Abstract
AIM: To detect the expression of p33ING1b protein and the change of p33ING1b gene in pancreatic carcinoma and to evaluate the significance of p33ING1b in pancreatic cell carcinogenesis.
METHODS: Pathological specimens from pancreatic carcinoma and matched non-tumor pancreatic tissues were examined for p33ING1b expression and mutation by immunohistochemistry, polymerase chain reaction single-strand conformation polymorphisms (PCR-SSCP) and loss of heterozygosity (LOH).
RESULTS: The rate of p33ING1b protein expression was 85% (34/40). A single germline missense mutation was detected in 1 of 40 tumors located at codon 215: TGC-TCC (Cys-Ser). Fourteen (60.9%) of 23 tumor samples showed LOH in all of the informative markers tested, but no mutation was detected in these tumors and only two of the informative tumors lacked expressions of p33ING1b protein.
CONCLUSION: Mutation and loss of expression are not the main reasons for the disfunction of p33ING1b in pancreatic carcinoma, an abnormality at the level of chromosome and/or transcription may inhibit their normal functions, potentially contributing to pancreatic cell carcinogenesis.
INTRODUCTION
A candidate tumor suppressor gene, ING1, has recently been cloned and mapped on human chromosome 13q34, encoding p33ING1b mainly, a nuclear protein which physically interacts with p53[1,2] and cooperates with it in many ways[3,4]. Forced overexpression of the ING1 gene could lead to cell arrest in the G1 phase of the cell cycle and induce apoptosis in several cell types[5,6]. Conversely, inhibition of ING1 expression by antisense constructs could promote the transformation of mouse breast epithelial cells and increase the frequency of focus formation with NIH3T3 cells and protect cells from apoptosis. Recently, p33ING1b protein has been found to contain many specific structures, such as plant homeodomain (PHD)-like finger, nuclear localization sequence (NLS), nucleolar targeting sequence (NTS), proliferating cell nuclear antigen (PCNA) interacting protein (PIP) domain, Sin3-associated polypeptide (SAP30) interaction region and bromodomain which implicate that the p33ING1b protein might play a critical role with other genes in oncogenesis, apoptosis, DNA repair and cell cycle regulation[7-9].
High rates of 13q loss of heterozygosity (LOH) have been detected in cancer of the esophagus, ovary, breast and head and neck[10]. And several gene mutations and reduced protein expression of p33ING1b were found in head and neck, esophageal, lung, bladder, ovary, kidney, breast, and liver cell cancers[7]. Major sequence features and biological effects and genetic alterations of p33ING1b indicate that p33ING1b may be a tumor suppressor and that functional loss of p33ING1b might contribute to tumorigenesis. To the author’s knowledge, only one study showed that p33ING1b expression was increased in pancreatic adenocarcinoma cell line Mia PaCa-2 after incubation with COX-2 inhibitors for specific times[11]. But little is known about its role and changes in pancreatic cancer. Accordingly, we examined whether genetic alterations, such as allelic imbalance or mutations of p33ING1b gene, as well as altered protein expression of p33ING1b, might be responsible for the emergence and progression of human pancreatic carcinoma using immunohistochemical study, LOH analysis, and PCR-SSCP.
MATERIALS AND METHODS
Materials
Fourty tumor and 23 matched normal tissues were obtained from 40 patients with pancreatic carcinoma undergoing surgical resection as primary therapy for their disease after informed consent was obtained. All patients were from the Department of Pathology, Changhai Hospital. Another four pancreatitis and 5 normal samples were also from the department. Histological studies were performed and all tumors were confirmed as exocrine pancreatic carcinoma. Among these patients whose age ranged from 31 to 86 years, 24 were males and 16 were females, and the average age was 60.5 years. Four types of tumor were involved namely, adenocarcinoma, adeno-squamous cell carcinoma, cystadenocarcinoma, and acinar cell carcinoma. The number of cases was 36, 2, 1, and 1, respectively. Thirteen cases of adenocarcinoma were well differentiated, and the other 23 cases were moderately or poorly differentiated.
DNA Extraction
Genomic DNAs were isolated from formalin-fixed paraffin-embedded sections and frozen by proteinase K treatment (method D)[12], phenol-choroform extraction, and ethanol precipitation following the manufacturer’s instructions.
Immunohistochemistry
The expression of p33ING1b protein in paraffin-embedded histological sections was determined using the EnVision method. Histological sections (4 µm thick) on 0.2 g/L poly-L-lysine-coated slides were deparaffinized and rehydrated, and the endogenous peroxidase activity was blocked by incubation with 20 mL/L H2O2 in phosphate buffer, followed by pre-treatment with proteinase K. Non-specific binding was blocked with normal goat serum, and sections were incubated with p33ING1b antibody (supplied by Dr. Riabowol, Canada)[1]. After washed with phosphate buffer, the sections were incubated with secondary antibody and enhanced labelled polymers for 3 h and washed with phosphate buffer, followed by incubation with DAB. Then the sections were washed and counterstained with methyl green. Sections of normal pancreas tissue were used as positive control for the expression of p33ING1b, and the sections incubated with PBS instead of the corresponding primary antibody were used as negative controls. To quantitate the p33ING1b expression in various samples, a scoring method was adopted[13]. A mean percentage of positive tumor cells was determined in at least five areas at × 400 magnification and assigned to one of the five following categories: 0, < 5%; 1, 5%-25%; 2, 25%-50%; 3, 50%-70%; and 4, > 75%. The intensities of immunostaining were scored as follows: weak, 1+; moderate, 2+; and intense, 3+. For tumors showing heterogeneous staining, the predominant pattern was taken into account for scoring. The percentage of positive tumor cells and the staining intensity were multiplied to produce a weighted score for each case. Cases with weighted scores of less than 1 were defined as negative, otherwise as positive.
SSCP Analysis
The coding region of exon1 was amplified by PCR with primers on the flanking regions, primers S1 (5’-ATGTTGAGTCCTGCCAACGGGGA) and AS1 (5’-CTCTTGGTATTTCGCGTCGAT), 138 bp; exon2 was amplified as four: overlapping fragments with four primer sets: (a) S2 (5’-ATCCTGAAG GAGCTAGACGAGTGC) and AS2 (5’-CTTGCCGCTGTTGCCCGCTG), 246 bp; (b) S3 (5’-TTCGAGGCGAGCAGGAGCT) and AS3 (5’-CTTGGCCTTCTTCTCCTTGGG), 210 bp; (c) S4 (5’-CAGCAACCACGACCACGACG) and AS4 (5’-TGAGCCCCACGCACGAGAAG), 240 bp; (d) S5 (5’-CCTCCCCATCGACCCCAACG) and AS5 (5’-CTACCTGTTGTAAGCCCTCTC), 235 bp. The PCR mixture contained 400 ng of DNA, 5 µL of 25 mmol/L MgCl2, 5 µL of 10 × PCR buffer, 5 µL of 2.5 mmol/L each deoxynucleotide triphosphate, 25 pmoL of each primer, and 1.25 unit of Taq DNA polymerase (Takara, Kyoto, Japan) in a 50-µL volume. Initial denaturation at 94 °C for 4 min was followed by 35 amplification cycles, each consisting of a denaturation at 94 °C for 1 min, an annealing at 55 °C for 1 min, and an extension at 72 °C for 1 min. A final extension step at 72 °C for 10 min was carried out. Fifteen µL of PCR products as mixed with 8 µL of 3 × loading dye (95% formamide, 20 mmol/L EDTA, 0.5 g/L bromphenol blue, and 0.5 g/L xylene cyanol), heat denatured, chilled on ice, applied onto an 400 g/L polyacrylamide gel with 50 g/L glycerol, and run at variable temperatures. The bands were detected by silver staining[14]. Aberrantly migrating bands on the gels were excised, reamplified with the same sets of primers. Reproduction of the result was confirmed by sequencing the samples at least twice.
LOH analysis using a microsatellite marker on 13q
To study allelic deletion in pancreatic cancer, we examined DNA for LOH at D13S261, D13S1047, D13S1315 and DS42490[15], which are located close to the ING1 locus. Primers sets were available through the Internet genome database. PCR was performed in 25-µL reaction mixtures containing 200 ng of template genomic DNA, 5 pmoL of each oligonucleotide primer, 0.75 unit of Taq DNA polymerase (Takara, Kyoto, Japan), 2.5 µL of 10 × buffer, and 2.5 µL of 2.5 mmol/L deoxynucleotide triphosphate. After predenaturation for 3 min at 94 °C, PCR was carried out for 35 amplification cycles, each consisting of denaturation for 50 s at 94 °C, annealing for 50 s at 55 °C (D13S261, D13S1047, D13S1315) or at 60 °C (DS42490), and extension for 1 min at 72 °C. A final extension step at 72 °C for 10 min was performed. After amplification, 8 µL of the reaction mixture was mixed with 8 µL of 2 × loading dye as described above, heat denatured, chilled on ice, and then electrophoresed on 80 g/L polyacrylamide gel containing 8 mol/L urea. DNA bands were visualized by silver staining. LOH was scored if one of the heterozygous alleles showed at least 50% reduced intensity in tumor DNA as compared with the corresponding normal DNA.
Statistical analysis
The SPSS 11.0 software package for Macintosh (SPSS Inc., Chicago, IL) was used for all statistical analyses. Variables associated with p33ING1b expression were analyzed by χ2 test. Differences in expression of p33ING1b mRNA between groups were checked by independent t test. P < 0.05 was considered statistically significant.
RESULTS
p33ING1b protein expression detected by IHC
p33ING1b protein expression was examined in paraffin-embedded specimens from 40 pancreatic cancer patients (Figure 1). Normal and benign lesion sections were examined for p33ING1b expression as controls. All of the non-tumor tissues were weak positive for p33ING1b, and reduced p33ING1b protein expression was noted only in 6 tumor samples (15%). The positive staining in cancer tissues was significantly stronger than that in matched normal tissues and other non-tumor samples (Figure 1). Statistical analysis showed no obvious correlation in p33ING1b expression between tumor and non-tumor tissues, and also no correlation between expression of p33ING1b protein and clinicopathological factors of all the patients.
Figure 1.
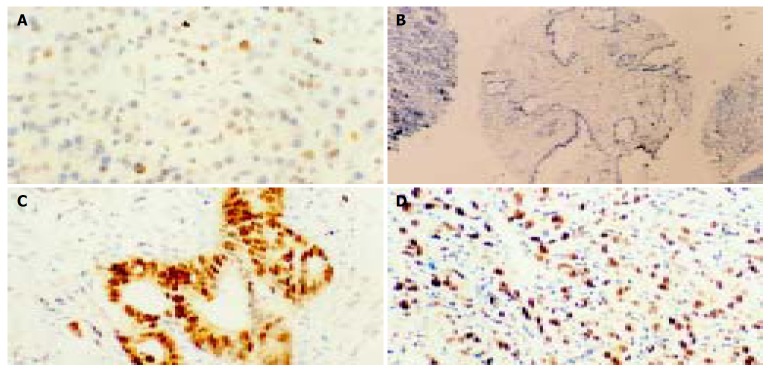
Representative examples of IHC staining for p33ING1b (× 200) on normal pancreatic tissues and cancer tissues. A: Weak positive pancreatic cells for p33ING1b; B: No p33ING1b; C and D: Extremely strong staining in pancreatic cancer samples.
Detection of p33ING1b gene mutation
To investigate whether the p33ING1b gene was the target of functional loss in tumors, we searched for mutations in the coding regions of the gene in all the 40 samples of pancreatic carcinoma. Only one possible mutation was identified by SSCP and DNA sequencing (Figure 2). The sample showed a substitution of cysteine (TGC) with serine (TCC) at codon 215, which located in plant homeodomain (PHD) finger. This change might affect the PHD finger and break the three-dimensional structure of ING1 protein, leading to the loss of function. But this sample still showed p33ING1b protein expression, implicating that the protein expressed in tumor could not exert its normal function.
Figure 2.

SSCP analysis of p33ING1b gene in genomic DNA from pancreatic carcinomas. N: normal DNA; T: tumor DNA. Arrow point indicates the altered band in tumor DNA as compared with the corresponding normal DNA. The amino acid substitution of p33ING1b are shown to the right.
LOH analysis
We examined DNA from 23 pairs of matched pancreatic carcinomas and normal tissues for losses at four microsatellite markers (D13S261, D13S1047, D13S1315 and DS42490) on the chromosome 13q33-34 region. Figure 3 shows the summary and representative examples. Fourteen of 23 informative tumors (60.9%) showed LOH of at least one marker. The rates of LOH at D13S261, D13S1047, D13S1315, and DS42490 loci was 8.7%, 21.5%, 13.0% and 26.1%, respectively. The most frequent loss was seen at marker DS42490, which was located in the ING1 region. These markers were less than 3 cM. Only two of these 23 pancreatic carcinomas showed no p33ING1b protein expression and none had a single mutation.
Figure 3.
LOH analysis on chromosome 13q33-34 in PCs. A: Submatic representation of LOH distribution;●LOH○ without LOH; a: cancer; b: matched cancer; B: Analysis of D13S1315; C: LOH analysis of D13S261; D: LOH analysis of DS42490; E: LOH analysis of D13S1047.
DISCUSSION
Several studies showed that reduced p33ING1 expression was found in esophageal squamous cell cancer (100%)[16] and hepatocellular carcinoma (66.3%)[17]. In our study, positive p33ING1b staining was present in 85.0% of the pancreatic carcinoma tissues. No significant difference was found between cancer and non-cancer tissues, indicating that loss of p33ING1b protein expression was not a major cause of the cancerization of pancreatic cells. But p33ING1b protein was overexpressed in tumors compared with matched normal tissues in the patients with positive p33ING1b stainings. Our results are in agreement with two previous studies[18,19]. The functions and states of these overexpressed p33ING1b proteins still need further study.
Limited analysis of ING1 gene demonstrated rearrangement of the gene in one neuroblastoma cell line and reduced expression in primary breast cancers and cell lines[1]. Then the following experiments found several missense mutations and silent changes in cancers of esophagus[16], head and neck[20], esophagogastric junction[21], gastrointestine[22], skin[18,19] and breast[23]. These mutations were all located in PHD finger and/or nuclear localization signal (NLS) in the COOH-terminal half of ING1. The PHD finger was found in several proteins, including transcription factors and proteins that could regulate chromatin structure. Mutations in PHD finger may lead to loss of the functions of ING1 through inhibiting the transcriptional activity of the gene. Mutations in NLS might interfere with the accumulation of ING1 protein in nuclei[7]. But the rate of mutations was very low and no missense mutations were found in oral squamous cell carcinoma[24], lymphoid malignancies[25], colorectal carcinoma[26], and haematological malignancies[27]. We only detected one missense mutation at codon 215: TGC-TCC (Cys-Ser) on PHD finger among 40 samples of pancreatic carcinomas. All of these results suggested that mutations in certain types of tumor could lead to loss of the functions of p33ING1b, but they were not the main reason.
Several studies have been reported that high rates of LOH were found in human chromosome 13q33-34 region[10,16,20]. The rate of LOH in esophageal/head and neck squamous cell cancers was 58.9% (20/34) and 68% (23/34), respectively[16,20]. Another study found that the rate of LOH in head and neck squamous cell cancers was 48% (21/44)[10]. But the range between every two markers was more than 1 cm, far from ING1 locus, and these markers could not reflect the particular information about ING1. So we selected four markers, D13S261, D13S1047, D13S1315, and DS42490, to detect LOH in 23 cancer tissues. D13S261 and D13S1047 located on the upstream of ING1, D13S1315 on the downstream, while DS42490 in the ING1 locus. The results were described as above. As compared with the LOH (60.9%), the loss of p33ING1b protein expression (8.7%) was very low in pancreatic carcinomas and no missense mutation of p33ING1 gene was found. The lack of mutations suggests that the ING1 gene is not the suppressor gene target of tumors in which frequent LOH was observed at 13q. The high frequency of LOH at ING1 locus and the vicinity may affect the function of p33ING1b but not the expression of p33ING1 protein. The reason is very complex. An alternative target of inactivation may be involved in this tumor type which can affect the function of p33ING1b protein. In addition, alterations at the transcriptional or post-transcriptional level may be another reason for the abnormal expression of p33ING1b protein, for several experiments also detected reduced expression of p33ING1b mRNA in primary breast cancers and cell lines[23,28], human gastric cancer[22], myeloid leukaemia[29], head and neck squamous cell carcinoma[20] and human diploid fibroblast[30]. More detailed studies are needed to elucidate this possibility.
Footnotes
Supported by the “258” Project Foundation of Changhai Hospital, Shanghai, China
Edited by Kumar M and Wang XL Proofread by Xu FM
References
- 1.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 2.Cheung KJ, Li G. The tumor suppressor ING1: structure and function. Exp Cell Res. 2001;268:1–6. doi: 10.1006/excr.2001.5258. [DOI] [PubMed] [Google Scholar]
- 3.Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature. 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 4.Leung KM, Po LS, Tsang FC, Siu WY, Lau A, Ho HT, Poon RY. The candidate tumor suppressor ING1b can stabilize p53 by disrupting the regulation of p53 by MDM2. Cancer Res. 2002;62:4890–4893. [PubMed] [Google Scholar]
- 5.Helbing CC, Veillette C, Riabowol K, Johnston RN, Garkavtsev I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997;57:1255–1258. [PubMed] [Google Scholar]
- 6.Shimada H, Liu TL, Ochiai T, Shimizu T, Haupt Y, Hamada H, Abe T, Oka M, Takiguchi M, Hiwasa T. Facilitation of adenoviral wild-type p53-induced apoptotic cell death by overexpression of p33(ING1) in T.Tn human esophageal carcinoma cells. Oncogene. 2002;21:1208–1216. doi: 10.1038/sj.onc.1205176. [DOI] [PubMed] [Google Scholar]
- 7.Feng X, Hara Y, Riabowol K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002;12:532–538. doi: 10.1016/s0962-8924(02)02391-7. [DOI] [PubMed] [Google Scholar]
- 8.Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP, Riabowol K. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci. 2001;114:3455–3462. doi: 10.1242/jcs.114.19.3455. [DOI] [PubMed] [Google Scholar]
- 9.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Cespedes M, Okami K, Cairns P, Sidransky D. Molecular analysis of the candidate tumor suppressor gene ING1 in human head and neck tumors with 13q deletions. Genes Chromosomes Cancer. 2000;27:319–322. doi: 10.1002/(sici)1098-2264(200003)27:3<319::aid-gcc13>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 11.Tseng WW, Deganutti A, Chen MN, Saxton RE, Liu CD. Selective cyclooxygenase-2 inhibitor rofecoxib (Vioxx) induces expression of cell cycle arrest genes and slows tumor growth in human pancreatic cancer. J Gastrointest Surg. 2002;6:838–843; discussion 844. doi: 10.1016/s1091-255x(02)00061-6. [DOI] [PubMed] [Google Scholar]
- 12.Frank TS, Svoboda-Newman SM, Hsi ED. Comparison of methods for extracting DNA from formalin-fixed paraffin sections for nonisotopic PCR. Diagn Mol Pathol. 1996;5:220–224. doi: 10.1097/00019606-199609000-00012. [DOI] [PubMed] [Google Scholar]
- 13.Lu CD, Altieri DC, Tanigawa N. Expression of a novel antiapoptosis gene, survivin, correlated with tumor cell apoptosis and p53 accumulation in gastric carcinomas. Cancer Res. 1998;58:1808–1812. [PubMed] [Google Scholar]
- 14.Bassam BJ, Caetano-Anollés G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 15.Borodina TA, Ivanov DV, Khusnutdinova EK, Spitsyn VA, Baranova AV, Iankovskiĭ NK. [A new pentanucleotide STR-marker, located in the intron of the ING1 tumor suppressor gene and its allelic polymorphism] Genetika. 2001;37:117–119. [PubMed] [Google Scholar]
- 16.Chen L, Matsubara N, Yoshino T, Nagasaka T, Hoshizima N, Shirakawa Y, Naomoto Y, Isozaki H, Riabowol K, Tanaka N. Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Res. 2001;61:4345–4349. [PubMed] [Google Scholar]
- 17.Ohgi T, Masaki T, Nakai S, Morishita A, Yukimasa S, Nagai M, Miyauchi Y, Funaki T, Kurokohchi K, Watanabe S, et al. Expression of p33(ING1) in hepatocellular carcinoma: relationships to tumour differentiation and cyclin E kinase activity. Scand J Gastroenterol. 2002;37:1440–1448. doi: 10.1080/003655202762671332. [DOI] [PubMed] [Google Scholar]
- 18.Campos EI, Cheung KJ, Murray A, Li S, Li G. The novel tumour suppressor gene ING1 is overexpressed in human melanoma cell lines. Br J Dermatol. 2002;146:574–580. doi: 10.1046/j.1365-2133.2002.04641.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen B, Campos EI, Crawford R, Martinka M, Li G. Analyses of the tumour suppressor ING1 expression and gene mutation in human basal cell carcinoma. Int J Oncol. 2003;22:927–931. [PubMed] [Google Scholar]
- 20.Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 2000;60:3143–3146. [PubMed] [Google Scholar]
- 21.Hara Y, Zheng Z, Evans SC, Malatjalian D, Riddell DC, Guernsey DL, Wang LD, Riabowol K, Casson AG. ING1 and p53 tumor suppressor gene alterations in adenocarcinomas of the esophagogastric junction. Cancer Lett. 2003;192:109–116. doi: 10.1016/s0304-3835(02)00635-3. [DOI] [PubMed] [Google Scholar]
- 22.Oki E, Maehara Y, Tokunaga E, Kakeji Y, Sugimachi K. Reduced expression of p33(ING1) and the relationship with p53 expression in human gastric cancer. Cancer Lett. 1999;147:157–162. doi: 10.1016/s0304-3835(99)00288-8. [DOI] [PubMed] [Google Scholar]
- 23.Toyama T, Iwase H, Watson P, Muzik H, Saettler E, Magliocco A, DiFrancesco L, Forsyth P, Garkavtsev I, Kobayashi S, et al. Suppression of ING1 expression in sporadic breast cancer. Oncogene. 1999;18:5187–5193. doi: 10.1038/sj.onc.1202905. [DOI] [PubMed] [Google Scholar]
- 24.Krishnamurthy J, Kannan K, Feng J, Mohanprasad BK, Tsuchida N, Shanmugam G. Mutational analysis of the candidate tumor suppressor gene ING1 in Indian oral squamous cell carcinoma. Oral Oncol. 2001;37:222–224. doi: 10.1016/s1368-8375(00)00081-6. [DOI] [PubMed] [Google Scholar]
- 25.Ohmori M, Nagai M, Tasaka T, Koeffler HP, Toyama T, Riabowol K, Takahara J. Decreased expression of p33ING1 mRNA in lymphoid malignancies. Am J Hematol. 1999;62:118–119. doi: 10.1002/(sici)1096-8652(199910)62:2<118::aid-ajh11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 26.Sarela AI, Farmery SM, Markham AF, Guillou PJ. The candidate tumour suppressor gene, ING1, is retained in colorectal carcinomas. Eur J Cancer. 1999;35:1264–1267. doi: 10.1016/s0959-8049(99)00104-5. [DOI] [PubMed] [Google Scholar]
- 27.Bromidge T, Lynas C. Relative levels of alternative transcripts of the ING1 gene and lack of mutations of p33/ING1 in haematological malignancies. Leuk Res. 2002;26:631–635. doi: 10.1016/s0145-2126(01)00185-0. [DOI] [PubMed] [Google Scholar]
- 28.Tokunaga E, Maehara Y, Oki E, Kitamura K, Kakeji Y, Ohno S, Sugimachi K. Diminished expression of ING1 mRNA and the correlation with p53 expression in breast cancers. Cancer Lett. 2000;152:15–22. doi: 10.1016/s0304-3835(99)00434-6. [DOI] [PubMed] [Google Scholar]
- 29.Ito K, Kinjo K, Nakazato T, Ikeda Y, Kizaki M. Expression and sequence analyses of p33(ING1) gene in myeloid leukemia. Am J Hematol. 2002;69:141–143. doi: 10.1002/ajh.10031. [DOI] [PubMed] [Google Scholar]
- 30.Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]



