Abstract
AIM: In vivo induced genes are thought to play an important role during infection of host. AlkA was identified as an in vivo-induced gene by in vivo expression technology (IVET), but its virulence in Shigella flexneri was not reported. The purpose of this study was to identify the role of alkA gene in the pathogenesis of S. flexneri.
METHODS: PCR was used to amplify alkA gene of S. flexneri 2a and fragment 028pKm. The fragment was then transformed into 2457T05 strain, a S flexneri 2a strain containing Red recombination system, which was constructed with a recombinant suicide plasmid pXLkd46. By in vivo homologous recombination, alkA mutants were obtained and verified by PCR and sequencing. Intracellular survival assay and virulence assay were used to test the intracellular survival ability in HeLa cell model and the virulence in mice lung infection model respectively.
RESULTS: Deletion mutant of S. flexneri 2a alkA was successfully constructed by λ Red recombination system. The mutant exhibited significant survival defects and much significant virulence defects in mice infection assay.
CONCLUSION: AlkA gene plays an important role in the infection of epithelial cells and is a virulent gene of Shigella spp.
INTRODUCTION
Shigella spp. is a Gram-negative facultative pathogen, which causes bacillary dysentery, a world endemic bloody diarrhea, particularly in developing countries. The disease is caused by invasion of the colorectal mucosa by Shigella spp., replicating within epithelial cells and moving between cells. The interaction between epithelial cells and Shigella spp. plays an important role in the pathogenicity of Shigella spp.[1-3]. During infection of epithelial cells, genes with inducible expression are important for Shigella spp. to replicate and survive in the cells. Hereby, these genes are generally thought to be related to the virulence of Shigella spp.. Many methods could be used to isolate in vivo expressed genes[4].Using in vivo expression technology (IVET) to identify the virulence-related genes of pathogens is a flourishing field in the world[5-11]. We have employed in vivo expression technology with asd gene as a reporter to screen S. flexneri 2a fusion gene library. The result indicated that alkA gene is an in vivo-induced gene for Shigella flexneri 2a.
AlkA gene or its homologous genes have been cloned from many organisms, such as Escherichia coli, Helicobacter pylori, Bacillus subtilis, Saccharomyces cerevisiae, human, etc[12-14]. They encode 3-methyladenine DNA glycosylase, whose function is excising hypoxanthine, demethylating, and mainly taking part in the repair of damaged DNA[15,16]. Up to now researches on alkA have mainly focused on regulation of its expression and its role in inducing repair after DNA alkylation damage. As for its relation to bacterial pathogenesis, it is a noteworthy issue. In this experiment, based on the sequence of E.coli alkA, alkA gene of S. flexneri 2a 2457T was cloned. Its mutant was constructed, and its role in pathogenesis was analyzed by a HeLa cell model and a mice infection model. This study perhaps would provide insights into the pathogenicity of this pathogen.
MATERIALS AND METHODS
Materials
The strains and plasmids used in this study are listed in Table 1. HeLa cell line was maintained in our laboratory. BALB/c mice were bought from the Laboratory Animal Center in the Academy of Military Medical Sciences, Beijing. All mice used in this study were female, specific pathogen free animals, with an age of 7-8 weeks and weight of 18-22 g. DNA endonucleases, DNA marker, T4 DNA ligase, T4 DNA polymerase, Ex Taq DNA polymerase, and CIAP were purchased from Takara Company. Newborn calf sera and RPMI1640 media were from HyClone, and deoxycholate sodium from Sigma. Primers (P1, P2, P3, and P4) were synthesized in our laboratory.
Table 1.
Strains and plasmids
| Strains and plasmids | Characteristics | Source (reference) |
| Strains | ||
| DH5α | endA1 hsdR17(rk-mk+) supE44 thi-1 recA1 gyrA(Nalr) | Our lab |
| RelA1 △ lacIZYA-argF) U169 deoR (φ80dlac △(lacZ)M15) | ||
| S17-1λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu, Smr, λpir | Guzman CA |
| MC1061 | hsdR2 mcrB araD139 △ (ara ABC-leu)7679 △ lacX74 galU galK rpsL thi, Smr | Our lab |
| 2457T | S. flexneri 2a wild type, Nalr | Maurelli AT |
| 2457T05 | a derivative strain of 2457T, contained araBp-gam-bet-exo, Nalr | This study |
| 2457T028D | △ alkA, derivative strain of 2457T, Nalr, Kmr | This study |
| Plasmids | ||
| pMD18-T | a derivative constructed from pUC18, Apr | TaKaRa |
| pXL275 | a mobile and suicide plasmid, ori R6K, mob RK2, sacBR, Cmr | Rui et al[17] |
| pKD46 | oriR101, repA101(ts), araBp-gam-bet-exo, Apr | CGSCa |
| pXLkd46 | pXL275 derivative, inserted a fragment containing araBp-gam-bet-exo, Cmr | This study |
| pMDKm05 | pMD18-T derivative, inserted Kmr gene, Kmr, Apr | Our lab |
| pMD028 | pMD18-T derivative, inserted alkA gene, Apr | This study |
| pMD028pKm | pMD18-T derivative, inserted a cassette (5’ alkA end-Kmr-3’alkA end), Kmr, Apr | This study |
CGSC: E. coli Genetic Stock Center.
Methods
Culture and maintenance of strains and HeLa cells Luria-Bertani (LB) broth and agar plate were used for the growth of S. flexneri and Escherichia coli strains at 37 °C. SOC culture medium was applied to the restoration of bacteria after electroporation. When appropriate, antibiotics were added in media as follows: 100 μg ampicillin (Ap), 100 μg streptomycin (Sm), 50 μg kanamycin (Km), 25 μg chloramphenicol (Cm), and 25 μg naladixic acid (Nal) per ml. HeLa cells were maintained in the RPMI-1640 medium supplemented with 10% fetal bovine serum, 200 mM L-glutamine, 2 mg sodium hydrogen carbonate per ml and 100 μg penicillin-streptomycin per ml. The cells were cultured in 37.5 cm2 or 10 cm2 flasks at 37 °C in a humidified atmosphere of 5% CO2. Confluent monolayers were split by treatment with sterile phosphate-buffered saline (PBS) and trypsin-EDTA.
Genetic techniques Plasmid DNA extraction was carried out using a Qiagen plasmid kit. Digestion, ligation, transformation, and other conventional methods of molecular biology were performed as previously described[18].
DNA amplifications For the amplification of S. flexneri 2a alkA gene, PCR was performed in a standard 100 μl reaction volume containing 2.5 mM Mg Cl2, 0.25 mM of each dNTP, 100 pmol of P1 and P2 primers, 10 μl boiled S. flexneri 2a 2457T, and 5 U Taq DNA polymerase. Reactions were allowed to proceed in a Perkin-Elmer 2400 thermal cycler programmed for 10 min at 94 °C, 30 cycles (for 45 s at 94 °C, for 40 s at 55 °C, for 3 min at 72 °C) and an additional extension reaction for 10 min at 72 °C. For the amplification of fragment 028pKm, PCR was carried out in 100 μl reaction volume containing 2.5 mM MgCl2, 0.25 mM of each dNTP, 100 pmol of P3 and P4 primers, 2 μl plasmid pMD028pKm (about 10 ng), and 5 U Taq DNA polymerase. The program of this PCR was at 94 °C for 10 min, 30 cycles (for 30 s at 94 °C, for 40 s at 58 °C, for 1.5 min at 72 °C) and for 10 min at 72 °C.
Bacterial mating The donor and recipient strains were grown in LB medium containing appropriate antibiotics overnight. The liquid cultures were then washed in PBS, mixed at 1:1 ratio, and spreaded on LB agar plates. The plates were incubated at 37 °C for 6-8 h. After incubation, the conjugation mixture was washed in PBS and spread onto LB agar plates containing chloramphenicol (25 μg·mL-1) and naladixic acid (25 μg·mL-1). The plates were incubated at 37 °C until transconjugants were visible.
Disruption of S. flexneri 2a alkA gene S. flexneri 2a 2457T05 was grown in 5 ml LB cultures with chloramphenicol, naladixic acid and L-arabinose to an OD600 = 0.45 and then made electrocompetent by concentrating 100-fold and washing three times with ice-cold 10% glycerol. The gel-purified 028pKm PCR products were digested with DpnI, repurified, and suspended in elution buffer (10 mM Tris, pH 8.0). Electroporation was done using a gene pulser® II with a pulse controller plus and 0.1 cm chambers according to the manufacturer’s instructions (Bio-RAD) using 40 μl of competent cells and 100 ng of 028pKm fragments. The parameters for electro- transformation were resistance 200 Ω, capacitance 25 μF, and voltage 2500 V. Shocked cells were added to 1 ml SOC culture, incubated for 1 h at 37 °C, and then one-half was spread onto agar to select Kmr and Nalr transformants. If none grew within 24 h, the remainder was spread after standing overnight at room temperature.
Intracellular survival assay HeLa cell infection assay was routinely used to detect the intracellular replication or survival ability of the mutant[19-21]. S. flexneri 2a 2457T, 2457T05, mutant strain and E. coli MC1061 (noninvasive control) were grown in an appropriate medium overnight. The liquid cultures were then washed in PBS and resuspended in antibiotic-free medium. Approximately 106 HeLa cells were cultured in a 10 cm2 flask. HeLa cells were washed three times in PBS prior to incubation with about 108 CFU bacteria at 37 °C for 3 h. The medium was removed from infected cells after 2.5 h, and the cells were washed three times in PBS. Fresh medium containing gentamicin (20 μg·mL-1) was added and the flasks were incubated for 5 h to eliminate extracellular bacteria. After that, the medium was replaced by RPMI1640 containing gentamicin (20 μg·mL-1) and the infected cells were cultured for another 40 h. The supernatants of culture were tested for extracellular surviving bacteria by plating them on LB agar plates. The monolayers were then washed three times in PBS and lysed by addition of 0.1% deoxycholate sodium to liberate the intracellular bacteria. Dilutions of the lysates of HeLa cells infected with bacteria were plated on LB agar plates and cultured at 37 °C overnight. The CFU of the bacteria was then counted.
Competition assay To test the mutant strains for alterations in virulence relative to the wild type, a competition assay was carried out by using a murine intranasal infection model[22-24] with some modifications. The mutant or MC1061 (negative control) or 2457T (positive control) and 2457T05 grown overnight were mixed at 1:1 (v/v) ratio and washed in PBS. After concentration of the mixture was adjusted by dilutions, 20 μl mixtures containing about 106 CFU in PBS was used to introduce droplets into the nares of BALB/c mice that was anesthetized. The number of bacteria in each inoculum was determined by plating serial dilutions of the inoculum. After 24 h, the recovered bacteria from the lungs of mice were counted, and then the number of mutant strains and 2457T was counted. According to the method of Camilli et al[25], the competitive index of each mutant was obtained.
Bioinformatics analysis The sequence of S. flexneri 2a alkA gene was analyzed by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The identity and gene function were also analyzed by NCBI.
Statistical analysis
Data from the intracellular survival assay were analyzed by Dunnett t test, and P value less than 0.05 was considered as statistically significant. Data from the competition assay were analyzed by the sign test, and P value less than 0.0078 was considered as statistically significant. The Dunnet t test and the sign test were performed using the SAS (Statistical Analysis Systems Inc., USA) program.
RESULTS
Cloning and homologous analysis of S. flexneri 2a alkA gene
In our previous work, we identified S. flexneri 2a alkA as an in vivo-induced gene, and obtained the partial sequence of S. flexneri 2a alkA gene which had an alignment with other bacteria. Based on the sequence of E. coli alkA, the primers, P1 (CGAGGAACGATTTTGGTGAT) and P2 (CTCGCT-GAAAGCGAATATGG) (Figure 1B) were designed. Using S. flexneri 2a 2457T chromosome DNA as a template, PCR was performed and the PCR products were purified by agarose gel electrophoresis, and ligated into plasmid pMD18-T to produce plasmid pMD028 after the confirmation of EcoRV and HindIII digestion analysis. The recombinant plasmid pMD028 was subjected to DNA sequencing to obtain the whole length DNA sequence of alkA. Its open read frame was 849 bp. In the upstream there was a promoter sequence (AGCAAAGCGTAACGTCTGAATAACGTTTATGCT) and the binding site (AAAGCAAA) of Ada protein which was a regulator of alkA gene expression in E. coli. Based on the sequence of S. flexneri 2a alkA, homologous analysis was carried out on the NCBI website. The results are listed in Table 2. Interestingly, the sequence alignment revealed a Helix- hairpin-Helix (HhH) motif common to DNA glycosylases. E. coli alkA was identified as the helix-hairpin-helix DNA glycosylase[26]. Very possibly, alkA protein from S. flexneri 2a belonged to an HhH-GPD super family. Its hallmark was Helix-hairpin-Helix and Gly/Pro rich loop followed by a conserved aspartate and its function was presumably involved in DNA replication and repair.
Figure 1.
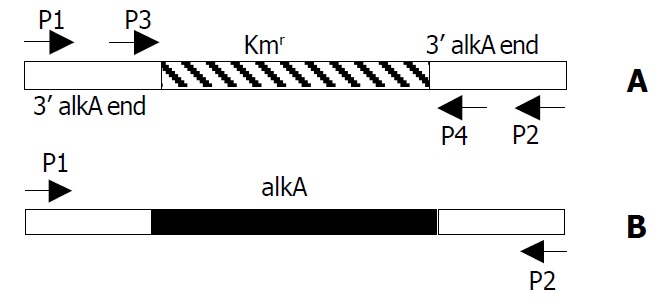
Location of primers P1, P2, P3 and P4 in PCR prod-ucts of alkA and 028pKm. A: 028pKm (about 1.4 kb); B: alkA (about 1.7 kb).
Table 2.
BLAST analysis of S. flexneri 2a alkA gene
| Bacteria | Gene product | Gene function | Identities (%) |
| Escherichia coli K12 | 3-methyl-adenine DNA glycosylase II | DNA - replication, repair, restriction/modification | 97% |
| Escherichia coli O157:H7 | 3-methyl-adenine DNA glycosylase II | Macromolecule synthesis, modification: DNA - replication, repair, restriction/modification | 97% |
| Salmonella typhimurium | 3-methyl-adenine DNA glycosylase II | Unknown | 74% |
| Ralstonia solanacearum | putative transcription regulator protein | Miscellaneous; not classified regulator | 38% |
| Pseudomonas aeruginosa | DNA-3-methyladenine glycosidase II | Unknown | 37% |
| Xanthomonas axonopodis | DNA methylation and regulatory protein | Unknown | 35% |
| Mycobacterium bovis | Methylated-DNA—protein-cysteine methyltransferase | Adaptative response | 34% |
| Mycobacterium tuberculosis | alkA protein | Unknown | 34% |
| Vibrio cholerae | ada regulatory protein | Unknown | 32% |
| Bacillus anthracis | DNA-3-methyladenine glycosidase | Unknown | 32% |
| Archaeoglobus fulgidus | 3-methyladenine DNA glycosylase | DNA repair at suboptimal and maybe even mesophilic temperatures | 30% |
Construction of S. flexneri 2a engineering strain
To delete alkA gene of S. flexneri 2a, construction of S. flexneri 2a engineering strain expressing gam, bet, and exo genes, was required. The fragment (about 4 kb) containing gam, bet, and exo genes was obtained by digestion of pKD46 plasmid with Pvu I and Bstx I and ligated into the Sal I site of suicide plasmid pXL275. Then the ligation products were then transformed into S17-1λpir. The recombinant plasmid was confirmed by BamHI digestion and known as pXLkd46. After bacterial mating of the donor (S17-1λpir/pXLkd46) and recipient (2457T), pXLkd46 plasmid was integrated into chromosome of S. flexneri 2a by homologous recombination. The process of pXLkd46 plasmid construction and chromosomal integration is showed in Figure 2. The resulting strain was verified with antibiotic selection and serum agglutination and designated as 2457T05.
Figure 2.
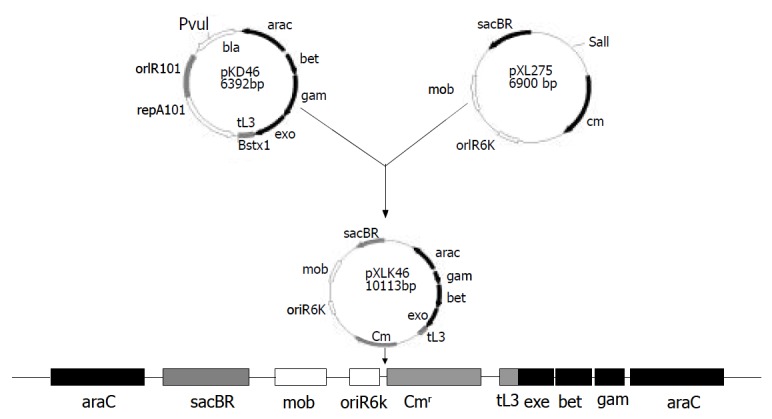
Construction and chromosomal integration of pXLkd46 plasmid.
Construction of deletion mutant of S. flexneri alkA gene
After 2457T05 was successfully constructed, the linear targeting DNA, 028pKm, was required for disruption of S. flexneri 2a alkA gene with λ Red recombination system. To obtain the fragment 028pKm, recombinant plasmid pMD028pKm was firstly to be constructed. Kmr gene fragment was obtained from plasmid pMDKm05 digested by HincI I and Sma I and inserted into the alkA gene of plasmid pMD028 digested by EcoRV and StuI and dephosphorized by CIAP. The ligation products were then transformed into E. coli DH5α. The recombinant plasmids were isolated from the transformants, confirmed by EcoR I digestion, and designated as pMD028pKm (Figure 3). Using pMD028pKm as a PCR template, P3 (TGTGCCAGTGAGGAAAGACC) and P4 (GAGAGAGCGTTTGCCCATTG) (Figure 1A) as primers, PCR was carried out. In order to reduce the interference of plasmid pMD028pKm, the second PCR was carried out at the same experimental condition except that the template was the first PCR products diluted by 1000 times. The second PCR products (028pKm) did not contain the template plasmid pMD028pKm that would lead to false positive colonies in the latter electroporation experiment. The 028pKm fragment was a cassette, 5’ alkA end-Kmr-3’alkA end (Figure 1A). The concentrated 028pKm was electroporated into S. flexneri 2a engineering strain 2457T05. The alkA gene was then replaced by kanamycin resistance gene through homologous recombination mediated by λ Red system. The positive transformants were selected on LB agar plates containing Km and Nal.
Figure 3.
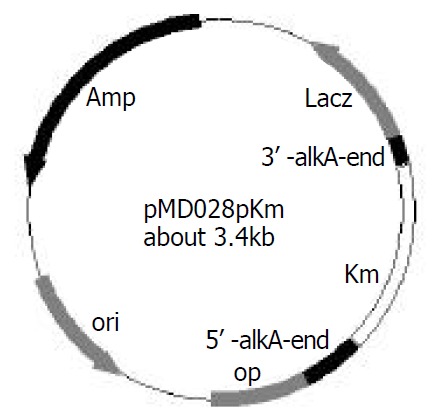
Construction of recombinant plasmid pMD028pkm.
To verify the replacement of S. flexneri 2a alkA gene, PCR and sequencing were used. PCR was performed in which 2457T05 (negative control), the transformant and pMD028pKm plasmid (positive control) were used as templates. The reaction conditions were the same as the amplification of S. flexneri alkA gene, and the primers were also P1 and P2. The PCR products from the transformant and pMD028pKm plasmid were about 1.4 kb and 1.7 kb from 2457T05 respectively (Figure 1, Figure 4). Then the PCR products were sequenced and analyzed by BLAST (data not shown). The result indicated that alkA gene of S. flexneri 2a was replaced by Kmr gene. Hereby, the deletion mutant of alkA gene was successfully constructed and designated as 2457T028D.
Figure 4.
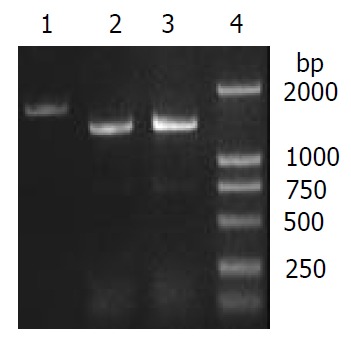
Verification of alkA gene deletion of 2457T028D mutant by PCR. 1: alkA PCR product (amplified from 2457T05), 2: 028pKm PCR product (amplified from 2457T028D), 3: 028Kkm PCR product (amplified from pMD028pKm), 4: DL2000 Marker.
Functional analysis of deletion mutant of S. flexneri 2a alkA gene
In order to detect the role of alkA in the pathogenesis of S. flexneri, intracellular survival assay and virulence assay were respectively carried out in HeLa cells and BALB/c mice.
Mutant 2457T028D was tested for its survival ability in HeLa cells relative to the wild-type strain. Equal volume of each strain (2457T, 2457T05, 2457T028D and MC1061) was respectively used to infect the HeLa cell monolayer. Within 48 h, the integrity of the infected HeLa cell monolayer was good and the configuration of HeLa cells had no significant alterations relative to normal cells. But the growth rate of the infected cells became slow. After 48 h, CFU of bacteria recovered from HeLa cells was counted. The results of the infection assay are summarized in Figure 5. Noticeably, CFU levels of the mutant recovered from HeLa cells were five-fold lower than that of the wild type (P < 0.01), indicating that the mutant had a lower capability of survival or replication. The survival probability of 2457T and 2457T05 showed no significant difference (Figure 5). In order to further confirm the survival probability of the mutant, competition assay was carried out with the HeLa cell infection model. A 1:1 (v/v) mixture of S. flexneri 2a 2457T05 and the mutant or 2457T or MC1061 was used to infect HeLa cells. The number of bacteria in each inoculum was determined by plating serial dilutions of the inoculum. After 48 h, recovered bacteria from HeLa cells were counted, and the number of bacteria was counted respectively. The experiment was separately repeated 3 times. Therefore, the competitive index of each strain obtained is shown in Table 3. The mutant 2457T028D whose survival probability was significantly lower in the infection assay, exhibited significant survival defects in this experiment (P < 0.0078). The data strongly indicated that the mutant was much less able to survive in HeLa cells.
Figure 5.
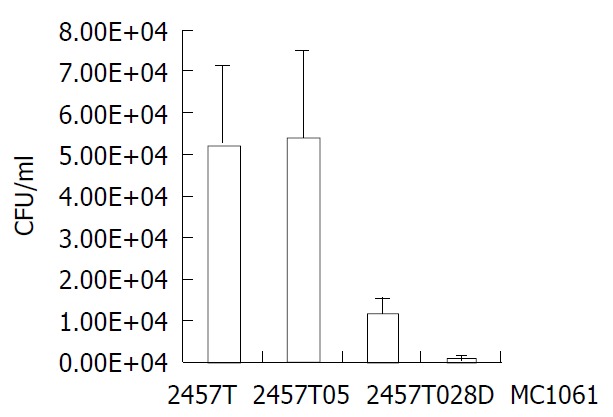
Comparison of HeLa cells infected by 2457T, 2457T05, 2457T028D and MC1061.
Table 3.
Competitive analysis of different strains of S. flexneri 2a
| Strains |
Competitive index (CI)a |
|
| BALB/c mouse infection | HeLa cells infection | |
| 2457T05 | 0.96 ± 0.27 | 1.07 ± 0.35 |
| 2457T028D | 0.21 ± 0.09b | 0.25 ± 0.11b |
| MC1061 | 0.03 ± 0.02b | 0.02 ± 0.01b |
Competitive index (CI) is the ratio of mutant CFU to wild-type CFU after correcting the ratios for deviations of the in-oculum ratio from a value of 1.
P < 0.0078.
The mutant was also tested in a mice lung infection model for alterations in virulence relative to the wild-type parental strain. Mice that were challenged only with S. flexneri 2457T showed early acute bronchiolitis at 24 h, followed by severe pneumonia at 48 h. Five mice were used for each group in the murine lung infection model. After 24 h, recovered bacteria from the lungs of mice were counted, and the number of each strain was counted. Then the competitive index of each strain obtained is summarized in Table 3. The mutant also exhibited significant colonization defects (P < 0.0078). The data further indicated that alkA gene was potentially related to the virulence of S. flexneri 2a 2457T.
DISCUSSION
It has been reported that in vivo-induced gene played an important role in the process of interaction between pathogen and host[27]. In vivo-induced genes are those whose expression is induced when pathogens infect their hosts. Their inducible expression is a molecular-level genetic adaptive response to special environments of host. Many virulent genes have been identified by mutational analysis of in vivo-induced genes. Heithoff et al[28] used purA-lacZY as a reporter to identify in vivo-induced genes of Salmonella typhimurim utilizing macrophages or BALB/c mice as a model. They discovered some in vivo-induced genes, including regulatory genes (phoP, pmrB, cadC, etc.) and metabolic genes (recD, hemA, mgtA, entF, etc.). Furthermore, insertion mutants of these genes were constructed, and their virulence was detected. Seven of them exhibited significant virulence defects. In an another research on Pseudomonas aeruginosa, 22 genes were in vivo-induced during infecting BALB/c mice, including np20, which has been proved to be a virulence gene, and known as virulent factor FptA[29]. In our previous study, we identified alkA as an in vivo-induced gene. However, it is unknown if alkA gene is related to the virulence of S. flexneri.
In order to detect the role of alkA gene in the virulence of S. flexneri 2a, it is required to construct the deletion mutant of alkA gene. The mutant is conventionally constructed by twice homologous recombination mediated by suicide plasmid. Although the asd gene of S. flexneri 2a was successfully disrupted in our previous study[30], the efficiency of this method is very low and the experimental period is quite long. Recently, a new method, which depends on Red recombination system of λ phage, has been successfully established and used to speed up the knockout of genes[31,32]. However, its application was limited to E. coli[33-35]. It was not reported in other bacteria except that the asd gene was deleted with λ Red system in our laboratory[36]. In this study, the deletion mutant of S. flexneri 2a alkA gene was successfully constructed also with λ Red system. Importantly, an engineering strain 2457T05 of S. flexneri 2a was constructed, and it was confirmed that the strain could be used to study the function of S. flexneri 2a genes.
After the mutant of S. flexneri 2a alkA gene was constructed, intracellular survival and competition assays were carried out. The results showed that alkA mutant of S. flexneri could exhibit a low intracellular survival ability and a significant virulence defect, indicating that alkA was a virulence-related gene in S. flexneri 2a. However, it has not been reported before whether alkA was associated with the virulence of pathogens. AlkA is an expression-induced gene and its product, 3-methyladenine DNA glycosylase II, is involved in the SOS-dependent adaptive response. Expression of alkA is regulated by Ada protein. When alkylation damages bacterial DNA, ada gene would be induced by alkyl-DNA. The produced Ada finishes directly-repairing damage of alkyl-DNA by transferring the methyl group from alkyl-DNA to its cysteine residues. At the same time Ada loses its activity. The methyl-Ada turns into a positive regulator of alkA, aidB, alkB, and itself as well. Methyl-Ada could recognize and bind onto the special region (AAAGCAAA) of alkA promoter, start transcription of alkA, and further complete repairing damage of other type alkylation, avoiding bacterial death due to damage of DNA alkylation[37-40]. The base excision repair could protect against the deleterious effects of DNA alkyl lesions. However, the activities of alkA gene must be balanced for optimal protection against the biological consequences of damaged DNA bases because inappropriate expression of this activity might have a detrimental consequence[41]. During infection of host, Shigella spp. probably suffers strong damage of alkyl in host. But alkylated DNA activates the adaptive response of Shigella spp. to host. Expression of alkA effectively repairs damage of DNA alkylation so that the killing-effect resulted from DNA damage could not carry out. Therefore, from this point of view, in vivo-induced-expression of alkA provides a significant safeguard for infection of Shigella spp. and is an essential gene for exhibiting Shigella spp. virulence.
Although there has been no report about the relationship between alkA and virulence of pathogens, it is known that a close relation lies between DNA methylation and bacterial virulence. Heithoff et al[42] discovered that DNA adenine methylase (Dam) could regulate expression of at least 20 in vivo-induced genes and that Dam- S. typhimurium as a live vaccine had a protective role with no side-effect. S. typhimurium with over-expressing Dam also exhibited a significant virulence defect and a protective effect as an oral vaccine[43]. Similar results have also been obtained in Yersinia pseudotuberculosis and Vibrio cholerae[44]. Hereby, during infection expression of Dam could induce the expression of in vivo-induced genes, but its over-expression could also lead to damage of methylation and attenuation of pathogens. Thus a suitable level of DNA methylation might play a key role for pathogens to keep the virulence. From this point of view, alkA may be a virulence-related gene of pathogens. The hypothesis illustrating the relationship between DNA methylation damage and alkA gene is shown in Figure 6. Whether a regulatory relation exists between alkA and dam remains to be further confirmed. However, we believe that alkA is a new target for studying on molecular pathogenesis mechanism of Shigella spp. and construction of attenuated live vaccines.
Figure 6.
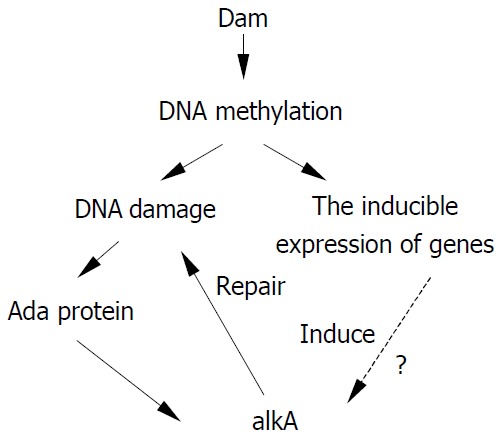
Hypothesis illustrating the relationship between alkA and damage of DNA methylation.
ACKNOWLEDGEMENTS
We thank Prof. Qi-Nong Ye and Dr Bao-Chang Fan for critical review of this manuscript. We also thank Miss Run-Yan Liu and Miss Hui-Ping Zhang for their help with experiments.
Footnotes
Supported by the Major State Basic Research Development Program of China (973 Program), No. G1999054103; National High-technology Research and Development Program of China (863 Program), No. 2001AA215211; and Capital “248” Key Innovation Program of China, No. H010210360119
Edited by Zhang JZ and Wang XL
References
- 1.Sansonetti PJ. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks. FEMS Microbiol Rev. 2001;25:3–14. doi: 10.1111/j.1574-6976.2001.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez MI, Sansonetti PJ. Shigella interaction with intestinal epithelial cells determines the innate immune response in shigellosis. Int J Med Microbiol. 2003;293:55–67. doi: 10.1078/1438-4221-00244. [DOI] [PubMed] [Google Scholar]
- 3.Yao X, Wang HL, Shi ZX, Yan XY, Feng EL, Yang BL, Huang LY. Identification of RanBMP interacting with Shigella flexneri IpaC invasin by two-hybrid system of yeast. World J Gastroenterol. 2003;9:1347–1351. doi: 10.3748/wjg.v9.i6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handfield M, Levesque RC. Strategies for isolation of in vivo expressed genes from bacteria. FEMS Microbiol Rev. 1999;23:69–91. doi: 10.1111/j.1574-6976.1999.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 5.Mahan MJ, Slauch JM, Mekalanos JJ. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 6.Young GM, Miller VL. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 7.Staib P, Kretschmar M, Nichterlein T, Köhler G, Michel S, Hof H, Hacker J, Morschhäuser J. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol Microbiol. 1999;32:533–546. doi: 10.1046/j.1365-2958.1999.01367.x. [DOI] [PubMed] [Google Scholar]
- 8.Gahan CG, Hill C. The use of listeriolysin to identify in vivo induced genes in the gram-positive intracellular pathogen Listeria monocytogenes. Mol Microbiol. 2000;36:498–507. doi: 10.1046/j.1365-2958.2000.01869.x. [DOI] [PubMed] [Google Scholar]
- 9.Lai YC, Peng HL, Chang HY. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect Immun. 2001;69:7140–7145. doi: 10.1128/IAI.69.11.7140-7145.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Lee SW, Hillman JD, Progulske-Fox A. Identification and testing of Porphyromonas gingivalis virulence genes with a pPGIVET system. Infect Immun. 2002;70:928–937. doi: 10.1128/IAI.70.2.928-937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartoleschi C, Pardini MC, Scaringi C, Martino MC, Pazzani C, Bernardini ML. Selection of Shigella flexneri candidate virulence genes specifically induced in bacteria resident in host cell cytoplasm. Cell Microbiol. 2002;4:613–626. doi: 10.1046/j.1462-5822.2002.00216.x. [DOI] [PubMed] [Google Scholar]
- 12.Morohoshi F, Hayashi K, Munkata N. Bacillus subtilis alkA gene encoding inducible 3-methyladenine DNA glycosylase is adjacent to the ada operon. J Bacteriol. 1993;175:6010–6017. doi: 10.1128/jb.175.18.6010-6017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Rourke EJ, Chevalier C, Boiteux S, Labigne A, Ielpi L, Radicella JP. A novel 3-methyladenine DNA glycosylase from Helicobacter pylori defines a new class within the endonuclease III family of base excision repair glycosylases. J Biol Chem. 2000;275:20077–20083. doi: 10.1074/jbc.M001071200. [DOI] [PubMed] [Google Scholar]
- 14.Saparbaev M, Mani JC, Laval J. Interactions of the human, rat, Saccharomyces cerevisiae and Escherichia coli 3-methyladenine-DNA glycosylases with DNA containing dIMP residues. Nucleic Acids Res. 2000;28:1332–1339. doi: 10.1093/nar/28.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Privezentzev CV, Saparbaev M, Sambandam A, Greenberg MM, Laval J. AlkA protein is the third Escherichia coli DNA repair protein excising a ring fragmentation product of thymine. Biochemistry. 2000;39:14263–14268. doi: 10.1021/bi001337g. [DOI] [PubMed] [Google Scholar]
- 16.Mansfield C, Kerins SM, McCarthy TV. Characterisation of Archaeglobus fulgidus AlkA hypoxanthine DNA glycosylase activity. FEBS Lett. 2003;540:171–175. doi: 10.1016/s0014-5793(03)00257-6. [DOI] [PubMed] [Google Scholar]
- 17.Rui X, Xu Y, Wan H, Su G, Huang C. Construction of a stable and non-resistant bivalent vaccine candidate strain against Shigella flexneri 2a and Shigella sonnei. Chin J Biotechnol. 1996;12:89–97. [PubMed] [Google Scholar]
- 18.Sambrook J, Friston E F, Maniatis T. Molecular Cloning: a Labo-ratory Manual. The 2end edition. New York: Cold Spring Harbor Laboratory Press. 1989:34–56, 237-261. [Google Scholar]
- 19.Roy S, Biswas T. Murine splenocyte proliferation by porin of Shigella dysenteriae type 1 and inhibition of bacterial invasion of HeLa cell by anti-porin antibody. FEMS Microbiol Lett. 1996;141:25–29. doi: 10.1111/j.1574-6968.1996.tb08358.x. [DOI] [PubMed] [Google Scholar]
- 20.Niesel DW, Chambers CE, Stockman SL. Quantitation of HeLa cell monolayer invasion by Shigella and Salmonella species. J Clin Microbiol. 1985;22:897–902. doi: 10.1128/jcm.22.6.897-902.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong QP. Pathogenic effects of Opolysaccharide from Shigella flexneri strain. World J Gastroenterol. 1999;5:245–248. doi: 10.3748/wjg.v5.i3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallett CP, VanDeVerg L, Collins HH, Hale TL. Evaluation of Shigella vaccine safety and efficacy in an intranasally challenged mouse model. Vaccine. 1993;11:190–196. doi: 10.1016/0264-410x(93)90016-q. [DOI] [PubMed] [Google Scholar]
- 23.van de Verg LL, Mallett CP, Collins HH, Larsen T, Hammack C, Hale TL. Antibody and cytokine responses in a mouse pulmonary model of Shigella flexneri serotype 2a infection. Infect Immun. 1995;63:1947–1954. doi: 10.1128/iai.63.5.1947-1954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Way SS, Borczuk AC, Dominitz R, Goldberg MB. An essential role for gamma interferon in innate resistance to Shigella flexneri infection. Infect Immun. 1998;66:1342–1348. doi: 10.1128/iai.66.4.1342-1348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camilli A, Mekalanos JJ. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang SL, Mekalanos JJ, Holden DW. In vivo genetic analysis of bacterial virulence. Annu Rev Microbiol. 1999;53:129–154. doi: 10.1146/annurev.micro.53.1.129. [DOI] [PubMed] [Google Scholar]
- 28.Heithoff DM, Conner CP, Hanna PC, Julio SM, Hentschel U, Mahan MJ. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Mushegian A, Lory S, Jin S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HL, Feng EL, Lin Y, Liao X, Su GF. Construction of asd mutant of Shigella flexneri 2a strain T32. Bull Acad Mil Med Sci. 2000;24:81–87. [Google Scholar]
- 31.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou S, Chen X, Wang H, Tao M, Hu Z. Efficient method to generate homologous recombinant baculovirus genomes in E. coli. Biotechniques. 2002;32:783–74, 786, 788. doi: 10.2144/02324st04. [DOI] [PubMed] [Google Scholar]
- 34.Murphy KC, Campellone KG, Poteete AR. PCR-mediated gene replacement in Escherichia coli. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- 35.Loh T, Murphy KC, Marinus MG. Mutational analysis of the MutH protein from Escherichia coli. J Biol Chem. 2001;276:12113–12119. doi: 10.1074/jbc.M007935200. [DOI] [PubMed] [Google Scholar]
- 36.Wang HL, Feng EL, Shi ZX, Yao X, Su GF, Huang LY. Quick knockout of Shigella flexneri asd gene with Red system. Bull Acad Mil Med Sci. 2002;26:172–175. [Google Scholar]
- 37.Furuichi M, Yu CG, Anai M, Sakumi K, Sekiguchi M. Regulatory elements for expression of the alkA gene in response to alkylating agents. Mol Gen Genet. 1992;236:25–32. doi: 10.1007/BF00279639. [DOI] [PubMed] [Google Scholar]
- 38.Landini P, Busby SJ. Expression of the Escherichia coli ada regulon in stationary phase: evidence for rpoS-dependent negative regulation of alkA transcription. J Bacteriol. 1999;181:6836–6839. doi: 10.1128/jb.181.21.6836-6839.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landini P, Busby SJ. The Escherichia coli Ada protein can interact with two distinct determinants in the sigma70 subunit of RNA polymerase according to promoter architecture: identification of the target of Ada activation at the alkA promoter. J Bacteriol. 1999;181:1524–1529. doi: 10.1128/jb.181.5.1524-1529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saget BM, Walker GC. The Ada protein acts as both a positive and a negative modulator of Escherichia coli's response to methylating agents. Proc Natl Acad Sci USA. 1994;91:9730–9734. doi: 10.1073/pnas.91.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD. 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays. 1999;21:668–676. doi: 10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 42.Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 43.Dueger EL, House JK, Heithoff DM, Mahan MJ. Salmonella DNA adenine methylase mutants elicit protective immune responses to homologous and heterologous serovars in chickens. Infect Immun. 2001;69:7950–7954. doi: 10.1128/IAI.69.12.7950-7954.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Julio SM, Heithoff DM, Provenzano D, Klose KE, Sinsheimer RL, Low DA, Mahan MJ. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect Immun. 2001;69:7610–7615. doi: 10.1128/IAI.69.12.7610-7615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


