Abstract
During apoptosis Bid and Bax are sufficient for mitochondrial outer membrane permeabilization, releasing pro-apoptotic proteins such as cytochrome c and Smac/Diablo into the cytoplasm. In most cells, both Bid and Bax are cytoplasmic but bind to mitochondrial outer membranes to exert pro-apoptotic functions. Binding to membranes is regulated by cleavage of Bid to truncated Bid (tBid), by conformation changes in tBid and Bax, and by interactions with other proteins. At least at the peripherally bound stage, binding is reversible. Therefore, regulation of apoptosis is closely linked with the interactions of tBid and Bax with mitochondria. Here we use fluorescence techniques and cell-free systems containing mitochondria or liposomes that faithfully mimic tBid/Bax-dependent membrane permeabilization to study the dynamic interactions of the proteins with membranes. We confirm that the binding of both proteins to the membrane is reversible by quantifying the binding affinity of proteins for the membrane. For Bax, both peripherally bound (inactive) and oligomerized (active) proteins migrate between membranes but much slower than and independent of tBid. When re-localized to a new membrane, Bax inserts into and permeabilizes it only if primed by an activator. In the case of tBid, the process of transfer is synergetic with Bax in the sense that tBid ‘runs' faster if it has been ‘kissed' by Bax. Furthermore, Mtch2 accelerates the re-localization of tBid at the mitochondria. In contrast, binding to Bcl-XL dramatically impedes tBid re-localization by lowering the off-rate threefold. Our results suggest that the transfer of activated tBid and Bax to different mitochondria is governed by dynamic equilibria and potentially contributes more than previously anticipated to the dissemination of the permeabilization signal within the cell.
Keywords: Bcl-2 family, Bax, tBid, liposomes, mitochondria, fluorescence
Mitochondrial outer membrane permeabilization (MOMP) is widely regarded as the commitment step in apoptosis and is controlled by the Bcl-2 family of proteins.1, 2, 3 MOMP requires an activator such as truncated Bid (tBid) or Bim to relay the apoptotic signal to the effector members, Bax and Bak, which oligomerize in and permeabilize membranes. Anti-apoptotic proteins such as Bcl-2 or Bcl-XL counter this process by binding to both the activators and Bax/Bak.4 In recent years, a consensus has emerged regarding the core MOMP mechanisms.2, 5, 6 In particular, the crucial role had by membranes in modulating the complex network of binding interactions between Bcl-2 family proteins has been recognized.7, 8, 9, 10 Moreover, it has been recently demonstrated that the interactions between key proteins in the family and the mitochondrial outer membrane (MOM) are reversible and dynamic.11, 12 Intermittent interactions of Bcl-2 family proteins with membranes may set up transient loci from which apoptotic events are regulated. Furthermore, increasing evidence suggests that all the mitochondria in the cell need to be permeabilized for efficient apoptosis to occur.13 Bcl-2 pro-apoptotic family members are heavily accumulated at sites of mitochondrial fission during apoptosis.14 However, with limited pool of proteins in the cell, it is easy to imagine a situation after the onset of apoptosis where the available protein pool is quickly used up on limited number of mitochondria and the cell has to reuse the available protein to permeabilize all the mitochondria to proceed with apoptosis. Indeed, in certain circumstances it appears that MOMP is initiated at one precise location and then propagates through the rest of the cell as a wave over the ensuing 5 min.15, 16 The diffusion of soluble apoptotic factors between mitochondria has been proposed as one potential mechanism to explain this phenomenon.15
Bax and Bak are composed of nine α-helices and contain a C-terminal tail-anchor region that targets them to the MOM and the endoplasmic reticulum.17, 18 In the death receptor pathway, the apoptotic signal is transmitted to Bax and Bak via tBid,19, 20 which, similar to Bax, is cytoplasmic in most dividing cells and localizes to target membranes only at the onset of apoptosis.21, 22, 23 After the engagement of death receptors on the cell surface, Bid undergoes a proteolytic cleavage by caspase 8 and the larger C-terminal fragment of cleaved Bid (cBid), termed tBid, binds the MOM and undergoes a conformational change facilitated by the MOM protein Mtch2, which renders tBid capable of activating Bax.10, 24 Interaction of Bax with tBid at the membrane also causes a conformational change in Bax, which is a necessary step for its activation.25 As soluble Bid is structurally homologous to soluble Bax and both proteins require a conformational change at the N-terminus before activation, we have proposed that Bid should be considered a Bax-like protein distinct from other Bcl-2 homology 3 (BH3) proteins.26
Another similarity between Bid and Bax that is relevant to the issue of propagation of an apoptotic signal is that both proteins interact dynamically with lipid membranes. Although tBid binds to liposomes rapidly,8 it dissociates from them with an apparent dissociation constant (KD) of 0.5 nM (lipids KD 44 μM).10 tBid has also been reported to have lipid-transfer activity.27 Consistent with this observed capacity of tBid to ‘retro-translocate' from the membrane, electron paramagnetic resonance experiments,24 NMR experiments28 and Monte Carlo simulations,29 all suggest that tBid binds only to the outer leaflet of the membrane. By contrast, fully activated Bax adopts a transmembrane topology with three α-helices (5, 6 and 9) inserted in the membrane30 that is carbonate resistant,17 which suggests a tight and possibly non-reversible interaction with the membrane. However, other conformations with only α9 inserted into membranes are possible. In the absence of activator BH3 proteins, Bax binds reversibly to artificial membranes in vitro,11, 31 a process that involves a reversible conformational change of its N-terminal region that exposes an epitope recognized by the monoclonal antibody called 6A7.11, 25 Accordingly, in growing cells, inactive Bax is normally in equilibrium between the cytoplasm and is peripherally bound to the MOM.12, 32 When Bcl-XL is present at the MOM, reversible binding of Bax favors the cytoplasmic form. This shift in localization has been referred to as retro-translocation and is an important aspect of the anti-apoptotic function of Bcl-XL.4, 12
Furthermore, the interaction between tBid and Bax is itself dynamic. It has been suggested that tBid activates Bax (and Bak) by a ‘kiss-and-run' mechanism,33, 34, 35 in which the interaction between the two proteins is transient allowing tBid to activate a series of different Bax (or Bak) molecules.15 Consistent with this notion, we have shown that the tBid–Bax interaction, which occurs only in or on lipid membranes, is reversible.8 Therefore, both tBid and/or Bax have the potential to propagate the apoptotic signal from one membrane to another.
Here we report that in an in vitro system consisting of fluorescently labeled tBid or Bax, and mitochondria or liposomes, both activated and membrane-bound tBid and Bax migrate between membranes. The transfer of activated Bax, however, proceeds very slowly and an activator at the destination membrane is required for permeabilization to occur. Surprisingly, the transfer of tBid is enhanced by Bax and is repressed by Bcl-XL. Overall, our work suggests that by regulating tBid transfer, interaction with Bax and Bcl-XL provides an unexpected mechanism for regulating the intracellular propagation of apoptotic signals.
Results
Mitochondria initially lacking tBid undergo Bax-mediated permeabilization in the presence of mitochondria pre-incubated with tBid
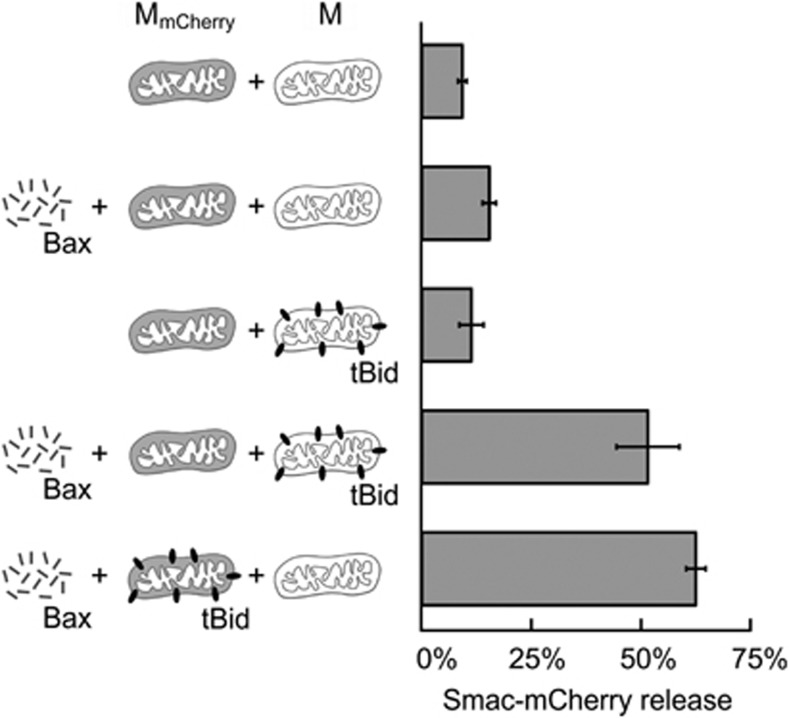
To determine whether tBid and Bax can mediate the transmission of apoptotic signaling between mitochondria without any other factors, we performed the experiment shown in Figure 1a. The first set of mitochondria (M) was isolated from bax −/− bak −/− BMK cells and incubated with tBid, and then the unbound tBid was removed by centrifugation. A second set of mitochondria (MmCherry) was isolated from bax −/− bak −/− BMK cells that express a fluorescent Smac-mCherry fusion protein localized to the mitochondrial intermembrane space. The two sets of mitochondria were mixed in a 1 : 1 ratio, incubated with Bax and collected by centrifugation to evaluate the amount of MOMP in the MmCherry by comparing the quantities of Smac-mCherry in the supernatant and pellet fractions (Figure 1). As expected, no MOMP was observed in control reactions lacking either tBid or Bax. Surprisingly, when the incubation contained MmCherry not pre-incubated with tBid, ∼50% of them underwent MOMP after addition of Bax, as long as mitochondria pre-incubated with tBid were also present. In a positive-control incubation containing Bax and MmCherry pre-incubated with tBid, the amount of Smac-mCherry released was similar (∼60%).
Figure 1.
Bax releases Smac-mCherry from bax−/− bak−/− mitochondria in incubations containing mitochondria pre-incubated with tBid. Bars indicate the percentage of Smac-mCherry release on addition of 20 nM Bax (as indicated) to incubations containing both bax−/− bak−/− mitochondria (M) and bax−/− bak−/− mitochondria expressing Smac-mCherry (MmCherry), where one of the mitochondria populations has been pre-incubated with 2 nM tBid (as indicated). Release of Smac-mCherry was detected 2 h after adding Bax by measuring the fluorescence in the supernatant after mitochondria were pelleted by centrifugation (mean±S.E.M., n=3). For each bar, the schematic on the left recapitulates the content of the incubation
The fact that Bax permeabilizes mitochondria with and without tBid pre-bound to the MOM demonstrates that apoptotic signaling is propagated between mitochondria in this simplified system. This transmission may be due to several mechanisms, either alone or in combination: (I) tBid originally bound to one mitochondrion can transfer to another mitochondrion, where it recruits soluble Bax; (II) after activation by tBid bound to a specific mitochondrion, activated Bax can insert into the membrane of another mitochondrion, irrespective of whether it contains tBid; (III) only mitochondria with membrane-bound tBid are directly permeabilized by Bax, but these fuse to other mitochondria, leading to MOMP. As explained in the (Supplementary Figure S1), this third possibility was discarded in control experiments and will not be discussed further.
Bax-mediated permeabilization of membranes initially lacking tBid is as efficient as and only marginally slower than that of membranes pre-incubated with tBid
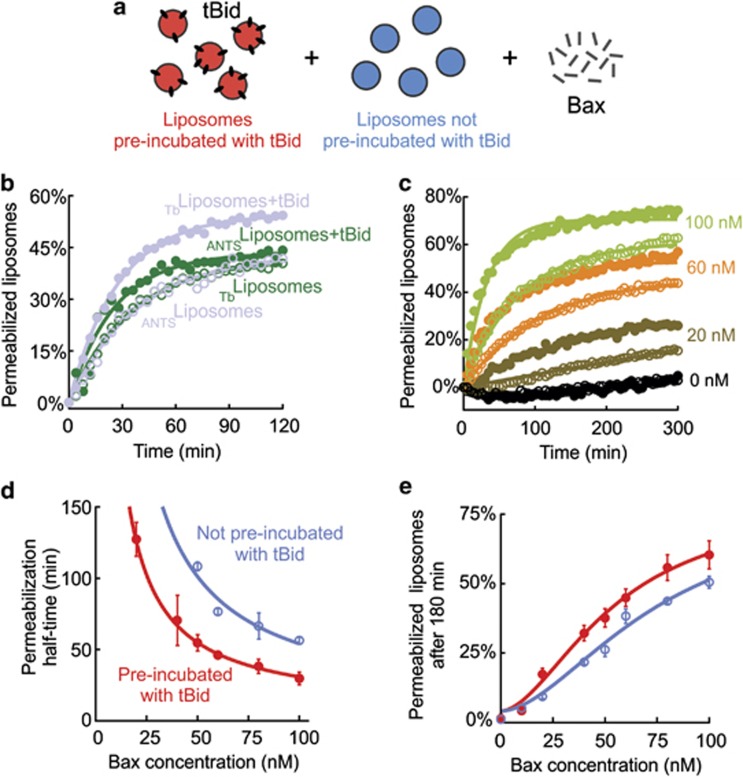
To determine which of the other two mechanisms mediate MOMP propagation, we used a liposome system that allowed precise measurements of reagent concentrations and reaction kinetics. Two distinct sets of liposomes containing spectrally distinct fluorophores (ANTSliposomes and Tbliposomes) made possible the concurrent evaluation of permeabilization of both sets after addition of Bax (Figure 2a).
Figure 2.
Bax permeabilizes liposomal membranes when liposomes pre-incubated with tBid are also present. (a) Principle of the protein transfer experiment: liposomes pre-incubated with tBid (black ovals) and encapsulating one type of fluorophore (red – color arbitrary) are mixed with liposomes not pre-incubated with tBid and filled with a spectrally different fluorophore (blue – color arbitrary). Liposome permeabilization on the addition of Bax can then be monitored separately. (b) Permeabilization of a 1 : 1 ratio of liposomes either pre-incubated with 20 nM tBid (filled symbols) or not (empty symbols) when mixed together and incubated with 100 nM Bax. Data represented by the same color symbols were recorded simultaneously in the same solution, using spectrally different encapsulated fluorophores (ANTS and Tb) as shown in a. (c) Permeabilization of liposomes either pre-incubated with tBid or mixed with an equal amount of liposomes not pre-incubated with tBid (filled symbols) or vice versa (empty symbols) when incubated with the indicated amount of Bax. Curves with the same color have been recorded for the same Bax concentration but in two different solutions (one set of representative data). In b and c, lines represent exponential fits of the data. (d) Permeabilization half-time as a function of Bax concentration and (e) percentage of permeabilized liposomes after 3 h of incubation (mean±S.E.M., n=3). The fit of the data with either a power law (d) or a sigmoidal function (e) is shown for both liposome populations
To reconstitute the observation with mitochondria in liposomes, we measured liposome permeabilization after mixing liposomes with or without membrane-bound tBid. First, liposomes were incubated with tBid, and then unbound tBid was removed by gel filtration chromatography to obtain liposomes with bound tBid (confirmed by immunoblotting, data not shown), denoted ANTSliposomes+tBid and Tbliposomes+tBid. Exactly as observed with mitochondria, after the addition of Bax to a 1 : 1 mixture of ANTSliposomes+tBid and Tbliposomes, both sets of liposomes were permeabilized with a comparable efficiency (Figure 2b, green symbols). To ensure that our observations were independent of the fluorescent molecules encapsulated in the liposomes, we performed the experiment with the reverse combination of fluorophores using 1 : 1 mixture of Tbliposomes+tBid and ANTSliposomes. We obtained the same results where both sets of liposomes were permeabilized to a similar extent (Figure 2b, light blue symbols). As expected, in control incubations without Bax, co-incubation of the different liposomes did not lead to permeabilization even when external tBid was added (data not shown).
To more closely examine the possible differences between the permeabilization of liposomes with and without membrane-bound tBid, we evaluated the influence of varying Bax concentrations on permeabilization kinetics and end points (Figure 2c). Similar to the results above, ANTSliposomes with or without pre-incubated tBid were permeabilized on addition of Bax. However, the permeabilization half-time for liposomes pre-incubated with tBid was about half that of liposomes not pre-incubated with tBid (Figure 2d), suggesting that the process is more complicated than simple diffusion. Nevertheless, permeabilization eventually proceeded to roughly the same extent in a Bax concentration-dependent manner for both types of liposomes at end point (Figure 2e).
Transfer of tBid between membranes is efficient and is accelerated by Bax
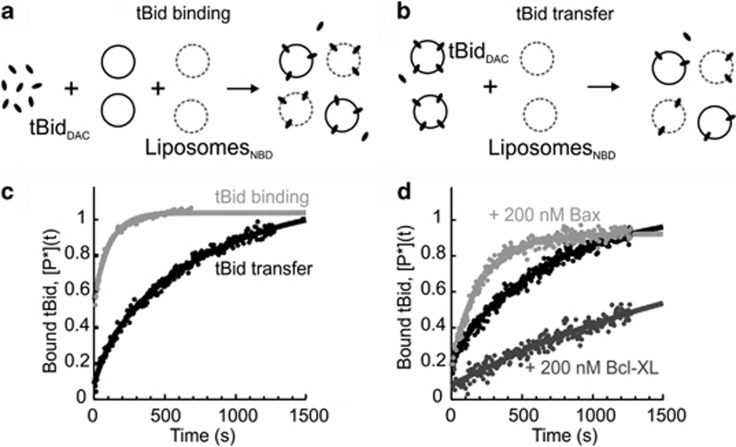
For tBid to transfer between liposomes, tBid binding to membranes must be reversible. We therefore used gel filtration chromatography to confirm our previous observation that binding of tBid to the membrane is reversible (Supplementary Figure S2).10 To determine the extent and the rate of tBid transfer between liposomes, we used a FRET (Förster resonance energy transfer)-based assay. For these experiments, tBid was fluorescently labeled with the donor DAC (tBidDAC) and the membrane contained the acceptor NBD-PE (liposomesNBD). FRET is observed only when tBidDAC migrates to liposomesNBD8, 10 As a positive control, we added tBidDAC to a 1 : 1 mixture of liposomesNBD and unlabeled liposomes (Figure 3a), and observed rapid FRET between tBidDAC and liposomesNBD (Figure 3c, gray symbols). A single-exponential fit of the data as predicted by the kinetic model obtained by considering a simple binding reaction between tBid and the membranes equation (3), gives an average relaxation rate k=(0.013±0.005)/s (mean±S.D., n=3) at this lipid concentration. Next, to determine the transfer of tBid between liposomes, we incubated tBidDAC with liposomes and used gel filtration to obtain a solution with liposomes+tBidDAC, which was then added to liposomesNBD in a 1 : 1 ratio (Figure 3b). We observed efficient FRET between tBidDAC and liposomesNBD that reached the same level of completion as in the control experiment, indicating that tBidDAC transferred from one population of liposomes to the other such that at equilibrium it was equally distributed between the two populations of liposomes (Figure 3c, black symbols). We fitted this data with a double exponential function equation (4) to account for the fact that tBid is initially partitioned between the membrane of the liposomes and the solution, and that both these fractions eventually interact with the membrane of liposomesNBD, but with different rates (see Materials and Methods). In this way, the retro-translocation rate of tBid was found to be on average: koff=(1.6±0.5) 10−3/s. This means that the half-time associated with the release of a molecule of tBidDAC from a liposome is ∼7 min, considerably longer than the average time necessary for a tBidDAC to bind to a liposome in our experimental conditions (∼40 s). These results demonstrate that tBid binding to mitochondrial-like membranes is reversible and the difference in rates is consistent with our previous observations that tBid co-fractionated with liposomes during gel filtration chromatography. Together, our results suggest that tBid spontaneously transfers between two populations of liposomes, leading to permeabilization of the destination liposome.
Figure 3.
The binding of tBid to membranes is reversible. (a) Schematic of the kinetic experiments performed to measure the binding and unbinding rates of tBid to liposomes. tBid labeled with the donor DAC (tBidDAC) is mixed with unlabeled liposomes (black ovals) and liposomes labeled with the acceptor NBD-PE (dotted ovals) in a 1 : 1 ratio. (b) Unlabeled liposomes pre-incubated with tBidDAC were mixed with liposomes labeled with the acceptor NBD-PE. (c) A tBid binding experiment (gray symbols, as shown in a) and a tBid transfer experiment (black symbols, as shown in b), showing the fraction of tBid bound to fluorescent liposomes as a function of time, as obtained by normalizing the signal obtained from the donor fluorescence. Lines are fit to the data according to equations (3) and (4). (d) Fraction of tBid transferred to fluorescent liposomes as a function of time in the presence of either 200 nM Bax (light gray symbols) or 200 nM Bcl-XL (dark gray symbols). Data in the absence of Bax or Bcl-XL are also shown for comparison (black symbols, as in c). Lines are fit to the data with equation (4)
As tBid interacts with other Bcl-2 family members at the MOM, we next examined the effect of Bax and Bcl-XL on the dynamic equilibrium of tBid transfer between liposomes. We incubated liposomes+tBidDAC with liposomesNBD in a 1 : 1 ratio (Figure 3b) with Bax or Bcl-XL. Consistent with their opposing functional roles in apoptosis, Bax and Bcl-XL had opposing effects on tBid transfer between liposomes. Although Bax accelerated tBid transfer between liposomes, increasing the retro-translocation rate threefold to koff= (4.5±0.2) 10−3/s (corresponding to a half-time of ∼2.5 min), Bcl-XL reduced the retro-translocation rate threefold, to koff=(0.5±0.2) 10−3/s (corresponding to a half-time of ∼25 min) (Figure 3d). This result suggests that regulation of retro-translocation may be a general property of Bcl-2 family proteins.
Although these experiments demonstrate that no other proteins are required for tBid membrane binding and transfer, an integral MOM protein Mtch2 facilitates tBid binding to membranes and MOMP by accelerating the conformational change in tBid required for Bax activation in cells.10, 36 Consistent with this function, Mtch2 at the recipient membrane increased the efficiency of tBid transfer (Supplementary Figure S3).
Bax transfers between membranes but requires an activator at the recipient membrane for oligomerization
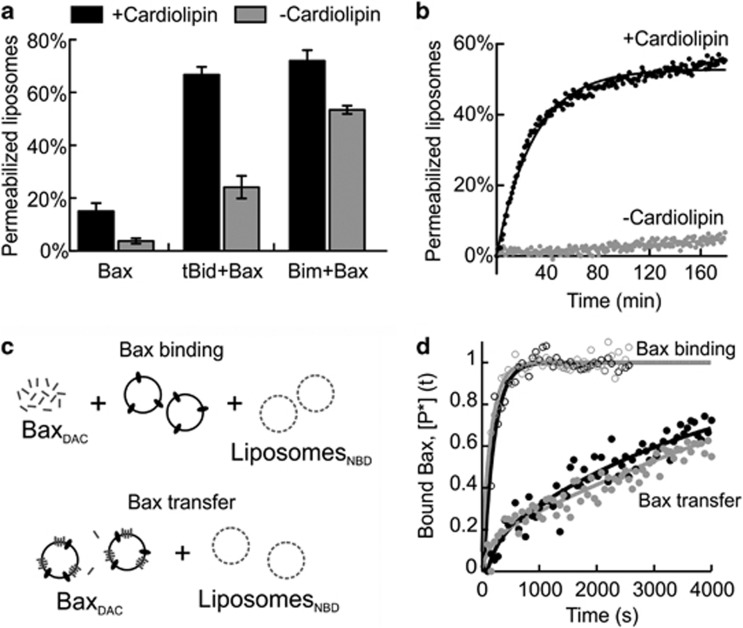
To determine whether activated Bax transfers between liposomes independent of tBid, we used liposomes that support Bax oligomerization but not tBid binding. We therefore compared a lipid composition without cardiolipin (−CL) to mitochondria-like liposomes with higher negative surface charge density (+CL). As expected, tBid was unable to bind to −CL liposomes (data not shown). Consequently, while tBid mediated Bax liposome permeabilization of +CL ANTSliposomes was very efficient, permeabilization was greatly reduced in −CL ANTSliposomes (Figure 4a). However, when using another BH3 protein BimL as an activator for Bax, both sets of ANTSliposomes were efficiently permeabilized, although −CL ANTSliposomes were permeabilized to a lesser extent (Figure 4a). Thus, once activated with BimL, Bax permeabilizes −CL ANTSliposomes (>50% release) much more efficiently than when tBid is the activator (<25% release).
Figure 4.
Bax transfers between membranes but requires an activator at the recipient membrane to form pores. Permeabilization of liposomes with either +CL or −CL lipid composition. (a) −CL liposomes were poorly permeabilized using 100 nM Bax and 20 nM tBid, but were efficiently permeabilized by 100 nM Bax and 20 nM BimL. (b) Permeabilization of liposomes by Bax as a result of transfer of tBid between two liposome populations. Non-fluorescent +CL liposomes pre-incubated with 20 nM tBid were mixed with equal concentration of fluorophore-encapsulated liposomes with indicated lipid composition, followed by the addition of 100 nM Bax. (c) Illustration of the assays used to measure Bax binding and transfer. (d) Bax binding: 100 nM Bax labeled with the donor, DAC, 20 nM tBid and liposomes containing the acceptor NBD-PE with the indicated lipid composition were incubated for 30 min. Line is fit to the data according to equation (3). Bax binds to both +CL (open symbols, black) and −CL (open symbols, gray) liposomes. Normalized FRET efficiency is indicative of binding of BaxDAC to liposomesNBD. Bax transfer: tBid activated, membrane-bound Bax transfers to +CL (filled symbols, black) and –CL (filled symbols, gray). Unlabeled liposomes pre-incubated with 200 nM BaxDAC and 20 nM tBid were passed over a gel-filtration column to remove free Bax and tBid. The fraction containing the liposomes and bound proteins was added to liposomesNBD in a 1 : 1 ratio to observe Bax transfer. Line is fit to the data according to equation (4). FRET efficiency was calculated as explained10 (mean±S.E.M., n=3)
To exploit this difference, we examined whether Bax activated at one set of liposomes can permeabilize another by performing the experiment analogous to that shown in Figure 2a. For this experiment, tBid was incubated with +CL liposomes and the liposomes containing bound tBid were isolated by gel filtration and mixed in an equal ratio to either +CL or −CL ANTSliposomes. As expected, Bax efficiently permeabilized +CL ANTSliposomes that promoted tBid transfer. However, −CL ANTSliposomes were not permeabilized (Figure 4b). As Bax is able to permeabilize these membranes when activated by BimL, this indicates that Bax either cannot transfer between membranes or it cannot permeabilize membranes that are not ‘primed' with an activator after binding.
To investigate the transfer of soluble Bax between liposomes, we used a protein–liposome FRET assay similar to the one described above for tBid (Figure 4c). When BaxDAC (donor) was added to a reaction containing either +CL or −CL liposomesNBD (acceptor), the efficiency of FRET was not only similar in both cases (Figure 4d, open circles) but was also independent of the presence of tBid (Supplementary Figure S4). This demonstrates that Bax binds to membranes independent of negative surface charge and that FRET cannot distinguish between the peripheral binding of Bax to membranes (that occurs in the absence of tBid) and the membrane-embedded form of Bax that mediates permeabilization (that occurs in the presence of tBid). Thus, there must be very little difference in the distance between the DAC dye on Bax and the NBD dye in the membrane for these two conformations. In addition, these results demonstrate that unlike tBid, the interaction of Bax with membranes is not dependent on the presence of CL in the membrane.
Next, we investigated whether the active, stably membrane-bound Bax transfers between liposomes. BaxDAC and tBid were incubated with liposomes and the liposomes containing membrane-bound proteins (liposomes+tBid+BaxDAC) were collected by gel filtration. These liposomes were then incubated with +CL or −CL liposomesNBD, and the transfer of BaxDAC was measured by FRET (Figure 4c). Surprisingly, membrane-bound BaxDAC re-equilibrated between the two populations of liposomes, although at a slower rate than tBid, with an average koff=(0.2±0.1) 10−3/s (binding half-time ∼60 min) (Figure 4d, filled circles) equation (4). Furthermore, membrane-bound BaxDAC transferred to −CL liposomes as efficiently as to +CL liposomes, demonstrating that transfer of both the active and the inactive conformers of Bax is independent of both tBid and negative membrane surface charge.
Discussion
The fact that soluble and membrane-bound forms of Bcl-2 family proteins are in reversible equilibrium is increasingly recognized as an important feature of these proteins.10, 31 The retro-translocation of Bax from mitochondrial membranes in transformed cells that has been highlighted in several recent publications12, 32 is a natural consequence of this equilibrium partition. The present study shows that the binding of both tBid and Bax to liposomal membranes is dynamic, with measurable off-rates on the order of 10−3/s similar to that measured for Bax in cells,12, 32 and that this permits redistribution of these proteins to other membranes. Surprisingly, even though both Bax and tBid have multiple membrane conformations, binding was reversible in all cases, even for membrane-bound Bax resistant to gel filtration. It remains speculative whether one Bax from an oligomer transfers or exclusively monomeric membrane-bound Bax retro-translocates, or if a combination of both processes occurs simultaneously. This result is consistent with the concept of multiple conformational changes between inactive and fully active tBid and Bax (Figure 5).8, 10
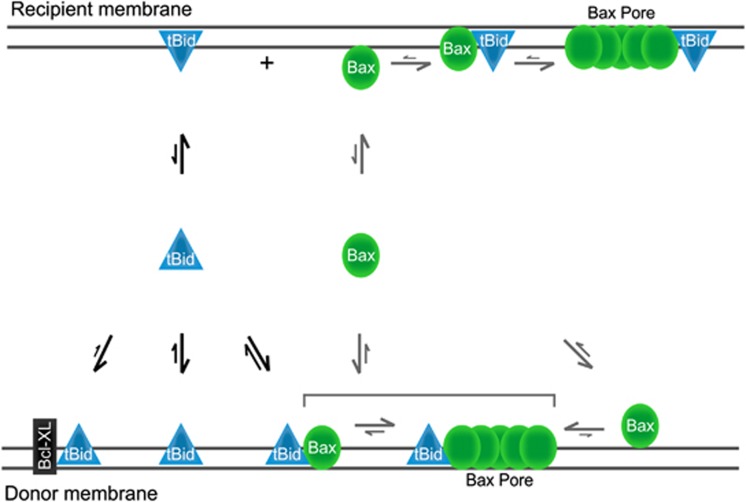
Figure 5.
Bax and tBid transfer between membranes and Bax permeabilizes recipient membranes primed by tBid. Binding of tBid and Bax to membranes is reversible and is regulated by dynamic equilibria as indicated by equilibrium arrows. Binding of Bcl-XL to tBid decreases the transfer rate of tBid between the donor and the recipient membranes by decreasing the koff threefold. Interaction of tBid with Bax increases the transfer rate of tBid threefold either by increasing the koff or by decreasing the kon. Both soluble and membrane-bound Bax are also constantly retro-translocating but the transfer rate of Bax is independent of tBid. Moreover, the koff of membrane bound Bax is eight-fold less than that of membrane bound tBid. Once at the recipient membrane, Bax requires re-activation by tBid for insertion and pore formation, suggesting that the active conformation of Bax is lost during retro-translocation
Consistent with redistribution of tBid being functionally important, it was modulated positively and negatively by pro- and anti-apoptotic Bax and Bcl-XL, respectively. For Bax, oligomerization in the recipient membrane is strictly dependent on the presence of a BH3 activator (tBid or BimL) (Figure 4). Bcl-2 family activators therefore ensure that Bax permeabilizes the correct subcellular membrane, similar to the way that Mtch2 may act to trigger the activating conformational change in tBid, thereby restricting its activity to mitochondrial membranes.
One implication of the finding that an activator needs to be present on the recipient membrane for pore formation to occur is that retro-translocated Bax likely reverts to its inactive conformation. Our results also suggest that the physical migration of the activator throughout the cell may be required to ensure that all mitochondrial membranes are permeabilized, a requirement for apoptosis to kill the cell.13 Consistent with this notion, in certain circumstances where the apoptosis signal is localized, a rapidly spreading wave is observed.16 Moreover, quantitative kinetic analysis in model cellular systems indicates that the duration of the lag phase (initiation) of apoptosis is long and is variable between different cells, but the execution phase is rapid and with little variation between cells,15, 37 consistent with the diffusion of an apoptotic factor across the cell.15 Our study suggests that BH3 activators have a bigger role in transmission of the apoptotic signal compared with effectors such as Bax, as the koff for tBid is more than eight times faster than that of Bax, in line with the different KD we previously measured for tBid (44 μM)10 and for Bax in the presence of tBid (∼1 μM).31 In addition, our results clearly demonstrate that both Bax and Bcl-XL regulate migration of tBid between membranes, further illustrating total integration of the system.
The mechanism we propose here is a slight revision of the ‘kiss-and-run' model, in which tBid ‘kisses' not only Bax but also a membrane, and then ‘runs' to other locations in order to propagate the apoptotic signal (Figure 5). Such a mechanism would explain how cells can achieve rapid full-scale apoptosis activation, as Bax continuously cycles on and off mitochondrial membranes and is primed to act, while requiring the signal provided by a fast-moving activator before pore formation can occur. Furthermore, the migration of tBid and Bax between different mitochondria may also be involved in regulating mitochondrial dynamics possibly by promoting mitochondrial fragmentation.38
Our study has identified a number of factors that affect membrane binding by tBid, and which we propose should affect its rate of transfer between mitochondria and the propagation of apoptotic signals throughout the cell. First, Bcl-XL not only inhibits tBid binding to Bax, our results demonstrate that it also slows down tBid migration between membranes. This is a logical but not necessarily obvious result of mutual sequestration due to Bcl-XL binding to tBid in the membrane with higher affinity than in solution.9 The p7 fragment of Bid slightly shifts the equilibrium in the other direction, towards the soluble form of tBid10 through a similar mechanism: it binds tBid in solution exclusively and with a high affinity (KD∼40 nM, Supplementary Fig. S2). Second, and more surprising is our observation that Bax enhances tBid retro-translocation and therefore the rate of transfer of tBid between liposomes (Figure 3d). The effect of Bax on tBid migration is harder to understand, as unlike the p7 fragment it binds tBid only in the membrane, and therefore would be expected to slow down retro-translocation. We speculate that the molecular mechanism by which Bax may accelerate retro-translocation of tBid is that it has an effect opposite to that of Mtch2. Mtch2 promotes tBid binding to MOM by enhancing a conformational change of tBid that leads to deeper membrane insertion and is required for its Bax activation capability (Supplementary Figure S3 and Shamas-Din et al.10), whereas Bax may be reversing this conformational change. In this way, after the ‘kiss' between tBid and Bax, tBid can ‘run' because the interaction with Bax (or the initiation of Bax oligomerization) pushes tBid out of the membrane. Regardless of the mechanism, it is interesting to note that the opposite effects of Bax and Bcl-XL on tBid retro-translocation are congruous with their opposite roles in apoptosis. This work therefore underlines the notion that a series of competitive equilibria between Bcl-2 family members and membranes lies at the heart of apoptotic regulation.
Materials and Methods
Protein purification and labeling
All experiments used recombinant full-length proteins or single-cysteine mutants. The nomenclature for the single-cysteine mutants is to indicate the residue number of the single cysteine introduced into variants without endogenous cysteine. Recombinant full-length human wild-type (WT) and single-cysteine mutant C126 of Bax were purified and labeled as described.8, 31 Recombinant full-length N-terminal 6 × -His-tagged murine WT and single-cysteine mutant 126C or 190C of Bid were purified, labeled and cleaved to isolate cBid or tBid as described.10 Recombinant full-length WT Bcl-XL was purified as described.4 Recombinant full-length N-terminal 6 × -His tagged human BimL mutant lacking all endogenous lysine and cysteine residues was purified as described.39 Dyes for labeling, DACM ((N-(7-dimethylamine-4-methylcoumarin-3-yl) maleimide) and Alexa Fluor 488 were purchased from Life Technologies (Waltham, MA, USA).
Mitochondria permeabilization assay
Mitochondria from bax−/− bak−/− BMK (baby mouse kidney) cells (either expressing Smac-mCherry or not) were isolated as described.30 To assay MOMP, 0.2 mg/ml mitochondria (in 20 mM HEPES pH 7.5, 250 mM sucrose, 150 mM KCl, 1 mM EDTA) were incubated with tBid (2 nM) for 30 min at room temperature. Mitochondria with membrane-bound tBid were then isolated by centrifugation (13 000 × g for 10 min at 4 °C). To assess MOMP, mitochondria expressing Smac-mCherry were incubated with mitochondria pre-targeted with 2 nM tBid and with 20 nM Bax for 2 h at 37 °C. Samples were then centrifuged at 13 000 × g for 10 min. The relative amount of mCherry present in the supernatant and pellet fractions was measured by fluorescence. Samples were excited at 580 nm and monitored at 610 nm, with excitation and emission bandwidths of 5 nm. The percentage of Smac-mCherry release was calculated as the fraction of the total fluorescence coming from the supernatant.
Liposome preparation
Liposomes with a composition resembling that of MOMs (PC, PE, PI, DOPS and TOCL in a 48 : 28 : 10 : 10 : 4 molar ratio, were prepared from lipids purchased from Avanti Polar Lipids, Alabaster, AL, USA), in assay buffer as described.10 Liposomes indicated as −CL are with low surface charge and contain PC, PE and PI in a 62 : 28 : 10 molar ratio. −CL liposomes have a net negative charge/lipid of 0.10 compared with 0.28 for liposomes with a mitochondria-like composition. All liposome preps used in this work included a small fraction (0.008 mol%) of lipophilic dye (DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt), Life Technologies) to monitor the liposomes concentration. The fluorescence of DiD was assessed by excitation at 644 nm and emission at 665 nm with 5 nm bandwidths for both.
Liposome permeabilization assay
Liposomes containing the fluorophore ANTS and the quencher DPX (ANTSliposomes) were prepared as described.4 Permeabilization of ANTSliposomes was measured by the increase in ANTS fluorescence that results from dilution of the liposome contents. The other set of liposomes contained terbium (Tb) and dipicolinic acid (DPA) (Tbliposomes). TbLiposomes were prepared by adding TbC13 (0.8 mM) and DPA (2.4 mM) in the assay buffer (without EDTA). The Tb/DPA complex is highly fluorescent when encapsulated in liposomes but the fluorescence is reduced dramatically when the liposomes are permeabilized in a solution containing EDTA, due to chelation of Tb. Therefore, the permeabilization curves from Tbliposomes were later normalized inversely to directly compare them with the curves generated from ANTSliposomes. In both cases, non-encapsulated molecules were removed by gel filtration chromatography on a CL-2B Sepharose column before use in permeabilization assays.
ANTS and Tb/DPA were excited concurrently at 355 and 276 nm, respectively, with 5 nm bandwidths. In order to separate the short-lifetime fluorescence emission of ANTS from the long-lifetime emission of the Tb/DPA complex, ANTS emission was collected at 520 nm with a 12-nm bandwidth for 20 μs without any delay, while Tb/DPA emission was collected at 545 nm with a 12-nm bandwidth for 2 ms after a 40-μs delay. The extent of dye release was calculated as a percentage of maximum possible release: P(t)=[F(t)−FB(t)]/[FM−FB(t)], where F(t) refers to either the ANTS or the Tb/DPA fluorescence signal, FB(t) refers to the background fluorescence of a control sample containing liposomes but no protein and FM refers the fluorescence measured for each sample after lysis of the liposomes with 0.5% w/v CHAPS or 0.2% v/v Triton X-100. P(t) was then fitted with an exponential function, P(t)= PM−(PM−P0)exp(−kt), which returned the percentage of dye release measured immediately after the addition of Bax at t=0 (P0), the extrapolated final percentage of dye release (PM) and the initial rate of release, k.
To prepare liposomes pre-incubated with tBid, 20 nM tBid was incubated with 0.3 mg/ml liposomes in assay buffer at 37 °C for 30 min. To collect liposomes with bound tBid, the reaction was passed over a CL-2B Sepharose column. Liposomes without tBid were mixed in 1 : 1 ratio, and indicated amount of Bax was added. The reactions were carried out at 37 °C.
Liposome-binding assay
For FRET experiments, 200 nM tBid 126C labeled with DAC was incubated with 0.3 mg/ml liposomes in assay buffer at 37 °C for 30 min, or 200 nM Bax 126C labeled with DAC was incubated with 0.3 mg/ml liposomes at 37 °C for 2 h. Liposomes containing bound protein were isolated by gel filtration on a CL-2B Sepharose column and mixed with 0.3 mg/ml liposomes containing the acceptor, NBD-PE (1-palmitoyl-2-{12-((7-nitro-2-1,3-benzoxadiazol-4-yl) amino]dodecanoyl}-sn-glycero-3-phosphoethanolamine)) at 1 mol% (Avanti) in 1 : 1 ratio. The detailed protocol for liposome preparation is previously published.8, 10 The reactions were carried out at 37 °C for 30 min to 2 h without stirring.
For gel filtration experiments, 200 nM cBid 190C labeled with Alexa Fluor 488 was incubated with 0.3 mg/ml liposomes at 37 °C for 30 min. Free protein was separated from liposome-bound protein using CL-2B Sepharose column and % bound protein was assessed by fluorescence (excitation at 495 nm, emission at 519 nm, 5 nm bandwidths) and by immunoblotting for Bid as described previously.4
Equilibrium binding of protein to liposomes
If a protein (e.g., tBid) binds reversibly to lipid membranes, the interaction can be represented by the equilibrium P+L↔P* (where P represents the protein in solution, P* the protein bound to the membrane and L is the lipid), with a KD=[P][L]/[P*]. In the case where the lipids are in excess and the available lipid concentration is equal to the total lipid concentration, [L], the equilibrium fraction of bound protein becomes (free ligand approximation)
KD is the effective dissociation constant that may vary with the lipid composition.
In the presence of a competitor, such as the p7 fragment of Bid, which can bind to the soluble protein to prevent membrane binding, a second equilibrium needs to be taken into account, P+P'↔C, with the associated KD'=[P][P']/[C]. Assuming that the lipids are in excess, and that [P]=[P']=cP (i.e. that the concentration of both Bid fragments is the same), this simple competition model predicts that the equilibrium fraction of bound protein is:
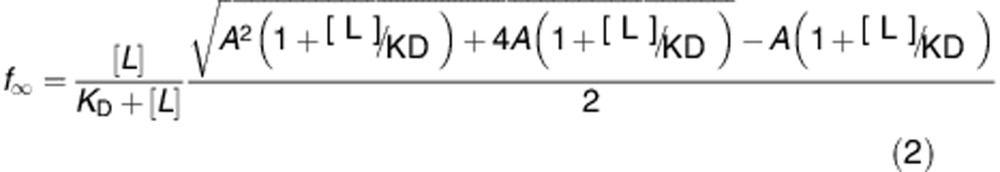 |
where A=KD'/cP.
Kinetics of protein transfer between liposomes
We consider again the simple reaction P+L↔P* to analyze the kinetics of the reaction by considering the on-rate, kon, and the off-rate, koff. Thus, d[P]/dt=kon[P][L]−koff[P*], leading to KD=koff/kon and to single exponential kinetics for the fraction of bound protein, f(t)=f∞(1−e−kt) when soluble proteins are mixed with liposomes at time t=0, where f∞ is given by equation (1) and where the relaxation rate is k=koff+kon[L]. In the case when soluble proteins are incubated with a 1 : 1 mix of two types of liposomes (with total lipid concentration [L]) starting at t=0, assuming that the affinity of the protein for both types of liposomes is the same, the fraction of bound protein to one type of liposome (a quantity accessible by FRET if the protein is labeled with a donor fluorophore and this set of liposomes is labeled with an acceptor) also increases with single exponential kinetics:
 |
with a rate k=koff+kon[L].
We now consider the more complicated case where protein is initially at equilibrium with a first population of liposomes (total lipid concentration [L]/2) and a second population of liposomes is introduced at time t=0 (also with total lipid concentration [L]/2). The kinetics of the system is defined by a set of two differential equations, which can be solved to calculate the fraction of protein bound to the second set of liposomes (the quantity of which is measurable using FRET):
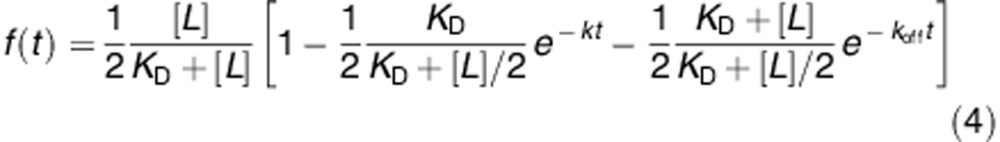 |
This time, the fraction of bound protein increases with double-exponential kinetics. The first exponential kinetics with rate k=koff+kon[L] as before corresponds to the binding of soluble protein to the second set of liposomes, while the second exponential kinetics with a slower rate koff corresponds to the binding of the protein molecules originally bound to the first set of liposomes, and which have to detach before they can bind to the second set of liposomes.
Acknowledgments
This work was supported by grant FRN86657 to CF and grant FRN12517 to DWA and BL from the Canadian Institute of Health Research (CIHR). CF was the recipient of Canada Research Chair funded by the Natural Sciences and Engineering Council (NSERC).
Glossary
- MOMP
mitochondrial outer membrane permeabilization
- MOM
mitochondrial outer membrane
- BH
Bcl-2 homology
- cBid
cleaved Bid
- tBid
truncated Bid
- KD
dissociation constant
- FRET
Förster resonance energy transfer
- CL
cardiolipin
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by M Campanella
Supplementary Material
References
- 1Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 2002; 111: 331–342. [DOI] [PubMed] [Google Scholar]
- 2Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev 2008; 9: 47–59. [DOI] [PubMed] [Google Scholar]
- 3Shamas-Din A, Kale J, Leber B, Andrews DW. Mechanisms of action of Bcl-2 family proteins. Cold Spring Harb Perspect Biol 2013; 5: a008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Billen LP, Kokoski CL, Lovell JF, Leber B, Andrews DW. Bcl-XL inhibits membrane permeabilization by competing with Bax. PLoS Biol 2008; 6: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Leber B, Lin J, Andrews DW. Embedded together: The life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 2007; 12: 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Leber B, Lin J, Andrews DW. Still embedded together binding to membranes regulates Bcl-2 protein interactions. Oncogene 2010; 29: 5221–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Dlugosz PJ, Billen LP, Annis MG, Zhu W, Zhang Z, Lin J et al. Bcl-2 changes conformation to inhibit Bax oligomerization. EMBO J 2006; 25: 2287–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 2008; 135: 1074–1084. [DOI] [PubMed] [Google Scholar]
- 9Garcia-Saez AJ, Ries J, Orzaez M, Perez-Paya E, Schwille P. Membrane promotes tBID interaction with BCL(XL). Nat Struct Mol Biol 2009; 16: 1178–1185. [DOI] [PubMed] [Google Scholar]
- 10Shamas-Din A, Bindner S, Zhu W, Zaltsman Y, Campbell C, Gross A et al. tBid undergoes multiple conformational changes at the membrane required for Bax activation. J Biol Chem 2013; 288: 22111–22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. Interaction with a membrane surface triggers a reversible conformational change in bax normally associated with induction of apoptosis. J Biol Chem 2003; 278: 48935–48941. [DOI] [PubMed] [Google Scholar]
- 12Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C et al. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell 2011; 145: 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Tait SW, Green DR. Mitochondrial regulation of cell death. Cold Spring Harb Perspect Biol 2013; 5: a008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A et al. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol 2002; 159: 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Rehm M, Huber HJ, Hellwig CT, Anguissola S, Dussmann H, Prehn JH. Dynamics of outer mitochondrial membrane permeabilization during apoptosis. Cell Death Differ 2009; 16: 613–623. [DOI] [PubMed] [Google Scholar]
- 16Garcia-Perez C, Roy SS, Naghdi S, Lin X, Davies E, Hajnoczky G. Bid-induced mitochondrial membrane permeabilization waves propagated by local reactive oxygen species (ROS) signaling. Proc Natl Acad Sci 2012; 109: 4497–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Valentijn AJ, Upton JP, Bates N, Gilmore AP. Bax targeting to mitochondria occurs via both tail anchor-dependent and -independent mechanisms. Cell Death Differ 2008; 15: 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Schinzel A, Kaufmann T, Borner C. Bcl-2 family members: integrators of survival and death signals in physiology and pathology [corrected]. Biochim Biophys Acta 2004; 1644: 95–105. [DOI] [PubMed] [Google Scholar]
- 19Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 1998; 94: 481–490. [DOI] [PubMed] [Google Scholar]
- 20Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998; 94: 491–501. [DOI] [PubMed] [Google Scholar]
- 21Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol 1997; 139: 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: a novel BH3 domain-only death agonist. Genes Dev 1996; 10: 2859–2869. [DOI] [PubMed] [Google Scholar]
- 23Hsu YT, Youle RJ. Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J Biol Chem 1998; 273: 10777–10783. [DOI] [PubMed] [Google Scholar]
- 24Oh KJ, Barbuto S, Meyer N, Kim RS, Collier RJ, Korsmeyer SJ. Conformational changes in BID, a pro-apoptotic BCL-2 family member, upon membrane binding. J Biol Chem 2005; 280: 753–767. [DOI] [PubMed] [Google Scholar]
- 25Hsu YT, Youle RJ. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem 1997; 272: 13829–13834. [DOI] [PubMed] [Google Scholar]
- 26Billen LP, Shamas-Din A, Andrews DW. Bid: a Bax-like BH3 protein. Oncogene 2009; 27(Suppl 1): S93–S104. [DOI] [PubMed] [Google Scholar]
- 27Esposti MD, Erler JT, Hickman JA, Dive C. Bid, a widely expressed proapoptotic protein of the Bcl-2 family, displays lipid transfer activity. Mol Cell Biol 2001; 21: 7268–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Wang Y, Tjandra N. Structural insights of tBid, the caspase-8-activated Bid, and its BH3 domain. J Biol Chem 2013; 288: 35840–35851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Veresov VG, Davidovskii AI. Monte Carlo simulations of tBid association with the mitochondrial outer membrane. Eur Biophys J 2007; 37: 19–33. [DOI] [PubMed] [Google Scholar]
- 30Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B et al. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J 2005; 24: 2096–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Satsoura D, Kucerka N, Shivakumar S, Pencer J, Griffiths C, Leber B et al. Interaction of the full-length Bax protein with biomimetic mitochondrial liposomes: a small-angle neutron scattering and fluorescence study. Biochim Biophys Acta 2012; 1818: 384–401. [DOI] [PubMed] [Google Scholar]
- 32Schellenberg B, Wang P, Keeble JA, Rodriguez-Enriquez R, Walker S, Owens TW et al. Bax exists in a dynamic equilibrium between the cytosol and mitochondria to control apoptotic priming. Mol Cell 2013; 49: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Eskes R, Desagher S, Antonsson B, Martinou JC. Bid induces the oligomerization and insertion of bax into the outer mitochondrial membrane. Mol Cell Biol 2000; 20: 929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Perez D, White E. TNF-alpha signals apoptosis through a bid-dependent conformational change in Bax that is inhibited by E1B 19K. Mol Cell 2000; 6: 53–63. [PubMed] [Google Scholar]
- 35Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev 2000; 14: 2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 36Zaltsman Y, Shachnai L, Yivgi-Ohana N, Schwarz M, Maryanovich M, Houtkooper RH et al. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat Cell Biol 2010; 12: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cell 2011; 144: 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Martinou JC, Youle RJ. Mitochondria in apoptosis: bcl-2 family members and mitochondrial dynamics. Dev Cell 2011; 21: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Sarosiek KA, Chi X, Bachman JA, Sims JJ, Montero J, Patel L et al. BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol Cell 2013; 51: 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.