN variants in affected children | 2,462 | 637 | 380 | 1,657 | 136 | 0.597 | 2.601 | 0.214 | 2.87 |
The first two rows show the number of UR synonymous (syn), LGD, missense (mis), and nonsense (non) variants found in parents from the SSC and EVS cohorts. Also shown are ratios of UR LGD, missense and nonsense variants to UR synonymous variants and the transition to transversion ratio (Ti/Tv) for UR substitutions. The EVS and SSC datasets are very similar, both in absolute numbers of UR variants and the observed ratios. For comparison, the last two rows show the numbers and the ratios of DN synonymous, LGD, missense, and nonsense variants reported in children affected with autism and unaffected siblings.
Genes Targeted by DN Mutation in Affected Individuals Have Low Mutational Load.
We next examined the load of LGD variation in genes that are targets of DN mutation in affected individuals and then compared these genes with targets of DN mutation in unaffected siblings (Table 2). These two target classes were discovered on the same sequencing platforms, sequenced to the same depth, and processed through identical informatics pipelines, and they arise in children matched for germ-line background by being full siblings. Moreover, the target genes in affected and unaffected individuals have length distributions that are closely matched (10). Their differential tolerance notwithstanding, the two target classes would have the same expectation of load for deleterious mutations. We further parse the DN targets among affected individuals into those DN targets occurring in lower and higher IQ individuals. This separation by IQ is made because our previous studies showed that the DN target set for higher IQ males has little overlap with DN targets in females, as well as in males of lower IQ (10). By contrast, DN targets from the latter two classes have extensive recurrence, higher ascertainment differentials, and similar functional class enrichments.
Table 2.
Burden of UR variants in the targets of DN LGD mutation
| DN LGD targets | Gene count | Proportion expected to be causal | No. of UR syn mutations | No. of UR LGD mutations | LGD/syn mutations | P value | Expected no. of UR LGD mutations | Class vulnerability | Causal class vulnerability |
| sib | 173 | 0.02 | 7,372 | 881 | 0.12 | 0.9842 | 881 | 1.00 | 1.00 |
| rec in aut | 39 | 0.90 | 2,568 | 79 | 0.03 | <0.0001 | 307 | 0.26 | 0.17 |
| autL | 204 | 0.44 | 10,244 | 790 | 0.08 | 0.0011 | 1,224 | 0.65 | 0.19 |
| autH | 151 | 0.30 | 7,039 | 678 | 0.10 | 0.1066 | 841 | 0.81 | 0.35 |
| aut | 509 | 0.36 | 24,758 | 2,062 | 0.08 | 0.0009 | 2,959 | 0.70 | 0.15 |
Target classes of DN LGD mutations occurring in “sib” (unaffected sibling), “autL” (affected, lower nonverbal IQ half), “autH” (affected, higher nonverbal IQ half), “aut” (all affected), and “rec in aut” (targets hit in >1 affected) are shown. For each class, we report the gene count, and the proportion expected to be causal, as determined by the ascertainment differential (Methods). In successive columns, the numbers of observed UR synonymous and LGD variants are reported, as well as the ratio of the latter to former for each class. Based on permutations of labels, we compute the P value of the observed ratios on the assumption that they arise from a gene class similar to the sibling targets. Target classes from affected children show a markedly lower load for UR LGD variants than the class from siblings, although this difference is not significant for the affected children of higher IQ. We also derived the expected loads in the gene classes by multiplying the fourth column (number of UR syn) by 0.12, the ratio of LGD mutations to synonymous mutations in the unaffected sibling class. The expected loads allow us to compute class vulnerability as the ratio of observed to expected and, more importantly, to use a linear model to compute vulnerability of causal genes within classes (Methods). The estimates of the causal class vulnerability for the true autism genes based on the rec in aut, autL, and aut values are close (0.17, 0.19 and 0.15, respectively) and quite low.
For each DN target class, we count variants in the WES data, combining EVS and parents from the SSC, and divide the number of UR LGD variants by the number of UR synonymous variants to yield the LGD/synonymous variant ratio. All DN target gene sets derived from affected children have lower LGD ratios than the DN target gene sets derived from unaffected siblings. To determine if these lower ratios were significant, we performed 10,000 permutations, randomly swapping genes between sets while keeping the size of sets fixed, and computed the LGD ratio. Judging by this measure, DN target gene sets from affected children with lower IQ have significantly lower LGD ratios than DN targets from siblings (P = 0.0011). On the other hand, the lower LGD ratio of the DN targets of higher IQ males does not reach standard statistical significance (P = 0.1066).
The above analysis does not provide us with a measure of the tolerance for causal target genes, because each set is composed of causal targets as well as “bystanders” that have DN mutation by chance, presumably the same random process by which the unaffected siblings acquire DN mutations. The ratio of causal to bystander targets can be estimated from the ascertainment differential, which is based on the excess of the frequency of events in affected children compared with unaffected siblings. Adjusting for recurrently hit genes (Methods), we estimate that 44% of the gene set for lower IQ affected individuals and 30% for higher IQ individuals are causal targets. On the assumption that there are two classes of targets, causal and bystander, and that the tolerance of bystander targets in affected children is the same as for their siblings, we can estimate the relative tolerance of the causal targets in each class. First, we compute a tolerance index for the set as the observed number of UR LGD variants divided by the expected number, under the assumption that the LGD variation is the same as observed in unaffected siblings. Then, using the proportion of causal to bystander targets, we compute the tolerance index for the causal component. The indices are 1.0 for unaffected siblings, by definition, and 0.15 for affected children. By class, we calculate 0.19 for causal target genes in lower IQ affected individuals, 0.17 for recurrent genes, 0.16 over all affected individuals, and 0.35 for higher IQ affected individuals. Thus, causal targets in higher IQ affected individuals seem to be less vulnerable as a class.
Differential Transmission of Alleles from Vulnerable Genes.
Although the SSC is a simplex collection that is enriched for low-risk families in which DN mutation would be a more prominent contributor to the affected state, we estimate that nearly half of simplex families are actually of the high-risk class in which causation by transmission predominates (9). Therefore, we reason that causal transmission might be observed even in this collection. Given that DN targets in affected children have a reduced load of LGD variants, we sought evidence that deleterious alleles of genes with lower LGD variant loads are preferentially transmitted to affected children. We considered 1,866 families for which both affected and unaffected siblings have been whole-exome sequenced (quads) so that we could compare the frequency of transmission only to affected children with the frequency of transmission only to unaffected siblings. We determined significance by permuting “affected” labels. Although we see bias for transmission of all UR LGD variants to affected rather than unaffected children (4,921 vs. 4,813 variants, respectively), it is not significant (P = 0.1398). On the other hand, we see clear significance in transmission (Table 3) for the set of genes with only a single LGD variant (809 vs. 708; P = 0.0101). Genes hit exactly twice by LGD variants show no bias in transmission (Table 3), indicating that virtually all signal comes from the UR LGD variants in genes with the lightest LGD loads. The ascertainment differential of transmission of UR LGD variants in the genes with a single LGD variant is 101, contributing to diagnosis in perhaps 5.4% of families from the SSC.
Table 3.
Transmission patterns for UR LGD mutations from SSC parents
| Set | Gene count | LGD mutations in quads | Transmission pattern | Delta (Aut only − Sib only) | Delta P value | |||
| None | Both | Aut only | Sib only | |||||
| All | 18,455 | 19,602 | 4,671 | 5,197 | 4,921 | 4,813 | 108 | 0.1398 |
| Genes with two UR LGD | 2,624 | 3,602 | 848 | 954 | 900 | 900 | 0 | 0.4981 |
| Genes with one UR LGD | 4,538 | 3,114 | 757 | 840 | 809 | 708 | 101 | 0.0101 |
| Split by IQ | ||||||||
| Lower IQ | 1,586 | 1,586 | 383 | 437 | 418 | 348 | 70 | 0.0071 |
| Higher IQ | 1,528 | 1,528 | 374 | 403 | 391 | 360 | 31 | 0.1489 |
| Split by parent | ||||||||
| Mother | 1,590 | 1,590 | 395 | 415 | 428 | 352 | 76 | 0.0031 |
| Father | 1,524 | 1,524 | 362 | 425 | 381 | 356 | 25 | 0.1890 |
| Split by length | ||||||||
| Long | 1,557 | 1,557 | 384 | 412 | 427 | 334 | 93 | 0.0005 |
| Short | 1,557 | 1,557 | 373 | 428 | 382 | 374 | 8 | 0.4034 |
| Transmission in genes with one UR LGD mutation by functional category | ||||||||
| FMRP | 128 | 128 | 32 | 33 | 38 | 25 | 13 | 0.0654 |
| Chromatin | 62 | 62 | 15 | 18 | 18 | 11 | 7 | 0.1299 |
| Embryonic | 298 | 298 | 59 | 82 | 94 | 63 | 31 | 0.0073 |
| PSD | 245 | 245 | 57 | 64 | 69 | 55 | 14 | 0.1195 |
| Essential | 258 | 258 | 69 | 63 | 75 | 51 | 24 | 0.0248 |
The first row shows transmission patterns for all of the 19,602 UR LGD mutations identified in parents of the 1,866 families for which both affected and unaffected siblings (quads) have been whole-exome sequenced (covering 18,455 genes). The four columns under the heading “Transmission pattern” give the numbers of UR LGD mutations transmitted to “None” of the children, to “Both” children, only to the affected child (“Aut only”), or only to the unaffected child (“Sib only”). We use the difference (delta) between the number of UR LGD mutations transmitted only to affected children and the number of UR LGD mutations transmitted only to unaffected children as a measure of overtransmission to the affected child. We test the significance of the delta against an empirically derived distribution through 10,000 iterations, randomly swapping the affected status of the two children within each family. Although there is a delta of 108 for all LGD mutations, it is not statistically significant (P = 0.1398). We then analyzed the subset of UR LGD mutations that occur in the 4,538 genes with exactly one UR LGD mutation in the SSC parents: 3,114 of these UR LGD mutations are in quads, and the delta in this smaller set of UR LGD mutations is 101; 809 are transmitted only to affected children, whereas 708 are transmitted only to the unaffected child. This delta is almost as large as the delta from all UR LGD mutations, and is significant (P = 0.0101). In contrast, the delta is 0 when we consider the UR LGD mutations in genes with exactly two UR LGD mutations. We then split the UR LGD mutations into roughly two equal halves independently, based on nonverbal IQ of the affected child, on the parent who carried the variant, and on the length of the gene measured as the load of UR synonymous variants. The overtransmission in each half is presented under the “split by IQ,” “split by parent,” and “split by length” subsections of the table. The most extreme difference is observed between long genes (delta = 93, P = 0.0005) and short genes (delta = 8, P = 0.4034), with the majority of overtransmission observed in the long genes. Most of the overtransmission is found in variants carried by the mother (delta = 76) relative to variants carried by the father (delta = 25), and in families with a lower-IQ affected child (delta = 70) relative to those families with a higher-IQ child (delta = 31). The lower section of the table shows the overtransmission of the UR LGD mutations in genes with one UR LGD mutation that are members of five functional classes: FMRP-associated, chromatin modifiers, embryonic, PSD (post synaptic density), and essential (10).
We next examine this transmission signal (to affected only vs. unaffected only) in greater detail, refining its source. We first compare variants in the less tolerant to more tolerant gene class by separating these variants equally in into “longer” and “shorter” genes (using the load of synonymous variants as a surrogate for length). Despite this even split, nearly all the signal in differential transmission comes from the longer genes: the differential is 93 in the longer genes and 8 in the shorter genes. We separately calculate the P value of the signal from both sources, and observe great significance only from the longer genes. We observe differential transmission of 76 from the mother compared with 25 from the father with P = 0.0031 and P = 0.1890, respectively. This result is in line with expectation from theory that mothers should be preferential carriers, as well as from other evidence: for example, that there is a fourfold greater concordance in half siblings sharing the maternal rather than paternal germ-line (17). Almost as striking is the decomposition by affected IQ: A differential of 70 comes from precisely half of the families with an affected child of lower IQ, and 30 come from families with an affected child of higher IQ (P = 0.0071 and P = 0.1489, respectively). This result is in keeping with finding more gene vulnerability in the DN targets of the lower IQ affected individuals than in the targets from the higher IQ affected individuals.
Likelihood of Being an Autism Gene, Given Its Vulnerability.
We can prioritize candidate autism genes by their recurrence as targets of DN mutation, the type of mutation, the ascertainment differential of the affected population, and now the load of disruptive variation. Although we are working with a small database of about 11,000 individuals, and much larger databases would be needed to determine precise tolerance to disruption, there are enough data to rerank ASD target genes as causal targets using tolerance scores. To compare LGD ratios between genes in different classes, such as the class of LGD targets in affected individuals with low IQ or the class of recurrent missense mutations in affected individuals, we compute posterior probabilities using the ascertainment differential for that class as a prior.
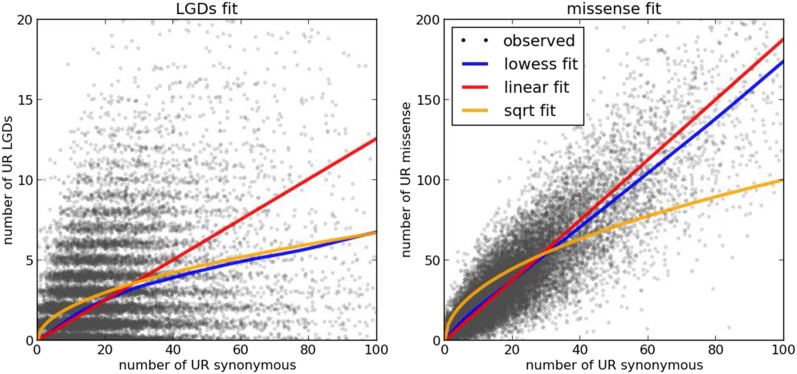
To realize this plan, we need a model of expected LGD ratio per gene. We developed a model of expectation based on synonymous variation, with the reasoning that gene length, coverage, ethnicity, and base composition are likely to be reflected in the load of UR synonymous mutations in a given gene in a given population. The load of UR LGD variants is not linear with the load of UR synonymous variants. The nonlinearity is seen most clearly by fitting the data with a locally weighted scatterplot smoothing (LOWESS) function (Fig. 1A). By contrast, UR missense accumulation proportionately follows UR synonymous accumulation (Fig. 1B).
Fig. 1.
Numbers of UR variants per gene. (Left) Numbers of UR synonymous variants (x axis) and UR LGD variants (y axis) found in the parents of the SSC and the EVS database for each of ∼18,000 protein-coding genes that were successfully captured by whole-exome sequencing. (Right) Similar, with the exception that the y axis represents the number of UR missense variants. Random noise is added to the integer counts for better visibility. In addition to the observed counts, the panels show fits to the number of UR synonymous variants: a linear function fit (red line), L = a * S; a square root fit (yellow line), L = b * sqrt(S); and a nonparametric LOWESS fit (blue line). The LOWESS fit agrees closely with the square root model for the number of UR LGD mutations as a function of the number of UR synonymous variants, but it aligns better with the linear fit for the number of UR missense as a function of UR synonymous variants.
The nonlinearity of the accumulation of LGD variants can be explained by theory. On the simple expectation that negative selection would most often act on the homozygous or compound heterozygous state, the accumulation of UR LGD variants would follow the square root of the gene length, and hence the square root of the number of synonymous mutations at the steady state. The reasoning is that for recessive mutations, which will be the majority, selection acts most strongly when both alleles of a gene are destroyed. So, the rate of elimination of LGD variants in that gene will be proportional to the square of the abundance of the LGD variation in the population. At the steady state, the rate of acquisition equals the rate of elimination, and because acquisition is proportional to the length of the gene and elimination is proportional to the square of abundance, the abundance is proportional to the square root of the length. Even for a dosage-sensitive gene, which will be eliminated through dominance or codominance, the strength of selection may be a function of the number of interactions of its protein, which could increase as the surface area of the protein, and hence as the square of gene length. Indeed, LGD accumulation in a population follows roughly the square root of synonymous accumulation as revealed by the similarity to the LOWESS fit. Because the great majority of missense mutations will be under weak to neutral selection, we expect accumulation of missense will be proportional to synonymous mutation (Fig. 1B). As a practical matter, to develop an expectation of mutational load for individual genes, we use the LOWESS function.
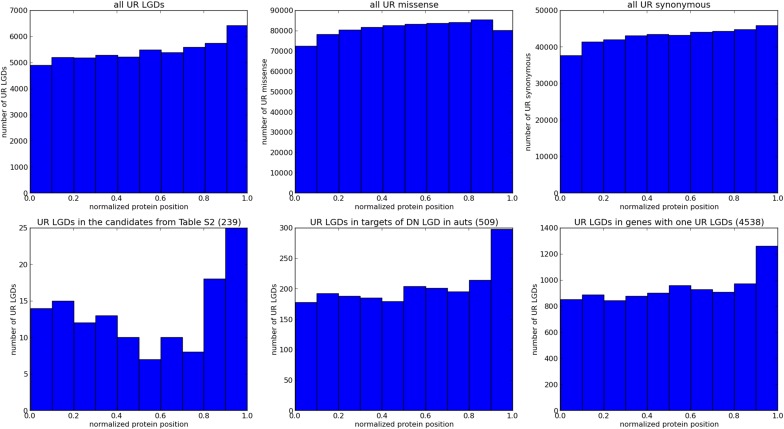
Our heuristic for prioritizing is to develop a discriminant for whether a gene is a vulnerable autism target or a typical gene (Methods). We begin with a prior that a gene is typical or vulnerable based on the expectation that a DN target gene in a class is causal (i.e., based on the ascertainment differential of the class). Then, we compare the observed mutational load for each gene in our database against the prediction based on the LOWESS fit to the load of UR synonymous mutations, obtaining an expected load for a typical gene. We use a Poisson distribution based on that expectation to derive the likelihood for the observed LGD load. We assign the expectation for highly vulnerable genes somewhat arbitrarily as 10% of the LGD expectation for a typical gene, given its synonymous load. It is unreasonable to expect that vulnerable genes are completely devoid of UR LGD mutations, because there can be error in the sequence or reference sequence and certain mutations might not destroy function. For example, we see an abundance of LGD mutations occurring at the end of the coding region of vulnerable genes relative to typical genes (Fig. S1). Moreover, penetrance need not be complete for a variant of strong effect, and so a variant can persist in the population occasionally for a few generations.
Fig. S1.
Amino acid positions. Shown are the normalized amino acid codon positions for six classes of UR variants. The normalized positions are computed as the position of the affected amino acid divided by the length of the encoded protein, using as a reference the isoform that encodes the longest protein. (Top) Histograms of the normalized positions for all UR LGD, missense, and synonymous variants. (Bottom) Histograms of the positions for three subsets of the UR LGD variants: the set of UR LGD variants found in the top 239 genes based on the heuristic posterior score (Dataset S2), the set of UR LGD variants found in the targets of DN LGD mutation in affected autistic (auts) children (509 genes), and the set of UR LGD variants found in genes with exactly one UR LGD variant (4,538 genes). Whereas the normalized amino acid positions of the UR missense and synonymous variants are fairly uniformly distributed across the length of the protein, there is a marked increase in UR LGD variants toward the regions encoding the C-terminal ends of the proteins. This increase is more pronounced for the sets of vulnerable genes shown in the bottom row.
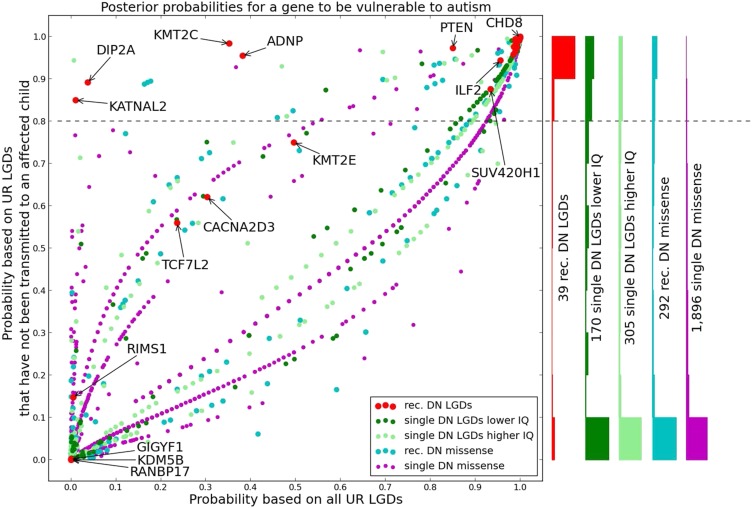
Combining the prior with the likelihood of the observed load of UR LGD mutations under the two models provides a global ranking of DN target genes for the classes of recurrent LGD targets, recurrent missense targets, and LGD targets in lower and higher IQ affected individuals (Fig. 2, x axis). We perform this procedure for both total observed burden of LGD mutations and for the LGD burden adjusted for transmission to an affected individual within the SSC portion of the database (Fig. 2, y axis). After posterior probabilities are calculated, including adjustment for transmission, the scores of target genes in the various classes become bimodal (Fig. 2, Right). This result gives us some assurance that the weight of the modes agrees with the expectation of the proportion of targets within a class that are estimated from the ascertainment differential to be causal. We can take a posterior probability of 0.8 as a good dividing line for the score. The posterior probabilities for all genes can be found in Dataset S1, and the 239 autism candidates with a posterior probability above 0.8 are summarized in Dataset S2.
Fig. 2.
Posterior probabilities for a gene to be a vulnerable autism target. A decreased load of rare LGD mutations is used to prioritize targets of DN LGD mutation. We have different confidence (priors) in targets of DN LGD mutations from affected children, based on the number of recurrent hits as well as nonverbal IQ. The diagram shows each of 2,702 targets of LGD mutation or missense DN mutation in ASD (10, 18); the symbol color and size depend on the prior confidence. The degree of UR LGD depletion is then used to update our confidence (posteriors) for all genes, and these results are displayed on the x axis. We use parents from the SSC, in addition to a collection of individuals from the EVS, to measure the degree of LGD depletion. It is possible that some SSC parents carry UR LGD mutations that have been ascertained because they caused autism in their affected children upon transmission. To address the ascertainment of causative variants in parents, we repeated the posterior computation after removing all UR LGD mutations that have been transmitted to an affected child. Readjusted posteriors are shown on the y axis. (Right) Bar graph represents histograms of the readjusted posteriors (y axis), split by the priors. The dotted line at a score of 0.8 represents an approximate threshold for candidacy as a causal autism gene.
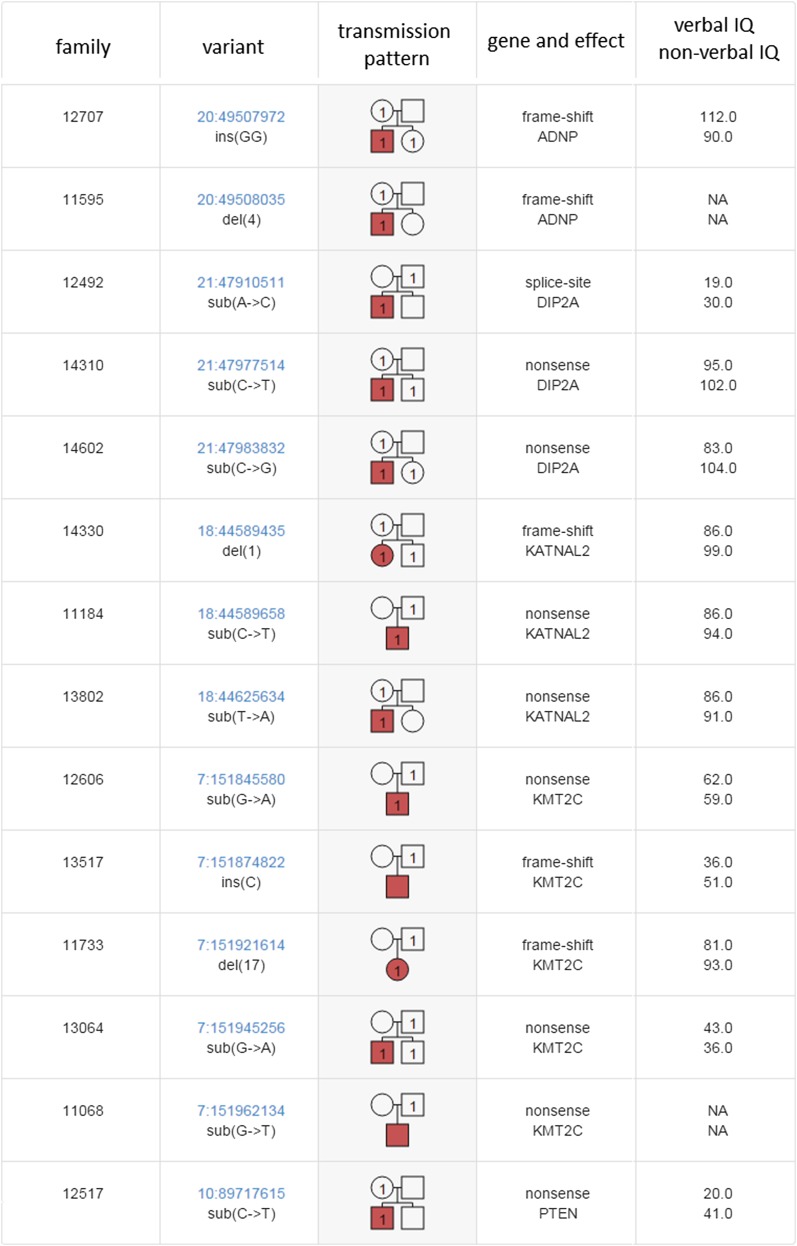
As an example, we consider the class of genes hit by recurrent DN LGD mutation in affected children. There are 39 such targets (10, 18), and we previously estimated that ∼90% of these targets are true causal targets. The majority of these genes, such as ANKRD11, ASH1L, CHD8, GRIN2B, MED13L, and SCN2A, are long but have not accumulated LGD mutations despite expectations. Eleven DN LGD targets are “demoted” by virtue of having LGD variants, including RIMS1 and KDM5B. On the other hand, five of these 12 genes (KMT2C, KATNAL2, ADNP, DIP2A, and PTEN) are “promoted” to be causal, because we observe that LGD variants found in these genes are also transmitted to the affected child. For four of these genes, the transmissions are mainly from the mother, but for one, the histone methyltransferase KMT2C, each of the four transmissions is from the father (Fig. S2). Overall, 32 (>80%) of the original 39 recurrent gene targets remain as excellent causal candidates.
Fig. S2.
Transmission patterns of UR LGD variants in five recurrently hit genes. Shown are the 14 UR LGD variants found in ADNP, DIP2A, KATNAL2, KMT2C, and PTEN, all of which have been targeted by multiple DN LGD mutations and have a decreased load of UR LGD variants after removing variants transmitted to affected children. The table lists the family identification in which the UR LGD variant was identified (family), the description of the genomic variant and its position (variant), and the gender (square for male, circle for female) and affected status (red for affected, white for unaffected) of each family member. Family members who carry the variant are indicated by “1.” In the next column, the targeted gene and the effect on the encoded protein (“gene and effect”) are shown, and the verbal and nonverbal IQs of the affected child are displayed in the final column.
LGD Load and Transmission for Various Gene Sets.
We examined the load for LGD variation more broadly in classes of genes, including those genes that are enriched as DN targets for mutation in affected children (Dataset S1): FMRP-associated genes that specify transcripts bound with the Fragile X mental retardation protein; chromatin-associated genes that encode transcription factors or proteins known to bind chromatin; and embryonic genes that are highly expressed in fetal brain but for which expression is rapidly turned down upon birth (10). All gene classes enriched for autism targets show a decreased load of UR LGD variants, especially the FMRP-associated class, as previously noted (10). In strong contrast, the targets of DN mutation in unaffected siblings have a greater accumulation of LGD variants (Table 4). Finally, we show transmission of the LGD variants within these gene categories for genes with a single LGD variant in the SSC (Table 3). The embryonically expressed category shows a surprising differential transmission of UR LGD variants to affected children over siblings. With 298 opportunities, 31 more are transmitted only to the affected child rather than to the unaffected sibling among quad families (P = 0.0073 by permutation test).
Table 4.
Mutational load of classes of genes
| Set | Gene count | No. of UR synonymous | No. of UR LGD mutations | Expected UR LGD mutations | Class vulnerability | Z-score |
| Rec. DN LGD in aut | 39 | 2,568 | 79 | 181 | 0.4 | 3.2 |
| DN LGD in sib | 173 | 7,372 | 881 | 694 | 1.3 | −2.7 |
| FMRP | 795 | 46,230 | 1,521 | 3,681 | 0.4 | 15.7 |
| Chromatin | 408 | 14,299 | 807 | 1,467 | 0.6 | 8.2 |
| Embryonic | 1,865 | 52,261 | 4,673 | 5,990 | 0.8 | 8.6 |
| PSD | 1,398 | 43,183 | 2,638 | 4,607 | 0.6 | 13.9 |
| Essential | 1,732 | 52,243 | 3,571 | 5,769 | 0.6 | 13.2 |
UR LGD mutational loads are shown for seven classes of genes: the gene targets of recurrent DN LGD mutations in children with autism (Rec. DN LGD in aut), the targets of DN LGD mutations in unaffected siblings (DN LGD in sib), FMRP-associated genes, the genes encoding chromatin modifiers, embryonic genes, the genes encoding postsynaptic density proteins (PSD), and essential genes (10). For each gene class, we list the observed number of UR synonymous and LGD variants, the expected number of UR LGD variants, and class vulnerability (the ratio of the observed to expected UR LGD variants). Expectations are computed with a simple permutation approach that addresses the nonlinear dependence of the UR LGD variants to gene length. We perform 10,000 random permutations in which each gene in the class is replaced with a random gene with the same number of UR synonymous mutations, and in each permutation, we record the number of UR LGD mutations in the randomly selected genes. We take the mean of the random UR LGD class loads as an expected number of UR LGD mutations for the gene class. We then use the SD of the 10,000 random UR LGD class loads to compute a Z-score as the number of SDs separating the observed and expected class loads, with positive Z-scores when the observed is smaller than the expected and negative otherwise. With the exception of the targets of DN mutation in unaffected children, all classes have a significantly decreased UR LGD mutational load.
Discussion
In our previous work, we made the prediction that the target genes of DN mutation that contribute to autism would be highly “vulnerable” genes, in the sense that disruptive mutation in these genes would be of strong effect and have a very high likelihood of causing the disorder. Because individuals with ASD have drastically reduced fecundity (11), the net prediction is that these target genes should be under severe purifying selection, and hence have a reduced load of disruptive variants in the human gene pool. Here, we validate this prediction. Moreover, the proportion of genes with a reduced load matches expectations based on the ascertainment differential in DN mutation frequency between affected and unaffected siblings, with target genes falling into two classes: those genes with reduced mutational load and those genes without (Fig. 2). With a likelihood model based on tolerance for disruption, we obtain a clear bimodal distribution of likelihood among all DN missense and LGD targets in affected individuals (Fig. 2), and we list the 239 most likely causal genes (Dataset S2).
The association of load with causation provides us with a tool by which we can study transmission of causative factors. Although the SSC is composed mainly of simplex families, we have previously calculated that about 40% of families are “high risk,” in which transmission genetics play a strong causative role (9). Indeed, we observe that there is biased transmission of UR disruptive variants within genes with very low load to the affected sibling only, compared with variants transmitted to the typical sibling only (P = 0.01, by permuting labels). The excess comprises about 5.4% of the families, but this estimate is very conservative for a number of reasons. First, we count only genes with LGD variants, yet we expect as much (or more) signal from disruptive missense variants. Second, we do not consider transmission of copy number variants, although they were observed at a similar magnitude in previous studies (3). Third, variants that fall outside the exome are not presently counted. Fourth, we do not count cases of transmission to both affected and unaffected siblings that might be causal because of incomplete penetrance, such as transmission seen when an affected child is male and the unaffected sibling is female. Overall, we estimate causation from transmission to be roughly equal to causation from DN mutation within the SSC (and ASD more generally), in line with an earlier prediction (1). By this theory, UR variants of vulnerable genes would be short-lived in the population, and therefore not detectable by population association studies. Previous theory also predicted a preferred role for the mother in transmission, based largely on the reduced incidence of autism in females. Earlier population studies have indicated the importance of a shared maternal bloodline in sibling risk (19, 20). In support of these reports, we find that the majority of the signal of biased transmission of UR LGD variants in vulnerable genes comes from the maternal line.
Based on this work, we make several additional observations. First, a substantial signal from transmission is seen in the set of “embryonic” genes (10). These genes are strongly expressed during prenatal development but have sharply lower expression upon birth. Although our previous work had shown that embryonic genes were enriched as DN targets (10), their involvement in transmission is greater than we would have predicted. Second, the targets of DN mutation in higher IQ affected individuals show higher mutational load than the targets in affected individuals with lower IQ. There are, of course, exceptions to this general rule (Dataset S2). Moreover, most of the biased transmission of disruptive mutation in vulnerable genes occurs in children of lower IQ. Overall, we can say there is an inverse correlation between phenotypic severity and the tolerance of the genetic target for disruptive mutation. It is worthwhile to speculate on the genetics of higher IQ autism. We know that transmitted and DN events in highly vulnerable genes occur in lower IQ affected children. We do not know if these transmitted and DN events act in combination with other factors; however, for now, we propose that each incidence of a lower IQ ASD is caused mainly by one such event. We refer to such events as “monodromal.” Clearly, more of the higher IQ disorders do not appear to be monodromal, and so must be the result of combinations of more equal genetic factors, or else not genetic. If the former, these variants should accumulate in the child mainly by transmission. This line of reasoning leads us to predict that endophenotypes will be seen more often in both parents of children with higher IQ, whereas in the parents of children with lower IQ, one expects endophenotypes only in one parent, if at all. Interestingly, endophenotypes in high-functioning children are mainly seen in their mothers (21).
That autism candidate genes have a reduced load of damaging mutation has been reported earlier by others using different methods for measuring tolerance of mutation based largely on missense mutation and/or overlapping autism sample sets (13, 14, 16). Our results strengthen this finding with somewhat stronger statistics by using a larger set of target genes divided by severity, by using a larger control population to hone the tolerance score, or both. Moreover, we show a bimodal distribution in mutational load in the proportion predicted by theory for candidate autism genes and bystander target genes. The inverse correlation of phenotypic severity with mutational load is very clear in our study. Recently published findings using the RVIS (16) present evidence for maternal transmission within the SSC of “private” LGD mutations for the lower half of RVIS-tolerant genes, and our result on maternal transmission based purely on genes with UR LGD mutations is very similar (statistically significant with a P = 0.003 for all, and P > 0.0005 for the more vulnerable long genes). Moreover, the signal is mainly seen in severely affected children.
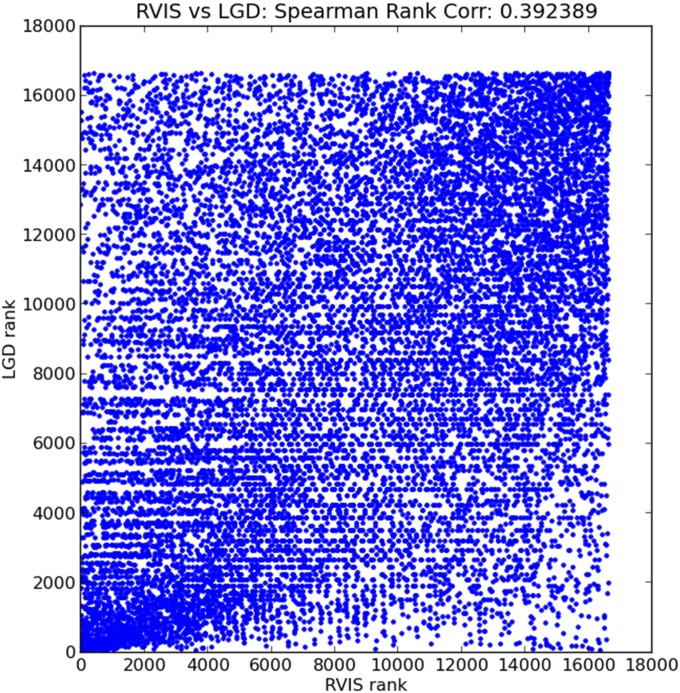
Our method of determining tolerance, based on UR LGD mutations and normalized by UR synonymous mutations, and the RVIS method are strongly but far from perfectly correlated (Fig. S3). Our method differs from the other method by including frame shifts among rare mutations, avoiding computation of load based on missense entirely, and using larger population databases. Counting LGD variants and synonymous variants is simpler computationally, by measuring loads of rare disruptions relative to loads of synonymous mutations. It is easily recomputed as new databases emerge, after appropriate treatment for the population size by repetitive down-sampling. We have relied solely on LGD variants because, unlike missense variants that are hard to interpret, their presence in the interior of a well-annotated gene (Fig. S1) will almost certainly cause disruption of gene function. Moreover, at least in theory, the LGD load should be less dependent on ethnic bias, and to the extent that such a bias was seen at particular genes, it would be of great interest. A direct comparison by gene of the tolerance rankings of the two methods is shown in Dataset S3.
Fig. S3.
Correlation between RVIS and LGD score. We created a per-gene score that reflects the extent to which a gene has a reduced load of UR LGD variants. This score is defined based on the Poisson distribution as the cumulative distribution function of the observed number of UR LGD variants, given the expected number of UR LGD variants, where the expectation is calculated using the LOWESS fit (main text). We ordered the 16,642 genes that had also been assigned an RVIS (13). The figure is a scatterplot of the rank of the UR LGD score (y axis) vs. the rank of RVIS (x axis). The Spearman rank correlation is 0.39.
The power of using gene tolerance for disruptive mutation should be of general value in the analysis of genetic disorders that reduce fecundity. Having a large universally accessible database of human variation, carefully annotated for coverage and ethnicity and searchable per individual genome, should be a community priority. This database should yield a tolerance score that is a property of each gene, and such data could provide ways to measure the contribution of genes to genetic disease more generally. Such databases should not be built “on the cheap” by combining control groups from small studies or borrowing from corporations or countries that keep private databases, but rather as a comprehensive mission specifically designed for this purpose and freely available to all.
Methods
Dataset.
We used the multinomial genotyper to identify transmitted variants from the WES for 2,471 families from the SSC (10). In addition, we used publicly available variants from the Exome Variant Server (evs.gs.washington.edu/EVS/) identified through exome sequencing of ∼6,000 neurotypical individuals.
Proportion of Causal Gene Targets of DN Mutation.
The increase in the rate of DN LGD mutation in affected children compared with the rate in their unaffected siblings was used to estimate the proportion (0.42) of 391 observed DN variants in the affected children that contribute to affected status (10). Ninety percent of the 65 variants that fell within the 27 recurrently hit genes are expected to be causal. Taking this fact into account, we estimate that 36% of the genes hit by one or more DN LGD mutations are causal target genes. Similarly, we computed the proportion of causal genes in the targets of DN LGD mutations in affected children with lower (44%) and higher (30%) nonverbal IQs.
Calculation of Causal Gene Vulnerability.
The DN LGD mutations in the affected children can be split into two classes: those mutations that fall in causal target genes and contributed to the autism diagnosis and others that fall in a bystander target gene and did not contribute to the disorder. The rate of noncontributory DN LGD mutations per affected individual should match the rate of DN LGD mutations in unaffected individuals; moreover, the genes targeted by noncontributory DN LGD mutations in affected and unaffected siblings should have similar loads of UR LGD mutations.
We observe that the targets of all DN LGD mutations in affected children have a decreased load of UR LGD mutations compared with the targets of DN LGD mutations in siblings. To quantify that observation, we compute the expected number of UR LGD mutations assuming that all the DN LGD mutations in affected children are noncontributory (thus the targets of these mutations have the same load as the targets in the unaffected siblings). We then define the class vulnerability (C) as the ratio of observed UR LGD mutations to expected UR LGD mutations. Using class vulnerability, we compute the causal genes vulnerability (V) using a simple model in which we split the targets of DN LGD mutations in affected children into causal target genes and noncausal bystander target genes, with the proportion of causal (R) calculated as described in the above section. V is calculated by solving the equation C = R * V + (1 − R) * 1, where the vulnerability of noncausal genes is set to 1, reflecting the assumption that they have the same load as the targets of DN LGD mutation in unaffected siblings.
Heuristic Prioritization Score.
We use both DN mutations and rare parental LGD mutations to prioritize genes. We developed a simple heuristic gene score based on the naive assumption that the set of autism vulnerable genes and the set of intolerant (protected) genes are the same. We call this gene set autism genes and set the score equal to (A |,), which should read roughly as the probability that a gene is a target autism gene (), given the observed DN data () and rare parental LGD mutations (). Using the Bayes rule, and further assuming that the DN and rare parental data are independent, we can rewrite this conditional probability as:
where is the probability not to be a target (), given . The probability to be a target given only the DN data, , is treated here as a prior and has already been established on a gene class level (10); as discussed above, the probability that a recurrently hit gene is a target is estimated as 0.9, and the probability that a gene with a single DN LGD hit in an affected child of lower IQ is 0.44. We denote this class level probability as . We define , based on the observed () and expected () numbers of UR LGD mutations in the given gene, as:
For simplicity, we denote this probability as , the likelihood assuming the gene is not protected. is computed based on a LOWESS fit of UR synonymous to UR LGD mutations across all genes. To define , we assume that a protected gene has only 10% of the typical share of UR LGD mutations:
and denote this probability as , the likelihood assuming the gene is protected. The score for a gene g is computed as:
Such a score should not be treated as absolute probability for a gene to be an autism gene. It is a heuristic that allows us to prioritize genes and has several useful properties. First, it allows ordering of the genes within a class following the intuition that the protected genes are more likely to be autism genes than unprotected ones. The score splits the genes within a class into two groups of “good” and “not so good” candidates with sizes that match the expectation based on ascertainment differential. Second, it successfully “demotes” some of the recurrently hit genes on the basis of extensive variation within the population. Finally, it allows comparison of targets from different classes such that a protected gene from the class of higher IQ DN hits can compete with the genes in the class of lower IQ DN hits.
Glossary
De novo (DN) means present in the child’s genome and presumed to arise from a mutation in a parental germ line.
Likely gene-disruptive (LGD) is a disruptive mutation, either a frameshift, nonsense, or destroying a consensus splice site.
Vulnerability is a measure of the likelihood that a disruptive mutation in a specified gene produces a specified phenotype.
Load refers to the total amount of a specified type of mutational variation in a gene within the population. If unspecified, the mutational type is assumed to be disruptive or an LGD mutation.
Tolerance is the distinction between genes with high load vs. genes with a low load. In general, but not always, it can be assumed that genes with low tolerance are under negative selective pressure, and are hence vulnerable genes for low fecundity.
Tolerance scores are methods for measuring and ranking load.
Residual variation intolerance score (RVIS) is one method for measuring tolerance.
Ultrarare (UR) refers to variants occurring independently only once in a database. What is UR will depend on the size of the database.
LGD/synonymous variant ratio (or LGD ratio) is the ratio of the load of UR LGD mutations divided by the load of UR synonymous mutations, and is another method of measuring tolerance. It is assumed the database is not above 12,000 persons, or else we obtain the LGD ratio by taking the mean after repetitive down-sampling.
Autism gene is a gene with a nonzero vulnerability for autism.
Gene target or target gene is one hit by mutation, most commonly by a DN LGD mutation in a proband.
Candidate autism gene or candidate target is a gene that is a candidate to be an autism gene as evidenced by being a DN target gene in an affected child.
Bystander target genes are target genes that are imputed not to be autism genes.
Causal target gene is a target gene imputed to be an autism gene.
Ascertainment differential is the difference in the frequency of a specified mutation type in two populations, one ascertained for a specified condition and one a control, which is typically the unaffected sibling in our work.
Supplementary Material
Acknowledgments
We thank all the families at the participating SSC sites, as well as the principal investigators (A. L. Beaudet, R. Bernier, J. Constantino, E. H. Cook, Jr., E. Fombonne, D. Geschwind, D. E. Grice, A. Klin, D. H. Ledbetter, C. Lord, C. L. Martin, D. M. Martin, R. Maxim, J. Miles, O. Ousley, B. Peterson, J. Piggot, C. Saulnier, M. W. State, W. Stone, J. S. Sutcliffe, C. A. Walsh, and E. Wijsman) and the coordinators and staff at the SSC sites for the recruitment and comprehensive assessment of simplex families, and the Simons Foundation Autism Research Initiative (SFARI) staff for facilitating access to the SSC. This work was supported by SFARI Grants SF235988 (to M.W.) and SF362665 (to I.I.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516376112/-/DCSupplemental.
References
- 1.Zhao X, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci USA. 2007;104(31):12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10(2):74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70(5):886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet. 2014;15(2):133–141. doi: 10.1038/nrg3585. [DOI] [PubMed] [Google Scholar]
- 10.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Power RA, et al. Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry. 2013;70(1):22–30. doi: 10.1001/jamapsychiatry.2013.268. [DOI] [PubMed] [Google Scholar]
- 12.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9(8):e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46(9):944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischbach GD, Lord C. The Simons Simplex Collection: A resource for identification of autism genetic risk factors. Neuron. 2010;68(2):192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Krumm N, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47(6):582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risch N, et al. Familial recurrence of autism spectrum disorder: Evaluating genetic and environmental contributions. Am J Psychiatry. 2014;171(11):1206–1213. doi: 10.1176/appi.ajp.2014.13101359. [DOI] [PubMed] [Google Scholar]
- 18.De Rubeis S, et al. DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Constantino JN, et al. Autism recurrence in half siblings: Strong support for genetic mechanisms of transmission in ASD. Mol Psychiatry. 2013;18(2):137–138. doi: 10.1038/mp.2012.9. [DOI] [PubMed] [Google Scholar]
- 20.Sandin S, et al. The familial risk of autism. JAMA. 2014;311(17):1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skuse DH. Rethinking the nature of genetic vulnerability to autistic spectrum disorders. Trends Genet. 2007;23(8):387–395. doi: 10.1016/j.tig.2007.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.