Abstract
Receptor-interacting protein (RIP)3 is a critical regulator of necroptosis and has been demonstrated to be associated with various diseases, suggesting that its inhibitors are promising in the clinic. However, there have been few RIP3 inhibitors reported as yet. B-RafV600E inhibitors are an important anticancer drug class for metastatic melanoma therapy. In this study, we found that 6 B-Raf inhibitors could inhibit RIP3 enzymatic activity in vitro. Among them, dabrafenib showed the most potent inhibition on RIP3, which was achieved by its ATP-competitive binding to the enzyme. Dabrafenib displayed highly selective inhibition on RIP3 over RIP1, RIP2 and RIP5. Moreover, only dabrafenib rescued cells from RIP3-mediated necroptosis induced by the necroptosis-induced combinations, that is, tumor necrosis factor (TNF)α, TNF-related apoptosis-inducing ligand or Fas ligand plus Smac mimetic and the caspase inhibitor z-VAD. Dabrafenib decreased the RIP3-mediated Ser358 phosphorylation of mixed lineage kinase domain-like protein (MLKL) and disrupted the interaction between RIP3 and MLKL. Notably, RIP3 inhibition of dabrafenib appeared to be independent of its B-Raf inhibition. Dabrafenib was further revealed to prevent acetaminophen-induced necrosis in normal human hepatocytes, which is considered to be mediated by RIP3. In acetaminophen-overdosed mouse models, dabrafenib was found to apparently ease the acetaminophen-caused liver damage. The results indicate that the anticancer B-RafV600E inhibitor dabrafenib is a RIP3 inhibitor, which could serve as a sharp tool for probing the RIP3 biology and as a potential preventive or therapeutic agent for RIP3-involved necroptosis-related diseases such as acetaminophen-induced liver damage.
Necroptosis, also known as programmed necrosis, is a kind of programmed cell death that occurs at conditions that result in blocking the execution of apoptosis.1, 2 The protein kinase receptor-interacting protein (RIP)3 is a serine/threonine protein kinase that has recently been demonstrated to be the critical regulator that switches cells from apoptosis to necroptosis.3, 4, 5, 6 The death receptor ligands, such as tumor necrosis factor (TNF)α, Fas ligand and TNF-related apoptosis-inducing ligand (TRAIL), are classical inducers of apoptosis or necroptosis. By binding to their respective receptors, they lead to activation of functional caspase-8, which results in apoptosis by activating the effector caspases such as caspase-3 but inactivating the necroptic kinases such as RIP3. When caspase-8 is absent or inhibited by caspase inhibitors such as z-VAD, those death receptor ligands cause necroptosis, which can be augmented by Smac mimetic that promotes degradation of inhibitor of apoptosis proteins.3, 4, 5, 6
RIP3 is widely involved in physiological processes and pathological states.6 RIP3 deficiency not only rescues the lethality of caspase-8−/− and FADD−/− mice7 and restores normal proliferation of their T cells,6 but also protects hepatocytes from ethanol-induced injury and steatosis,8 rescues caspase-8 or FADD deficiency-induced massive inflammation in epithelium,9 prevents cerulean-induced acute necrotizing pancreatitis,3, 4 inhibits photoreceptor and cone cell death10, 11 and alleviates macrophage necrosis in advanced atherosclerosis lesions.12 Acetaminophen is an extensively used analgesic and antipyretic. When taken in overdose, its most frequent toxicity is hepatotoxicity including fatal centrilobular hepatic necrosis.13, 14 Acetaminophen overdose is the most common cause of acute liver failure in the United States and the United Kingdom.15 It also causes 11.86% of acute liver failure in China.16 Enhanced levels of high-mobility group box-1 and necrosis keratin-18 marked occurrence of hepatic necrosis.14 Necrosis has been considered as the predominant mode of cell death in this case, for which RIP3 has been shown to be responsible.17 In addition, RIP3 might also be associated with carcinogenesis and tumor drug resistance to chemotherapeutics.18, 19 These lines of evidence suggest potential extensive uses of small-molecule RIP3 inhibitors in medical prevention or therapy.
However, few RIP3 inhibitors have been reported20 and no small-molecule RIP3 inhibitors have been investigated for the potential medical uses. One possible cause is that there lacks a proper RIP3 kinase assay for screening for its inhibitors at molecular levels, which should be highly sensitive, free of radioisotopes, and high throughput. We thus established a non-radioactive luminescent RIP3 kinase assay in this study. By using this assay, we found that 6 B-Raf inhibitors inhibited the RIP3 enzymatic activity in vitro. But only dabrafenib could rescue cells from RIP3-mediated necroptosis induced by TNFα, TRAIL or Fas ligand plus Smac mimetic and the caspase inhibitor z-VAD. Dabrafenib directly and ATP-competitively bound to RIP3 protein and caused highly selective inhibition on RIP3 over RIP1, RIP2 and RIP5. Dabrafenib was demonstrated to ease acetaminophen-induced necrosis in normal human hepatocytes and to prevent acetaminophen-induced liver injury in mice. Our study raises a possibility that the medical indications of the B-RafV600E inhibitor dabrafenib might be extended from cancers to RIP3-involved diseases.
Results
Establishment of a non-radioactive luminescent RIP3 kinase assay
To evaluate the RIP3 inhibition of compounds at the molecular level, we established a non-radioactive luminescent RIP3 kinase assay (Supplementary Figure 1) by expressing the full-length human RIP3 protein with a glutathione S-transferase (GST) tag as shown schematically in Supplementary Figure 1a. To ensure the reproducibility of the assay and make it fit for high-throughput screening, we optimized the reaction conditions (Supplementary Figures 1b–f). One unit of the recombinant RIP3 enzymatic activity was defined as its amount required to consume 100 nM ATP in a 10-μl reaction system containing the substrate myelin basic protein (MBP, 25 ng/μl) at 30 °C for 90 min. The maximal signal-to-noise ratio of this assay could be achieved when RIP3 was used at between 50 U and 75 U (Supplementary Figure 1b). The ATP consumption reached its maximum at about 25 ng/μl of MBP (Supplementary Figure 1c). The ATP concentration and consumption relationship curve had its maximal slope at between 5 and 13 μM of ATP with the Km value of 12.58 μM for ATP (Supplementary Figure 1d). The reaction percentage reached a plateau at between 20 and 35 °C for 90 min (Supplementary Figures 1e and f). According to these results and considering the operability, the assay was set to be run at 30 °C for 90 min in a 10-μl system containing 50 U of the recombinant RIP3 enzyme, 25 ng/μl of the substrate MBP and 10 μM of ATP.
The activity of our recombinant RIP3 was similar to that of the enzyme from Abcam with the Km values of 28.83 and 30.59 ng/μl, respectively (Supplementary Figures 1c and 2a). Moreover, the sensitivity of this non-radioactive luminescent RIP3 kinase assay was also similar to that of the radioactive RIP3 kinase assay performed by the Reaction Biology Corporation (Malvern, PA, USA; Table 1).
Table 1. The inhibitory activity of the compounds against RIP3 and Rafs.
| Compounds |
IC50
for RIP3 (μM) |
IC50
for Rafs (nM) |
||||
|---|---|---|---|---|---|---|
| Luminescent assaysa | Radioactive assaysa | B-Raf | B-RafV600E | C-Raf | Refsb | |
| Dabrafenib | 0.25±0.16 | 0.002 | 3.2 | 0.8 | 5.0 | 35 |
| AZ628 | 0.90±0.15 | 4.48 | 100 | 34 | 29 | 36 |
| Regorafenib | 2.56±1.02 | 0.95 | 28 | 19 | —c | 37 |
| PLX4720 | 4.72±2.25 | 1.52 | — | 13 | — | 38 |
| Vemurafenib | 6.73±3.72 | 14.30 | 100 | 31 | 48 | 39 |
| Sorafenib | 14.52±0.51 | NAd | 22 | — | — | 40 |
| GDC-0879 | NA | ND | — | 0.13 | — | 41 |
| SB590885 | NA | ND | 0.16 | — | 1.72 | 42 |
| SU11248 | NA | ND | — | — | — | |
| Necrostatin-1 | NA | NA | — | — | — | |
Abbreviations: NA, not applicable; ND, not determined; RIP, receptor-interacting protein
IC50 values from luminescent assays were presented as mean±S.D. from three independent experiments in our laboratory, whereas the IC50 values from radioactive assays were presented as the mean of two independent experiments performed by the Reaction Biology Corporation
References (refs) reporting the IC50 values of Rafs inhibition by the corresponding compounds
No data reported
No IC50 available at the tested conditions
The B-Raf inhibitor dabrafenib selectively inhibits RIP3 kinase activity by its ATP-competitive binding to the enzyme protein
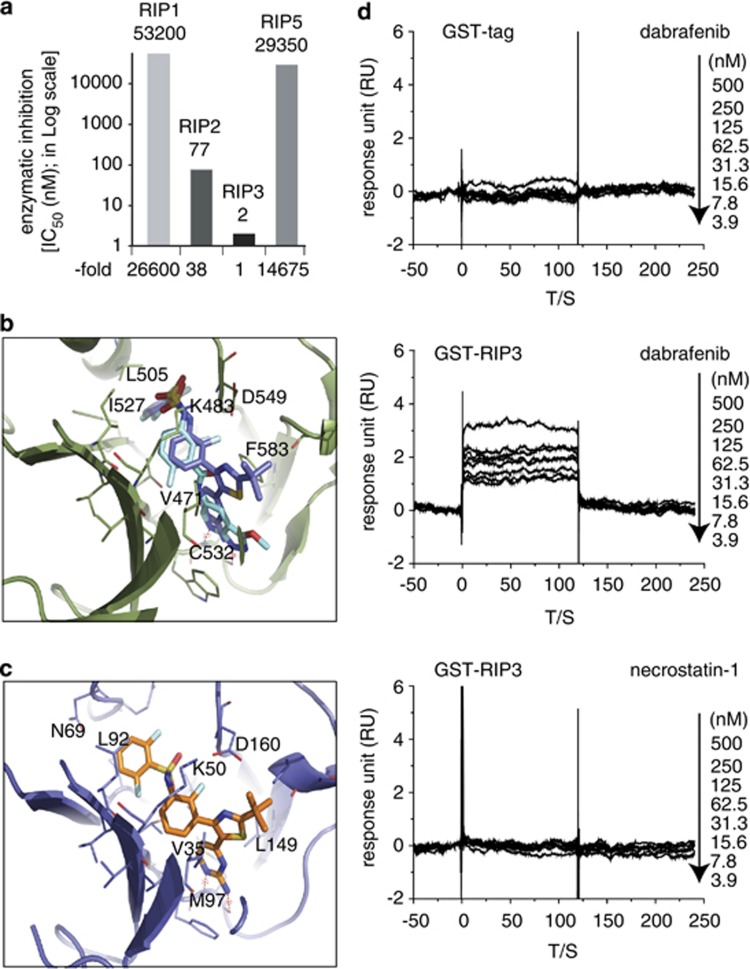
Based on the primary sequence similarity between RIP3 and RIP1 proteins and the conformational similarity between RIP1 and B-Raf proteins,21 we tested 10 commercially available protein kinase inhibitors in this assay, including eight B-Raf/B-RafV600E inhibitors, necrostatin-1 (RIP1 inhibitor)21, 22 and SU11248 (tyrosine protein kinase inhibitor; Table 1 and Supplementary Figure 2b). The result showed that the six tested B-Raf inhibitors (dabrafenib, AZ628, regorafenib, PLX4720, vemurafenib and sorafenib) inhibited the RIP3 activity with IC50s ranging from 0.25 to 14.52 μM, but the other two B-Raf inhibitors (GDC-0879 and SB590885), necrostatin-1 and SU11248 did not (Table 1 and Supplementary Figure 2b). The results were further confirmed by a classical radioactive assay, which gave the similar data of RIP3 inhibition (Table 1). Both assays revealed that dabrafenib had the most potent RIP3 inhibitory activity with the lowest IC50s among all those tested compounds. Selectivity analyses against the RIP family members revealed that dabrafenib inhibited RIP3 26 600-, 38- and 14 675-fold more potently than RIP1, RIP2 and RIP5, respectively (Figure 1a).
Figure 1.
Dabrafenib selectively inhibits RIP3 enzymatic activity by binding to the enzyme protein. (a) The inhibition of dabrafenib against RIP1∼RIP3 and RIP5 was examined by the radioactive assay (Reaction Biology Corporation). Column, the mean of the IC50 values from two independent experiments. The relative fold was obtained by normalizing the IC50 value for RIP3 as 1. (b) Molecular docking of dabrafenib binding to B-Raf. The B-Raf protein was shown as a cartoon style, the original ligand in 3SKC was colored cyan and dabrafenib was colored marine blue. (c) Molecular docking of dabrafenib binding to RIP3. RIP3 was shown in cartoon and dabrafenib was shown in the stick model. (d) The surface plasmon resonance assay showed that dabrafenib did not bind to the GST tag protein (upper) but bound to the recombinant RIP3 protein (middle). The RIP1 inhibitor necrostatin-1 did not bind to RIP3 (lower). The representative data were shown from two independent experiments
We next investigated the possibility of the direct interaction between dabrafenib and RIP3. For this purpose, we explored the interaction between dabrafenib and B-Raf. By checking the B-Raf crystal structures deposited in PDB database, we found the crystal structure 3SKC23 containing a ligand with high similarity to dabrafenib. The molecular docking study was conducted to obtain the binding mode of dabrafenib at the ATP site of B-Raf. Similar to the interaction seen in the crystal structure of B-Raf (3SKC), dabrafenib bound to the ATP-binding site of B-Raf by utilizing the pyrimidin-2-amine group forming hydrogen bonds with the hinge part. Its sulfonamide group extended to the sub-pocket in the vicinity of the C-alpha helix of B-Raf, and superimposed well with the original ligand in 3SKC (Figure 1b). For the docking study on the binding of dabrafenib to RIP3, we performed the homology modeling of human RIP3 based on the B-Raf crystal structure 3SKC. Then this theoretical model of RIP3 was used as the receptor to dock dabrafenib into its ATP-binding site. The result showed that dabrafenib could be fit into the binding pocket of RIP3 very well (Figure 1c). Although the interaction pattern of the pyrimidin-2-amine group with the hinge residue M97 is similar to B-Raf, the 2,6-difluorobenzene sulfonamide group of dabrafenib binds to a different sub-pocket when compared with the predicted B-Raf-dabrafenib complex structure (Figure 1b). The data suggest that dabrafenib could directly bind to RIP3.
To confirm the result, we then conducted the surface plasmon resonance biochemical analysis with necrostatin-1 as the negative control. Dabrafenib did not bind to the GST tag in the recombinant RIP3 protein (Figure 1d, upper panel), excluding its influence on the assay for the binding of dabrafenib to RIP3. Dabrafenib (Figure 1d, middle panel), but not necrostatin-1 (Figure 1d, lower panel), could bind to the recombinant RIP3 protein in a concentration-dependent manner with a dissociation constant (Kd) value of 26.5 nM. Lineweaver-Burk plots based on the substrate ATP competition assay revealed that dabrafenib was competitive with ATP to inhibit the RIP3 kinase activity (Supplementary Figure 2c), further supporting the conclusion about its direct binding to the protein.
Dabrafenib rescues cells from the RIP3-mediated necroptosis
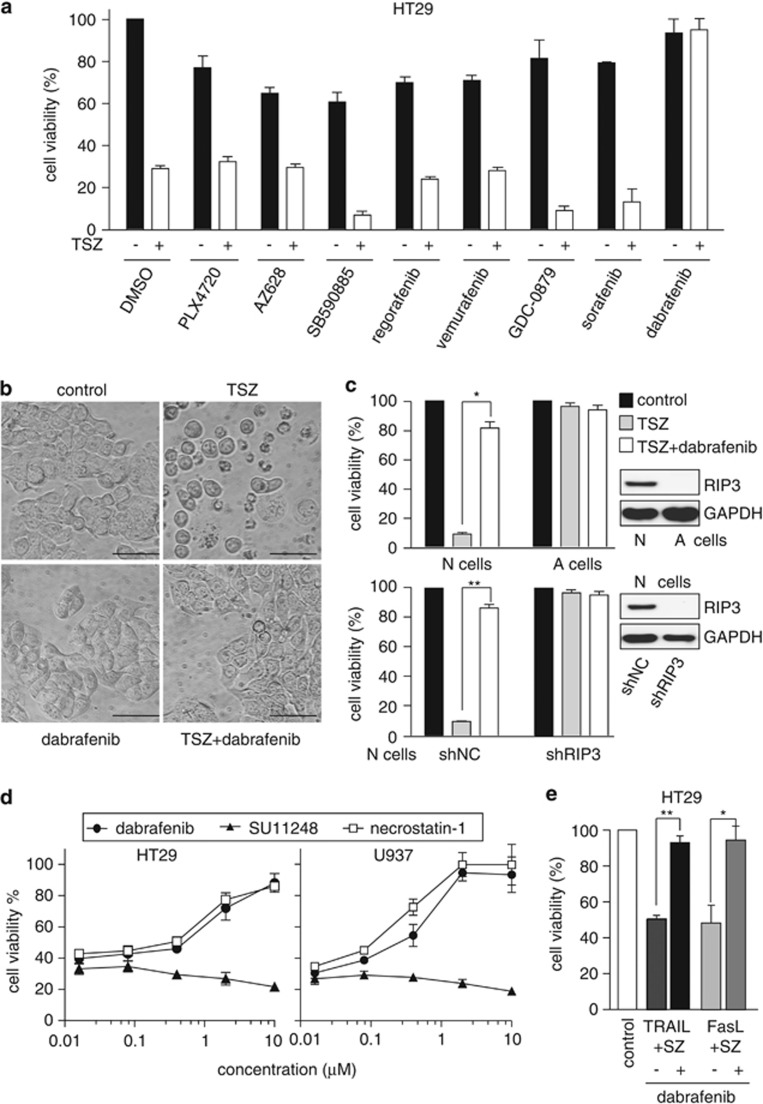
TNFα plus Smac mimetic and the caspase inhibitor z-VAD (TSZ) is a classical combination that is used to induce RIP3-mediated necroptosis.3, 4, 5 We treated RIP3-expressed HT29 cells (Supplementary Figure 3a) with this combination and eight compounds that inhibited the RIP3 kinase activity in vitro. Only dabrafenib was found to rescue TSZ-caused cell death significantly (Figure 2a). Dabrafenib also prevented the TSZ-induced necrotic morphology such as cell swelling, loss of membrane integrity and cell disruption in HT29 cells (Figure 2b). Either TSZ or TSZ plus dabrafenib did not obviously affect the cell viability of RIP3-deficient A cells3 or shRIP3 N cells; in contrast, TSZ apparently reduced the cell viability of RIP3-proficient N cells,3 which was effectively restored by dabrafenib (Figure 2c). The data further revealed the RIP3 dependence of dabrafenib in preventing TSZ-induced necroptosis. In RIP1- and RIP3-expressed HT29 and U937 cells4, 24 (Supplementary Figure 3a) exposed to TSZ, dabrafenib rather than the tyrosine protein kinase inhibitor SU11248,25 restored the cell viability as effectively as the RIP1 inhibitor necrostatin-1 did (Figure 2d). However, dabrafenib did not ease apoptosis induced by TNFα plus Smac mimetic (TS) in HT29 cells (Supplementary Figures 3b and c),4 further indicating that dabrafenib specifically inhibited the necroptosis pathway. Moreover, dabrafenib rather than another clinically used B-Raf inhibitor vemurafenib (with poor RIP3 inhibitory activity) reversed the loss of propidium iodide (PI)-negative HT29 cells caused by TSZ (Supplementary Figures 3d). In addition to TNFα, other cytokines such as TRAIL and Fas ligand have also been shown to be able to induce necroptosis.4 So we tested whether dabrafenib could block either TRAIL or Fas ligand-induced necroptosis. The result showed that dabrafenib reversed the reduction of the HT29 cell viability induced by TRAIL or Fas ligand plus Smac mimetic and z-VAD (SZ; Figure 2e). The data further strengthen the evidence that dabrafenib acts as a RIP3 inhibitor.
Figure 2.
Dabrafenib inhibits RIP3-mediated necroptosis. (a) Cell viability was examined in HT29 cells with or without TNFα (20 ng/ml) plus Smac mimetic (100 nM) and the caspase inhibitor z-VAD (20 μM; TSZ) in the presence of the indicated compounds (dabrafenib, 10 μM; all the others, 100 μM) for 24 h. (b) HT29 cells exposed to TSZ for 24 h in the presence or absence of 10 μM dabrafenib were observed under an inverted microscope at 20 × magnification. Scale bar, 50 μm. The data were representative of three independent experiments. (c) The cell viability of N and A cells (upper panel) or the RIP3-silenced N cells (lower panel) exposed to TSZ or TSZ plus dabrafenib for 24 h was determined. The protein levels of RIP3 in the cells were examined by western blotting. *P=0.002; **P=0.001. shNC and shRIP3, N cells transfected with the control shRNA and RIP3 shRNA, respectively. (d) The effects of the indicated compounds on cell viability were examined in HT29 cells or U937 cells treated with TSZ for 24 h. (e) The cell viability of HT29 cells treated with SZ plus 1 μg/ml Fas ligand or 200 ng/ml TRAIL was examined in the presence or absence of 10 μM dabrafenib. *P=0.003; **P=0.0003
Effects of dabrafenib on the Ser358 phosphorylation of mixed lineage kinase domain-like protein (MLKL) and the interaction between RIP3 and RIP1 or MLKL induced by TSZ
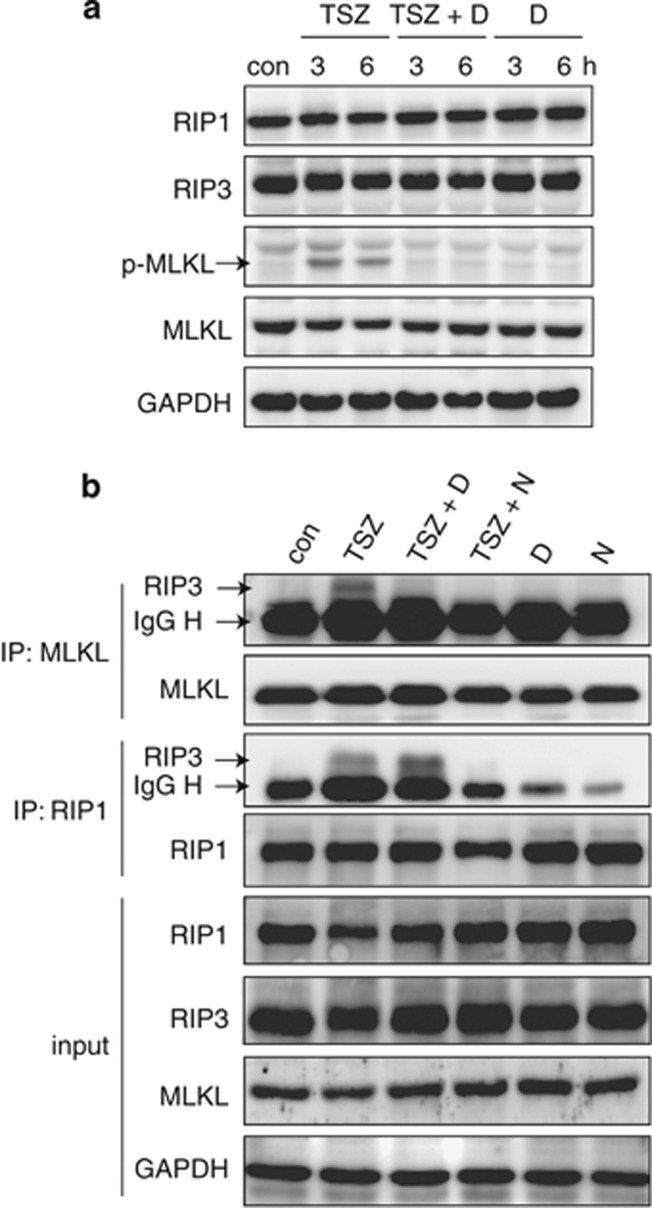
MLKL is a core component of the RIP1/RIP3 necrosome, and can be phosphorylated by RIP3 in responding to necrosis induction at the Ser358 site.26 Dabrafenib was shown to decrease the TSZ-caused-RIP3-mediated Ser358 phosphorylation of MLKL, but not to change the total protein levels of all RIP1, RIP3 and MLKL at this tested condition (Figure 3a). In addition, TSZ could induce the typical interaction between RIP3 and RIP1 or MLKL (Figure 3b). However, dabrafenib only disrupted the interaction between RIP3 and MLKL, but did not reduce the interaction between RIP3 and RIP1 (Figure 3b). The results suggest that MLKL rather than RIP1 mediates the biological effects of RIP3 inhibition by dabrafenib.
Figure 3.
Effects of dabrafenib on the Ser358 phosphorylation of MLKL and the interaction between RIP3, RIP1 or MLKL in HT29 cells. (a) Dabrafenib (10 μM) inhibited the Ser358 phosphorylation of MLKL induced by TSZ at the indicated times. (b) Effects of dabrafenib on the interaction between RIP3, RIP1 or MLKL in the cells exposed to the indicated combinations for 6 h. con, control; D, dabrafenib (10 μM); N, necrostatin-1 (10 μM); S, Smac mimetic (100 nM); Z, z-VAD-fmk (20 μM); T, TNF-α (20 ng/ml). The data were representative of three independent experiments
Dabrafenib inhibits RIP3 independently of its effect on the B-Raf family members
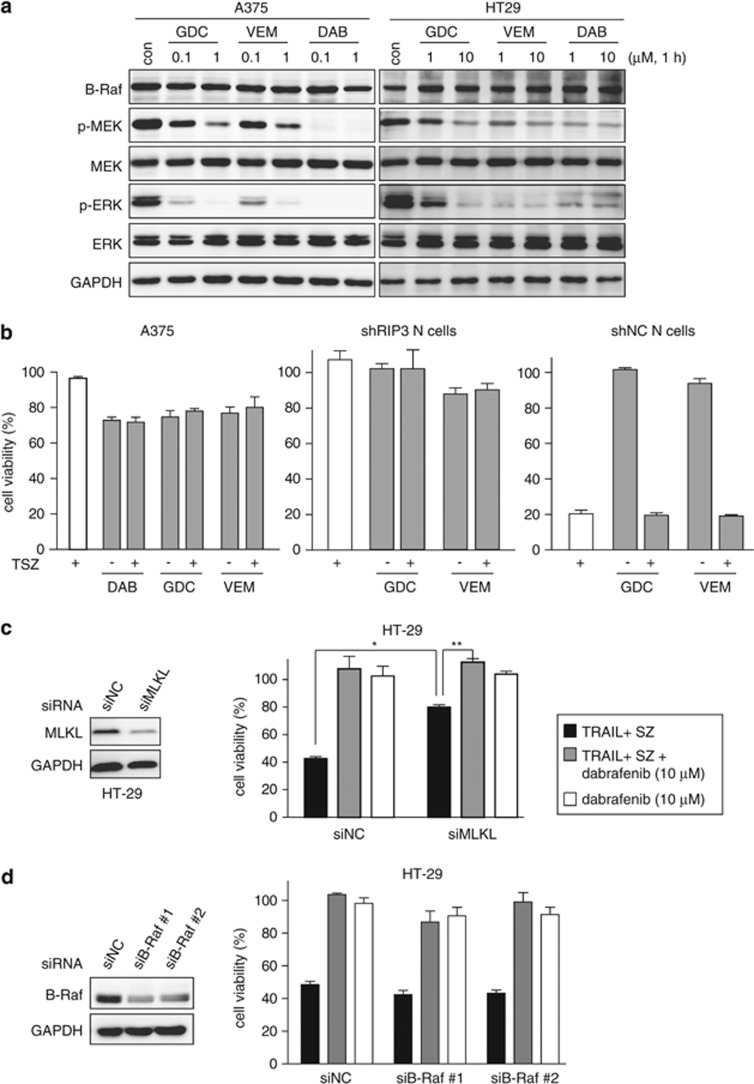
Table 1 showed that the inhibitory capability of those tested compounds including dabrafenib on RIP3 and other protein kinases, respectively, was not apparently correlative. In terms of proliferative inhibition, B-RafV600E-expressed colon HT29 cells show differential sensitivity to B-RafV600E inhibitors as evidenced by their reported sensitivity to vemurafenib27 but insensitivity to GDC-0879;28 in contrast, B-RafV600E-expressed melanoma A375 cells are similarly sensitive to B-RafV600E inhibitors including dabrafenib, vemurafenib and GDC-0879.28 However, all the three inhibitors caused similar phosphorylation reduction of the downstream signaling proteins MEK and ERK in both HT29 and A375 cells (Figure 4a). On the other hand, only dabrafenib reversed TSZ-induced necroptosis in RIP3-expressed HT29 cells, possibly because the other 2 B-RafV600E inhibitors have significantly weak RIP3 inhibitory activity (Table 1). A375 cells did not express RIP3 (Supplementary Figure 3a), and thus the treatment with TSZ did not affect their cell viability, which was not affected by adding dabrafenib, vemurafenib or GDC-0879 either (Figure 4b, left). Similar results were observed in RIP3-silenced (shRIP3) N cells (Figure 2c, lower and Figure 4b, middle). Similar in HT29 cells, dabrafenib rather than vemurafenib or GDC0879 prevented TSZ-induced necroptosis in RIP3-proficient N cells or shNC N cells (Figures 2c and 4b (right)). In addition, the reduced expression of MLKL partially prevented, but when combined with dabrafenib, completely reversed the loss of the HT29 cell viability caused by TRAIL+SZ, possibly because of the remaining MLKL (Figure 4c). In contrast, the reduced expression of B-Raf did not change the TRAIL+SZ-induced loss of the cell viability or the prevention of dabrafenib (Figure 4d). The data also showed that HT29 cells were resistant to dabrafenib alone, just as to GDC-0879.28 These data collectively indicate that (1) both RIP3 inhibition and B-RafV600E inhibition are separable, (2) the biological outcomes of their inhibition are mutually independent and (3) dabrafenib inhibits RIP3 independently of its effect on the B-Raf family members.
Figure 4.
Dabrafenib inhibits RIP3 independently of its effect on the B-Raf family members. (a) The B-RafV600E inhibitors GDC-0879 (GDC), vemurafenib (VEM) and dabrafenib (DAB) inhibited the phosphorylation of MEK and ERK in B-RafV600E-expressed A375 cells and HT29 cells. con, control. The data were representative of three independent experiments. (b) The cell viability of the cells exposed to TSZ in the presence or absence of the indicated B-RafV600E inhibitors for 24 h was examined. All the B-RafV600E inhibitors were used at 1 μM in A375 cells while at 10 μM in both shRIP3 and shNC N cells. (c) MLKL was silenced with the specific siRNA in HT29 cells. The protein levels were examined by western blotting. The cell viability of the cells exposed to the indicated combinations for 24 h was examined. Dabrafenib was used at 10 μM. *P=0.00002; **P=0.0002. (d) B-Raf was silenced with the specific siRNA in HT29 cells. The protein levels were examined by western blotting (left). The cell viability of HT29 cells exposed to the indicated combinations for 24 h was examined
Dabrafenib prevents acetaminophen-induced necrosis in normal human hepatocytes
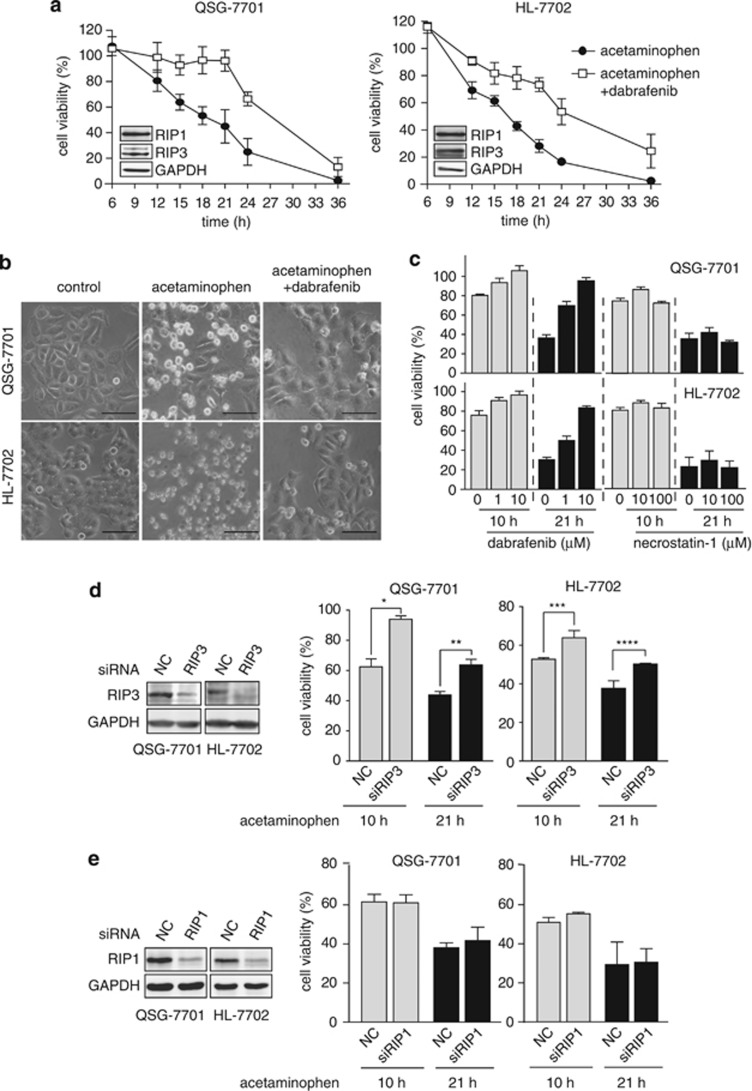
Acetaminophen taken in overdose can cause fatal hepatotoxicity including fatal centrilobular hepatic necrosis, for which RIP3 has been shown to be responsible.13, 14, 17 In RIP3-expressed normal human hepatocyte QSG-7701 and HL-7702 cells (Figure 5a), the treatment with 20 mM acetaminophen led to progressively reduced cell viability, which was alleviated by dabrafenib (Figure 5a). As expected, dabrafenib obviously attenuated the necrotic morphological changes of those cells exposed to acetaminophen (Figure 5b). This capability of dabrafenib was much more potent than that of the RIP1 inhibitor necrostatin-1 (Figure 5c). The human RIP3 inhibition by dabrafenib achieved similar effects to RIP3 deficiency in preventing acetaminophen-induced hepatocyte necrosis17 and ethanol-caused liver injury8 in mice. Moreover, RIP3 silencing partially reversed the acetaminophen-induced loss of the cell viability (Figure 5d). In contrast, necrostatin-1 or RIP1 silencing did not reduce acetaminophen-induced cell death in both human hepatocyte cells (Figures 5c and e) as necrostatin-1 did in mice,17 suggesting a differential role of RIP1 in human and mouse. In addition, z-VAD increased cell death induced by acetaminophen, which was prevented by dabrafenib partly (Supplementary Figure 4a); dabrafenib but not vemurafenib decreased the acetaminophen-induced loss of the cell viability (Supplementary Figure 4b). Together with the previous reports,8, 17 our results suggest a potential preventive or therapeutic use of dabrafenib as a RIP3 inhibitor in RIP3-mediated liver injury.
Figure 5.
Protection of dabrafenib from acetaminophen-induced cell viability reduction in human hepatocytes. (a) Human liver QSG-7701 and HL-7702 cells were treated with 20 mM acetaminophen in the presence or absence of 10 μM dabrafenib for the indicated times and the cell viability was examined. Insets, the levels of RIP1 and RIP3 proteins in the cells were detected by western blotting. (b) The cells exposed to 20 mM acetaminophen for the indicated times in the presence or absence of 10 μM dabrafenib were observed under an inverted microscope at × 20 magnification. Scale bar, 50 μm. (c) The effects of dabrafenib and necrostatin-1 on QSG-7701 and HL-7702 cells treated with 20 mM acetaminophen for 10 or 21 h were examined. (d and e) RIP3 (d) or RIP1 (e) was silenced with the specific siRNA in QSG-7701 cells and HL-7702 cells. The protein levels were examined by western blotting. The cell viability of the indicated cells exposed to 20 mM acetaminophen at the indicated times was examined. *P=0.004; **P=0.005; ***P=0.03; ****P=0.05. All the image data were representative of three independent experiments
Dabrafenib protects mice from acetaminophen-induced hepatotoxicity
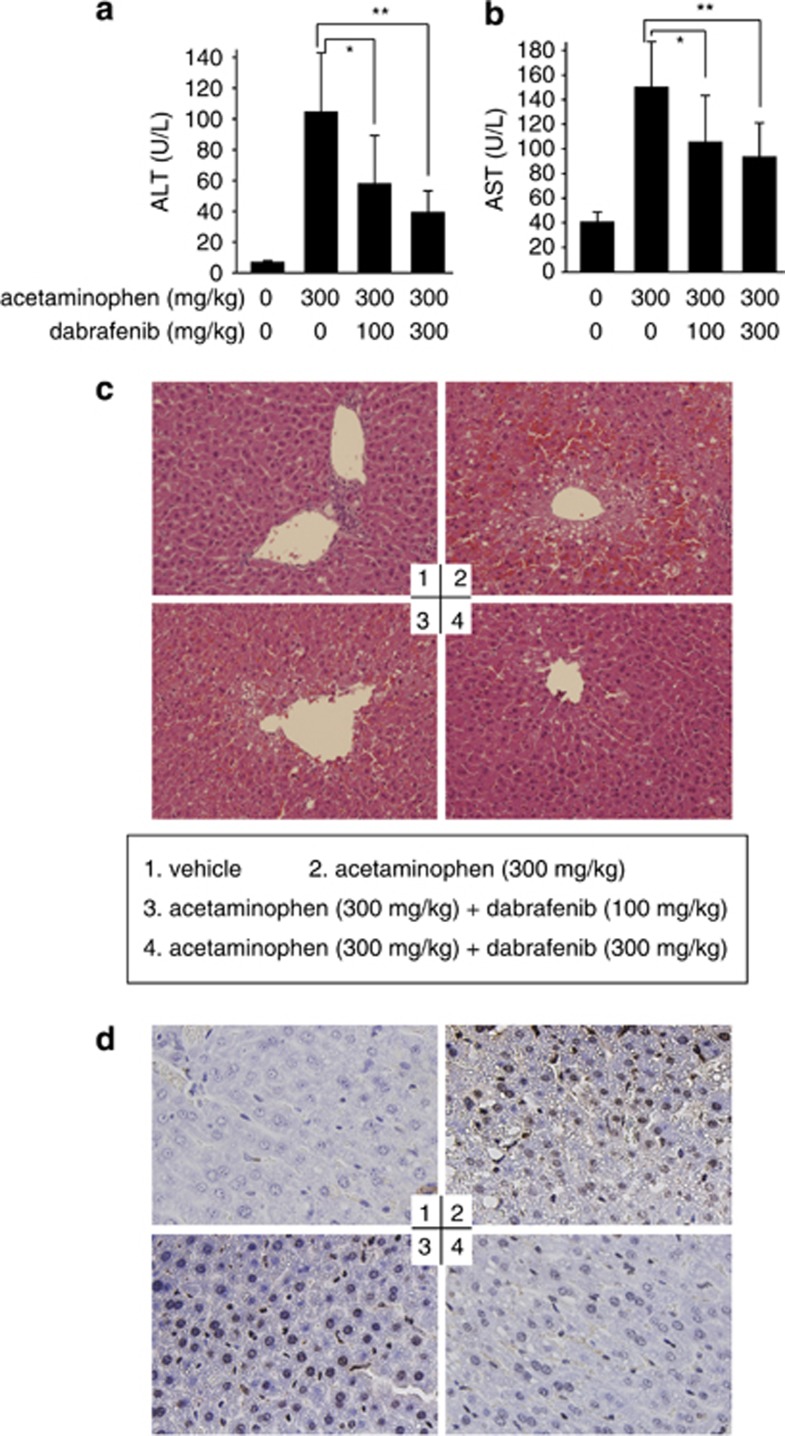
To validate the data from normal human hepatocyte QSG-7701 and HL-7702 cells, we examined the hepatoprotective effect of dabrafenib in acetaminophen-overdosed mouse models. The 6-h treatment of mice with acetaminophen (300 mg/kg) resulted in significant liver injury as indicated by the biochemical analyses showing the increase in the plasma levels of alanine aminotransferase, aspartate aminotransferase, interleukin (IL)-1β and TNFα and in the liver glutathione disulfide (GSSG) levels (Figures 6a and b; Supplementary Figure 5). This was further supported by its histological changes characteristic of centrilobular hepatic necrosis, peripheral hemorrhage and focal necrosis of hepatocytes (Figure 6c) and by the TUNEL staining revealing the increase in DNA breaks in the liver cells (Figure 6d). The pretreatment of mice with dabrafenib (100 mg/kg or 300 mg/kg) apparently eased the acetaminophen-caused liver injury (Figure 6 and Supplementary Figure 5), but another B-Raf inhibitor vemurafenib that has very weak RIP3 inhibitory activity did not (Supplementary Figure 5). Therefore, dabrafenib can protect acetaminophen-induced hepatotoxicity in mice in a dose-dependent manner.
Figure 6.
Dabrafenib alleviates acetaminophen-induced hepatotoxicity in mice. Mice were treated with 300 mg/kg acetaminophen (i.p.), with or without pretreatment with 300 mg/kg or 100 mg/kg dabrafenib (p.o.). Plasma alanine aminotransferase (ALT) (a) and aspartate aminotransferase (AST) (b) were determined. Data were expressed as mean±S.D.; n=10. In a, *P=0.015 and **P=0.0007; and in b, *P=0.03; **P=0.002. (c) The histological changes in mouse liver tissues shown by hematoxylin and eosin (H&E) staining. (d) TUNEL staining for nuclear DNA fragmentation in mouse liver cells. The typical images were captured under an optical microscope at × 20 (c) or × 40 (d) magnification
Discussion
In this study, we established a non-radioactive RIP3 kinase assay and with it, found that dabrafenib elicited the most potent inhibition on RIP3 among the eight tested Raf inhibitors. By competitively binding of ATP to the enzyme protein dabrafenib inhibited RIP3 with high selectivity relative to other RIP family members. Dabrafenib apparently rescued cells from RIP3-mediated necroptosis, which was independent of its B-Raf inhibition. Moreover, dabrafenib alleviated acetaminophen-induced RIP3-mediated necrotic injury in either human liver cells or mouse liver tissues.
RIP3-mediated necroptosis has been demonstrated to be involved in multiple pathological states.6 RIP3 knockout or silence could reverse or prevent some of those diseases or injuries, suggesting potential preventive or therapeutic uses of RIP3 inhibitors in the clinic. There have been only two RIP3 inhibitors (GSK'843 and GSK'872) reported as yet,20 but these two inhibitors were just used to probe the role of RIP3 in TLR3-induced necrosis; no data were shown about their preventive or therapeutic action in RIP3-associated diseases or injuries. Therefore, dabrafenib is the first reported small-molecule RIP3 inhibitor showing preventive or therapeutic action in RIP3-mediated pathological states, that is, the acetaminophen-induced liver injury. Dabrafenib has been used for other diseases in the clinic, indicating its acceptable human safety. This apparently raises the possibility of its preventive or therapeutic clinical uses for RIP3-involved diseases or injuries including acetaminophen-overdosed hepatotoxicity.
Although RIP1 and RIP3 proteins have similar primary sequences and RIP1, RIP3 and B-Raf proteins have similar conformation,21 our data showed that the RIP1 inhibitor necrostatin-1 did not inhibit RIP3. The tested B-Raf inhibitors also showed differential RIP3 inhibitory activities, which is obviously irrelevant to their Raf inhibitory activities. Dabrafenib showed potent inhibition against both RIP3 and Raf kinases. Moreover, our data showed that dabrafenib inhibited RIP3 independently of its effect on the B-Raf family members. Based on the structure of dabrafenib, therefore, careful structure-activity relationship studies help to differentiate the moieties that contribute to its RIP3 inhibition from the ones that contribute to its Raf inhibition. The results may lay an important basis for development of highly selective inhibitors of RIP3 or B-Raf in the future. Selective RIP3 inhibitors, except for their potential applications in RIP3-associated pathological states, can be used as probes to explore the roles of RIP3 in regulating cell death and the related signaling transduction processes.
In addition, both dabrafenib and vemurafenib are recently approved for the treatment of metastatic melanoma harboring B-RafV600E.29 Dabrafenib shows 39-fold more potent inhibition against B-RafV600E than vemurafenib. However, dabrafenib produces a little weaker clinical therapeutic effect and causes lower incidence of cutaneous squamous-cell carcinoma (incidence: 6% in a dabrafenib phase III trial versus 26% in a vemurafenib phase III trial), a common side effect of B-RafV600E inhibitors.29 Dabrafenib inhibited RIP3 also 27-fold more potently than vemurafenib as determined by the luminescent assay (Table 1). These differences between the 2 B-RafV600E inhibitors appear to suggest that RIP3 inhibition might have a role in their therapeutic effect and toxicity, which deserves clarifying. In this aspect, selective inhibitors of B-Raf against RIP3 may be also helpful.
Finally, the luminescent RIP3 kinase assay we established in this study has several advantages. It has high sensitivity equivalent to the widely used radioactive RIP3 kinase assay. It is non-radioactive, and thus free of potential radioactive contamination and risks. This assay can be run in a relatively small (10 μl) system in a 384-well-plate format that can meet the requirements for high-throughput screening for small-molecule compounds. The recombinant RIP3 used this assay is full-length human RIP3, which ensures the high relevance of the compound-screening results to human cells or tissues. In addition, all other materials or reagents used in this assay are common and easily commercially available. All the advantages indicate a possibility that this RIP3 kinase assay established in this study could be widely used in the future.
Materials and Methods
Protein expression and purification
The full-length human RIP3 cDNA was amplified from PMD 18-T-RIP3 plasmid (Jiran Bio-Technology Co., Ltd., Shanghai, China) by PCR with the following primers: forward primer (5′-TATGCGGCCGCTATGTCGTGCATCAAGTTAT-3′), which contains a NotI site; reverse primer (5′-CGCAAGCTTTTATTTCCCGC TATGATTATACC-3′), which contains a HindIII site. The PCR reaction (4 μl 5 × PrimeSTAR buffer, 3.2 μl of 2.5 mM dNTP, 0.5 μl of 100 μM forward primer, 0.5 μl of 100 μM reverse primer, 1 μl PrimeSTARHS DNA polymerase, 1 μl PMD 18-T-RIP3 (5 ng/μl) and 9.8 μl double distilled (dd)H2O) was run at 95 °C for 2 min, followed by 30 cycles (95 °C, 30 s; 60 °C, 30 s; 72 °C, 1 min) and 72 °C, 1 min.
The full-length human RIP3 cDNA was cloned into pFastBac HT B plasmids (Invitrogen, Grand Island, NY, USA), which was modified by inserting a fragment of DNA sequence encoding GST into the upstream of the multiple cloning sites of the primary plasmid. The digestion conditions were as follows: 2 μl 10 × NEB buffer II, 500 ng plasmid or PCR product, 1 μl Hind III, 2 μl NotI and ddH2O to 20 μl at 37 °C for 4 h. The ligation conditions were as follows: 1 μl T4 ligase, 1 μl pFastBac HT B, 10 μl PCR product, 2 μl 10 × T4 buffer and 6 μl ddH2O at 16 °C for 4 h. The ligation product was then transformed into DH5α cells to obtain the recombinant pFastBac HT B-RIP3 constructs. Then the amplified recombinant pFastBac HT B-RIP3 constructs were transformed into DH10Bac cells to get the recombinant Bacmid by ‘white-blue plaque selection'. The sequence of the PCR primer used to verify the RIP3-Bacmid clones was as follows: 5′-TTTGGCCTGTCCACATTTCAG-3′ (forward) and 5′-GGTTGGCAACTCAACTTCTCTT-3′ (reverse). The PCR assay was performed with PrimeSTARHS DNA polymerase according to the manufacturer's instructions (TaKaRa, Dalian, China).
The recombinant RIP3 protein with the GST tag at the N terminal was expressed in Sf9 cells according to the protocol of the Bac-to-Bac baculovirus expression system (Invitrogen) and was purified by using glutathione-sepharose beads according to the manufacturer's instructions (GE Healthcare, Pittsburgh, PA, USA). The protein was eluted in the buffer (PH 7.5) containing 50 mM Tris-HCl, 150 mM NaCl, 25% glycerol and 10 mM reduced glutathione. The eluted protein was stored at −80 °C.
The GST-tag protein was expressed and purified using the similar methods as shown above.
Quantitation of the recombinant RIP3 protein
The recombinant RIP3 protein was quantified by using the GST tag ELISA detection kit according to the manufacturer's instructions (GenScript, Piscataway, NJ, USA).
Non-radioactive luminescent RIP3 kinase assays
The Kinase-Glo Luminescent Kinase Assays Kit (Promega, Madison, WI, USA) was used to establish the non-radioactive luminescent RIP3 kinase assay. The purified recombinant RIP3 protein was incubated with its substrate MBP (Invitrogen) in the kinase reaction buffer (5 mM MOPS, pH=7.2, 2.5 mM β-glycerol-phosphate, 5 mM MgCl2, 2.5 mM MnCl2, 1 mM EGTA, 0.4 mM EDTA, 0.05 mM DTT) and ATP at the indicated reaction conditions, followed by adding the corresponding reagents from the kit according to the manufacturer's instructions. Luminescence was recorded with a PerkinElmer EnVision Multilabel Reader (Perkin-Elmer, Waltham, MA, USA).
In this assay, ATP consumption due to phosphorylation of the substrate MBP catalyzed by RIP3 was defined as signal or reaction percentage, which was calculated as [1-luminescence (+MBP)/luminescence (−MBP)] × 100%. In contrast, ATP consumption due to RIP3-mediated phosphorylation of non-MBP proteins (no MBP in the reaction system) was defined as noise, which was calculated as [1-luminescence (+RIP3)/luminescence (−RIP3)] × 100%. When evaluating the inhibitory activity of the tested compounds on RIP3, we defined the inhibition rate as [1-signal (+compounds)/signal (−compounds)] × 100%. The compound concentration required for 50% inhibition (IC50) of the RIP3 enzymatic activity was calculated with the Logit method.
Compounds
Dabrafenib, AZ628, regorafenib, PLX4720, vemurafenib, sorafenib, GDC-0879, SB590885 and SU11248 were purchased from Selleck (Houston, TX, USA). zVAD.fmk was obtained from Abcam (Cambridge, UK), necrostatin-1 was from Tocris (Bristol, UK) and acetaminophen was from Sigma-Aldrich (St. Louis, MO, USA). Smac mimetic was synthesized and purified as previously described.30
Cell culture
Human colon cancer HT29 cells, melanoma A375 cells, leukemia U937 cells and insect Sf9 cells were obtained from American Type Culture Collection (Manassas, VA, USA). Human normal liver QSG-7701 and HL-7702 cell lines were purchased from the Cell Bank of the Chinese Academy of Science Type Culture Collection (Shanghai, China). A cells and N cells were kind gifts from Professor Jiahuai Han (Xiamen University, Xiamen, China). All of the human cells were maintained in RPMI1640 medium except HT29 cells that were maintained in McCoy's 5A medium, supplemented with 10% fetal bovine serum. A cells and N cells were maintained in DMEM medium. Sf9 cell was maintained in SF-III SFM medium. All the cell lines were cultured according to the suppliers' instructions. All the cells were also periodically authenticated by morphologic inspection and tested Mycoplasma contamination.
Proteins, antibodies and other reagents
TNFα and MBP were purchased from Invitrogen. Fas ligand was obtained from Sino Biological Inc. (Beijing, China). TRAIL was purchased from Novoprotein (Summit, NJ, USA). Commercially available human RIP3 protein was from Abcam (Cambridge, UK). Human RIP1 and B-Raf antibodies were purchased from BD Biosciences (Franklin Lakes, NJ, USA). Human MEK, p-MEK, ERK and p-ERK antibodies were purchased from the Cell Signaling Technology (Danvers, MA, USA). Human RIP3 antibody, mouse RIP3 antibody, mouse GAPDH antibody and human GAPDH antibody were purchased from Santa Cruz (Dallas, TX, USA). Human MLKL and phospho-MLKL (Ser358) antibodies were obtained from Merck Millipore (Darmstadt, Germany). The glutathione-sepharose was purchased from GE Healthcare (Piscataway, NJ, USA) and other common reagents were from Sigma-Aldrich.
In vitro cell viability assays
Cells were seeded in 96-well plates at proper density. For necroptosis induction assays, TNFα (T, 20 ng/ml), Fas ligand (1 μg/ml) or TRAIL (200 ng/ml) plus Smac mimetic (S, 100 nM) and z-VAD (Z, 20 μM) were added to induce necroptosis. The tested compounds were incubated with the cells exposed to one of the above combinations at the indicated concentrations for 24 h. For acetaminophen hepatotoxicity assays, dabrafenib or necrostatin-1 at the indicated concentrations was incubated with the cells exposed to acetaminophen (20 mM) for the indicated times. Cell viability was then examined by using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega). Luminescence was recorded with an EnVision Multilabel reader (Perkin-Elmer).
Western blotting
Cellular levels of proteins were determined by the standard western blotting using appropriate antibodies. Proteins were visualized with an ImageQuant LAS 4000 mini system (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer's instructions.
Real-time PCR
Total RNA of the cells was extracted using the RNeasy Mini Kit (QIAGEN Inc., Valencia, CA, USA). One microgram of total RNA was reverse-transcribed using a RT reagent kit (TaKaRa). The resulting cDNA was amplified by real-time PCR using the SYBR PrimeScript RT-PCR kit (TaKaRa) in the Applied Biosystems 7500 Fast real-time PCR System (Applied Biosystem, Grand Island, NY, USA). The primer sequences were as follows: 5′-AATTCGTGGCTGCGCCTAGAAG-3′ (forward) and 5′-TCGTGCAGGTAAAACATCCCA-3′ (reverse) for RIP3; 5′-GGATGCAGAAGGAGATCACTG-3′ (forward); and 5′-CGAT CCACACGGAGTACTTG-3′ (reverse) for β actin. The relative quantification of RIP3 mRNA levels was automatically analyzed by the software of the PCR system.
Cell staining
For Hochest33342/PI staining, cells were incubated with Hochest33342/PI at 37 °C for 30 min and analyzed in INCell Analyzer 2000 (GE Healthcare) according to the manufacturer's instructions. For FACS, cells were trypsinized, collected by centrifugation, washed once with PBS buffer and then resuspended in PBS containing 5 μg/ml PI. The levels of PI incorporation were quantified with BD FACSCalibur (BD, Franklin Lakes, NJ, USA).
Small interfering RNA (siRNA)
Cells were transfected with 100 nM siRNA using RNAiMax (Invitrogen) according to the manufacturer's instructions. siRNA sequences were as follows: 5′-UGCAGUCUCUUCAACUUGAATT-3′ for RIP1;5 (#1) 5′-AGAAUUGGAUCUGGAUCAU-3′, (#2) 5′-AAAGAAUUGGAUCUGGAUCAU-3′ for B-Raf; 5′-TGAGTTACCAGGAAGTTTGTT-3′ for MLKL;31 5′-UAACUUGACG CACGACAUCAGGCUGUU-3′ for RIP3;5 and 5′-UUCUCCGAACGUGUCACGUTT-3′ as negative control. The siRNAs were obtained from Shanghai GenePharma Co. Ltd (Shanghai, China).
RIP3-silenced N cells
The expression of RIP3 was silenced in N cells with the specific short-hairpin RNAs (shRNAs) on a lentivirus vector (pLV-H1-EF1α-puro-RIP3-shRNA, a kind gift from Professor Jiahuai Han, Xiamen University, Xiamen, China). The sequence of RIP3 shRNA is 5′-AAAAGCTGAGTTGGTAGACAAGATTGGATCCAACGACTCAACCATCTGTTCTTTTTTTTT-3′. The sequence of the control shRNA is 5′-AAAAGCAGTTATCTGGAAGATCAGGTTGGATCCAACCTGATCTTCCAGATAACTGC-3′. The shRNA-contained lentivirus was packaged in HEK293FT cells and was used to infect N cells. The RIP3-silenced N cells were selected by adding the culture medium containing 1 μg/ml puromycin.3
Surface plasmon resonance assay
The binding affinity of dabrafenib or necrostatin-1 to RIP3 was determined by using Biacore T200 (GE Healthcare, Piscataway, NJ, USA). The purified recombinant RIP3 protein was immobilized to the CM5 chip surface (GE Healthcare) according to a standard amine-coupling procedure in 10 mM sodium acetate (pH 4.5) using HBS-EP (10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.05% (v/v) surfactant P20) as the running buffer. Dabrafenib and necrostatin-1 were serially diluted and injected onto the sensor chip at a flow rate of 30 μl/min for 120 s (contact phase), followed by 120 s of the buffer flow (dissociation phase). The Kd value of dabrafenib was determined with the BIA evaluation software (GE Healthcare).32
Homology modeling and molecular docking
Based on the similarity of dabrafenib to the ligand in B-Raf structure (PDB ID: 3SKC), we selected the 3SKC as the template to build the three-dimensional structure of human RIP3. The software Modeller (version 9.12)33 was used to perform the homology modeling and refinement with the default parameters. And the amino-acid residues were numbered in this study according the sequence NP_006862.
The ligand dabrafenib was built in Maestro (Schrödinger software, http://www.schrodinger.com/), and refined with the MMFF94 force field. The B-Raf (3SKC) and modeled human RIP3 structures were prepared with autodock tools to add the hydrogens and assign charges and saved as the pdbqt file format. Dabrafenib was also prepared with the autodock tool. Then the software Vina (version 1.1)34 was used to dock dabrafenib into the ATP-binding sites of B-Raf and RIP3. Totally, 10 runs were performed for each protein, and the lowest energy conformation was selected for the detailed analysis of interaction patterns.
Immunoprecipitation
HT-29 cells at 90% confluence grown were washed once with PBS and lysed for 30 min on ice in the NP-40 lysis buffer (Beyotime, Jiangsu, China). Cell lysates were then centrifuged at 12 000 × g for 20 min. The soluble fraction was collected, and the protein concentration was determined by using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Next, 1 mg of the extracted protein in the lysis buffer was immunoprecipitated overnight with anti-MLKL or anti-RIP1 antibody at 4 °C and then with protein A+G agarose beads (Beyotime) for another 4 h. After that, the protein A+G agarose beads were washed three times with the lysis buffer. The beads were then boiled in 1% SDS loading buffer for western blotting with the indicated antibodies.
Animal experiments
Specific pathogen-free male C57Bl/6J mice (weight 18–22 g) were supplied from the Shanghai Laboratory Animal Center, Chinese Academy of Sciences. The animals were housed. All experiments abided by institutional ethical guidelines of the Animal Care and Use Committee (Shanghai Institute of Materia Medica, Chinese Academy of Sciences, China). For drug administration, acetaminophen was dissolved in normal saline, whereas dabrafenib and vemurafenib were dissolved in pH 8.0 distilled water containing 0.5% HPMC and 0.2% Tween 80. Overnight-fasted mice were given dabrafenib (300 mg/kg or 100 mg/kg) or vemurafenib (300 mg/kg) by oral gavage 1 h before the intraperitoneal injection of 300 mg/kg acetaminophen. The mice were killed 6 h post the acetaminophen treatment. Plasma alanine aminotransferase and aspartate aminotransferase were measured with a Cobas C501 Chemistry Analyzer (Roche, Basel, Switzerland). Plasma IL-1β and TNF-α were measured by using the mouse IL-1β ELISA kit and the mouse TNF-α ELISA kit (MultiSciences, Bellingham, WA, USA), respectively. Liver GSSG was measured by using a GSSG Assay kit (Beyotime). To evaluate the changes in liver histology, sections of paraformaldehyde-fixed liver tissues were stained with hematoxylin and eosin and photographed under an optical microscope. The standard TUNEL staining was carried out to evaluate the DNA breaks in liver cells.
Statistical analysis
All the data, if applicable, were expressed as mean±S.D. Comparison between two groups was performed with the Student's t-test. P<0.05 was considered statistically significant. All the data expressed as mean±S.D. were from three independent experiments unless otherwise specified.
Acknowledgments
We thank Professor Jia-Huai Han (Xiamen University, Xiamen, China) for his kind gifts of A cells, N cells and the plasmids pLV-H1-EF1α-puro-RIP3-shRNA (shRIP3) and pLV-H1- EF1α-puro-NC (shNC). This work was supported by grants from the National Natural Science Foundation of China (No. 81373446, No. 81025020 and No. 81321092), the National Basic Research Program of China (No. 2012CB932502), the National Science and Technology Major Project of China (No. 2012ZX09301-001-002), the ‘Interdisciplinary Cooperation Team' Program for Science and Technology Innovation of the Chinese Academy of Sciences and the State Key Laboratory of Drug Research (No. SIMM1203ZZ-0103).
Glossary
- RIP
protein kinase receptor-interacting protein
- MLKL
mixed lineage kinase domain like protein
- TNF
tumor necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
- FADD
Fas-associated protein with death domain
- MBP
myelin basic protein
- IL-1β
interleukin-1β
- GST
glutathione S-transferase
- GSSG
glutathione disulfide
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Oberst
Supplementary Material
References
- 1Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol 2010; 22: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714. [DOI] [PubMed] [Google Scholar]
- 3Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336. [DOI] [PubMed] [Google Scholar]
- 4He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 1100–1111. [DOI] [PubMed] [Google Scholar]
- 5Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Moriwaki K, Chan FK. RIP3: a molecular switch for necrosis and inflammation. Genes Dev 2013; 27: 1640–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Roychowdhury S, McMullen MR, Pisano SG, Liu X, Nagy LE. Absence of receptor interacting protein kinase 3 prevents ethanol-induced liver injury. Hepatology 2013; 57: 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 2011; 477: 330–334. [DOI] [PubMed] [Google Scholar]
- 10Trichonas G, Murakami Y, Thanos A, Morizane Y, Kayama M, Debouck CM et al. Receptor interacting protein kinases mediate retinal detachment-induced photoreceptor necrosis and compensate for inhibition of apoptosis. Proc Natl Acad Sci USA 2010; 107: 21695–21700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Murakami Y, Matsumoto H, Roh M, Suzuki J, Hisatomi T, Ikeda Y et al. Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc Natl Acad Sci USA 2012; 109: 14598–14603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Lin J, Li H, Yang M, Ren J, Huang Z, Han F et al. A role of RIP3-mediated macrophage necrosis in atherosclerosis development. Cell Rep 2013; 3: 200–210. [DOI] [PubMed] [Google Scholar]
- 13James LP, Mayeux PR, Hinson JA. Acetaminophen-induced hepatotoxicity. Drug Metab Dispos 2003; 31: 1499–1506. [DOI] [PubMed] [Google Scholar]
- 14Antoine DJ, Dear JW, Lewis PS, Platt V, Coyle J, Masson M et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology 2013; 58: 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS et al. Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology 2005; 42: 1364–1372. [DOI] [PubMed] [Google Scholar]
- 16Zhao P, Wang C, Liu W, Chen G, Liu X, Wang X et al. Causes and outcomes of acute liver failure in china. PLoS One 2013; 8: e80991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Ramachandran A, McGill MR, Xie Y, Ni HM, Ding WX, Jaeschke H. Receptor interacting protein kinase 3 is a critical early mediator of acetaminophen-induced hepatocyte necrosis in mice. Hepatology 2013; 58: 2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Vucur M, Reisinger F, Gautheron J, Janssen J, Roderburg C, Cardenas DV et al. RIP3 inhibits inflammatory hepatocarcinogenesis but promotes cholestasis by controlling caspase-8- and JNK-dependent compensatory cell proliferation. Cell Rep 2013; 4: 776–790. [DOI] [PubMed] [Google Scholar]
- 19He JX, Wang YQ, Feng JM, Li JX, Xu L, Li XH et al. Differential sensitivity of RIP3-proficient and deficient murine fibroblasts to camptothecin anticancer drugs. Acta Pharmacol Sin 2012; 33: 426–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Kaiser WJ, Sridharan H, Huang C, Mandal P, Upton JW, Gough PJ et al. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J Biol Chem 2013; 288: 31268–31279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Xie T, Peng W, Liu Y, Yan C, Maki J, Degterev A et al. Structural basis of RIP1 inhibition by necrostatins. Structure 2013; 21: 493–499. [DOI] [PubMed] [Google Scholar]
- 23Wenglowsky S, Ahrendt KA, Buckmelter AJ, Feng B, Gloor SL, Gradl S et al. Pyrazolopyridine inhibitors of B-RafV600E. Part 2: structure-activity relationships. Bioorg Med Chem Lett 2011; 21: 5533–5537. [DOI] [PubMed] [Google Scholar]
- 24Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 2005; 1: 112–119. [DOI] [PubMed] [Google Scholar]
- 25Motzer RJ, Hutson TE, Cella D, Reeves J, Hawkins R, Guo J et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med 2013; 369: 722–731. [DOI] [PubMed] [Google Scholar]
- 26Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227. [DOI] [PubMed] [Google Scholar]
- 27Yang H, Higgins B, Kolinsky K, Packman K, Bradley WD, Lee RJ et al. Antitumor activity of BRAF inhibitor vemurafenib in preclinical models of BRAF-mutant colorectal cancer. Cancer Res 2012; 72: 779–789. [DOI] [PubMed] [Google Scholar]
- 28Hoeflich KP, Herter S, Tien J, Wong L, Berry L, Chan J et al. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer Res 2009; 69: 3042–3051. [DOI] [PubMed] [Google Scholar]
- 29Luke JJ, Hodi FS. Ipilimumab, vemurafenib, dabrafenib, and trametinib: synergistic competitors in the clinical management of BRAF mutant malignant melanoma. Oncologist 2013; 18: 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science 2004; 305: 1471–1474. [DOI] [PubMed] [Google Scholar]
- 31Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012; 148: 228–243. [DOI] [PubMed] [Google Scholar]
- 32Wen WW, Xie S, Xin XL, Geng MY, Ding J, Chen Y. Oligomannurarate sulfate inhibits CXCL12/SDF-1-mediated proliferation and invasion of human tumor cells in vitro. Acta Pharmacol Sin 2013; 34: 1554–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Eswar N, Webb B, Marti-Renom MA, Madhusudhan M, Eramian D, Shen My et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2006: 1–30 Chapter 5, Unit 5.6. [DOI] [PMC free article] [PubMed]
- 34Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010; 31: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Laquerre S, Arnone M, Moss K, Yang J, Fisher K, Kane-Carson L et al. A selective Raf kinase inhibitor induces cell death and tumor regression of human cancer cell lines encoding B-RafV600E mutation. Mol Cancer Ther 2009; 8: B88. [Google Scholar]
- 36Khazak V, Astsaturov I, Serebriiskii IG, Golemis EA. Selective Raf inhibition in cancer therapy. Expert Opin Ther Targets 2007; 11: 1587–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Wilhelm SM, Dumas J, Adnane L, Lynch M, Carter CA, Schutz G et al. Regorafenib (BAY 73-4506): a new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int J Cancer 2011; 129: 245–255. [DOI] [PubMed] [Google Scholar]
- 38Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA 2008; 105: 3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 2010; 467: 596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 2004; 64: 7099–7109. [DOI] [PubMed] [Google Scholar]
- 41Wong H, Belvin M, Herter S, Hoeflich KP, Murray LJ, Wong L et al. Pharmacodynamics of 2-[4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridine-4-yl)-1H-pyraz ol-1-yl]ethan-1-ol (GDC-0879), a potent and selective B-Raf kinase inhibitor: understanding relationships between systemic concentrations, phosphorylated mitogen-activated protein kinase kinase 1 inhibition, and efficacy. J Pharmacol Exp Ther 2009; 329: 360–367. [DOI] [PubMed] [Google Scholar]
- 42King AJ, Patrick DR, Batorsky RS, Ho ML, Do HT, Zhang SY et al. Demonstration of a genetic therapeutic index for tumors expressing oncogenic BRAF by the kinase inhibitor SB-590885. Cancer Res 2006; 66: 11100–11105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.