Significance
Translation is a fundamental biochemical process in which ribosomes use an mRNA’s nucleotide sequence as a template to synthesize a protein with a specific amino acid sequence. Errors in this process are deleterious because they can alter a protein’s structure. Yet such errors are surprisingly frequent. Here we ask whether and how evolution can affect the ability of proteins to cope with these errors. In principle, evolution could reduce the rate of such errors, or it could leave this rate unchanged but reduce the damaging effects of errors. We find that populations of proteins evolving in the laboratory pursue the second route, increasing their robustness to translation errors. Evolution may preferentially mitigate damage to a biological system than reduce the source of this damage.
Keywords: molecular evolution, mutational robustness, phenotypic mutations, protein stability, antibiotic resistance
Abstract
How biological systems such as proteins achieve robustness to ubiquitous perturbations is a fundamental biological question. Such perturbations include errors that introduce phenotypic mutations into nascent proteins during the translation of mRNA. These errors are remarkably frequent. They are also costly, because they reduce protein stability and help create toxic misfolded proteins. Adaptive evolution might reduce these costs of protein mistranslation by two principal mechanisms. The first increases the accuracy of translation via synonymous “high fidelity” codons at especially sensitive sites. The second increases the robustness of proteins to phenotypic errors via amino acids that increase protein stability. To study how these mechanisms are exploited by populations evolving in the laboratory, we evolved the antibiotic resistance gene TEM-1 in Escherichia coli hosts with either normal or high rates of mistranslation. We analyzed TEM-1 populations that evolved under relaxed and stringent selection for antibiotic resistance by single molecule real-time sequencing. Under relaxed selection, mistranslating populations reduce mistranslation costs by reducing TEM-1 expression. Under stringent selection, they efficiently purge destabilizing amino acid changes. More importantly, they accumulate stabilizing amino acid changes rather than synonymous changes that increase translational accuracy. In the large populations we study, and on short evolutionary timescales, the path of least resistance in TEM-1 evolution consists of reducing the consequences of translation errors rather than the errors themselves.
Protein synthesis or translation is a key step in genetic information processing. Despite being fundamental to all cellular life, translation is remarkably error prone. Mistranslation events are estimated to occur once per 102–104 codons (1–3). When mRNA is mistranslated, synthesized proteins carry phenotypic mutations in positions where ribosomes incorrectly decoded the mRNA. A pool of proteins with such phenotypic mutations, also called statistical proteins (4), can differ from error-free proteins in sequence, structure, and function.
Comparative genomics studies show that mistranslation can help explain why highly expressed proteins evolve slowly (5–8). All else being equal, highly expressed genes experience more translation events, and thus give rise to a higher number of mistranslated proteins (6, 7). Mistranslation is costly because it can destabilize proteins, and increases proteotoxic stress by promoting protein misfolding and aggregation (2, 9, 10).
Natural selection can reduce mistranslation costs via two non-mutually-exclusive groups of mechanisms. The first increases translational accuracy, i.e., it reduces the rate at which translational errors occur. A global increase in accuracy, for example through hyper-accurate ribosomes, affects all proteins but comes with high energetic and kinetic costs (11). Alternatively, translational accuracy can increase locally at amino acid sites where (phenotypic) mutations would cause the largest fitness defects (12). This increase is possible through synonymous mutations toward codons with a low propensity for mistranslation (1, 13, 14).
A second group of mechanisms does not reduce mistranslation itself, but mitigates its deleterious consequences, and thus increases the translational robustness of proteins. Some error-mitigation mechanisms are global and affect many proteins. They include chaperones that can help proteins fold even if they harbor destabilizing mutations (15). In contrast to such global mechanisms, which can become overwhelmed when mistranslation rates are high, local mechanisms are less costly. They rely on (genetic) mutations called suppressors, which increase the stability of a single protein, and thus buffer destabilizing effects of other (phenotypic) mutations (16–18).
The only experimental study of protein evolution under phenotypic mutations focused on errors in transcription (19). It could not answer whether proteins evolve to reduce the mutational load of mistranslation by increasing their translational accuracy, or by mitigating the effect of errors resulting from such accuracy. Here, we address this question by evolving genes encoding the antibiotic resistance protein TEM-1 β-lactamase in strains of E. coli with different rates of mistranslation. We also ask whether relaxed and stringent selection for antibiotic resistance affect the adaptation to elevated mistranslation in different ways. Under relaxed selection, mistranslating populations adapt by reducing TEM-1 expression through inefficient initiation codons, which lowers the cost of mistranslation. Under stringent selection, where reducing gene expression would be detrimental, populations increase translational robustness by accumulating stabilizing and purging destabilizing single nucleotide polymorphisms (SNPs).
Results
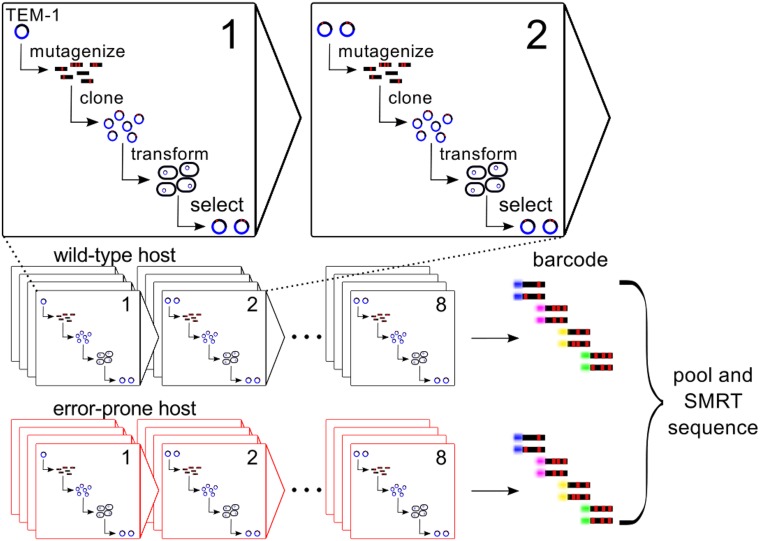
To study how proteins adapt to mistranslation in evolving laboratory populations, we experimentally evolved TEM-1 β-lactamase independently in two E. coli strains, the wild type, and the mistranslating, or error-prone rpsD12 strain (Fig. 1). The error-prone strain carries a mutation in the ribosomal protein S4, which results in increased missense, readthrough, and frameshift (phenotypic) mutations during protein synthesis (1). Specifically, we evolved TEM-1 in populations of 108–109 individuals for eight cycles of PCR-based mutagenesis and selection (Fig. 1), with four replicate populations for each of the two host strains and each of the two selection conditions (relaxed: 25 μg mL−1 ampicillin; stringent: 250 μg mL−1 ampicillin). We also evolved two replicate control populations per strain. These populations were mutagenized in the same way as evolved populations, but experienced no selection for β-lactamase activity. We sequenced more than 500 evolved molecules per population using single molecule real-time (SMRT) sequencing (20) (SI Appendix, Table S1).
Fig. 1.
Experimental evolution of TEM-1 under mistranslation. In each round of evolution, we subjected TEM-1 to mutagenic PCR and recloned the resulting mutant alleles into a fresh plasmids backbone, thus ensuring that only the TEM-1 evolves in our experiments. We transformed plasmids with mutagenized TEM-1 into host cells (wild type or error prone), and exerted relaxed and stringent selection for antibiotic resistance by growing 108–109 transformed hosts in liquid LB media with ampicillin (25 or 250 μg mL−1, respectively). Subsequently, we isolated plasmids and used them as templates for the next round of evolution. We evolved 4 replicate populations per host and per selection regime, for a total of 16 populations. After eight cycles of evolution, we subjected evolved TEM-1 populations to SMRT sequencing.
From the sequenced library of ancestral TEM-1 (395 sequences), we estimated the compound sequencing and variant calling error rate as per nucleotide. In control libraries (4,567 variants), we observed an average of 0.73 mutations per variant, implying a mutation rate of per nucleotide. Consistent with reports from previous studies (21, 22), our mutagenesis protocol is AG and TC biased (SI Appendix, Table S2).
Mistranslation Slows down TEM-1 Evolution.
The TEM-1 protein has two parts: the N-terminal signal peptide (the first 25 residues in Ambler numbering; ref. 23), and the mature enzyme. The signal peptide guides the translocation of TEM-1 to the periplasmic space. Once translocation is complete, the signal sequence is cleaved and the mature TEM-1 folds into its active conformation. The signal peptide controls the expression, and the localization of TEM-1. We observe that SNPs in the signal peptide have frequencies up to (SI Appendix, Fig. S1 and Table S7). In contrast, SNPs in the mature part of TEM-1 all have frequencies below 5% (SI Appendix, Fig. S1 and Tables S5 and S6).
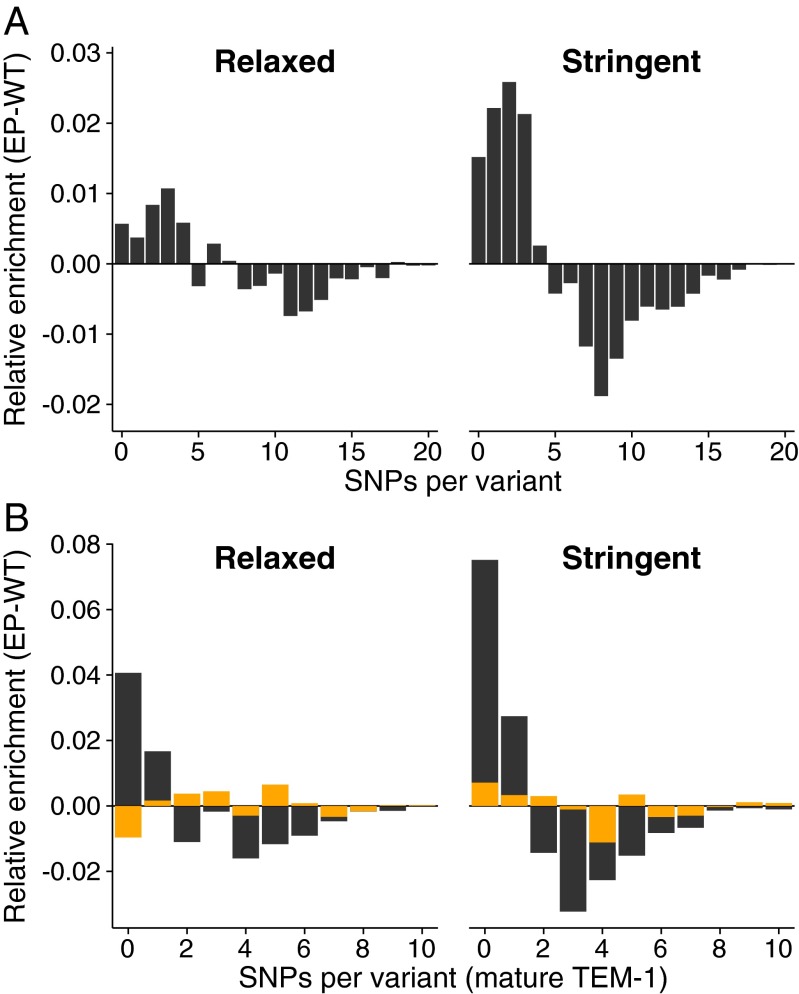
Next, we compared the average number of SNPs per TEM-1 variant among the two strains. Wild-type populations have a higher relative frequency of variants with more SNPs than error-prone populations (Fig. 2A), and selection makes this difference more pronounced. Specifically, under relaxed selection, wild-type and error-prone populations have different estimated means of 5.41 and 5.08 SNPs per variant, respectively [estimates and comparisons are based on general linear models (GLMs) with a Wald test of the corresponding estimate, ]. Under stringent selection the estimated mean number of SNPs per variant was also different between wild-type (5.28) and error-prone (4.59) populations (Wald test, GLM, ).
Fig. 2.
SNP enrichment in evolved populations for relaxed and stringent selection. (A) The relative enrichment of variants with a given number of SNPs (horizontal axis) for the whole protein, calculated by subtracting the frequency (SI Appendix, Fig. S2) of variants with this number of SNPs in the wild-type host from that in the error-prone host. The data show that error-prone strains have more variants with fewer SNPs, and fewer variants with more SNPs, relative to the wild type. For this analysis, we binned variants according to the number of observed SNPs, and calculated the frequency of alleles in a bin relative to the pooled data from all replicates for a given host. (B) As in A, but only for synonymous (orange) and nonsynonymous (black) SNPs in the mature part of TEM-1.
The accumulation of nonsynonymous SNPs in mature TEM-1 proteins (Fig. 2B, black) shows that error-prone populations are under stronger purifying selection. Specifically, under relaxed selection, error-prone populations accumulated significantly fewer nonsynonymous SNPs per variant than wild-type populations (1.76 vs. 2.01 nonsynonymous SNPs per variant; Wald test, GLM, ). Under stringent selection, this difference became even more pronounced (1.51 vs. 1.90 nonsynonymous SNPs per variant; Wald test, GLM, ). In addition, the ratio of nonsynonymous to synonymous SNPs was significantly lower in error-prone lines, indicating stronger purifying selection under both selection regimes (relaxed selection: 0.83 vs. 0.95 for mistranslating and for wild-type populations, respectively; Wald test, GLM, ; stringent selection: 0.71 vs. 0.87; Wald test, GLM, ). Further evidence for stronger selection under mistranslation comes from the distribution of fitness effects of nonsynonymous SNPs, where error-prone populations are depleted of deleterious, and enriched in neutral and beneficial nonsynonymous SNPs (SI Appendix, Fig. S4).
As opposed to these indicators of purifying selection, the mean number of synonymous SNPs per variant did not differ significantly between error-prone and wild-type populations (Fig. 2B in orange; relaxed selection: 2.12 vs. 2.14 syn. SNPs, Wald test, GLM, ; stringent selection: 2.18 vs. 2.13, Wald test, GLM, ). Taken together, our observations show that the nonsynonynmous SNPs we observe are subject to purifying selection associated with error-prone translation. In contrast, synonymous SNPs accumulate neutrally with respect to mistranslation, contrary to what one would expect if there was strong selection for increased translational accuracy through high-fidelity synonymous SNPs in our experimental system.
TEM-1 Adapts to Mistranslation through Increased Stability and Changes in Expression.
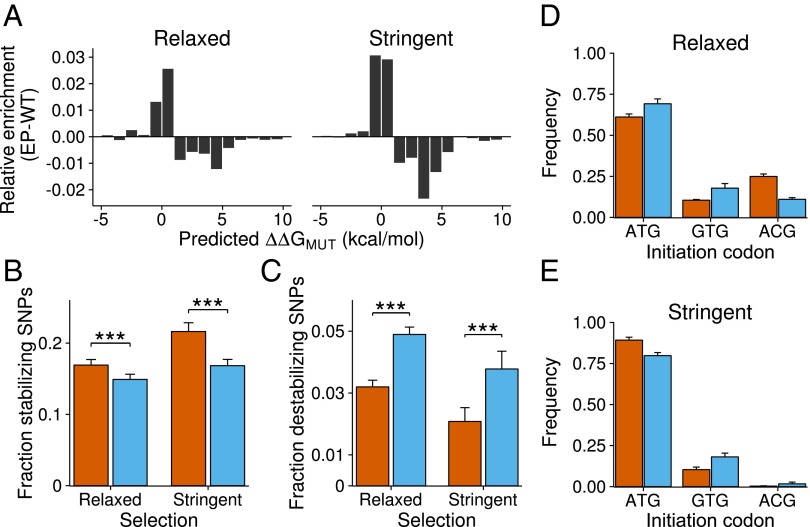
To see how mistranslation affects the robustness of evolved proteins, we first predicted the stability effects of observed SNPs using FoldX (24). FoldX can compute the thermodynamic impact of a mutation, expressed as G, where a mutation with G < 0 is stabilizing. Error-prone populations accumulated more stabilizing SNPs and fewer destabilizing SNPs, a difference that is once again more pronounced under stringent selection (Fig. 3A; Wilcoxon rank-sum test, two-sided for both relaxed and stringent selection).
Fig. 3.
Stability and expression changes in evolved populations. (A) Thermodynamic impact of enriched SNPs as predicted by FoldX (24). SNPs are binned according to G. Positive values of enrichment (vertical axis) correspond to a larger number of mutations with a given G in error-prone populations. (B) Fraction of experimentally validated stabilizing SNPs among all nonsynonymous SNPs. (C) Fraction of validated destabilizing SNPs among all nonsynonymous SNPs. (D) Frequencies of sequences with each of three initiation codons (horizontal axis) under relaxed selection. (E) Like D, but for stringent selection. Only codons that appear with a frequency greater than 5% in at least one of the populations are included in D and E. Error bars represent SDs across four replicate populations.
To validate computational predictions of FoldX, we compiled a list of mutations known from experiment to either increase or decrease the stability of TEM-1 (16–18, 22, 25–32). TEM-1 in error-prone populations accumulated significantly more stabilizing SNPs than in wild-type populations, and it did so for both selection regimes (Fig. 3B; relaxed selection: 17.1% vs. 14.8%, Wald test, GLM, ; stringent selection: 21.6% vs. 16.7%, Wald test, GLM, ). At the same time, error-prone populations accumulated fewer destabilizing SNPs compared with wild-type populations (Fig. 3C; relaxed selection: 3.2% for error-prone, 4.8% for wild-type, Wald test, GLM, ; stringent selection: 2.1% for error-prone populations, 3.9% for wild-type populations, Wald test, GLM, ). Furthermore, the well-known stabilizing mutation M182T is the nonsynonymous SNP with the highest frequency in two populations under mistranslation and stringent selection (SI Appendix, Table S5). No significant differences exist in the accumulation of synonymous SNPs at sites where mutations are known to affect stability (SI Appendix, Fig. S5).
SNPs found at the initiation codon (within the signaling peptide), have the highest frequencies in our dataset (Fig. 3 D and E; SI Appendix, Table S7). Under relaxed selection, sequences evolved in error-prone hosts are more likely to have non-ATG initiation codons (Fig. 3D, 38.6% vs. 31.1%, Wald test, GLM, ), which reduce efficiency of translation initiation (33, 34). In contrast, under stringent selection, sequences evolved in wild-type hosts are more likely to have non-ATG initiation codons (Fig. 3E; 20.0% vs. 10.7%, Wald test, GLM, ).
Mistranslating Populations Accumulate Nonsynonymous SNPs in Surface Residues.
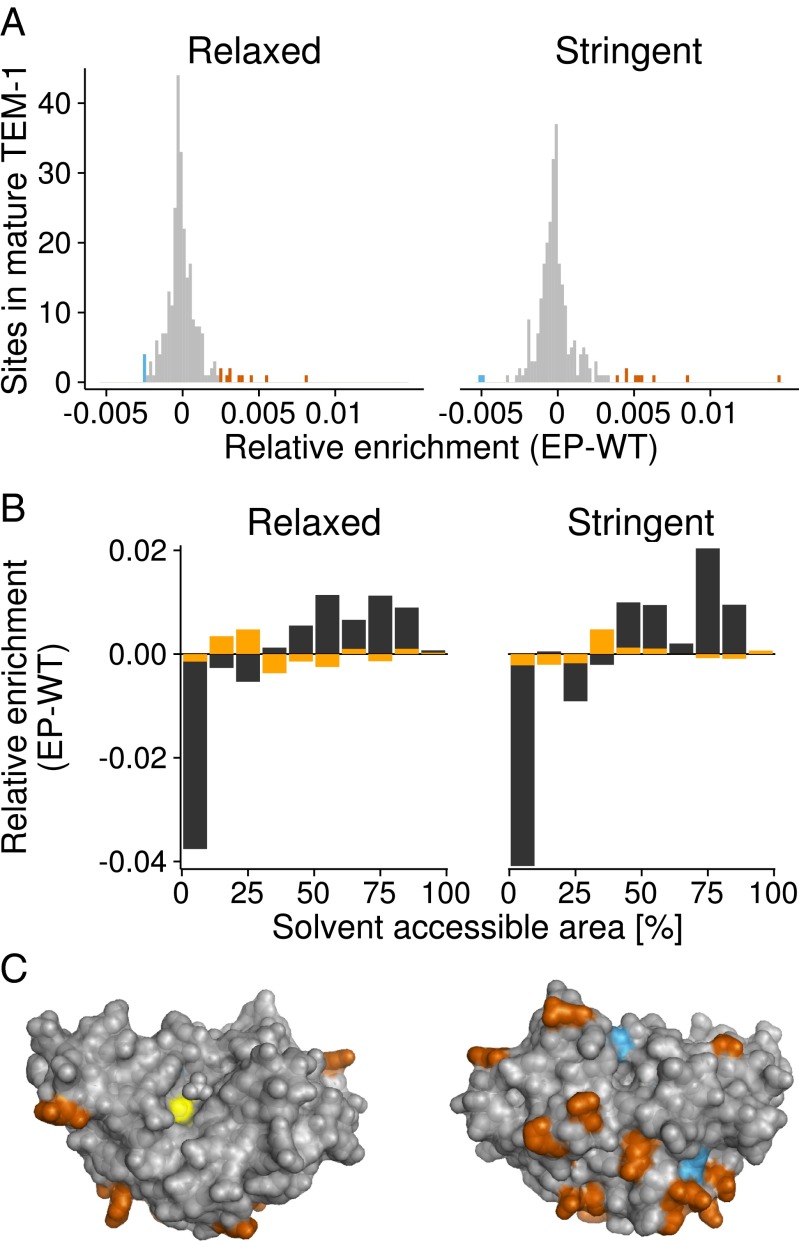
We next examined where in the TEM-1 tertiary structure SNPs accumulated during evolution (Fig. 4). In general, mutations that affect a protein’s core tend to be more destabilizing than mutations of surface residues (35). Core residues are buried and have a low solvent-accessible surface area (SASA), and surface residues have high SASA. We computed SASA for each of the residues affected by SNPs, and found that residues with higher SASA tend to be enriched in nonsynonymous SNPs in error-prone populations, relative to wild-type populations (relaxed selection: Wilcoxon rank-sum test, two-sided ; stringent selection: Wilcoxon rank-sum test, two-sided ; Fig. 4 B in black and C). In contrast, distributions of SASA for residues harboring synonymous SNPs did not differ between error-prone and wild-type strains (relaxed selection: Wilcoxon rank-sum test, two-sided ; stringent selection: Wilcoxon rank-sum test, two-sided ; Fig. 4B in orange).
Fig. 4.
Accumulating SNPs and where they occur on the TEM-1 structure. (A) Histogram of sites that harbor host-specific enriched nonsynonymous SNPs. Strongly enriched bins (more than two SD) are shown in red (error prone) and blue (wild type). (B) Relative enrichment of synonymous (orange) and nonsynonymous (black) SNPs in areas with a given solvent accessibility (horizontal axis). (C) TEM-1 structure with the sites of polymorphic nonsynonymous SNPs colored as in A according to their relative enrichment. The active site is shown in yellow. Left and right structures are rotated horizontally by 180 degrees relative to each other.
Discussion
We find first that mistranslation can indeed affect the rate of protein evolution. Specifically, error-prone populations accumulate fewer nonsynonymous changes, and show a lower ratio of nonsynonymous to synonymous changes. This pattern of evolution is readily explained through the observation that most nonsynonymous mutations destabilize proteins (35, 36). In populations subject to high rates of mistranslation, proteins harboring such destabilizing mutations suffer additional destabilizing effects from phenotypic mutations, and thus become even more destabilized. In such populations, a greater fraction of nonsynonymous mutations should thus get eliminated by natural selection, just as we observed. Also consistent with our observation is that those nonsynonymous changes that survive high rates of mistranslation occur preferentially on the TEM-1 surface (Fig. 4 B and C) where they are less likely to be destabilizing (35).
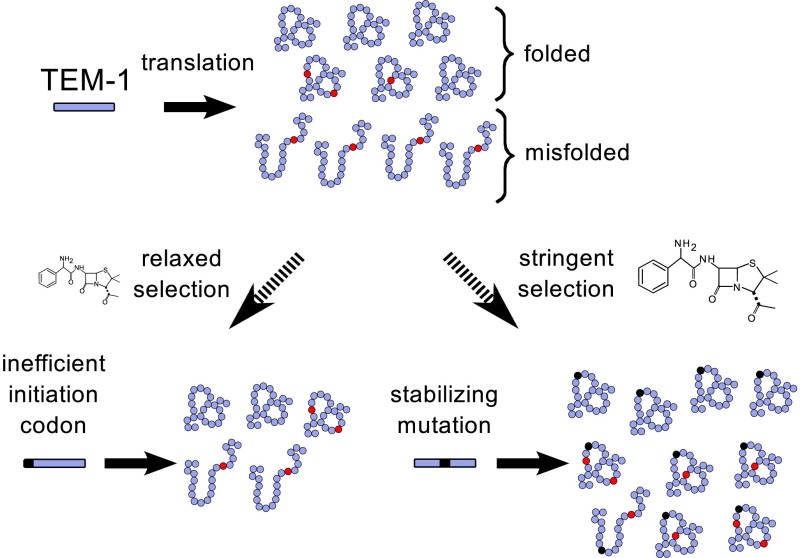
Second, several lines of evidence show that cells adapt to mistranslation by reducing TEM-1 mistranslation costs (Fig. 5). They do so with two different strategies, depending on whether selection for antibiotic resistance is relaxed or stringent. Under relaxed selection (low ampicillin concentrations), error-prone populations reduce TEM-1 expression by adopting inefficient non-ATG initiation codons, which reduce the cost of misfolding by reducing TEM-1 expression (Fig. 5). Changes in initiation codons have the highest frequency among all SNPs we observed (Fig. 3 D and E; SI Appendix, Table S7). Such changes are known to reduce TEM-1 expression and with it the cost of mistranslation. Specifically, the GTG initiation codon, which is used in about 14% of E. coli genes (37) has a 1.5–3 times lower initiation efficiency than ATG (33). Similarly, ACG can serve as an initiation codon (37), but its initiation efficiency is only 1–3% of that of ATG (34). In addition, both GTG and ACG initiation codons are frequently observed in comprehensive TEM-1 mutagenesis libraries selected at low levels of ampicillin (38). Reducing the concentration of TEM-1 is a simple strategy to mitigate mistranslation costs, but it is only viable where amounts of ampicillin are so low that TEM-1 expression can be reduced without adverse effects.
Fig. 5.
A model for the evolution of translational robustness under error-prone translation. Phenotypic mutations are shown in red, genotypic mutations in black. The strength of selection affects adaptation. Under relaxed selection (low ampicillin concentration), only small amounts of β-lactamase are needed, and populations reduce the cost of mistranslation by reducing expression. Under stringent selection (high ampicillin concentration), larger amounts are needed, and populations reduce the cost of misfolding by accumulating stabilizing changes and purging destabilizing changes.
In contrast, under stringent selection (high antibiotic concentration), a high concentration of active TEM-1 is needed to degrade ampicillin. In this condition, mistranslating lines, where a fraction of functional TEM-1 is already lost to mistranslation, should not be able to reduce TEM-1 expression much further. This prediction is borne out by our observation that error-prone lines are more likely to use the standard ATG initiation codon under stringent selection. A related observation was made in experiments with elevated mistranscription rates (19), where transcription with error-prone RNA polymerase reduces the effective expression of TEM-1, and populations with increased mistranscription adapted to higher concentrations of ampicillin by increasing TEM-1 expression.
Error-prone populations subject to stringent selection (high ampicillin concentration) mitigate the effects of mistranslation not by reducing TEM-1 expression, but by accumulating stabilizing and depleting destabilizing mutations in TEM-1 (Fig. 3 A and B). Remarkably, the change with the highest frequency in our mistranslating populations is the well-known M182T substitution (SI Appendix, Table S5). Frequently observed in natural TEM-1 isolates and in laboratory evolution experiments (30), M182T increases the stability of TEM-1, making it more robust to genetic mutation and denaturation (36). In addition, error-prone populations are impoverished relative to wild type in predicted and known destabilizing SNPs in TEM-1 (Fig. 3 A and C). Furthermore, they have especially few nonsynonymous SNPs in the TEM-1 core, where amino acid changes would be strongly destabilizing (Fig. 4). In other words, mistranslation causes efficient purging of destabilizing mutations.
In sum, we find that under laboratory conditions evolving proteins adapt to mistranslation by mitigating the damage it causes. Our observations are consistent with previous experiments showing that increased mistranscription rates (19) can lead to increases in protein stability (32).
Our third observation pertains to whether evolution alters the robustness or the accuracy of translation. Specifically, does TEM-1 evolve increased translational accuracy, which can occur by synonymous changes toward high-fidelity codons? Our observations suggest that, at least in our experimental system, the answer is no. First, synonymous SNPs do not generally accumulate at a higher rate in error-prone populations. Second, the incidence of synonymous SNPs at sites where mutations are known to have stabilizing effects is not greater in error-prone populations. Third, the incidence of synonymous SNPs at sites where mutations have destabilizing effects is not lower in error-prone populations. Finally, the incidence of synonymous SNPs in codons adjacent to those with known stability effects does not differ between error-prone and wild-type populations (SI Appendix, Fig. S5). These findings are consistent with theoretical predictions that adaptation to mistranslation may predominantly occur through increased translational robustness, because robustness provides bigger benefits and is thus easier to evolve (7, 8). They are also rendered plausible by two further observations. First, the number of known stabilizing and destabilizing amino acid changes is large (SI Appendix, Supplementary Methods S3.13), which implies that evolutionary modulation of protein stability is easily achieved. Second, once a stabilizing SNP reduces the destabilizing effects of mistranslation, further selection for high fidelity synonymous SNPs will be less effective (39).
The conditions of our experimental evolution differ from those experienced by many natural populations. For example, our experimental design imposed strong selection (high antibiotic concentrations), a high mutation rate, and large populations, as well as few () cell generations. The last condition is especially important, because the evolution of synonymous changes may require many more generations (40). These differences may help explain why selection for translational accuracy can be effective in some natural populations, even though it was not effective in our experiments (7, 41–43).
Error-prone protein translation has occurred since life’s earliest days (4), and it has contributed to the evolution of a robust genetic code (44). Our observations demonstrate that it can still influence the structure of modern proteins. The path of least evolutionary resistance in laboratory-evolved TEM-1 reduces the consequences of errors rather than the errors themselves.
Materials and Methods
For detailed description of experimental procedures, see SI Appendix.
Strains and Plasmids.
The wild-type and the error-prone hosts were derived from E. coli strain MG1655, and were isogenic except for the rpsD allele. That is, the error-prone host carried an rpsD12 allele (45), and the wild-type had a normal rpsD allele. We used the high-copy number plasmid pHS13T (SI Appendix, Fig. S6B), derived from pHSG396 (46), which carries a chloramphenicol resistance marker, as the vector for TEM-1 evolution.
Directed Evolution.
To mutagenize the TEM-1 population, we used error-prone PCR with nucleoside analogs (21). After PCR, we digested, purified, and ligated the mutagenized TEM-1 sequences into fresh plasmid backbones. Subsequently, we transformed the ligation product into electrocompetent DH5α cells to ensure plasmid methylation, which resulted in library sizes between 105 and 106 sequences. After recovering the transformed cells, we grew them overnight in LB media supplemented with 34 μg/mL of chloramphenicol, purified plasmids (preselection libraries) from these overnight cultures, and transformed these libraries into electrocompetent rpsD12 or wt cells (library sizes: 108–109 sequences). We allowed recovered transformants to grow for approximately six generations in LB media supplemented with 34 μg/mL chloramphenicol (for plasmid maintenance), as well as either 25 or 250 μg/mL of ampicillin, for relaxed and stringent selection regime, respectively. After purifying plasmids from the resulting postselection libraries, we used them as templates in the next round of evolution, as well as for single molecule real-time sequencing.
Primary Data Analysis.
We assembled consensus reads (referred to as variants in the manuscript) of TEM-1 sequences from subreads (13.5 passes per consensus TEM-1 variant) with the SMRTAnalysis v2.3 package. We mapped reads to the reference (ancestral) TEM-1 sequence using BLASR (47), and filtered mapped reads that spanned the entire TEM-1 coding region to an average Phred quality above 20. We considered a mismatch of a TEM-1 variant sequence to the TEM-1 reference sequence a true SNP only if its Phred quality score was above 20.
Statistical Methods.
Unless specified otherwise, we used GLMs (48) to compare the four groups given by the selection regimes (relaxed, stringent) and the host strains (wild-type, error-prone). We report the estimated means of quasi-Poisson models for count data (number of SNPs) and estimated proportions (such as dN/dS ratios) of quasi-binomial models. For comparisons involving GLMs, we indicate the z value of the corresponding Wald test statistic and the corresponding P value, which we adjusted for multiple testing with the Holm–Bonferroni procedure. We took the grouping of the data in four replicate populations into account via an extension to generalized linear mixed models (49). However, based on model diagnostics, we decided to report the estimates of the GLMs. Further details on the statistical methods are given in SI Appendix, Supplementary Methods S3.12.
Supplementary Material
Acknowledgments
We thank M. O’Connor for advice on recombineering, M. Ackermann and L. Keller for helpful discussions, and the Functional Genomics Center Zurich for generating the sequence data. A.W. acknowledges support through Swiss National Science Foundation Grant 31003A-146137, as well as through the University Priority Research Program in Evolutionary Biology at the University of Zurich. F.G. acknowledges the University of Zurich Research Priority Program on Global Change and Biodiversity.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences reported in this paper have been deposited in the GenBank database (accession codes KT391064–KT423096).
See Commentary on page 12553.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510071112/-/DCSupplemental.
References
- 1.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13(1):87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10(10):715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyerovich M, Mamou G, Ben-Yehuda S. Visualizing high error levels during gene expression in living bacterial cells. Proc Natl Acad Sci USA. 2010;107(25):11543–11548. doi: 10.1073/pnas.0912989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woese CR. On the evolution of the genetic code. Proc Natl Acad Sci USA. 1965;54(6):1546–1552. doi: 10.1073/pnas.54.6.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158(2):927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA. 2005;102(40):14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134(2):341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilke CO, Drummond DA. Population genetics of translational robustness. Genetics. 2006;173(1):473–481. doi: 10.1534/genetics.105.051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiler-Samerotte KA, et al. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci USA. 2011;108(2):680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro S, Villar-Piqué A, Ventura S. Selection against toxic aggregation-prone protein sequences in bacteria. Biochim Biophys Acta. 2014;1843(5):866–874. doi: 10.1016/j.bbamcr.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 11.Ruusala T, Andersson D, Ehrenberg M, Kurland CG. Hyper-accurate ribosomes inhibit growth. EMBO J. 1984;3(11):2575–2580. doi: 10.1002/j.1460-2075.1984.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou T, Weems M, Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol. 2009;26(7):1571–1580. doi: 10.1093/molbev/msp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Precup J, Parker J. Missense misreading of asparagine codons as a function of codon identity and context. J Biol Chem. 1987;262(23):11351–11355. [PubMed] [Google Scholar]
- 14.Manickam N, Nag N, Abbasi A, Patel K, Farabaugh PJ. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014;20(1):9–15. doi: 10.1261/rna.039792.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459(7247):668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 16.Huang W, Palzkill T. A natural polymorphism in beta-lactamase is a global suppressor. Proc Natl Acad Sci USA. 1997;94(16):8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marciano DC, et al. Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase. J Mol Biol. 2008;384(1):151–164. doi: 10.1016/j.jmb.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown NG, Pennington JM, Huang W, Ayvaz T, Palzkill T. Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM β-lactamases. J Mol Biol. 2010;404(5):832–846. doi: 10.1016/j.jmb.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldsmith M, Tawfik DS. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc Natl Acad Sci USA. 2009;106(15):6197–6202. doi: 10.1073/pnas.0809506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 21.Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J Mol Biol. 1996;255(4):589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 22.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444(7121):929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 23.Ambler R, Coulson A. 1991. A standard numbering scheme for the class A β-lactamases. Biochem J 276(1):269–272.
- 24.Schymkowitz J, et al. The FoldX web server: An online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382-8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raquet X, et al. Stability of TEM beta-lactamase mutants hydrolyzing third generation cephalosporins. Proteins. 1995;23(1):63–72. doi: 10.1002/prot.340230108. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320(1):85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 27.Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23(1):160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kather I, Jakob RP, Dobbek H, Schmid FX. Increased folding stability of TEM-1 beta-lactamase by in vitro selection. J Mol Biol. 2008;383(1):238–251. doi: 10.1016/j.jmb.2008.07.082. [DOI] [PubMed] [Google Scholar]
- 29.Guthrie VB, Allen J, Camps M, Karchin R. Network models of TEM β-lactamase mutations coevolving under antibiotic selection show modular structure and anticipate evolutionary trajectories. PLOS Comput Biol. 2011;7(9):e1002184. doi: 10.1371/journal.pcbi.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salverda MLM, De Visser JAG, Barlow M. Natural evolution of TEM-1 β-lactamase: Experimental reconstruction and clinical relevance. FEMS Microbiol Rev. 2010;34(6):1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- 31.Abriata LA, Salverda ML, Tomatis PE. Sequence-function-stability relationships in proteins from datasets of functionally annotated variants: The case of TEM β-lactamases. FEBS Lett. 2012;586(19):3330–3335. doi: 10.1016/j.febslet.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol. 2008;379(5):1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Reddy P, Peterkofsky A, McKenney K. Translational efficiency of the Escherichia coli adenylate cyclase gene: Mutating the UUG initiation codon to GUG or AUG results in increased gene expression. Proc Natl Acad Sci USA. 1985;82(17):5656–5660. doi: 10.1073/pnas.82.17.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sussman JK, Simons EL, Simons RW. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol Microbiol. 1996;21(2):347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 35.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369(5):1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 36.Bloom JD, et al. Thermodynamic prediction of protein neutrality. Proc Natl Acad Sci USA. 2005;102(3):606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 38.Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol Biol Evol. 2014;31(6):1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajon E, Masel J. Evolution of molecular error rates and the consequences for evolvability. Proc Natl Acad Sci USA. 2011;108(3):1082–1087. doi: 10.1073/pnas.1012918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bull JJ, Molineux IJ, Wilke CO. Slow fitness recovery in a codon-modified viral genome. Mol Biol Evol. 2012;29(10):2997–3004. doi: 10.1093/molbev/mss119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akashi H. Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics. 1994;136(3):927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porceddu A, Zenoni S, Camiolo S. The signatures of selection for translational accuracy in plant genes. Genome Biol Evol. 2013;5(6):1117–1126. doi: 10.1093/gbe/evt078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaborske JM, et al. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 2014;12(12):e1002015. doi: 10.1371/journal.pbio.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freeland SJ, Hurst LD. The genetic code is one in a million. J Mol Evol. 1998;47(3):238–248. doi: 10.1007/pl00006381. [DOI] [PubMed] [Google Scholar]
- 45.Ballesteros M, Fredriksson A, Henriksson J, Nyström T. Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001;20(18):5280–5289. doi: 10.1093/emboj/20.18.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 47.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelder JA, Wedderburn RWM. Generalized Linear Models. J R Stat Soc [Ser A] 1972;135(3):370–384. [Google Scholar]
- 49.Bolker BM, et al. Generalized linear mixed models: A practical guide for ecology and evolution. Trends Ecol Evol. 2009;24(3):127–135. doi: 10.1016/j.tree.2008.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.