Abstract
Intracellular accumulations of altered, misfolded proteins in neuronal and other cells are pathological hallmarks shared by many neurodegenerative diseases including amyotrophic lateral sclerosis (ALS). Mutations in several genes give rise to familial forms of ALS. Mutations in Sigma receptor 1 have been found to cause a juvenile form of ALS and frontotemporal lobar degeneration (FTLD). We recently described altered localization, abnormal modification and loss of function of SigR1 in sporadic ALS. In order to further elucidate the molecular mechanisms underlying SigR1-mediated alterations in sporadic and familial ALS, we extended our previous studies using neuronal SigR1 knockdown cell lines. We found that loss of SigR1 leads to abnormal ER morphology, mitochondrial abnormalities and impaired autophagic degradation. Consistent with these results, we found that endosomal trafficking of EGFR is impaired upon SigR1 knockdown. Furthermore, in SigR1-deficient cells the transport of vesicular stomatitis virus glycoprotein is inhibited, leading to the accumulation of this cargo protein in the Golgi apparatus. Moreover, depletion of SigR1 destabilized lipid rafts and associated calcium mobilization, confirming the crucial role of SigR1 in lipid raft and intracellular calcium homeostasis. Taken together, our results support the notion that loss of SigR1 function contributes to ALS pathology by causing abnormal ER morphology, lipid raft destabilization and defective endolysosomal pathways.
ER stress causes alterations in protein quality control, autophagy, calcium imbalance and mitochondrial dysfunction.1, 2, 3 It is central to the pathogenesis of many neurodegenerative diseases.4, 5 Altered proteins are targeted by molecular chaperones for protein repair or refolding. However, failure of this first line of defense leads to abnormal aggregates of such proteins that then form ubiquitinated cellular inclusions and compromise UPS function.6, 7, 8, 9 Macroautophagy, the major lysosomal degradative pathway in cells, is responsible for degrading long-lived cytoplasmic constituents; it is the principal mechanism for turning over cellular organelles and protein aggregates too large to be degraded by the proteasome.10, 11, 12, 13 Macroautophagy, hereafter referred to as autophagy, is a multistep process, initiated primarily by sequestration of portions of the cytoplasm in double-membrane-bound vesicles to form an autophagosome. Autophagosomes along with their cargoes are then degraded upon fusing with late endosome- or lysosome-containing cathepsins, other acid hydrolases, and vacuolar [H+] ATPase.14 Even though autophagy is a selective and efficient mechanism for the degradation of misfolded and mutant proteins related to neurodegeneration, recent evidence indicates that the alterations in certain disease-related genes may actually impair autophagic activity at different levels, including accumulation of autophagic vacuoles,15 substrate recognition, lysosomal acidity and autophagosome membrane nucleation.16, 17
SigR1 interacts with a variety of ligands and is involved in a broad array of biological functions that have only been partially defined so far. They include regulation of neuronal survival, neuritogenesis, ion channel activity, IP3R-mediated Ca2+ signaling, memory and drug addiction. SigR1 is an ER chaperone that is located at the mitochondria–ER interface and is normally bound to another chaperone, BiP/GRP78.18 Upon IP3 receptor stimulation or Ca2+ depletion within the ER, SigR1 dissociates from BiP and stabilizes IP3 receptors, leading to prolonged Ca2+ signaling into mitochondria.18 Recently, a mutation in SigR1, E1O2Q, has been reported to cause a juvenile form of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD).19 SigR1 protein is decreased significantly in human sporadic ALS spinal cord.20 In addition, functional relevance of SigR1 in ALS pathogenesis is demonstrated by SigR1 knockout mice that display motor deficits with a shorter latency period in rotarod experiments, and by another study showing that lack of SigR1 exacerbates ALS progression in SOD1 tg mice.21, 22 Furthermore, a recent study showed improvement in motor function and survival of motor neurons in SOD1 mice treated with a SigR1 agonist.23 Recently, we reported altered localization and abnormal modification of SigR1 in sporadic ALS and showed that loss of SigR1 function leads to deformities in ER structure, formation of ER-derived autophagic vacuoles and induction of ER stress.20 Loss of function of SigR1 also leads to aberrant calcium homeostasis and cell death.20 Together, these data suggest a crucial role of SigR1 in neuronal function and survival.
Autophagy has been known to be tightly linked to ER function and is decisive in neurodegeneration mediated by ER stress.24, 25, 26 Therefore, we hypothesized that loss of SigR1 function contributes to a vicious circle including ER stress, defective autophagy and altered calcium signaling that causes multifactorial ALS pathology. In the present study, we show that depletion of SigR1 leads to the accumulation of numerous autophagic vacuoles often filled with nondegraded autophagic substrates, and severe deformities in ER ultrastructure including loss of ER tethering. Biochemical analysis in SigR1-deficient cells revealed the accumulation of various autophagic substrates and defects in endosomal trafficking, suggesting an impairment of endolysosomal pathways. Finally, loss of SigR1 destabilized lipid rafts causing both impaired calcium mobilization and altered endosomal trafficking. Altogether, our results support the notion that loss of SigR1 contributes to ALS pathogenesis by causing abnormal ER morphology, lipid raft destabilization and defective autophagy.
Results
Loss of function of SigR1 leads to structural abnormalities of the ER/Golgi and to accumulation of autophagic material
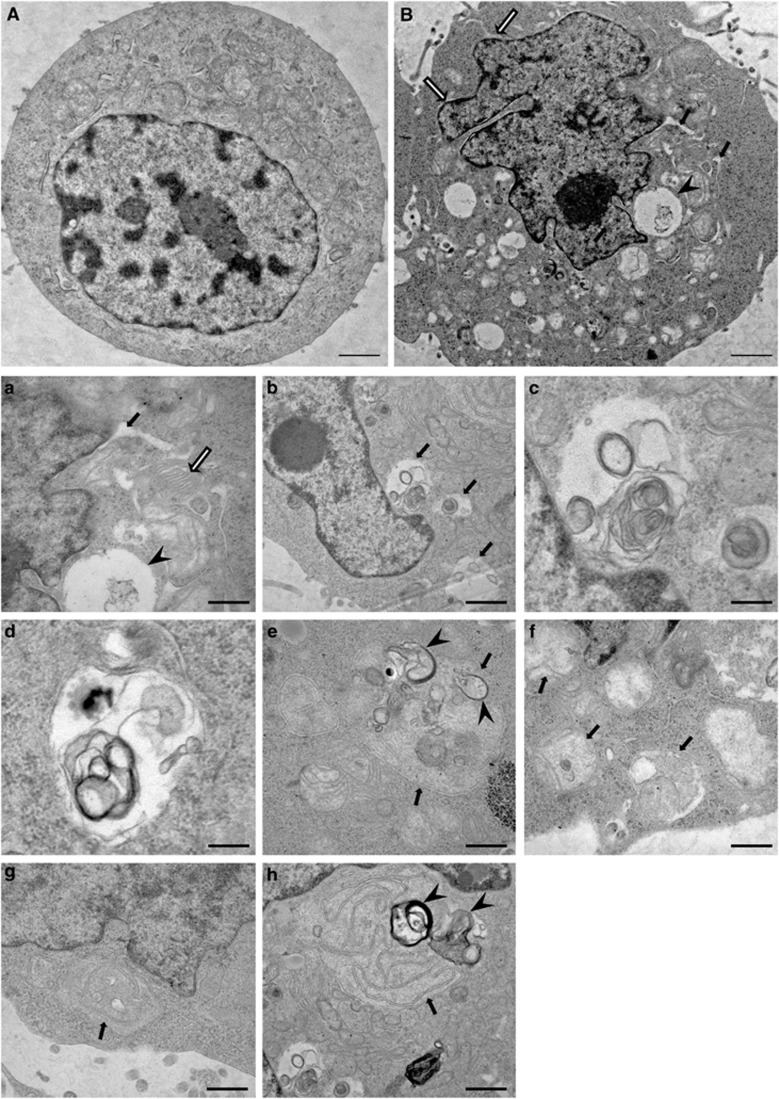
We previously reported that knockdown of SigR1 in HEK 293 and NSC34 cells caused ER widening and induced the formation of numerous ER-derived vacuoles often filled with membranous autophagic material.20 In the present study, we confirmed our previous findings and further characterized these vacuolar structures in HEK 293 and NSC34 motor neuron-like cells lacking SigR1. EM studies revealed that depletion of SigR1 leads to the accumulation of numerous autophagic vacuoles15 often filled with nondegraded autophagic substrates (Figure 1B and Supplementary Figures 2A–D) and deformities of ER ultrastructure (Figure 1B). Furthermore, knockdown of SigR1 induced mitochondrial abnormalities as well as mitophagy and led to ER untethering and proliferation, suggesting a crucial role of SigR1 in maintaining structural integrity of the ER and stabilizing mitochondria at the MAM (Figure 1Be–h).
Figure 1.
Depletion of SigR1 leads to ER deformities and accumulation of autophagic vacuoles. HEK293 cells were either transfected with control shRNA or SigR1 shRNA. After 48 h, the transfected cells were fixed with 2.5% buffered glutaraldehyde and processed for electron microscopy. (A) HEK293 cell transfected with control shRNA shows normal mitochondria and ER and no evidence of defective autophagy. Scale bar=1 μm. (B) HEK293 cell transfected with SigR1 shRNA shows widened ER profiles (black arrows) and membrane-bound vacuolar structures, some corresponding to autophagosomes (arrowhead). Note the focal widening of the nuclear envelope (white arrows). Scale bar=1 μm. (a) Higher magnification of the widened ER depicted in (B), which is now seen to be connected to the nuclear envelope (arrow), and stacked membranes (white arrow) probably derived from the Golgi. Arrowhead indicates incipient autophagosome. Scale bar=0.5 μm. (b) Multiple lamellar bodies in large autophagic vacuoles (arrows) of another HEK293 cell transfected with SigR1 shRNA. Scale bar=1 μm. (c) Enlarged view of one of the autophagic vacuoles depicted in (b), probably a mature double membranous autophagosome containing phospholipids and other undigested substrates. Scale bar=0.3 μm. (d) Large, double membrane-bound autophagic vacuole containing membranous and other osmiophilic accumulations. Scale bar=0.3 μm. (e) Enlarged mitochondrion (arrows) containing autophagic material indicating mitophagy. Scale bar=0.4 μm. (f) Mitochondria (arrows) showing dissolution of cristae architecture. Scale bar=0.5 μm. (g and h) Untethered ER (arrows) with several multilamellar bodies (arrowheads, only in h). Scale bar=0.5 μm
Knockdown of SigR1 leads to the accumulation of various components of the lysosomal pathway
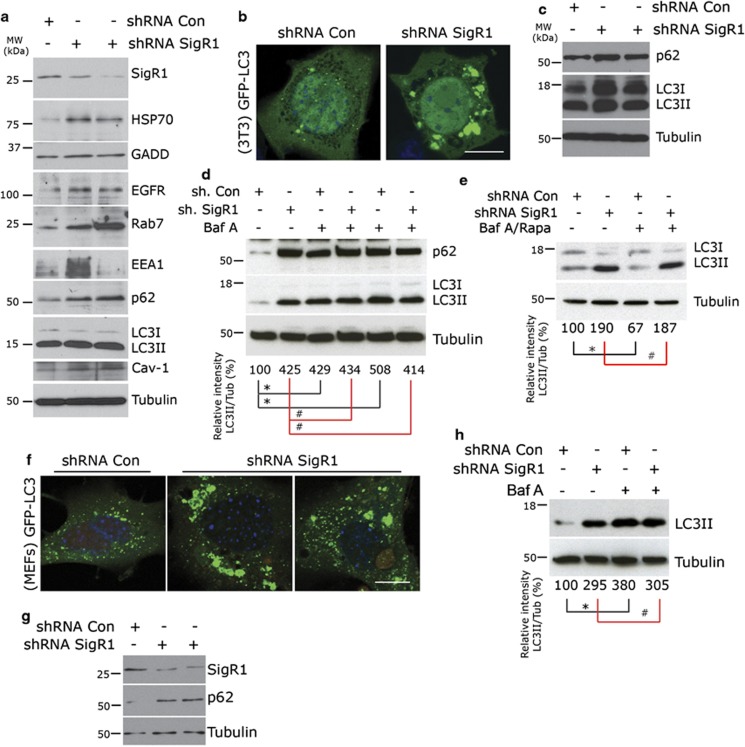
Based on our observation that SigR1 deficiency induces the formation of defective ER and of ER-derived autophagic vacuoles, we hypothesized that the autophagic process is impaired in SigR1-deficient cells. Autophagy is characterized by the engulfment of cytoplasmic components26 in double-membrane-bound structures25 that are then delivered to lysosomes/vacuoles for degradation. Autophagic activity can thus be analyzed by determining the levels of LC3II and p62 by western blotting as well as the number of LC3II-positive cytoplasmic punctae by immunofluorescence.10, 27, 28 NSC-34 cells lacking SigR1 showed evidence of increased ER and misfolded protein stress (Figure 2a), confirming and extending our previous results,20 along with the accumulation of p62, LC3II and several other known markers for endolysosomal pathways such as epidermal growth factor receptor (EGFR), the early endosomal antigen EEA1 and Rab7 (Figure 2a and Supplementary Figure 1a). In order to further confirm that autophagy is defective, we generated a mouse fibroblast (NIH-3T3) cell line stably expressing GFP-LC3 with normal basal autophagic activity (Supplementary Figure 1d). Under basal conditions, knockdown of SigR1 in these cells led to abnormal accumulation of GFP-LC3 (Figure 2b). Abnormal autophagy was also evident by p62 and LC3 accumulation already in the absence of autophagy inhibitors (Figure 2c). Autophagy inhibition due to loss of SigR1 was further confirmed by the disproportionate increase in LC3II and of p62 levels in the presence of the known autophagy inhibitor bafilomycin A and of the autophagy inducer rapamycin (Figures 2d and e). Parallel experiments confirmed these results in HeLa cells transfected with a GFP-LC3 construct and obtained analogous results (Supplementary Figures 1b and c). In order to further confirm our results at a physiological expression level of LC3 in a primary cell culture system, we used MEFs from GFP-LC3 tg mice.29 Knockdown of SigR1 in MEFs confirmed the above-described results of disproportionate GFP-LC3 and p62 accumulation (Figures 2f–h), again consistent with the inhibition of autophagic degradation after SigR1 knockdown. Taken together, these results demonstrate that loss of SigR1 protein leads to a defective endolysosomal autophagy pathway.
Figure 2.
Accumulation of various components of the lysosomal pathway in SigR1-depleted cells. (a) NSC34 cells were transiently transfected with two different SigR1 shRNA constructs, achieving different levels of suppression of SigR1 expression. Cell lysates were subjected to immunoblot analysis with antibodies indicated. (b) NIH-3T3 cells stably expressing GFP-LC3 were transiently transfected either with control or SigR1 shRNA. At 48 h after transfection, cells were processed for immunofluorescence. Representative images depict the accumulation of GFP-LC3 as a marker for autophagy inhibition. Scale bar=8 μm. (c) NIH-3T3 cells stably expressing GFP-LC3 were transiently transfected with two different SigR1 shRNA constructs, achieving different levels of suppression of SigR1 expression. Immunoblot analysis was performed using p62 and LC3 as markers for autophagy. (d) NIH-3T3 cells stably expressing GFP-LC3 were transiently transfected either with control or SigR1 shRNA. At 48 h after transfection, cells were treated with or without the autophagy inhibitor bafilomycin A at increasing concentration (lanes 3 and 4: 10 nM, lanes 5 and 6: 20 nM) for 3 h. Immunoblot analysis was performed using p62 and LC3 as markers for autophagy. (e) NIH-3T3 cells stably expressing GFP-LC3 were transiently transfected as described above. After 48 h, the cells were treated with or without the autophagy inhibitor bafilomycin A (20 nM) for 3 h, and then replaced by the autophagy inducer rapamycin (20 nM) for additional 3 h. Cell lysates were prepared and subjected to immunoblot analysis using the LC3 antibody. (f) MEFs obtained from GFP-LC3 tg mice were transiently transfected either with control or SigR1 shRNA. At 48 h after transfection, cells were processed for GFP staining. Scale bar=6 μm. (g) Western blot analysis of these MEFs transiently transfected either with control or two different SigR1 shRNAs. At 48 h after transfection, cell lysates were subjected to p62 and LC3 immunoblots. (h) MEFs obtained from GFP-LC3 tg mice were transiently transfected either with control or SigR1 shRNA. At 48 h after transfection, cells were treated with or without the autophagy inhibitor bafilomycin A (20 nM) for 3 h. Cell lysates were prepared and subjected to immunoblot analysis using the LC3 antibody as a marker for autophagy. For all the experiments, values are means±S.D. of three independent experiments. *P<0.05, **P<0.005, #not significant
Depletion of SigR1 leads to defective endosome-mediated EGFR degradation/fusion of autophagosome to lysosomes
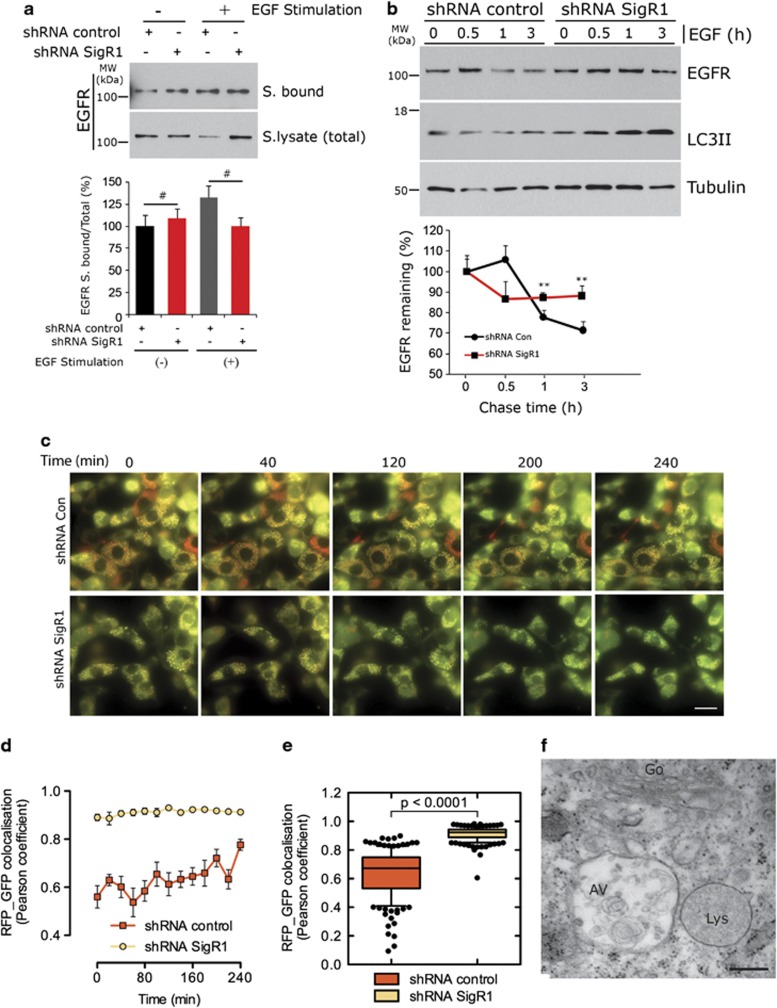
Because the impaired degradation and accumulation of autophagic substrates was indicative of a deficient autophagic process, we hypothesized that the transport and fusion of endosomes/autophagosomes to lysosomes was impaired. We took advantage of the widely studied internalization and degradation of the EGF–EGFR complex in A431 cells expressing high levels of EGFR and showing normal autophagic activity (Supplementary Figure 1e). In this system, EGFR can be activated by both EGF and TGF-α,30, 31 leading to clustering and internalization of the EGF–EGFR complex into early endosomes or membrane compartments characterized by tubular vesicular morphology and a mild acidic pH. From early endosomes, EGF receptors can be either recycled back to the plasma membrane (recycling pathway) or into multivesicular bodies (MVBs) for degradation in lysosomes.32 Suppression of SigR1 expression by three different shRNA constructs caused a significant accumulation of EGFR along with p62 and LC3II in A431 cells, confirming that deficiency of SigR1 leads to defective autophagy (Supplementary Figure 1f). Importantly, downregulation of SigR1 did not impair receptor internalization as indicated by similar biotinylation levels of plasma membrane-associated EGFR in SigR1-deficent cells and control cells after 10 min of EGF stimulation (Figure 3a). Next, we monitored the degradation of EGFR after internalization. Interestingly, we observed an efficient degradation of EGFR in control cells after 1 h of activation followed by almost 30% reduction after 3 h, whereas the levels of EGFR in SigR1-deficient cells were unaffected within 1 h after stimulation and only slightly reduced (10–15%) after 3 h. The differences in reduction of EGFR levels after 3 h between control transfected and SigR1-deficient cells were statistically significant (P<0.05). Consistent with these data, we found an increased accumulation of LC3II in the SigR1-deficient cells as compared with the controls, again confirming the above-described impairment of lysosomal degradation.
Figure 3.
Defective endosome-mediated EGFR degradation/fusion of autophagosomes to lysosomes in SigR1 shRNA knockdown cells. (a) A431 cells were transiently transfected with either control shRNA or SigR1 shRNA. At 48 h after transfection, cells were processed for surface biotinylation assays. Immunoblot analysis using EGFR antibody revealed no significant alteration in EGFR internalization. Values in the quantification (lower panel) are means±S.D. of three independent experiments. #not significant. (b) A431 cells were transiently transfected as described above and proceeded for EGFR degradation assays combined with LC3 immunoblots. Lower panel: densitometric analysis of EGFR immunoblots (Adobe PS CS5). Values are expressed as mean±S.D. from three independent experiments. **P<0.005. (c) NIH-3T3 cells stably expressing RFP-GFP-LC3 were transiently transfected either with control or SigR1 shRNA (without RFP tag, see Materials and Methods). At 48 h after transfection, cells were treated with bafilomycin A (20 nM) for 3 h to block autophagosome fusion; 1 h later, the rate of lysosome fusion with autophagosomes was monitored in control and SigR1 shRNA-transfected cells by live cell imaging. Scale bar=25 μm. (d and e) The rate of autophagosome maturation reflected by Pearson's coefficient (green/red fluorescence ratio) at each time point indicated after 1 h of bafilomycin A removal. Values are represented as means±S.E.M. of triplicate experiments. (f) Large, double membrane-bound autophagic vacuole (AV) containing membranous and other osmiophilic accumulations, not fused with lysosomes (Lys) in SigR1 shRNA-transfected cells. Scale bar=0.25 μm
To corroborate these findings, we assessed the fusion of endosomes or autophagosomes to lysosomes using NIH-3T3 cells stably expressing a tandem fluorescence-tagged autophagosomal marker in which LC3 was linked to both monomeric red fluorescent protein (mCherry) and GFP. Because GFP signal is lost upon degradation, this fusion protein serves as a reliable tool for monitoring autophagosome fusion with lysosomes by determining the extent of the colocalization of RFP and GFP signals.33 By using live cell imaging (Figure 3c and Supplementary Movie 1), we found that the fusion of autophagosomes to lysosomes was significantly impaired in SigR1-deficient cells as indicated by the minimal loss of GFP fluorescence and maximal colocalization of RFP and GFP signals (Figures 3d and e and Supplementary Movie 1). In most of the control cells, in contrast, we observed a clear reduction of GFP fluorescence and RFP–GFP colocalization. As a final confirmation of the above-described results, we next analyzed the autophagosome to lysosome fusion by EM. We consistently observed the accumulation of several double-membrane autophagosomes (AV) filled with cargos (Supplementary Figures 3a–d) that failed to fuse with lysosomes (Lys) in SigR1-deficient GFP–RFP cell lines (Figure 3f and Supplementary Figures 3b and c) and in SigR1-deficient NSC34 and HEK293 cell lines (Supplementary Figures 2B–D). Taken together, these findings support the notion that SigR1 regulates the endolysosomal pathway by controlling the delivery to and fusion of cargo vesicles with lysosomes.
Depletion of SigR1 leads to impaired ER to Golgi transport of VSVG-GFP
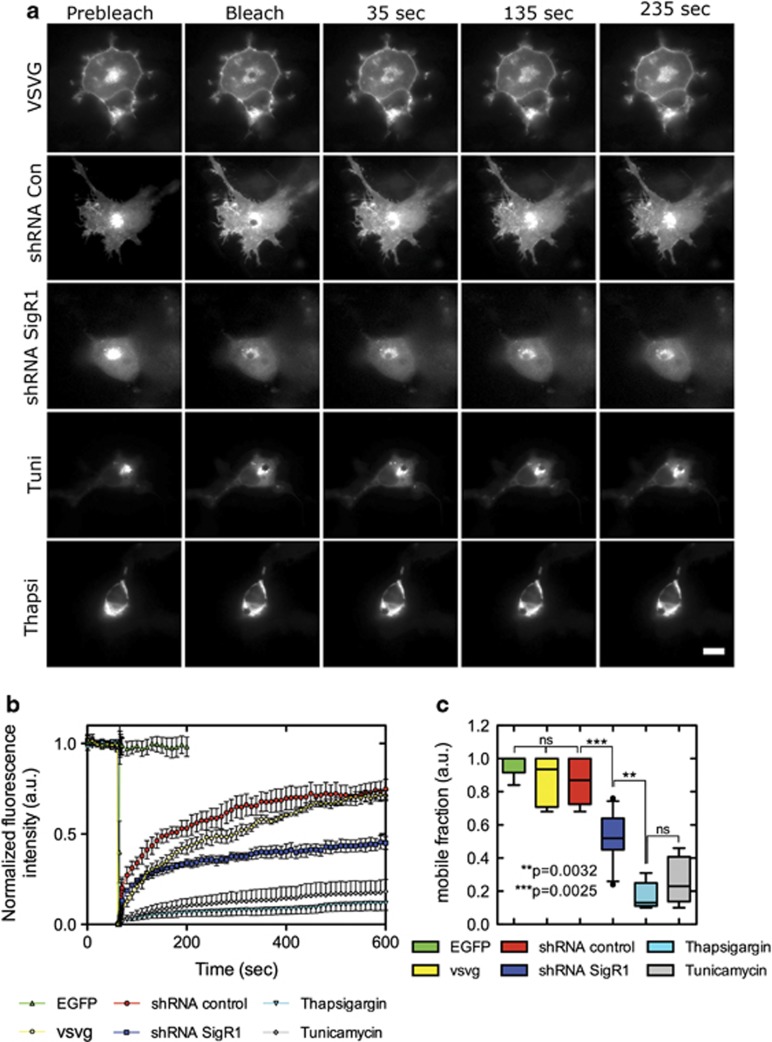
Nascent proteins that leave the ER are channeled through the Golgi complex before they are sorted for transport to their various final destinations. Having observed that ER ultrastructure was compromised considerably in SigR1-deficient cells, we hypothesized that the ER defects might be associated with alterations of vesicle transport from ER to Golgi. To test this hypothesis, we used the temperature-sensitive viral glycoprotein VSVG tagged with green fluorescent protein (VSVG-GFP). VSVG-GFP is misfolded and retained in the ER at 40 °C, whereas it acquires a normal conformation at 32 °C, thus being able to leave the ER and move to the Golgi complex along microtubules by using the microtubule minus-end-directed motor complex of dynein/dynactin (Supplementary Figure 4a).34, 35 We analyzed the delivery of VSVG-GFP from pre-Golgi intermediates to the Golgi complex by fluorescence recovery after photobleaching (FRAP) imaging in control cells and SigR1-deficient cells. In VSVG controls (COS-7 cells transfected with VSVG-GFP only) and shRNA control cells, the trafficking of VSVG-GFP vesicles was efficient as indicated by the fast recovery of VSVG-GFP after photobleaching (Figures 4a–c and Supplementary Movies 2 and 3). In contrast, in SigR1-deficient cells and cells treated with the known ER stressors Thapsigargin and Tunicamycin, the intracellular movement of VSVG-GFP vesicles was severely impaired, causing a decrease in VSVG-GFP fluorescence recovery (Figures 4a–c and Supplementary Movie 2). Thus, in agreement with our previous work showing ultrastructural abnormalities of the ER/Golgi complex in SigR1-deficient cells,20 these observations indicate that SigR1 is essential for the transport of vesicles from the ER to the Golgi complex. Moreover, these results are also consistent with previous studies showing that VSVG transport is impaired by ER stress.34, 35
Figure 4.
Loss of function of SigR1 leads to impaired ER to Golgi transport of VSVG-GFP. (a) Cos7 cells were transfected with VSVG-GFP alone (upper panel) or co-transfected with shRNA control or SigR1 shRNA (as indicated in the figure panels). At 48 h after transfection, the cells were either treated with ER stressor thapsigargin (5 μM) or tunicamycin (10 μM), or left untreated (as indicated in the figure panels). Fluorescence associated with the Golgi complex was photobleached with high-intensity laser light. Subsequent inward delivery of VSVG-GFP from pre-Golgi intermediates was monitored for the indicated periods of time. Scale bar=10 μm. (b) Fluorescence recoveries after photobleaching curves of SigR1 shRNA-transfected cells as well as cells treated with thapsigargin and tunicamycin show a clear decrease of the mobile fraction as compared with the shRNA and VSVG controls. Error bars indicate the S.E.M. (c) Comparison of the mobile fractions in controls and SigR1-deficient cells. In the box plots, the line in the middle of the box indicates the median; the top line indicates the 75th quartile, whereas the bottom line indicates the 25th quartile. Whiskers represent the 10th (lower) and 90th (upper) percentile, respectively. **P=0.0032; ***P=0.0025
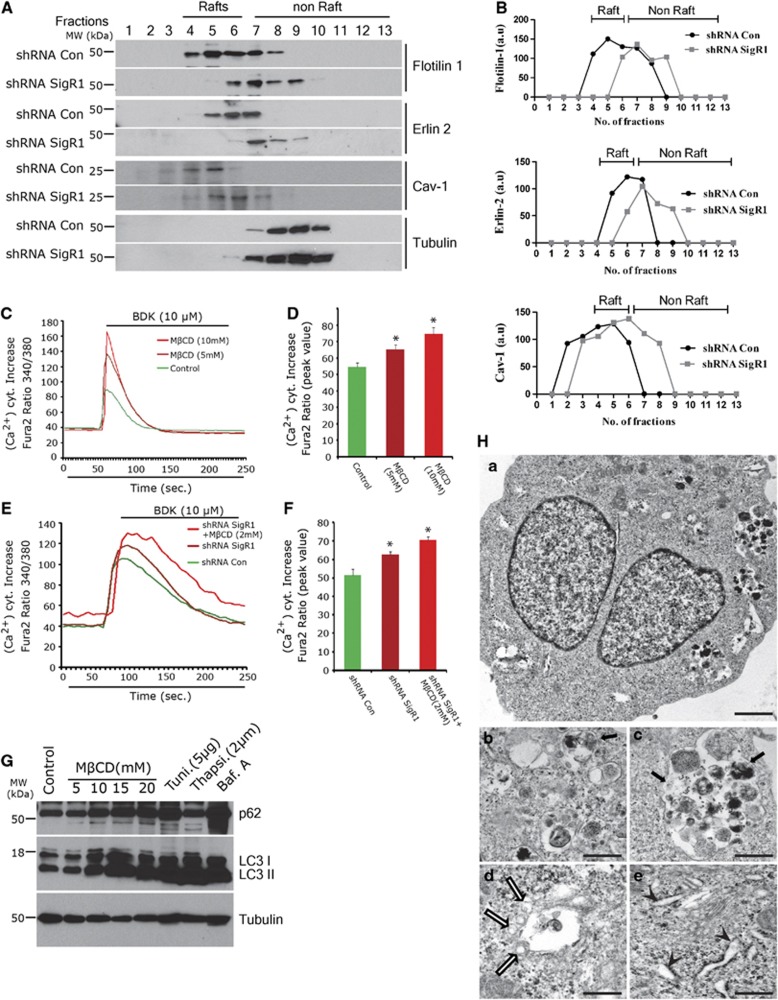
shRNA knockdown of SigR1 protein destabilizes membrane lipid rafts and associated calcium signaling
Lipid rafts are microdomains enriched in cholesterol and sphingolipids that – among other functions – orchestrate Ca2+ channels to maintain Ca2+ mobilization from intracellular stores via the IP3R as well as Ca2+ influx across membranes.36, 37 SigR1 has been shown to play an important role in modeling lipid rafts and regulating IP3R-mediated Ca2+ mobilization at the MAM.38 We investigated whether the loss of SigR1 function can affect these processes. Downregulation of SigR1 was found to disrupt lipid rafts by shifting the raft components toward the nonraft fractions. In particular, in SigR1-deficient cells, the raft markers Flotillin-1, Erlin-2 and Caveolin-1 became more enriched in the nonraft fractions (Figures 5A and B). To confirm these findings, we disrupted lipid rafts using mild concentrations of methyl-β-cyclodextrin (MβCD) that removes cholesterol from lipid rafts.39 Treatment with MβCD shifted the raft marker vesicle-associated membrane protein-associated protein B (VAPB) to nonraft fractions to a similar extent as in SigR1-deficient cells (Supplementary Figure 5c). Interestingly, treatment with bafilomycin A, which impairs the fusion of autophagosomes to lysosomes, caused alterations similar to the ones we observed after MβCD treatment or SigR1 depletion (Supplementary Figure 5c). Moreover, we observed that SigR1 (itself a raft marker) was shifted to and accumulated in nonraft fractions after MβCD or bafilomycin A treatment (Supplementary Figure 5c).
Figure 5.
Knockdown of SigR1 protein destabilizes membrane lipid rafts and raft-associated Ca2+ signaling. (A) NSC34 cells were transiently transfected either with control shRNA or SigR1shRNA as described above. At 48 h after transfection, cell lysates were subjected to sucrose density gradient centrifugation to isolate lipid rafts. Proteins from equal volumes of representative fractions (see also Supplementary Figures 2A and B) were resolved by SDS-PAGE and analyzed by western blotting using specific antibodies against raft markers. (B) Quantification of western blots of (A). (C and D) NSC34 cells were treated with MβCD for 24 h at the doses indicated. After the treatment, cells were loaded with Fura-2AM for 30 min, washed twice and were then stimulated with 10 μM BDK. Average traces of the BDK-induced increase in [Ca2+]i in the transient intracellular Ca2+ signaling evoked from the ER store through IP3R. (E and F) NSC34 cells were transiently transfected with SigR1 shRNA for 48 h and then treated with MβCD for 24 h. After the treatment, transient intracellular Ca2+ signaling evoked from the ER store through IP3R was measured as described above. Note the increase in basal levels of intracellular calcium in shRNA knockdown cells treated with MβCD. Average traces of the BDK-induced increase in [Ca2+]i in the knockdown cells and MβCD-treated cells as compared with control cells are shown in the pictograph. Results are expressed as mean±S.E.M. of ∼30 cells. Right panels in (E) and (F) indicate mean changes in peak [Ca2+]i measured. The asterisks denote a statistically significant difference (*P<0.05). (G) Western blot analysis of NSC34 cells either treated with different doses of MβCD or with the ER stressors tunicamycin and thapsigargin or the autophagy inhibitor bafilomycin A. (H) NSC34 cells were treated with 5 mM MβCD for 24 h, fixed with 2.5% buffered glutaraldehyde and processed for EM. (a) MβCD-treated cell showing prominent autophagic vacuoles and MVBs. Scale bar=2.5 μm. (b–e) Higher magnification of the autophagic vacuoles (arrows) of vesicles before fusing with multivesicular body (white arrows) and of the widened ER (arrowheads)
Given the crucial role of lipid rafts in maintaining intracellular calcium homeostasis, we examined the consequences of lipid raft disruption on calcium mobilization. We observed a dose-dependent increase of Ca2+ release from the ER mediated by the IP3R in cells treated with MβCD (Figures 5C and D) that was further increased in amplitude and duration in SigR1-deficient cells (Figures 5E and F).20 Given the similar defects in Ca2+ signaling and lipid raft structure caused by SigR1 deficiency and MβCD treatment and the altered autophagic function in SigR1-deficient cells, we speculated that lipid raft alterations may be associated with alterations of the endolysosomal pathway. We therefore analyzed the levels of autophagy markers in cells pretreated with MβCD. Treatment with MβCD led to a robust accumulation of the autophagic substrates p62 and LC3II comparable to the accumulation induced by the autophagy inhibitor bafilomycin A and the ER stressors tunicamycin and thapsigargin (Figure 5G) and to that induced by SigR1 deficiency (Figures 2c, d, f and g). Finally, we corroborated these observations by EM analysis. MβCD caused the accumulation of various double-membrane autophagosomes filled with autophagic substrates (Figure 5Hb–d), MVBs (Figure 5Hb and c) and of endosomes in the process of merging with autophagosomes (Figure 5Hd, white arrows) as well as widening of the ER (Figure 5He, arrowheads). Similar ultrastructural alterations were also found in SigR1-deficient cells (Figure 1).
Altogether, our findings indicate that SigR1 plays a crucial role in the regulation of the biochemical link between lipid rafts, calcium mobilization, endolysosomal pathways and autophagic processes.
Discussion
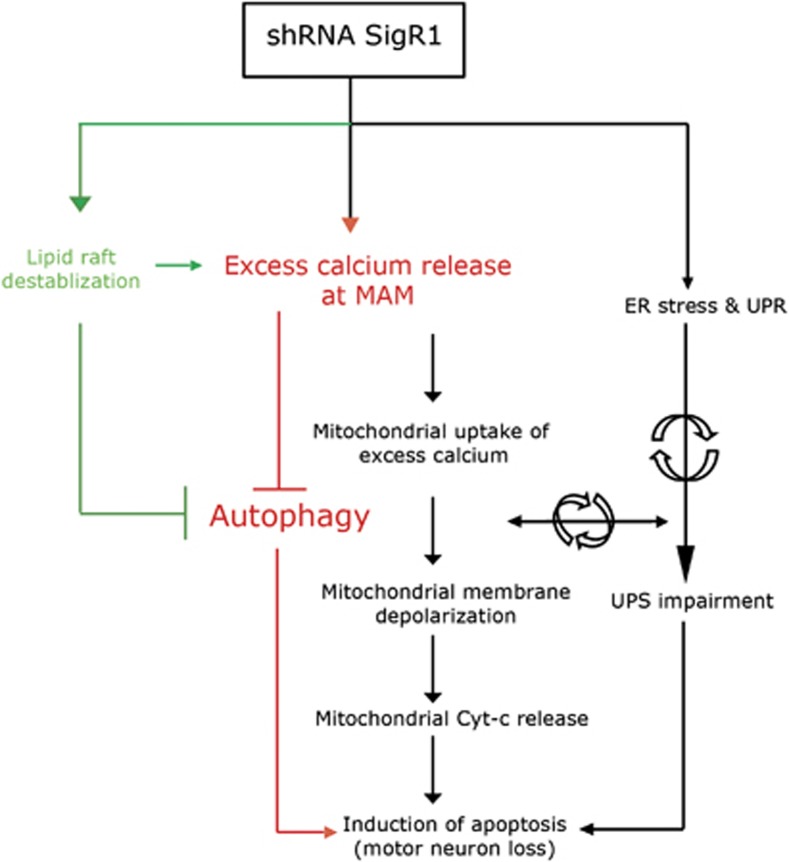
Recent studies have demonstrated that the SigR1 mutation (E102Q) causes ALS/FTLD in patients and reduces cell viability in cell culture model.19 Similarly, a truncated form of the SigR1 (a splice variant generated by splice site at residue E102 of full-length SigR1 protein) promotes mitochondrial energy depletion, ER stress and apoptosis in cell culture models.40 Furthermore, aberrant subcellular distribution of altered SigR1 protein was found in motoneurons of sporadic and familial ALS patients.20 However, whether mutant SigR1 exerts its pathological effects by a toxic gain- or a loss-of-function mechanism is still an open question. Importantly, SigR1 knockout mice display an ALS-like phenotype by showing motor deficits22 and ALS progression exacerbates in SOD1 transgenic mice lacking SigR1.21, 22 These findings strongly support loss of function of SigR1 as a causative mechanism of ALS pathogenesis. Consistent with the above-mentioned studies (loss-of-function hypothesis), we previously reported ER stress, mitochondrial abnormalities and apoptosis upon SigR1 depletion in various cell culture models.20 In the present study, we provide a more precise molecular mechanism of pathogenesis associated with loss of SigR1 function and propose that depletion of SigR1 involves a vicious circle composed of ER deformation, lipid raft disruption and deranged calcium homeostasis and a defective endolysosomal pathway (see Figure 6).
Figure 6.
Schematic representation of pathways of autophagy inhibition and apoptosis induction mediated by SigR1 knockdown. Molecular mechanisms corresponding to black lines and arrows were previously described.20 As described in the present paper, depletion of SigR1 leads to excess calcium release that induces autophagy inhibition and induction of apoptosis. Alternatively, knockdown of SigR1 leads to the destabilization of lipid rafts that in turn also induces excess calcium release and further autophagy inhibition leading to apoptosis. Lipid raft destabilization can also directly impair the autophagy that can further lead to induction of apoptosis
SigR1, autophagy and neurodegeneration
Autophagy is considered to be a selective and efficient mechanism for the degradation of misfolded and mutant proteins associated with neurodegenerative diseases including amyloid precursor protein (APP), α-synuclein, superoxide dismutase-1 (SOD1) and many others.41, 42, 43 Recent reports showed that defects in autophagic flux or in specific autophagy regulatory processes contribute to neurodegeneration and that autophagy is an essential protective mechanism in neurodegenerative disease.44, 45 Defective autophagy results in increased autophagosome content and inefficient cargo clearance, leading to the accumulation of damaged organelles and proteins. Moreover, disease proteins such as huntingtin, parkin, Pink-1 and α-synuclein alter autophagy by pathological interaction with autophagy regulators.41, 42, 43, 46
Consistent with the above studies, we showed by EM that depletion of SigR1 leads to accumulation of autophagosomes (Figure 1 and Supplementary Figures 2 and 3). Autophagy inhibition was confirmed biochemically by western blotting and immunostaining (Figure 2) and also by functional assays that demonstrated autophagosome accumulation, failed fusion with lysosomes and inhibition of EGF–EGFR complex degradation (Figure 3). These results are in line with previous studies showing similar accumulations of autophagosomes and LC3II in cells expressing a truncated form of SigR1 that led to ER stress, mitochondrial abnormalities and apoptosis.40 Recent studies in AD models suggest that presenilin-1 (PS1) expression may directly control the acidification of lysosomes through targeting the v-ATPase to these vesicle compartments, thus leading to defective lysosomal proteolysis.47 However, we did not observe any significant defects in lysosomal acidification in our preliminary staining experiments using Lysosensor yellow/blue DND-160 (data not shown).
SigR1 and defective endosomal/vesicle trafficking
Interestingly, knockdown of SigR1 also impairs vesicle trafficking from ER to Golgi as shown by VSVG transport analysis (Figures 4a–c). These results are reminiscent to the phenotypes seen after depletion of cellular proteins involved in endosomal sorting and trafficking, such as Hrs, TSG101 and Vps24.48, 49 Endosomal abnormalities are among the earliest pathological features of AD and Down syndrome.50 Mutations of Rab7,51 of the guanine nucleotide exchange factor alsin52 and of Vps5453 cause neuropathy and motor neuron diseases in mammals. In addition, mutations in the ESCRT-III complex subunit CHMP2B cause frontotemporal dementia54 and dysfunction of the ESCRT-III complex leads to autophagosome accumulation and neurodegeneration.55 Taken together, these results provide a link between the dysfunction of the endosomal sorting machinery and neurodegeneration and are consistent with our proposed role of SigR1 as a regulator of endosomal sorting, MVB formation and endosome-to-lysosome trafficking of membrane cargo.
Lipid raft alterations due to loss of SigR1
Lipid raft domains provide the spatial environment for direct protein–protein interactions and intramolecular cross-talk and function as a unique signal transduction platform.56 Lipid raft microdomains regulate several Ca2+ channels. Disruption of lipid raft domains inhibits the movement of the calcium sensor stromal interaction molecule 1 (STIM1) into lipid rafts and its association with TRPC1, thereby decreasing TRPC1-dependent store-operated Ca2+ entry (SOCE).57 We consistently observed that shRNA knockdown of SigR1 disrupts lipid rafts and leads to Ca2+ dysregulation as did treatment with MβCD (Figure 5). Moreover, in cells treated with MβCD, SigR1 protein is shifted to nonraft fractions (Supplementary Figure 5). Lipid rafts stabilize the association of GPI proteins with the ER membrane and are involved in other mechanisms essential for receptor-mediated signal transduction pathways and membrane trafficking.58, 59 The defect in ER to Golgi trafficking of VSVG in SigR1 KO cells can also be explained by the fact that SigR1 in lipid rafts plays an important role in vesicular trafficking. This is supported by a study showing similar defects after disruption of lipid rafts.60 SigR1 depletion, like MβCD treatment, leads to mislocalization of raft components and was accompanied by abnormal accumulation of internal vesicles in MVBs (Figures 5A and G).
Calcium and neurodegeneration
ER and mitochondria interact physically at the MAM. Ca2+ is transferred from the ER to mitochondria in order to stimulate oxidative metabolism; conversely, the metabolically energized mitochondria regulate ER Ca2+ homeostasis via the MAM.18 SigR1 regulates IP3-mediated Ca2+ homeostasis at the MAM. Neuronal intracellular Ca2+ signaling is abnormal in many neurodegenerative disorders such as AD, PD, ALS and HD.61 Given that neurons are very sensitive to alterations in Ca2+ levels, subtle abnormalities in Ca2+ signaling may cause neuronal damage in the long term. We previously reported that loss of function of SigR1 increases IP3-mediated ER calcium release and defective ER calcium storage.20 In the present study, we provide evidence for a defective ER morphology and altered lipid raft composition after the loss of SigR1 function. We also observed increased Ca2+ levels that were further increased by treatment with MβCD. These data confirm previous reports that lipid raft microdomains regulate Ca2+ mobilization57 and suggest that SigR1 is an important constituent of lipid rafts and a regulator of ER Ca2+ homeostasis.
Materials and Methods
Reagents
Fluorescent nucleic acid stain Hoechst 33258 was purchased from Molecular Probes (Life Technologies GmbH, Munich, Germany). Thapsigargin, tunicamycin, EGF, rapamycin, bafilomycin A1, Fura 2AM, NaCl, KCl, pluronic acid, CaCl2, MgCl2, Glucose, HEPES 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, MβCD and Bradykinin were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Antibodies
The antibodies used in this study and their dilutions are described in Supplementary Table 1.
Cell culture, transient transfection and treatments
Cell culture and treatment
Human epithelial cancer cells (HeLa) and motor neuron-like cells (NSC34) were maintained in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA), supplemented with 10% FBS and 1% antibiotic/anti-mycotic solution (Invitrogen). Cell cultures were maintained in a humidified incubator at 37 °C/5% CO2. To differentiate NSC34 cells, cells were plated onto poly-D-lysine-coated tissue culture plastic and grown in differentiation medium containing 1 : 1 DMEM/Ham's F12 (Invitrogen), supplemented with 1% FBS, 1% penicillin/streptomycin and 1% modified Eagle's medium with non-essential amino acids (Invitrogen) for 3 days. For the stable cell lines, all cells were grown in a humidified atmosphere at 37 °C with 5% CO2. NIH-3T3 cells as well as tandem mCherry/GFP NIH-3T3 cells were cultured in DMEM supplemented with 10% heat-inactivated FCS. Plat-E cells were grown in DMEM supplemented with 10% heat-inactivated FCS, 1% penicillin/streptomycin (Gibco, Life Technologies GmbH), 1 μg/ml puromycin (Sigma-Aldrich) and 10 μg/ml blasticidin (Invitrogen).
Generation of stable cell lines with retroviral infection
NIH-3T3 cells stably expressing GFP-LC3 and mCherry-EGFP-LC3B were generated by retroviral infection. Retroviruses were generated by transient transfection of the retroviral producer cell line Plat-E with 20 μg pBABEpuro GFP-LC3,62 and pBABEpuro mCherry-EGFP-LC3B,63 respectively. After two rounds of infection, retrovirus-expressing NIH-3T3 cells were selected with 3 μg/ml puromycin to generate stable cell pools. For the transfections the cells were seeded without antibiotics.
Retroviral infection
The day before transfection, Plat-E cells64 were seeded at 2 × 106 cells per 100 mm dish. The next day, pBABE-based retroviral vectors were introduced into Plat-E cells using the Ca2+ phosphate precipitation technique. At 6 h after transfection, the medium was replaced and cells were incubated overnight at 37 °C with 5% CO2. On the same day, NIH-3T3 were seeded at 8 × 105 cells per 100 mm dish. After 24 h, virus-containing supernatants derived from the Plat-E cultures were filtered through a 0.45 μm PVDF filter (Millipore, Merck Chemicals GmbH, Darmstadt, Germany) and supplemented with 4 μg/ml polybrene (Sigma-Aldrich). NIH-3T3 cells were incubated in the virus/polybrene-containing supernatants overnight. After infection, the cells were re-plated in 10 ml fresh medium. A second round of infection was performed after 24 h to maximize the rate of infection. At 48 h after the last infection, puromycin (3 μg/ml, Sigma-Aldrich) was added to select for stable clones for at least 2 weeks.
Plasmids and RNA interference
The HuSH shRNA (pRFP-C-RS) retroviral RFP vector containing four unique 29-mer shRNA SigR1 oligoneucleotides (four different shRNA SigR1 constructs) for optimal suppression of SigR1 was purchased from Origene (Rockville, MD, USA). Non-effective 29-mer scrambled shRNA cassette in the same vector was used as a shRNA control in all the experiments. For some experiments, shRNA SigR1 vector was modified to generate a stop codon in the RFP sequence to generate shRNA SigR1 without the RFP tag. Knockdown by transient transfections of shRNA SigR1 in all the cell lines used was performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's recommendations. Western blots were performed to verify the efficiency of the knockdown (see below). VSVG-GFP was a kind gift from Professor Jennifer Lippincott-Schwartz.34, 35
Immunocytochemistry
HeLa cells or NIH-3T3 cells were cultured on μ-dish (ibidi, GmbH, Am Klopferspitz, Germany) and transiently transfected either with shRNA SigR1 alone or together with GFP-LC3 constructs. The medium was changed after 4 h. After 48 h, cells were fixed in 4% PFA and processed for confocal microscopy. In further experiments after 48 h of knockdown, cells were permeabilized with 0.1% Triton X-100, fixed in 4% PFA and immunostained (see Supplementary Table 1 for antibody dilutions). Secondary Alexa488- or Alexa594-conjugated anti-mouse or anti-rabbit antibodies (Invitrogen) were used for visualization. Nuclei were stained with Hoechst 33342 (1 μg/ml). Samples were mounted with fluorescent mounting media (DAKO, Glostrup, Denmark) and visualized using a Zeiss LSM 700 confocal microscope (Zeiss, Göttingen, Germany). Resulting images were processed using the Zeiss LSM software and Adobe Photoshop CS5 (Adobe Systems GmbH, Munich, Germany).
Fura-2 calcium imaging
The intracellular calcium ion concentration, [Ca2+]i, was measured using a conventional Fura-2 technique.65 After 48 h of SigR1 knockdown, NSC34 cells in glass bottom μ-dishes (ibidi, GmbH) were loaded with the membrane-permeable AM form of Fura-2 (1.5 ng/μl; Invitrogen) in the presence of pluronic acid (25%) for 30 min at 37 °C. Emitted fluorescence at 530 nm (detected using a Sensicam; pco.imaging, PCO AG, Kelheim, Germany) in response to alternate excitation at 340 and 380 nm (using the Polychrome V monochromator; TILL Photonics, FEI Munich GmbH, Germany) was used to measure intracellular Ca2+ concentrations. Data were expressed as emission ratios in response to 340 nm/380 nm excitation. Whole-cell calcium measurements from SigR1-deficient and control NSC34 cells were obtained at room temperature (23–25 °C). During imaging, cells were kept in a bathing solution containing 100 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 10 MES, 5.5 mM glucose and pH was adjusted to 7.4.
Transmission electron microscopy
Hek293 cells were transfected with control, scrambled shRNA and SigR1 shRNA as described above. Cells were collected by scraping, washed in 0.1 M phosphate buffer and immediately fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer for 24 h followed by washing in buffer for further 24 h. Cell pellets were collected by centrifugation (1000 r.p.m., 5 min) and embedded in 2% agarose (at 60 °C; Fluka 05073, St. Louis, MO, USA). Small blocks of embedded cells were sliced and post-fixed in 2.5% glutaraldehyde for 24 h followed by washing in 0.1 M phosphate buffer for 24 h. Agarose blocks were then incubated in 1% OsO4 (in 0.2 M phosphate buffer) for 3 h, washed twice in distilled water and dehydrated using ascending alcohol concentrations (i.e., 25, 35, 50, 70, 85, 95 and 100% each step for 5 min). Dehydrated blocks were incubated in propylenoxide followed by subsequent 20 min of incubation in a 1 : 1 mixture of epon (47.5% glycidether, 26.5% dodenylsuccinic acid anhydride, 24.5% methylnadic anhydride and 1.5% Tris (dimethylaminomethyl) phenol) and propylenoxide. The samples were then incubated in epoxy resin for 1 h at room temperature followed by polymerization (28 °C for 8 h, 80 °C for 2.5 h and finally at room temperature for 4 h). Ultra-thin sections (70 nm) were mounted on grids, contrast enhanced with uranyl acetate and lead citrate and examined with a Philips EM 400 T electron microscope (Philips GmbH, Germany) as previously described.66 The images were captured using a Morada digital camera (Olympus, Japan).
Fluorescence recovery after photobleaching
For live cell imaging experiments, Cos7 cells were seeded onto homemade glass-bottomed dishes (6 cm), and co-transfected either with shRNA SigR1/VSVGts045-GFP or shRNA control VSVGts045-GFP. VSVGts045-GFP was allowed to accumulate in the ER at 42 °C for 8 h and cells were imaged on an Axio Observer Z1 inverted microscope equipped with heating stage and CO2 controller (Zeiss) maintained at a constant temperature of 32 °C. A portion of the ER (approximately ø 3.84 μm) in the periphery of the cell was photobleached using a 405 nm laser driven by the UGA-40 control unit (Rapp Opto Electronic GmbH, Wedel, Germany). The recovery of the fluorescent signal was monitored by imaging the cells every second for 15 min. Imaging was done using an Evolve EM-CCD camera driven by ZEN software (Zeiss). For all experiments the area bleached and the duration and intensity of the laser impulse was kept constant. The extent of recovery of the fluorescent signal was determined using ImageJ (developed by Rasband, W.S., National Institutes of Health, Bethesda, MD, USA, http://imagej.nih.gov/ij/) to measure the average pixel intensity values within three distinct regions of interest (ROIs): ROI1: bleached area; ROI2: unbleached area within the cell; and ROI3: background. Normalized FRAP recovery curves and the mobile fractions were calculated using the program easy FRAP.67
Live cell imaging to analyze RFP–GFP–LC3 fusion
To analyze the dynamics of the RFP–GFP–LC3 fusion protein, GFP and RFP channels were acquired every min for up to 4 h using the imaging system described above. The extent of autophagosome maturation was determined by measuring the colocalization of the GFP and RFP signals as expressed by Pearson's coefficient using ZEN software.
Western blot analysis
Cells were washed with ice-cold PBS twice, scraped off the culture plate and resuspended in lysis buffer (0.5% Triton X-100 in PBS, 0.5 mM PMSF and complete protease inhibitor mixture, Roche Applied Sciences, Roche Diagnostics GmbH, Mannheim, Germany). After incubation on ice for 30 min, lysates were briefly sonicated. Clear lysates were obtained after centrifugation for 5 min at 6000 r.p.m. Protein concentrations were determined using the BCA method (Molecular Probes). Equal amounts of protein were boiled for 5 min in 2 × SDS sample buffer and subjected to 10 or 12% SDS-PAGE before transfer to a polyvinylidenedifluoride (PVDF) membrane. The membranes were blocked in 5% skimmed milk in 0.05% Tween 20/Tris-buffered saline (TBS-T) for 30 min before incubation with primary antibody. Primary antibodies were used in a dilution of 1 : 1000. The membranes were incubated overnight at 4 °C, then washed 3 times in TBS-T and incubated for 1 h with appropriate horseradish peroxidase-conjugated secondary antibody (dilution 1 : 10 000, Thermo Scientific, Waltham, MA, USA). Immunoreactive proteins were detected by enhanced chemiluminescence (Amersham Biosciences, GE Healthcare, Life Sciences, Germany). Densitometric quantification of the band intensity was normalized to tubulin levels using Adobe Photoshop CS5.
EGFR endocytosis and degradation assay
For EGFR endocytosis and degradation analysis, A431 cells were transfected with either control or SigR1 shRNA, and 48 h later, the cells were starved in DMEM with 0.1% BSA for 4 h. After starvation, cells were treated with EGF (100 μg for 10 min) to stimulate EGFR endocytosis. Cells were then collected at various time points (as indicated in Figure 3b) and lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA and 0.1% Na deoxycholate) with protease inhibitor. Protein extracts were resolved by SDS-PAGE and immunoblotted using anti-EGFR antibody.
EGFR surface biotinylation assay
Surface biotinylation assays were performed as previously described68 with minor modifications. Briefly, A431 cells were transfected with either control or SigR1 shRNA, and 48 h later, the cells were starved in DMEM with 0.1% BSA for 4 h. After starvation, cells were treated with EGF (100 μg for 10 min) to stimulate EGFR endocytosis and then proceeded for surface biotinylation assay.
Membrane lipid raft analysis
Membrane lipid rafts were prepared as described previously57 with some modifications. Briefly, NSC34 cells transfected with SigR1 shRNA or control shRNA either treated with MβCD or left untreated were washed with phosphate-buffered saline, pH 7.4, and lysed for 30 min on ice in pre-chilled TNE buffer (1% v/v Triton X-100, 25 mMTris-HCl, 150 mM NaCl, and 5 mM EDTA pH 7.5) supplemented with protease and phosphatase inhibitors (Roche Applied Science). Lysates were homogenized using a Dounce homogenizer (VWR International GmbH, Langenfeld, Germany) followed by a brief centrifugation. Equal amount of proteins were quantified by BCA method (Supplementary Figure 2A). Then, 800 μl of the postnuclear supernatant (PNS) was mixed with an equal volume of 80% sucrose (w/v) and overlaid with 2 ml of 35% sucrose followed by 1 ml of 5% sucrose (in TNE buffer). A total of 5 ml of samples was centrifuged at 39 000 r.p.m. for 18 h at 4 °C in a SW 55 Ti rotor (Beckman Coulter GmbH, Krefeld, Germany). Then, 13 equal fractions (320 μl) were collected from the top of the tube and proteins were precipitated using ice-cold acetone. Samples were then mixed with SDS sample buffer and analyzed by western blotting. Coomassie blue staining showed equal loading of either control shRNA or SigR1 shRNA fractions (Supplementary Figures 4b and c).
Statistical analysis
Except for the FRAP and colocalization analyses, we used the unpaired Student's t-test for the comparison of the means of two samples. Samples were considered statistically different if P<0.05. In all graph, data are represented as mean±S.D. of three independent experiments. For Ca2+ measurements, results are expressed as mean±S.E.M. of ∼30 independent measurements.
For the FRAP and colocalization experiments (Figures 3 and 4), all graphs and statistical analyses were done using Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). A total of 5 to 6 independent experiments were analyzed. Differences among the various samples were determined using the two-tailed Mann–Whitney nonparametric U-test. The null hypothesis (the two groups have the same median values, i.e., they are not different) was rejected when P>0.05.
Acknowledgments
We thank A Knischewski, H Wiederholt, E Pascual, C Krude and H Mader (Institute of Neuropathology, RWTH Aachen University) and H Heidtmann (Institute of Physiology, RWTH Aachen University) for technical support. We also thank V Kumar (International Max Planck Research School, Tuebingen, Germany) for image analysis and S Gründer (Institute of Physiology, RWTH Aachen University) and his lab members for confocal microscopy and other generous help. Finally, we thank B Lüscher (Institute of Biochemistry and Molecular Biology, RWTH Aachen University) for his valuable comments and his lab members for other generous help. This work was supported by grants from the German research council (DFG) to JW (WE1406/13-1), of the Interdisciplinary Centre for Clinical Research (IZKF Aachen; N5-3) to JW and of the German Myopathy Society (DGM) to AG and JW. JW is a member of the German Motor Neuron Disease Network (MND-Net).
Glossary
- ALS
amyotrophic lateral sclerosis
- FTLD
frontotemporal lobar degeneration
- MVBs
multivesicular bodies
- EGFR
epidermal growth factor receptor
- FRAP
fluorescence recovery after photobleaching
- Lys
lysosomes
- MβCD
methyl-β-cyclodextrin
- APP
amyloid precursor protein
- SOD1
superoxide dismutase-1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Author contributions
AG raised the hypotheses and designed the experiments. Experimental work was performed by JTV, AG, AS, AD, DW, AC, EB, JP and CMJ. Calcium imaging was performed by AG and DW. Live cell imaging was done by AS. MEF cells from GFP-LC3 tg mouse and NIH-3T3 cell lines stably expressing GFP-LC3 and tandemly fused mCherry-GFP-LC3 were generated by JV and MD. Confocal imaging was done by AG. Electron microscopy was performed by IK. The manuscript was written by AG, TV and JW. All authors discussed results and commented on the manuscript.
Edited by G Raschellà
Supplementary Material
References
- 1Benbrook DM, Long A. Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp Oncol 2012; 34: 286–297. [PubMed] [Google Scholar]
- 2Chua CE, Tang BL. Linking membrane dynamics and trafficking to autophagy and the unfolded protein response. J Cell Physiol 2013; 228: 1638–1640. [DOI] [PubMed] [Google Scholar]
- 3Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ 2007; 14: 1576–1582. [DOI] [PubMed] [Google Scholar]
- 4Matus S, Glimcher LH, Hetz C. Protein folding stress in neurodegenerative diseases: a glimpse into the ER. Curr Opin Cell Biol 2011; 23: 239–252. [DOI] [PubMed] [Google Scholar]
- 5Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med 2004; 10(Suppl): S10–S17. [DOI] [PubMed] [Google Scholar]
- 6Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2001; 292: 1552–1555. [DOI] [PubMed] [Google Scholar]
- 7Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 2003; 40: 427–446. [DOI] [PubMed] [Google Scholar]
- 8Goswami A, Dikshit P, Mishra A, Mulherkar S, Nukina N, Jana NR. Oxidative stress promotes mutant huntingtin aggregation and mutant huntingtin-dependent cell death by mimicking proteasomal malfunction. Biochem Biophys Res Commun 2006; 342: 184–190. [DOI] [PubMed] [Google Scholar]
- 9Ross CA, Poirier MA. Opinion: What is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol 2005; 6: 891–898. [DOI] [PubMed] [Google Scholar]
- 10Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy 2007; 3: 181–206. [DOI] [PubMed] [Google Scholar]
- 11Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct 2002; 27: 421–429. [DOI] [PubMed] [Google Scholar]
- 12Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med 2013; 19: 983–997. [DOI] [PubMed] [Google Scholar]
- 13Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol 2006; 76: 89–101. [DOI] [PubMed] [Google Scholar]
- 14Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 1998; 23: 33–42. [DOI] [PubMed] [Google Scholar]
- 15Lee S, Sato Y, Nixon RA. Primary lysosomal dysfunction causes cargo-specific deficits of axonal transport leading to Alzheimer-like neuritic dystrophy. Autophagy 2011; 7: 1562–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 2004; 305: 1292–1295. [DOI] [PubMed] [Google Scholar]
- 17Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol 2010; 190: 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 2007; 131: 596–610. [DOI] [PubMed] [Google Scholar]
- 19Al-Saif A, Al-Mohanna F, Bohlega S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol 2011; 70: 913–919. [DOI] [PubMed] [Google Scholar]
- 20Prause J, Goswami A, Katona I, Roos A, Schnizler M, Bushuven E et al. Altered localization, abnormal modification and loss of function of Sigma receptor-1 in amyotrophic lateral sclerosis. Hum Mol Genet 2013; 22: 1581–1600. [DOI] [PubMed] [Google Scholar]
- 21Mavlyutov TA, Epstein ML, Andersen KA, Ziskind-Conhaim L, Ruoho AE. The sigma-1 receptor is enriched in postsynaptic sites of C-terminals in mouse motoneurons. An anatomical and behavioral study. Neuroscience 2010; 167: 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Mavlyutov TA, Epstein ML, Verbny YI, Huerta MS, Zaitoun I, Ziskind-Conhaim L et al. Lack of sigma-1 receptor exacerbates ALS progression in mice. Neuroscience 2013; 240: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Mancuso R, Olivan S, Rando A, Casas C, Osta R, Navarro X. Sigma-1R agonist improves motor function and motoneuron survival in ALS mice. Neurotherapeutics 2012; 9: 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Engedal N, Torgersen ML, Guldvik IJ, Barfeld SJ, Bakula D, Saetre F et al. Modulation of intracellular calcium homeostasis blocks autophagosome formation. Autophagy 2013; 9: 1475–1490. [DOI] [PubMed] [Google Scholar]
- 25Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2013; 2: e00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem 2002; 277: 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19: 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S et al. In search of an ‘autophagomometer'. Autophagy 2009; 5: 585–589. [DOI] [PubMed] [Google Scholar]
- 29Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15: 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Carpenter G. Receptors for epidermal growth factor and other polypeptide mitogens. Annu Rev Biochem 1987; 56: 881–914. [DOI] [PubMed] [Google Scholar]
- 31Derynck R. Transforming growth factor-alpha: structure and biological activities. J Cell Biochem 1986; 32: 293–304. [DOI] [PubMed] [Google Scholar]
- 32Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol 2009; 10: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol 2009; 452: 1–12. [DOI] [PubMed] [Google Scholar]
- 34Nehls S, Snapp EL, Cole NB, Zaal KJ, Kenworthy AK, Roberts TH et al. Dynamics and retention of misfolded proteins in native ER membranes. Nat Cell Biol 2000; 2: 288–295. [DOI] [PubMed] [Google Scholar]
- 35Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature 1997; 389: 81–85. [DOI] [PubMed] [Google Scholar]
- 36Brazer SC, Singh BB, Liu X, Swaim W, Ambudkar IS. Caveolin-1 contributes to assembly of store-operated Ca2+ influx channels by regulating plasma membrane localization of TRPC1. J Biol Chem 2003; 278: 27208–27215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Isshiki M, Anderson RG. Calcium signal transduction from caveolae. Cell Calcium 1999; 26: 201–208. [DOI] [PubMed] [Google Scholar]
- 38Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol 2009; 19: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Kilsdonk EP, Yancey PG, Stoudt GW, Bangerter FW, Johnson WJ, Phillips MC et al. Cellular cholesterol efflux mediated by cyclodextrins. J Biol Chem 1995; 270: 17250–17256. [DOI] [PubMed] [Google Scholar]
- 40Shioda N, Ishikawa K, Tagashira H, Ishizuka T, Yawo H, Fukunaga K. Expression of a truncated form of the endoplasmic reticulum chaperone protein, sigma-1 receptor, promotes mitochondrial energy depletion and apoptosis. J Biol Chem 2012; 287: 23318–23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Aguib Y, Heiseke A, Gilch S, Riemer C, Baier M, Schatzl HM et al. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy 2009; 5: 361–369. [DOI] [PubMed] [Google Scholar]
- 42Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet 2006; 15: 433–442. [DOI] [PubMed] [Google Scholar]
- 43Fornai F, Longone P, Cafaro L, Kastsiuchenka O, Ferrucci M, Manca ML et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 2008; 105: 2052–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441: 885–889. [DOI] [PubMed] [Google Scholar]
- 45Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441: 880–884. [DOI] [PubMed] [Google Scholar]
- 46Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem 2010; 285: 11061–11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 2010; 141: 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Bache KG, Stuffers S, Malerod L, Slagsvold T, Raiborg C, Lechardeur D et al. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol Biol Cell 2006; 17: 2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Doyotte A, Russell MR, Hopkins CR, Woodman PG. Depletion of TSG101 forms a mammalian ‘Class E' compartment: a multicisternal early endosome with multiple sorting defects. J Cell Sci 2005; 118: 3003–3017. [DOI] [PubMed] [Google Scholar]
- 50Jiang Y, Mullaney KA, Peterhoff CM, Che S, Schmidt SD, Boyer-Boiteau A et al. Alzheimer's-related endosome dysfunction in Down syndrome is Abeta-independent but requires APP and is reversed by BACE-1 inhibition. Proc Natl Acad Sci USA 2010; 107: 1630–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Saxena S, Bucci C, Weis J, Kruttgen A. The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. J Neurosci 2005; 25: 10930–10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Yamanaka K, Vande Velde C, Eymard-Pierre E, Bertini E, Boespflug-Tanguy O, Cleveland DW. Unstable mutants in the peripheral endosomal membrane component ALS2 cause early-onset motor neuron disease. Proc Natl Acad Sci USA 2003; 100: 16041–16046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Schmitt-John T, Drepper C, Mussmann A, Hahn P, Kuhlmann M, Thiel C et al. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat Genet 2005; 37: 1213–1215. [DOI] [PubMed] [Google Scholar]
- 54Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet 2005; 37: 806–808. [DOI] [PubMed] [Google Scholar]
- 55Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol 2007; 17: 1561–1567. [DOI] [PubMed] [Google Scholar]
- 56Anderson RG, Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 2002; 296: 1821–1825. [DOI] [PubMed] [Google Scholar]
- 57Pani B, Ong HL, Liu X, Rauser K, Ambudkar IS, Singh BB. Lipid rafts determine clustering of STIM1 in endoplasmic reticulum-plasma membrane junctions and regulation of store-operated Ca2+ entry (SOCE). J Biol Chem 2008; 283: 17333–17340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58Sarnataro D, Campana V, Paladino S, Stornaiuolo M, Nitsch L, Zurzolo C. PrP(C) association with lipid rafts in the early secretory pathway stabilizes its cellular conformation. Mol Biol Cell 2004; 15: 4031–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59Vinten J, Johnsen AH, Roepstorff P, Harpoth J, Tranum-Jensen J. Identification of a major protein on the cytosolic face of caveolae. Biochim Biophys Acta 2005; 1717: 34–40. [DOI] [PubMed] [Google Scholar]
- 60Shogomori H, Futerman AH. Cholesterol depletion by methyl-beta-cyclodextrin blocks cholera toxin transport from endosomes to the Golgi apparatus in hippocampal neurons. J Neurochem 2001; 78: 991–999. [DOI] [PubMed] [Google Scholar]
- 61Bezprozvanny I. Calcium signaling and neurodegenerative diseases. Trends Mol Med 2009; 15: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell 2008; 19: 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63N'Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 2009; 10: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Therapy 2000; 7: 1063–1066. [DOI] [PubMed] [Google Scholar]
- 65Usachev Y, Shmigol A, Pronchuk N, Kostyuk P, Verkhratsky A. Caffeine-induced calcium release from internal stores in cultured rat sensory neurons. Neuroscience 1993; 57: 845–859. [DOI] [PubMed] [Google Scholar]
- 66Schroder JM, Hoheneck M, Weis J, Deist H. Ethylene oxide polyneuropathy: clinical follow-up study with morphometric and electron microscopic findings in a sural nerve biopsy. J Neurol 1985; 232: 83–90. [DOI] [PubMed] [Google Scholar]
- 67Rapsomaniki MA, Kotsantis P, Symeonidou IE, Giakoumakis NN, Taraviras S, Lygerou Z. easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics 2012; 28: 1800–1801. [DOI] [PubMed] [Google Scholar]
- 68Brauers E, Dreier A, Roos A, Wormland B, Weis J, Kruttgen A. Differential effects of myopathy-associated caveolin-3 mutants on growth factor signaling. Am J Pathol 2010; 177: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.