The mechanisms guiding cells toward bone surfaces are generally unknown. Here, Nevius et al. show that the Gαi protein–coupled receptor EBI2 is expressed in mouse osteoclast precursors to guide these cells toward bone surfaces. Defective EBI2 signaling increased bone mass in male mice and protected female mice from age- and estrogen deficiency–induced osteoporosis.
Abstract
Bone surfaces attract hematopoietic and nonhematopoietic cells, such as osteoclasts (OCs) and osteoblasts (OBs), and are targeted by bone metastatic cancers. However, the mechanisms guiding cells toward bone surfaces are essentially unknown. Here, we show that the Gαi protein–coupled receptor (GPCR) EBI2 is expressed in mouse monocyte/OC precursors (OCPs) and its oxysterol ligand 7α,25-dihydroxycholesterol (7α,25-OHC) is secreted abundantly by OBs. Using in vitro time-lapse microscopy and intravital two-photon microscopy, we show that EBI2 enhances the development of large OCs by promoting OCP motility, thus facilitating cell–cell interactions and fusion in vitro and in vivo. EBI2 is also necessary and sufficient for guiding OCPs toward bone surfaces. Interestingly, OCPs also secrete 7α,25-OHC, which promotes autocrine EBI2 signaling and reduces OCP migration toward bone surfaces in vivo. Defective EBI2 signaling led to increased bone mass in male mice and protected female mice from age- and estrogen deficiency–induced osteoporosis. This study identifies a novel pathway involved in OCP homing to the bone surface that may have significant therapeutic potential.
Osteoclasts (OCs) are multinucleated cells that regulate skeletal development and integrity by actively resorbing excess or damaged bone produced by osteoblasts (OBs) and osteocytes. OBs and osteocytes differentiate from rare mesenchymal stem cells that reside in BM parenchyma (Méndez-Ferrer et al., 2010). In contrast, OCs differentiate from BM-resident and circulatory monocytic precursors that come into close contact with bone surfaces where the essential cytokines ligand for receptor activator of nuclear factor kappa binding (RANKL, encoded by Tnfsf11) and M-CSF (encoded by Csf1) are locally produced by OBs and osteocytes (Teitelbaum, 2000; Nakashima et al., 2011). Cellular interactions between OCs and OBs regulate the activity of both cell types, such that resorbed bone is accurately replaced by newly formed bone. Although the understanding of the molecular signals required for OC differentiation has increased significantly over the past several decades, very little is understood about the mechanisms controlling the migration and positioning of OC precursors (OCPs) near, or in contact with, the bone surface.
Monocytes and OCPs are dynamic within BM parenchyma (Ishii et al., 2009). OCPs recirculate between BM and peripheral organs via the action of sphingosine 1-phosphate receptors (S1PRs), which attract multiple hematopoietic cell subsets from BM parenchyma into blood circulation (Walzer et al., 2007; Ishii et al., 2009, 2010; Pereira et al., 2010b). In contrast, the signals promoting cell movement toward bone surfaces remain unknown. However, systemic RANKL administration has been shown to increase OCP homing back to BM and to promote local OC differentiation (Kotani et al., 2013). These studies suggest that OCP movement in and out of BM tissue is highly regulated and that balanced responsiveness to various chemoattractants regulates OCP movement and differentiation. Consistent with this hypothesis, bone resorption produces a milieu that contains potent chemoattractants for monocytes/OCPs (Mundy et al., 1978).
Here, we investigated the role played by EBI2, a Gαi protein–coupled receptor (GPCR) involved in dendritic cell and B lymphocyte migration in secondary lymphoid organs (Gatto et al., 2009, 2013; Pereira et al., 2009b; Hannedouche et al., 2011; Kelly et al., 2011; Liu et al., 2011; Yi and Cyster, 2013), in controlling monocyte and OCP movement and positioning within BM. We show that EBI2 is highly expressed in OCPs and mature OCs and promotes OCP motility in vitro and in vivo. Furthermore, we show that OCPs deficient in EBI2 migrate poorly toward bone surfaces, which reduces OC differentiation. In contrast, OCPs that overexpressed EBI2 preferentially localized at the bone surface and fused with preexisting OCs more efficiently than control OCPs. OBs expressed the enzymes CH25H and CYP7B1 that are required for the synthesis of the EBI2-ligand 7α,25-dihydroxycholesterol (7α,25-OHC) and secreted EBI2 ligands in vitro. Interestingly, OCPs also secreted 7α,25-OHC, which results in autocrine EBI2 signaling and tempers EBI2-mediated migration toward bone surfaces. Finally, EBI2 signaling–deficient mice exhibit increased bone mass at young and old ages, and EBI2 signaling deficiency significantly protects female mice from osteoporosis induced by estrogen deficiency.
RESULTS
EBI2 is expressed in OCPs and mature OCs and regulates bone mass
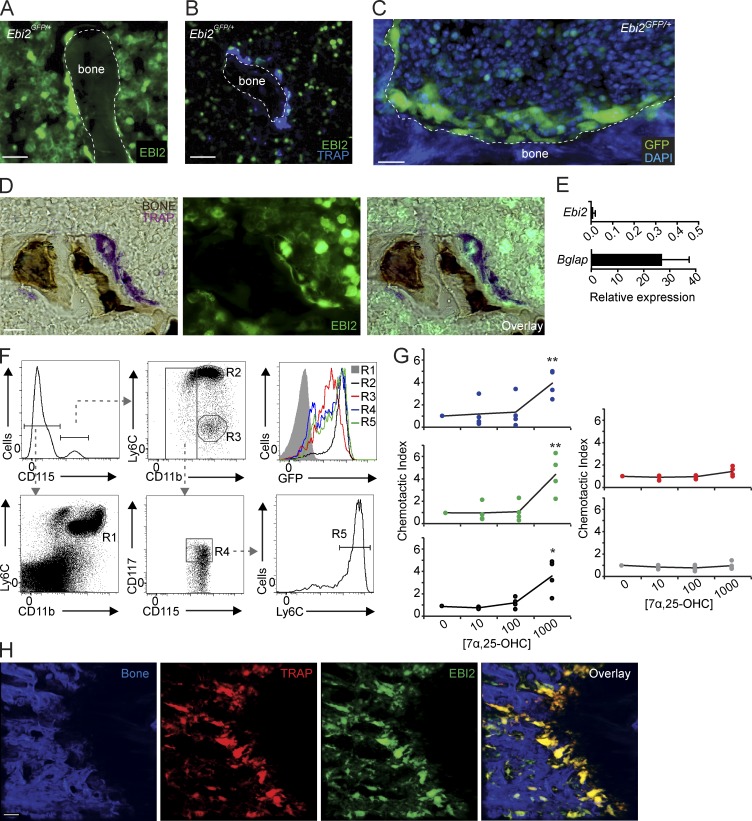
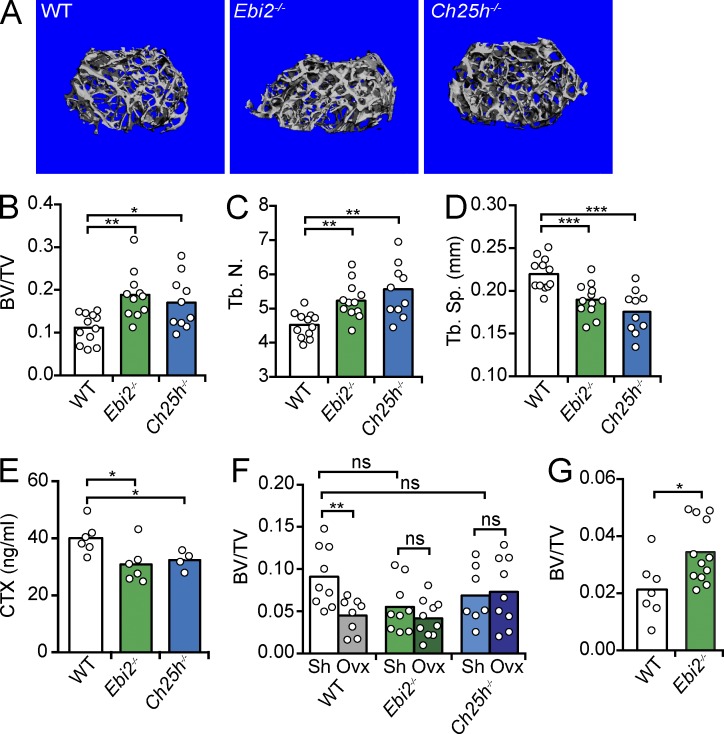
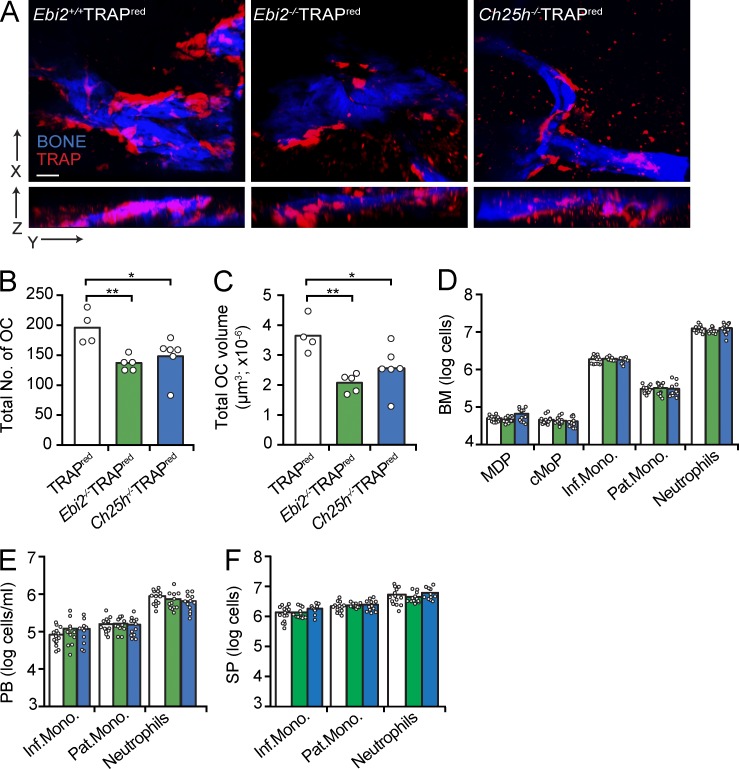
While examining EBI2-expressing cells in BM cavities of Ebi2GFP/+ mice (Pereira et al., 2009b), we detected abundant EBI2 expression in large and multinucleated bone-lining cells marked by tartrate-resistant acid phosphatase (TRAP) histochemistry (Fig. 1, A–D; Filgueira, 2004), suggesting that OCs express EBI2. In contrast, we could not detect EBI2 expression in OBs in vitro (Fig. 1 E) nor in vivo (Video 1). Using Ebi2GFP/+ mice, we examined EBI2 expression in hematopoietic cell subsets, particularly in monocyte/OCP subsets, by flow cytometry. Monocytic lineages differentiate from hematopoietic stem cells through sequential developmental stages, namely monocyte-macrophage and dendritic cell precursor (MDP) and common monocyte progenitor (cMoP) stages (Geissmann et al., 2010; Hettinger et al., 2013). MDPs and cMoPs expressed high amounts of EBI2, and its expression further increased in inflammatory monocytes, whereas it was reduced in patrolling monocytes and undetectable in neutrophils (Fig. 1 F). MDPs, cMoPs, and inflammatory monocytes migrated toward a concentration gradient of 7α,25-OHC, demonstrating that EBI2 is functional in these cells, whereas neutrophils and patrolling monocytes were unresponsive (Fig. 1 G). Using TRAP reporter mice (Kikuta et al., 2013), herein designated TRAPRed, we found that EBI2 was expressed in essentially all bone-lining TRAP+ OCs in vivo (Fig. 1 H and Video 2). To determine whether EBI2 signaling plays a role in bone mass homeostasis, we analyzed femurs and tibias of EBI2- and CH25H-deficient and -sufficient mice by microcomputed tomography (μCT). We found that EBI2 signaling–deficient male mice exhibited an increased ratio of bone volume to trabecular volume (Fig. 2, A and B), increased number of trabecular bones (Fig. 2 C), and reduced spacing between trabecular bones (Fig. 2 D), characteristic features of increased bone mass. Furthermore, we detected a significant reduction in the concentration of circulatory carboxy-terminal collagen cross-links (CTXs) by ELISA in EBI2- and CH25H-deficient mice when compared with littermate controls (Fig. 2 E), suggesting reduced OC resorptive activity in EBI2 signaling–deficient mice. Even though 16-wk-old sham-operated female mice did not show significant differences in bone mass (Fig. 2 F), EBI2-deficient females were significantly protected from ovariectomy-induced bone loss (Fig. 2 F). Furthermore, 1-yr-old EBI2-deficient female mice were significantly protected from age-induced reduction in bone mass (Fig. 2 G). These data showed that EBI2 is required for trabecular bone mass homeostasis in both sexes. We did not detect significant differences in cortical bone thickness between EBI2- or CH25H-deficient and control littermate mice. Histomorphometry of EBI2-deficient, CH25H-deficient, and control littermate femurs revealed a small but significant difference in OC numbers per tissue area (NOC/TAR) and showed no significant differences in OB numbers and bone surface area covered by OBs (Table 1). Furthermore, analyses of bone formation rate did not reveal significant differences between EBI2-deficient and -sufficient mice (Table S1). Combined, these data showed that EBI2 signaling is required for bone mass homeostasis, presumably as the result of a direct role in OC differentiation.
Figure 1.
EBI2 expression and activity in monocytes, OCPs, and mature OCs. (A and B) Fluorescence histochemistry of femur sections of Ebi2GFP/+ mice. (A and B) Distribution of Ebi2-expressing cells (A) and TRAP detection (B). (C) Femurs of Ebi2GFP/+ mice stained to detect nuclear DNA with DAPI detected by two-photon microscopy; Ebi2-expressing cells and bone are shown. (D) TRAP and bone histochemistry of Ebi2GFP/+ sectioned femurs. Images were visualized by light (left) and fluorescence (middle) microscopy. Right panel depicts overlay. (A–D) Data are representative of at least three mice independently analyzed. (E) Ebi2 and Bglap mRNA expression in OB differentiated in vitro. Bars indicate mean ± SD of triplicate measures; three independent experiments. (F) Flow cytometric analyses of EBI2 expression in BM myeloid cell subsets from Ebi2GFP/+ mice. R1, neutrophils; R2, inflammatory monocytes; R3, patrolling Ly6Clo monocytes; R4, MDP cells; and R5, cMoPs. Data are representative of three mice independently analyzed. (G) Transwell migration assay of MDPs (blue), cMoPs (green), inflammatory (black) and patrolling (red) monocytes, and neutrophils (gray) toward a gradient of 7α,25-OHC concentration (nM). Lines indicate mean, and circles depict individual experiments. Data are representative of four mice independently analyzed. (H) Analysis of Ebi2GFP/+ TRAPRed femur; left to right: bone, TRAP+, EBI2-expressing cells, and overlay. Imaged area is 500 × 500 × 100 µm. Data are representative of >10 mice independently analyzed. *, P < 0.05; **, P < 0.01 by unpaired Student’s t test. Bars: (A and B) 30 µm; (C, D, and H) 50 µm. See also Videos 1 and 2.
Figure 2.
Bone mass analyses in EBI2- and CH25H-deficient mice. (A) Representative μCT images of femurs from 14-wk-old male WT (left), EBI2-deficient (middle), and CH25H-deficient mice (right). (B) Ratio of bone volume (BV) and trabecular volume (TV). (C) Number of trabecular bones (Tb.N.). (D) Spacing between trabecular bones (Tb.Sp., mm). (A–D) Data were generated by μCT of femurs, n = 10–12; four independent experiments. (E) Concentration of CTXs in serum of 14-wk-old male EBI2-deficient, CH25H-deficient, and control littermate mice, n = 4–6; three independent experiments. (F) Percentage of bone loss 4 wk after ovariectomy. Control (white), EBI2-deficient (green), and CH25H-deficient (blue) mice. (G) Ratio of bone volume and trabecular volume in 1-yr-old female WT and EBI2-deficient mice. (F–G) Data were generated by μCT of femurs, n = 7–12; three independent experiments. (B–G) Bars indicate mean, and circles depict individual mice analyzed. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by unpaired Student’s t test. See also Table 1 and Table S1.
Table 1.
Histomorphometry of WT, Ebi2−/−, and Ch25h−/− mice
| Strain | n | BV/TV | Tb.N | Tb.Th | Tb.Sp | OCS/BS | NOC/TAR | NOC/BPm | OBS/BS | OBS/OS | NOB/TAR | NOB/BPm | NOB/Opm |
| % | mm−1 | µm | µm | % | % | % | |||||||
| WT | 8 | 19.80 ± 7.75 | 4.69 ± 0.99 | 41.20 ± 7.55 | 179.65 ± 45.65 | 9.59 ± 5.22 | 80.62 ± 22.67 | 4.28 ± 1.32 | 30.59 ± 7.55 | 11.74 ± 4.66 | 105.60 ± 40.20 | 11.41 ± 4.51 | 36.68 ± 7.14 |
| Ebi2−/− | 4 | 31.34 ± 11.01a | 5.56 ± 0.95 | 55.09 ± 13.51a | 131.04 ± 51.20a | 8.85 ± 4.12 | 59.90 ± 22.48a | 3.78 ± 1.15 | 32.37 ± 6.17 | 12.99 ± 4.72 | 131.36 ± 57.42 | 11.64 ± 4.45 | 36.01 ± 12.16 |
| Ch25h−/− | 10 | 23.57 ± 11.74a | 5.10 ± 1.17 | 44.50 ± 11.63 | 160.77 ± 53.42 | 6.17 ± 1.86b | 55.20 ± 11.28a | 4.21 ± 1.19 | 47.11 ± 9.46 | 14.54 ± 5.85 | 141.59 ± 83.40 | 13.20 ± 5.27 | 37.61 ± 7.19 |
Data are presented as mean ± SD. BV/TV, bone volume per total volume; Tb.N, trabecular number; Tb.Th, trabecular thickness; Tb.Sp, trabecular separation; OCS/BS, OC surface; NOC/TAR, OC number per total area; NOC/BPm, OC number per bone perimeter; OBS/BS, OB surface per bone surface; OB/OS, OB surface/osteoid surface; NOB/TAR, OB number per total area; NOB/BPm, OB number per bone perimeter; NOB/Opm, OB number per osteoid perimeter.
Significant difference from WT controls, P < 0.05.
P = 0.06.
EBI2 promotes OCP motility and enhances OC differentiation
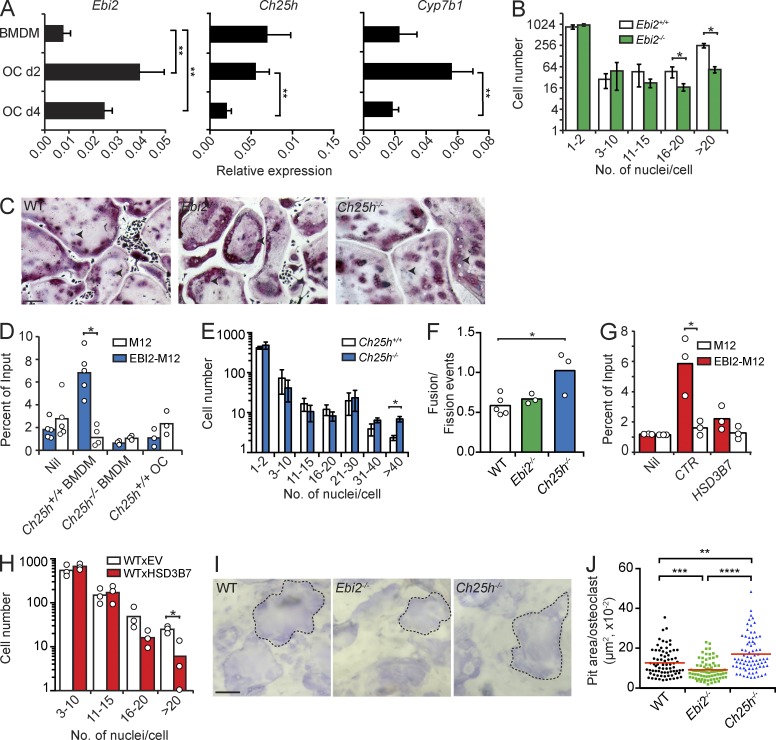
BMDMs and OCs differentiated in vitro with recombinant RANKL and M-CSF expressed Ebi2, as well as Ch25h and Cyp7b1 (Fig. 3 A). EBI2-deficient OCPs generated threefold less large multinucleated OCs containing >16 nuclei per cell (Fig. 3, B and C), even though EBI2 ligands were not added to the culture conditions. The cell-intrinsic requirement for EBI2 in the absence of exogenous EBI2 ligands suggested that OCPs secreted EBI2 ligands in vitro. To test this hypothesis, we measured chemotaxis of M12 cells overexpressing EBI2 toward WT or CH25H-deficient BMDM culture supernatants, as described previously (Hannedouche et al., 2011; Kelly et al., 2011). EBI2-transduced M12 cells migrated vigorously to WT BMDM supernatants but not to CH25H-deficient BMDM supernatants (Fig. 3 D), demonstrating that OCPs secrete EBI2 ligands in vitro. To our surprise, CH25H-deficient BMDMs differentiated efficiently into large OCs in vitro (Fig. 3 E). CH25H deficiency increases cellular cholesterol (Russell, 2003), and changes in cholesterol concentration affect OC differentiation in vitro (Grasser et al., 2003). OC sizes are not only dependent on the number of OCP fusion events, but can also be determined by the number of OC fission events (Jansen et al., 2012; Pavlos and Ng, 2012). To gain insight on how CH25H deficiency led to increased OC size, we visualized OC differentiation in vitro using time-lapse microscopy and quantified the number of cell fusions and fissions in WT and EBI2- and CH25H-deficient OCPs during OC differentiation. The ratio of fusion and fission events was significantly increased in CH25H-deficient precursors when compared with WT precursors (Fig. 3 F). To circumvent this problem, we measured OC differentiation from OCPs transduced with hydroxy-delta-5-steroid dehydrogenase, 3 β- and steroid delta-isomerase 7 (HSD3B7), an enzyme that metabolizes EBI2 ligands into downstream bile acid metabolic precursors (Russell, 2003). EBI2 ligand concentration was significantly reduced in HSD3B7-transduced OCPs (Fig. 3 G), which resulted in a significant reduction in the number of large multinucleated OCs (Fig. 3 H). Although EBI2-deficient OCPs differentiated into smaller OCs, these cells could resorb bone in vitro, as measured by bone-resorptive pit assays. However, the pit areas formed by EBI2-deficient OCs were significantly smaller than in OCs formed from WT and CH25H-deficient OCPs (Fig. 3, I and J).
Figure 3.
OC differentiation in EBI2 signaling deficiency in vitro. (A) Ebi2, Ch25h, and Cyp7b1 mRNA expression in BMDMs and OCs differentiated for 2 (d2) and 4 d (d4). Bars indicate mean ± SD of triplicate measures, representative of five independent experiments. (B) OC differentiation from EBI2-sufficient and -deficient BMDMs for 4.5 d. (C) Representative TRAP (pink) and nuclei (purple) staining of OCs differentiated from WT, Ebi2−/−, and Ch25h−/− precursors. Black arrowheads indicate individual nuclei (purple). Data are representative of five independent experiments. (D) Migration of M12-EBI2 or control cells toward BMDM culture supernatants. (E) OC differentiation from Ch25h+/+ and Ch25h−/− BMDMs for a period of 4.5 d. (F) Total fusion events relative to fission events after 3 h of time-lapse imaging of WT, Ebi2−/−, and Ch25h−/− OCs on day 4. (G) Migration of M12 cells overexpressing EBI2 or control cells toward supernatants of control (CTR) or HSD3B7-overexpressing BMDMs. (H) OC differentiation from control and HSD3B7-transduced BMDMs for 4.5 d. (I) Light microscopy of bone-resorptive pits formed by WT, Ebi2−/−, and Ch25h−/− OCs. Dashed lines indicate a pit from an individual OC. (J) Quantification of bone pit area/OC. Symbols depict individual pits formed by OCs, and red lines indicate mean. (I and J) Data are representative of three independent experiments. (B and E) OC subsets were distinguished by number of nuclei/cells. Bars indicate mean ± SD of triplicate wells. (D and F–H) Bars indicate the mean, and circles represent mean data from individual mice. (B and D–H) Data are representative of at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student’s t test. Bars: (C) 100 µm; (I) 50 µm.
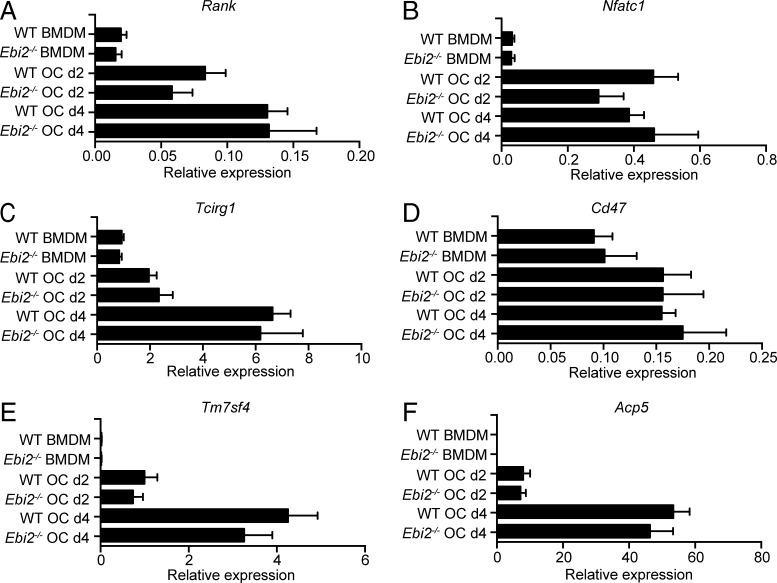
A recent study demonstrated an association between single nucleotide polymorphisms in Ebi2 that reduce EBI2 function and increased IRF7-driven inflammatory gene expression in BMDMs (Heinig et al., 2010). Therefore, we considered the possibility that IRF7 might be increased in Ebi2-deficient and Ch25h-deficient OCPs and BMDMs, which could lead to altered OC differentiation (Ji et al., 2009). To test this possibility, we analyzed Irf7 and Rank expression in BMDMs from WT and Ebi2-deficient mice stimulated for 6 h with 10 nM 7α,25-OHC. We did not detect significant differences in Irf7 or Rank expression upon EBI2 signaling (not depicted). Furthermore, RANK (encoded by Tnfrsf11a) expression was largely similar in EBI2-deficient and -sufficient OCPs (Fig. 4 A), and Nfatc1, Tcirg1 (encodes V-ATPase a3), Tm7sf4 (encodes DC-STAMP), Cd47, and Acp5 (Fig. 4, B–F) expression was also equally unaffected by EBI2 deficiency. These data suggest that the defect in the generation of large OC numbers was not caused by defective expression of genes controlling OC differentiation or cell fusion.
Figure 4.
RANK and RANK signaling–dependent gene expression in EBI2-deficient and -sufficient OCPs and mature OCs. (A–F) Rank (A), Nfatc1 (B), Tcirg1 (C), Cd47 (D), Tm7sf4 (E), and Acp5 (F) mRNA expression of BMDMs and OCs differentiated in vitro from WT and Ebi2−/− BM cells. Bars indicate mean ± SD of triplicate measures. Data are representative of five to nine independent experiments.
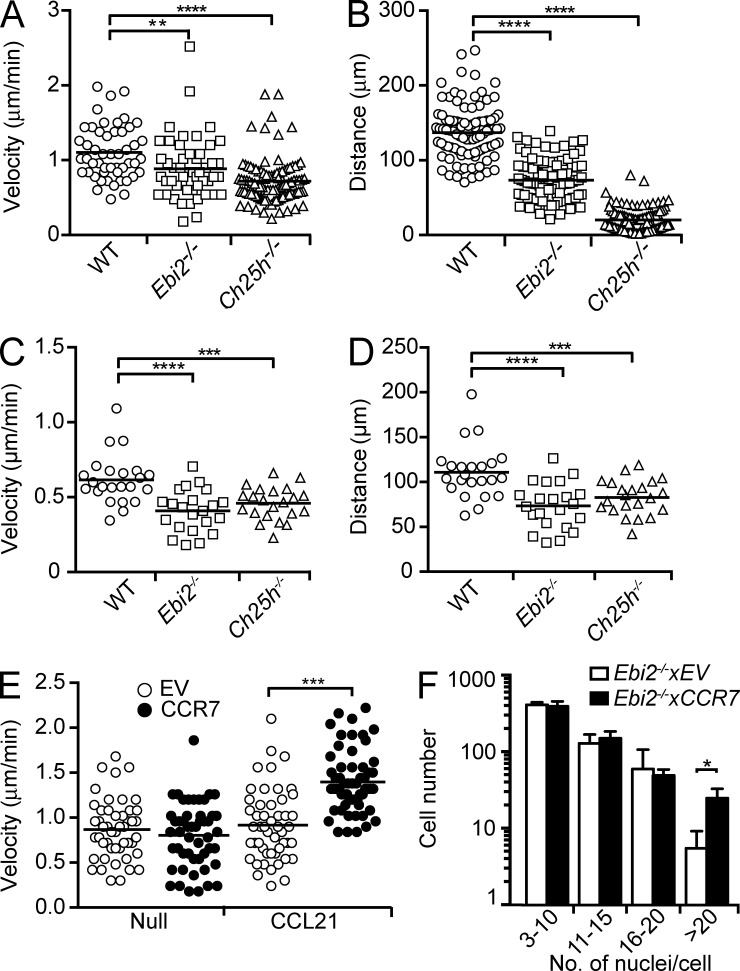
EBI2 and its oxysterol ligands control immune cell migration (Gatto et al., 2009; Pereira et al., 2009b, 2010a; Hannedouche et al., 2011; Liu et al., 2011). We hypothesized that EBI2 promotes BMDM and OCP migration, thereby influencing cell–cell interaction and cell fusion. To test this hypothesis, we visualized the dynamic behavior of BMDMs from Ebi2- and Ch25h-deficient and -sufficient mice by time-lapse microscopy and measured cell velocity and displacement for 3 h. We found that Ebi2- and Ch25h-deficient BMDMs and developing OCs moved with slower velocity and exhibited reduced displacement than WT cells imaged in similar conditions (Fig. 5, A–D; and Videos 3 and 4). These data suggested that random cell motility might be critical for the generation of large OCs. To test this hypothesis, we overexpressed CCR7, a GPCR that controls T lymphocyte migration (Okada and Cyster, 2007; Worbs et al., 2007), in Ebi2−/− OCPs and allowed them to differentiate into mature OCs in the absence or presence of the CCR7 ligand CCL21. CCR7-transduced Ebi2−/− OCPs were equally motile to control-transduced cells in the absence of CCL21, but their velocity was significantly increased in the presence of CCL21 (Fig. 5 E). Remarkably, CCR7-transduced precursors differentiated into large multinucleated OCs (>20 nuclei/cell) significantly better than control-transduced cells (Fig. 5 F), demonstrating that increased cell motility promotes the development of large OCs.
Figure 5.
OCP motility in EBI2 signaling deficiency in vitro. (A and B) WT, EBI2-deficient, and CH25H-deficient BMDM median velocity (µm/min; A) and displacement (µm; B). Data are representative of four independent experiments. (C and D) Median velocity (µm/min; C) and displacement (µm; D) of OCs differentiated from WT, Ebi2−/−, and Ch25h−/− OCPs. Each symbol represents the mean of five to eight tracks per OC, and lines indicate mean. Data are representative of three independent experiments. (E) Median velocity (µm/min) of EBI2-deficient BMDMs transduced with control or CCR7-encoding retroviruses, in the absence (null) or presence of 0.5 µg/ml CCL21. Data are representative of three independent experiments. (A–E) Data were collected by time-lapse imaging in vitro; symbols depict single cells, and lines indicate means. (F) OC differentiation from EBI2-deficient control and CCR7-transduced BMDMs for 4.5 d with 0.5 µg/ml CCL21. OC subsets were distinguished by number of nuclei/cell. Bars indicate mean ± SD of triplicate wells. Data are representative of three individual experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student’s t test. See also Videos 3 and 4.
To determine whether EBI2 signaling was required for the generation of large OCs in vivo, we crossed EBI2- and CH25H-deficient mice with TRAPRed reporter mice and examined the number and size (volume) of OCs in the femur primary spongiosa and trabecular bones by two-photon microscopy (Fig. 6 A). We found that the number and total cell volume of TRAP+ OCs in EBI2-deficient and in CH25H-deficient femurs was significantly reduced in a scanned volume of 5,000 × 5,000 × 100 µm3 (Fig. 6, B and C). We did not detect numerical differences in MDPs and cMoPs (cell gates that include OCPs; Muto et al., 2011; Charles et al., 2012; Jacome-Galarza et al., 2013) and in monocyte subsets in BM, blood, and spleen of Ebi2−/− and Ch25h−/− mice (Fig. 6, D–F). Therefore, the differences detected in OC numbers and volumes are likely not caused by differences in OCP retention and/or recirculation.
Figure 6.
OC numbers and volumes in EBI2- and CH25H-deficient mice. (A) Two-photon microscopy of WT, EBI2-deficient, and CH25H-deficient femurs. Representative of at least five independent experiments. Bar, 50 µm. (B and C) OC total number (B) and total volume (C) in WT, EBI2-deficient, and CH25H-deficient femurs quantified from two-photon microscopy. Data are representative of more than five independent experiments. (D–F) Flow cytometric analysis of cells isolated from BM (D), peripheral blood (PB; E), and spleen (SP; F) of the indicated mice genotypes. MDP cells were gated as CD11bloCSF1R+CD117+; cMoP cells were gated as Ly6ChiCD11bloCSF1R+CD117+; inflammatory monocytes (Inf.Mono.) were gated as CSF1R+Ly6ChiCD11b+; patrolling monocytes (Pat.Mono.) were gated as CSF1R+Ly6Cint/loCD11b+; and neutrophils were gated as CSF1R−Ly6C+CD11b+. Data are representative of at least three independent experiments. Bars indicate the mean of WT (white, n = 14), Ebi2−/− (green, n = 12), and Ch25h−/− (blue, n = 12) mice; circles depict individual mice. *, P < 0.05; **, P < 0.01 by unpaired Student’s t test.
EBI2 signaling controls OCP-directed migration toward bone surfaces
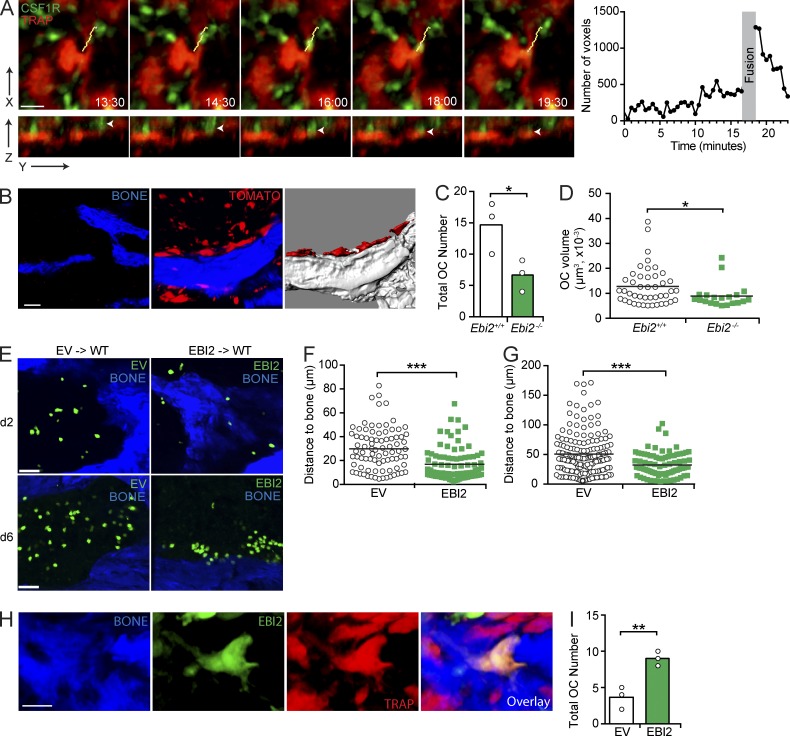
To gain insight on how OC differentiation occurs in vivo and on how EBI2 signaling contributes to this process, we visualized the dynamic behavior of OCPs and mature OCs in vivo by intravital two-photon microscopy. To discriminate between OCPs and mature OCs, we crossed CSF1R-GFP transgenic mice (Burnett et al., 2004) with TRAPRed reporter mice (Kikuta et al., 2013). After careful analysis of 24 datasets, each consisting of 30-min-long imaging periods obtained from three independent experiments, we could observe three examples of single GFP+ OCPs moving toward and fusing with bone-lining TRAP+ OCs (Fig. 7 A and Video 5). Even though TRAP+ OCs are GFPlo, we could detect an increase in GFP intensity during cell fusion, possibly because of GFP dilution throughout the OC volume, which reduced to basal GFP intensity between 1 and 4 min after cell fusion (Fig. 7 A). These data suggested that OCPs migrate in a directed manner toward bone-associated OCs. To examine the possibility that EBI2 guides OCPs to bone surfaces, we sorted EBI2-deficient and -sufficient CSF1R+ cells that overexpress tdTomato fluorescent proteins (see Materials and methods for details) and adoptively transferred 3 × 105 cells into sublethally irradiated C57BL/6 recipients. 1 wk after cell transfer, large tdTomato+ bone-lining cells, presumably OCs, were readily detected in femurs by two-photon microscopy (Fig. 7 B and Video 6). Interestingly, EBI2-deficient precursors differentiated poorly into bone-lining OCs in vivo, as indicated by a significant 2.5-fold reduction in the total number and volume of large tdTomato+ bone-lining cells when compared with EBI2-sufficient controls (Fig. 7, C and D). This defect was not caused by differences in BM homing between EBI2-deficient and -sufficient cells because similar numbers of transferred EBI2-deficient and -sufficient OCPs were detected in BM and spleen (not depicted). We next asked whether EBI2 was sufficient to guide OCPs toward bone surfaces and tested the effect of EBI2 overexpression in OCP positioning in BM. We adoptively transferred EBI2- or control-transduced OCPs into sublethally irradiated C57BL/6 mice and allowed cells to equilibrate in the host for 2 or 6 d. Remarkably, EBI2-overexpressing cells were positioned in closer proximity to bone surfaces 2 d after transfer in comparison with control-transduced OCPs (Fig. 7, E and F). EBI2-overexpressing cells had a propensity to form clusters near or at the bone surface 1 wk after transfer (Fig. 7 E), which was reflected in a significant change in their positioning relative to bone surfaces (Fig. 7 G). Using TRAPRed reporter mice as recipients of EBI2- or control-transduced OCPs, we observed that EBI2-transduced GFP+ cells fused more efficiently with TRAP+ OCs than control-transduced precursors (Fig. 7, H and I). These data demonstrate that EBI2 overexpression is sufficient for directed OCP migration toward bone surfaces and for enhancing OC differentiation in vivo. These data also suggest that EBI2 ligand gradients are formed in bone-proximal niches, perhaps as the result of restricted OB positioning at bone surfaces.
Figure 7.
EBI2 guides OCPs toward bone surfaces. (A, left) Time-lapse sequence of two-photon intravital microscopy of monocyte/OCP (green) fusion with an OC (red). Yellow lines indicate the cell track, and arrowheads indicate an OCP undergoing cell fusion. (right) GFP fluorescence in OCs before and after fusion. (B–D) Transfer of CSF1R+tdTomato+ cells into WT recipients analyzed by two-photon microscopy of femurs. (B, left) BM (black) and bone of control mice. (middle) tdTomato+ cell distribution after transfer. (right) tdTomato+ OCs. (C) OC numbers. (D) OC volumes. (E–G) Control (EV) and EBI2-transduced OCPs transferred into WT for 2 (E and F) or 6 d (E and G), and distance to bone was analyzed by two-photon microscopy of femurs. (D, F, and G) Symbols depict individual cells, and lines represent means. (H and I) Control (EV)- and EBI2-transduced OCPs transferred into TRAPRed mice for 6 d and analyzed by two-photon microscopy of femurs. (H) GFP+ cell fused with an OC. (I) Number of TRAP+GFP+ OCs. (C and I) Bars indicate the mean, and circles depict individual mice. (A–I) Data are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by unpaired Student’s t test. Bars: (A) 20 µm; (B and E) 50 µm; (H) 24 µm. See also Videos 5 and 6.
CH25H expressed by radiation-resistant cells promotes OC differentiation
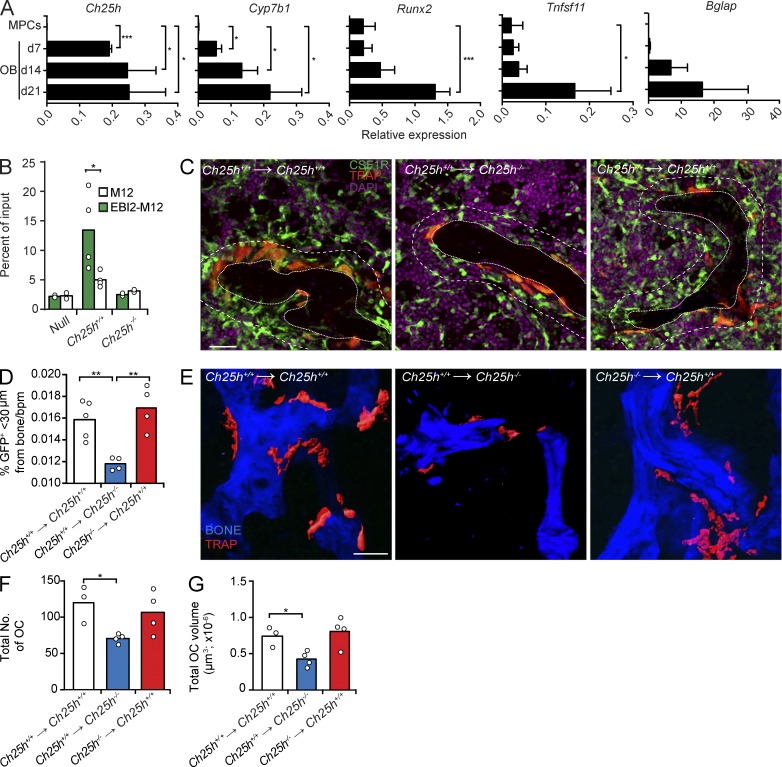
As mature OCs do not secrete measurable amounts of EBI2 ligands (Fig. 3 D), we considered the possibility that other bone-lining cells secrete EBI2 ligands. OC differentiation is critically dependent on signals provided by bone-forming OBs. Thus, we measured Ch25h and Cyp7b1 expression in FACS-sorted (CD45− Ter119− VE-Cadherin− CD31− LEPR+), freshly isolated mesenchymal progenitor cells (MPCs; Méndez-Ferrer et al., 2010; Zhou et al., 2014) and in OBs differentiated in vitro from CD45-depleted marrow cells from femurs and tibias of adult C57BL/6 mice or from neonatal calvaria. We found that Ch25h and Cyp7b1 are undetectable in MPCs, but expression is evident in pre-OBs and increases during mature OB differentiation (Fig. 8 A; data obtained from neonatal calvaria OBs are not depicted). Importantly, Ch25h and Cyp7b1 expression is highest at OB differentiation stages in which Runx2, Tnfsf11 (encodes RANKL), and Bglap (encodes osteocalcin) are also highest (Fig. 8 A). Furthermore, OBs secrete EBI2 ligands in vitro in a CH25H-dependent manner (Fig. 8 B). To determine whether oxysterols produced by radiation-sensitive hematopoietic cells (including OCs) are required for OCP positioning and differentiation in vivo, we performed BM chimeras using CH25H-deficient and -sufficient CSF1R-GFP;TRAPred double reporter mice as BM donors to reconstitute lethally irradiated WT recipients. We also analyzed the contribution of CH25H expressed by radiation-resistant cells (largely nonhematopoietic, mesenchymal-derived cells, including OBs) in OCP homing and differentiation by reconstituting lethally irradiated CH25H-deficient and -sufficient mice with BM cells isolated from WT CSF1R-GFP;TRAPred double reporter mice. Interestingly, CSF1R+ cells were distributed abnormally in the BM of CH25H-deficient recipient mice with fewer CSF1R+ cells positioned near bone surfaces (Fig. 8, C and D). Furthermore, the total number and volume of OCs that differentiated in CH25H-deficient recipient mice were significantly reduced (Fig. 8, E–G). In contrast, CH25H expressed by hematopoietic cells (of which the vast majority is radiation sensitive) played no detectable role in OCP positioning and OC differentiation in this experimental model (Fig. 8, C–G). Combined, these data support a model in which bone-associated radiation-resistant cells, possibly OBs, secrete oxysterol chemoattractants sensed by EBI2 expressed in OCPs and facilitates OC differentiation in vivo.
Figure 8.
CH25H expressed in radiation-resistant cells promotes OCP positioning near bone and OC differentiation. (A) Ch25h, Cyp7b1, Runx2, Tnfsf11, and Bglap mRNA expression in MPCs and in OBs differentiated in vitro for 7, 14, and 21 d. Bars indicate mean ± SD of triplicate measures. (B) Migration of M12 cells overexpressing EBI2 or control cells toward OB culture supernatants. Data are representative of four independent experiments. (C) Analysis of CSF1R+ cells and TRAP+ OCs by confocal microscopy in situ. Dashed lines delineate areas <30 µm distal from bone, and dotted lines delineate trabecular bones. (left and middle) Lethally irradiated Ch25h+/+ and Ch25h−/− mice were reconstituted with Ch25h+/+ CSF1R-GFP;TRAPRed BM cells. (right) Lethally irradiated Ch25h+/+ mice were reconstituted with Ch25h−/− CSF1R-GFP;TRAPRed BM cells. (D) Quantification of CSF1R+ cell proximity to bone surfaces: total number of CSF1R+ cells that are <30 µm distal from bone was divided by total number of CSF1R+ cells in the field of view and multiplied by 100 (%). The percentage of bone-proximal CSF1R+ cells was subsequently divided by the trabecular bone perimeter (bpm). Data are representative of at least four independent experiments. (E) Two-photon microscopy of femurs recovered from the BM chimeras described in C. (C and E) Bars, 50 µm. (F and G) OC total number (F) and total volume (G) in femurs from BM chimeras described in C. (A, F, and G) Data are representative of at least three independent experiments. (B, D, F, and G) Bars indicate mean, and circles indicate individual experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by unpaired Student’s t test.
EBI2 signaling promotes OCP motility in BM
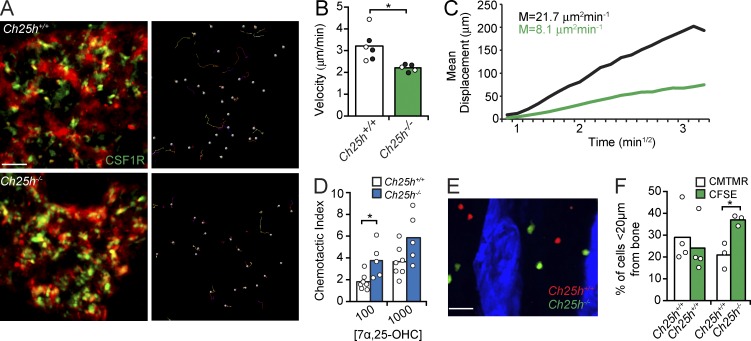
As autocrine EBI2 signaling promoted OCP motility in vitro, we considered the possibility that EBI2 also controls OCP movement in BM. To visualize EBI2 signaling–deficient and –sufficient OCPs in vivo, we crossed CSF1R-GFP reporter mice (Burnett et al., 2004) or Cx3cr1EGFP/+ mice (Jung et al., 2000) with Ch25h−/− mice. We found that the motility of Ch25h−/− GFP+ cells was significantly reduced when compared with WT cells (Fig. 9, A–C; and Video 7). Autocrine EBI2 signaling contributes to OCP motility in vitro, presumably because cells are not exposed to exogenous sources of EBI2 ligands. However, in vivo, OCPs presumably follow EBI2 ligand gradients formed by other cells, such as OBs. This led us to investigate the biological significance of autocrine EBI2 signaling in OCPs in vivo. EBI2 is sensitive to ligand-induced receptor internalization (Hannedouche et al., 2011). Thus, we considered the possibility that autocrine EBI2 signaling in OCPs attenuates the sensing of EBI2 ligand gradients in vivo. To test this hypothesis, we compared CH25H-deficient and -sufficient monocyte chemotaxis toward a gradient of 7α,25-OHC concentration established across transwells in vitro. We found that Ch25h−/− monocytes migrated significantly better toward 7α,25-OHC than Ch25h+/+ monocytes, suggesting that EBI2 function is more efficient when autocrine signaling is defective (Fig. 9 D). To test whether autocrine EBI2 signaling played a role in OCP positioning in vivo, we co-transferred CFSE- or CMTMR-labeled Ch25h−/− and Ch25h+/+ OCPs into C57BL/6 mice and analyzed their distribution relative to bone surfaces 48 h after transfer. We found that Ch25h−/− OCPs were significantly closer to bone surfaces than Ch25h+/+-sufficient cells (Fig. 9, E and F), suggesting that autocrine EBI2 signaling tempers the sensing of EBI2 ligand gradients formed in vivo.
Figure 9.
EBI2 signaling controls OCP motility and positioning in BM. (A) Intravital two-photon microscopy of CH25H-sufficient (top) and -deficient (bottom) monocytes/OCPs in vivo. (left) CSF1R+ cells and vasculature (red). (right) Cell tracks. Data are representative of six independent experiments. (B and C) Median velocity (B) and motility coefficient (M; C) of CH25H-sufficient and -deficient monocytes/OCPs. Circles indicate individual mice: CSF1R-GFP, white; and Cx3cr1GFP/+, black. Data are representative of six experiments. (D) CH25H-sufficient or -deficient monocyte/OCP migration toward 7α,25-OHC (nM). Data are representative of six experiments. (E) BM distribution of adoptively transferred CH25H-sufficient (red) and CH25H-deficient (green) monocytes/OCPs. (F) Monocyte/OCP frequency <20 µm distal from bone. Data were pooled from four experiments. (B, D, and F) Bars indicate mean, and circles represent individual experiments and/or mice. *, P < 0.05 by unpaired Student’s t test. Bars: (A) 50 µm; (E) 30 µm. See also Video 7.
DISCUSSION
The restricted positioning of OCs in direct contact with bone surfaces suggests that guidance cues attract OCPs to these locations, but their identity has not been defined for many decades now. Here we showed that EBI2 is expressed in mouse OCPs and mature OCs and that EBI2 signaling promotes basal OCP motility and guides OCPs to bone surfaces where OC differentiation is known to occur. Using conventional histomorphometry analyses and a novel two-photon microscopy–based method for measuring OCs in undecalcified whole-mount femurs and tibias, we showed that defects in EBI2 signaling caused a significant increase in bone mass and a significant reduction in OC numbers. Furthermore, the two-photon microscopy method that we used enabled measurements of OC volumes in situ and showed that OCs were significantly smaller in EBI2 signaling–deficient mice than in control mice. EBI2 signaling accelerated OCP movement in vitro and in vivo, which impacted OC differentiation and size. Likewise, the artificial manipulation of OCP motility by CCR7 overexpression also favored the development of large OCs. EBI2 is a chemoattractant receptor of the GPCR family with no known roles other than controlling cell migration and/or positioning. Specifically, EBI2 promotes the positioning of dendritic cells and activated B lymphocytes to specialized outer and interfollicular niches in secondary lymphoid organs and plays important roles in primary antibody responses (Gatto et al., 2009, 2013; Pereira et al., 2009b, 2010a; Yi et al., 2012; Yi and Cyster, 2013). Thus, the phenotypic similarity between EBI2- and CH25H-deficient mice favor a model in which oxysterols sensed by EBI2 expressed in OCPs regulate bone mass homeostasis predominantly by promoting OCP cell movement and positioning, which facilitates cell fusion and enhances the development of large OCs.
OBs did not express EBI2 but secreted measurable amounts of EBI2 ligands in a CH25H-dependent manner in vitro and are a likely source of EBI2 ligands at the bone surface in vivo. However, macrophages also express CH25H (Bauman et al., 2009) and CYP7B1 and can secrete EBI2 ligand activity. Therefore, our experiments do not rule out the possibility that endosteal macrophages may secrete EBI2 ligands and contribute to enforce OCP positioning near bone surfaces. In addition to OBs, OCPs also secrete EBI2 ligands, which enables autocrine EBI2 signaling in these cells. Autocrine chemoattractant receptor signaling has been described in OCPs and was shown to enhance OC differentiation in vitro by an unclear mechanism (Yang et al., 2006; Miyamoto et al., 2009; Hoshino et al., 2010). Our study revealed that autocrine EBI2 signaling accelerated OCP movement in vitro, which presumably maximized cell–cell interactions that precede cell fusion. In support of this possibility, we also showed that the size of OCs generated under in vitro conditions was sensitive to changes in OCP velocity. However, the positive effect of autocrine chemoattractant receptor signaling in OCP differentiation is likely an artifact of in vitro culture conditions because in these conditions OCPs are not exposed to other chemoattractants secreted by BM cells. In vivo, our findings suggest that autocrine EBI2 signaling attenuates the sensing of EBI2 ligand gradients presumably formed by bone-proximal cells. EBI2, like many other GPCRs, is internalized upon binding to cognate ligands (Hannedouche et al., 2011), although the internalization mechanism has not been defined. Autocrine EBI2 signaling and receptor internalization provide additional cell-intrinsic mechanisms for regulating OCP movement toward bone surfaces. Future studies will elucidate whether autocrine EBI2 signaling is constitutive or inducible in OCPs and whether it is altered during pathological conditions.
Although EBI2 contributes and is sufficient to promote OCP movement and differentiation in vivo, EBI2-deficient cells can still migrate toward bone surfaces and differentiate into multinucleated mature OCs, albeit less efficiently. GPCRs induce cell migration through the activation of small GTPases, particularly RAC1 and RAC2. Conditional inactivation of Rac1 and Rac2 in myeloid cells, including OCPs, led to a dramatic reduction in OC numbers in vivo (Wang et al., 2008). However, a more recent study examined RAC1 and RAC2 contribution in OC differentiation in vivo (using a similar genetic approach) and failed to detect significant roles in OC differentiation in vivo, but reported a dramatic RAC dependency in OCs for bone resorption (Croke et al., 2011), presumably caused by defective CSF1R signaling (Sakai et al., 2006). Although other chemoattractant receptors are likely involved in promoting OCP motility and directed migration to sites of OC differentiation, it is possible that GPCR signaling in these cells utilizes small GTPases other than RAC1 and RAC2.
Several chemokine receptors such as CCR1, CCR2, CCR3, CX3CR1, and CXCR4 have been proposed to control OCP trafficking within BM and toward bone surfaces (Ponomaryov et al., 2000; Lean et al., 2002; Petit et al., 2002; Grassi et al., 2003; Yu et al., 2003, 2004; Oba et al., 2005; Semerad et al., 2005; Wright et al., 2005; Kim et al., 2006; Yang et al., 2006; Li et al., 2007; Saitoh et al., 2007; Binder et al., 2009; Koizumi et al., 2009; Hoshino et al., 2010, 2013). However, these studies did not assess OCP migration in vivo, presumably because of technical difficulties with imaging OCPs in situ and/or in vivo. Instead, these studies relied on OCP migration assays in vitro and on analyses of chemokine expression by OBs differentiated in vitro. Furthermore, in the case of CCR2 and CXCR4, their cognate ligands CCL2 and CXCL12, respectively, are predominantly expressed by cells that are distal to bone surfaces, such as perivascular stromal cells and MPCs positioned in the parenchyma. In fact, CCL2 and CXCL12 are either not expressed or minimally expressed by OBs or other bone-associated cells in vivo (Sugiyama et al., 2006; Shi et al., 2011; Ding and Morrison, 2013). A recent study also demonstrated that CX3CR1 signaling reduced monocyte motility in parenchyma and controlled their BM retention (Jacquelin et al., 2013). These findings also imply that the CX3CR1 ligand CX3CL1 is abundant in bone-distal parenchymal locations.
In conclusion, our experiments reveal a novel role played by oxysterols and EBI2 in the guidance of hematopoietic monocytic precursors toward endosteal surfaces and in OC differentiation. Furthermore, our experiments showed that OBs secrete EBI2 ligands in vitro and may be the relevant cellular sources of EBI2 ligand production in BM under normal homeostatic conditions. Besides OBs, macrophages isolated from several organs have also been reported to express CH25H and to secrete 25 hydroxycholesterol and EBI2 ligands. Importantly, CH25H expression is sensitive to inflammatory signals including type I and II interferons (Bauman et al., 2009; Diczfalusy et al., 2009; Dennis et al., 2010; Park and Scott, 2010). Given the high EBI2 expression in monocyte-lineage cells, including inflammatory monocytes, it is possible that an EBI2–CH25H pathway is involved in inflammatory cell recruitment to inflamed tissues, such as inflamed synovial spaces that are typical in rheumatoid arthritis. Of note, antibodies recognizing citrullinated self-antigens are important drivers of inflammation in rheumatoid arthritis, and patients with antibodies against citrullinated antigens have a more erosive disease than patients without detectable autoantibodies. Given the role played by EBI2 in B cell differentiation into antibody-secreting plasma cells, and in OCP migration and differentiation, therapeutic strategies targeting EBI2 may have significant clinical potential for the treatment of skeletal disorders, including menopause-induced bone loss, and importantly, rheumatoid arthritis. Endosteal surfaces are also supporting environments for several bone metastatic cancers (Coleman, 2001). The identification of single nucleotide polymorphisms in EBI2 coding regions by whole-exome sequencing of acute myeloid leukemias in humans suggests that altered EBI2 signaling confers an advantage to some cancer cells. Oxysterols and EBI2 may thus be relevant therapeutic targets for the treatment of hematopoietic cancers and bone metastatic cancers.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were obtained from The Jackson Laboratory. The generation of Ebi2GFP/GFP mice was described previously (Pereira et al., 2009b). Nestin-cre mice were from the Jackson Laboratory. Ch25h−/− mice were provided by D.W. Russell (The University of Texas Southwestern Medical Center at Dallas, Dallas, TX; Bauman et al., 2009). TRAP-tdTomato (herein referred as TRAPRed) mice were provided by M. Ishii (Immunology Frontier Research Center, Osaka University, Osaka, Japan; Kikuta et al., 2013). TRAPRed mice were intercrossed with Ebi2GFP/GFP and with Ch25h−/− mice to generate TRAPRed Ebi2GFP/GFP and TRAPRed Ch25h−/− and littermate control mice. TRAPRed transgenic mice were also crossed with CSF1R-EGFP (Burnett et al., 2004) and Cx3cr1GFP/+ (Jung et al., 2000) mice to generate double reporter mice. Lyz2Cre/+ (The Jackson Laboratory) were crossed with mice carrying Rosa26(CAG)tdTomato/+ (The Jackson Laboratory) to generate Lyz2Cre/+Rosa26(CAG)tdTomato/+ and intercrossed with Ebi2GFP/GFP to generate Lyz2Cre/+Rosa26(CAG)tdTomato/+Ebi2GFP/GFP. All mice were cared for in accordance with institutional animal care and use committee–approved protocols at the Yale University School of Medicine animal facility.
Cell isolations, cell sorting, monocyte retroviral transduction, and adoptive transfers.
BM was prepared by flushing one femur and one tibia with 2 ml (1 ml per bone) of α-MEM (Gibco) containing 2% FBS (EMD Millipore), antibiotics (50 IU/ml of ampicillin and 50 mg/ml of streptomycin; CellGro), and 10 mM Hepes (CellGro) with a 1-cc insulin syringe (BD). Cell clumps were dissociated by gently pipetting up and down. Blood, BM, and spleen cells were counted with a Coulter Counter (Beckman Coulter). Cell populations were identified by staining with the following antibodies: CD11b PE-Cy7 (clone M1/70; BD), Ly6C PerCP-Cy5.5 (clone HK1.4; eBioscience), CSF1R APC (clone AF598; BioLegend), and CD117 APC-Cy7 (clone 2B8; BD). After staining, data were acquired on an LSRII (BD) and analyzed using FlowJo software (Tree Star). For cell sorting, RBC-lysed BM cells were plated overnight (∼16 h) on tissue culture–treated 10-cm Petri dishes, and the nonadherent fraction was collected and stained with CSF1R APC (clone AF598; BioLegend). Cells from Lyz2-cre.Rosa26(CAG)tdTomato/+ were sorted based on CSF1R+tdTomato+, and cells from Ebi2GFP/GFP.Lyz2-cre.Rosa26(CAG)tdTomato/+ were sorted based on GFP+CSF1R+tdTomato+ expression to >95% purity (FACSAria; BD). Sorted cells were adoptively transferred i.v. into sublethally irradiated (one dose of 3.35 Gy) C57BL/6 mice (3 × 105/mouse). For monocyte retroviral transduction, RBC-lysed BM cells were plated for 10–12 h on tissue culture–treated 10-cm Petri dishes (one dish/four long bones). The nonadherent fraction was collected and incubated in α-MEM containing 10% FBS, antibiotics (50 IU/ml of ampicillin and 50 mg/ml of streptomycin), and 10 mM Hepes + 10 ng/ml M-CSF (R&D Systems) on nontissue culture–treated 10-cm Petri dishes (Thermo Fisher Scientific; one dish/four long bones) for ∼16 h. Cells were then spin infected with a retroviral construct expressing Ebi2 with an (IRES)-GFP cassette as a reporter or empty construct as a control in nontissue culture–treated 6-well plates (Cyto-One) and immediately transferred i.v. into sublethally irradiated (one dose of 3.35 Gy) WT or TRAPRed mice (BM cells collected from two mice were transferred into one recipient mouse). For labeling of CSF1R+ cells sorted from Ch25h−/− or Ch25h+/+ before transfer, sorted cells were incubated in either 5 µM CMTMR or 5 µM CFSE for 20 min at 37°C, and then equal numbers were transferred i.v. into WT recipient mice (∼5 × 106/mouse). Femurs were analyzed 48 h after transfer. For MPC sorting, long bones recovered from three to four C57BL/6 mice (ages 6–12 wk) were flushed using HBSS supplemented with 2% of heat-inactivated FBS. Whole BM was digested with 200 U/ml Collagenase IV (Worthington Biochemical Corporation) at 37°C for 20 min. Cells clumps were dissociated by gentle pipetting, followed by a further incubation of 10 min at 37°C. Cells were washed with 5 ml HBSS/2% FBS, spun at 1,000 rpm for 7 min, and stained with anti-LEPR antibody for 1 h, on ice, followed by staining with biotin-conjugated anti–goat for 40 min. After washing, cells were incubated with anti-CD45, anti-Ter119, anti-CD31, and anti-CD144 antibodies, Streptavidin BV605, and DAPI for 30 min. Cells were acquired and sorted on a FACSAria II (BD) equipped with UV (355 nm), violet (405 nm), blue (488 nm), green (532 nm), and red (640 nm) lasers and with FACSDiva 7.
BMDM, OC, and OB in vitro differentiation.
For BMDM, BM cells were cultured in 70% DMEM (CellGro) containing 10% FBS, antibiotics, 10 mM Hepes, and 2 mM l-glutamine and 30% L929-conditioned media as a source of M-CSF, at 37°C with 5% CO2. L929-conditioned media were generated by culturing confluent L929 cells in DMEM containing 10% FBS, antibiotics, 10 mM Hepes, and 2 mM l-glutamine for 10 d and collecting supernatants and filtering through a 20-µm filter. For OC differentiation, BM cells were lysed with RBC lysis buffer for 1–2 min at room temperature and plated in 10-cm tissue culture–treated Petri dishes for ∼16 h. The nonadherent cells were recovered and plated at a concentration of 7.5 × 104 cells/well for 96-well plates, 3.0 × 105 cells/well for 24-well plates, and 106 for 35-mm glass-bottomed dishes in α-MEM containing 10% FBS, antibiotics, 10 mM Hepes, and M-CSF and RANKL at 100 ng/ml (R&D Systems). Media were changed to fresh media every other day. Typically, large multinucleated OCs became evident after 5 d of culture. For TRAP staining of OCs, the TRAP staining kit (Kamiya) was used according to the manufacturer’s instructions. OC nuclei were stained with hematoxylin (Sigma-Aldrich). OC nuclei were enumerated and stratified by total number of nuclei. For OC retroviral transduction, day 2 OC cultures were aspirated and media were replaced with supernatants from Phoenix 293T cells containing retroviral constructs expressing Hsd3b7 or Ccr7 with an (IRES)-hCD4 as a reporter or empty vector control. OC were spin infected one time, and media were replaced with α-MEM OC media. OCs were stained and enumerated at day 5. For OB differentiation from long bones, BM cells were flushed from femurs and the bones were incubated in DMEM containing 2% Collagenase A (Roche) and antibiotics at 37°C with agitation for 30 min (500 µl/bone) and then washed with DMEM. Bones were incubated a second time in DMEM containing 2% Collagenase A and antibiotics at 37°C with agitation for 30 min, and this fraction was collected. To remove contaminating hematopoietic cells, cells were incubated with biotin α-CD45 (clone 30-F11; BioLegend) at 1:100 dilution in 100 µl DMEM containing 2% FBS for 20 min at 4°C and then washed two times, and cells were then resuspended in 100 µl DMEM containing 10 µl streptavidin beads (Invitrogen) and incubated at 4°C with gentle shaking for 45 min. Labeled hematopoietic cells were separated using a magnetic field, and remaining cells were plated up to 106 per well on 6-well plates in α-MEM containing 15% FBS, 10 mM Hepes, 50 µg/ml l-ascorbic acid 2-phosphate (Sigma-Aldrich), and 10 nM glycerophosphate (Sigma-Aldrich). Media were changed every 3–4 d.
Bone resorption pit assay.
OC bone resorption was assayed by culturing BM cells in OC differentiation conditions on devitalized bovine cortical bone slices (4.4 × 4.4 × 0.2 mm; Hurley et al., 1998). In brief, BM cells were plated overnight, nonadherent cells were recovered, and 75,000 cells were plated on individual bone slices in a total volume of 200 µl of OC differentiation media. OCs were cultured on the bone slices for 10 d (media were changed every 2–3 d). Bone slices were then fixed and stained for TRAP to verify OC formation. OCs were removed from the bone slices by ultrasonication in water (5–10 min) and stained with a 1% toluidine blue in 1% sodium borate 10-hydate solution to visualize resorption pits. Pits were analyzed by light microscopy and quantified using OsteoMeasure software (OsteoMetrics).
In vitro time-lapse microscopy.
WT, Ebi2GFP/GFP, and Ch25h−/− BMDMs were cultured on 35-mm glass-bottomed dishes (MatTek) for 2 d before imaging in 30% L929-conditioned media at 37°C with 5% CO2. In vitro–differentiated OCs from the same mice were cultured on 35-mm glass-bottomed dishes for 4 d before imaging in OC differentiation media containing M-CSF and 100 ng/ml RANKL (R&D Systems) at 37°C with 5% CO2. For motility recovery experiments, WT or Ebi2GFP/GFP BMDM media were removed after 2 d and replaced with media containing titrated concentrations of CCL21 (R&D Systems), and cells were imaged 1 h after media change. Images were collected by time-lapse microscopy (Vivaview; Olympus) every 2 or 3 min for BMDMs or OCs, respectively, for 3 h total (objective 20×) at 37°C with 5% CO2. The images from four to five fields for each sample were collected and analyzed with Imaris software (Bitplane).
Chemotaxis assays.
Supernatants for chemotaxis assays were prepared from confluent cell cultures. The media were replaced with DMEM containing 0.5% fatty acid–free bovine serum albumin (MP Biomedicals), 10 mM Hepes, antibiotics, and 2 mM l-glutamine for 10–12 h. Supernatants were collected and spun down to clean off contaminating cells. Chemotaxis assays were performed using primary murine BM cells or M12 B cell line transduced with an Ebi2 (IRES)-GFP retroviral construct (Kelly et al., 2011). Cells were allowed to migrate through 5-µm–pore sized transwells (Corning) for 3 h at 37°C with 5% CO2 against supernatants collected from BMDM or OB cultures. Primary BM cells or M12 B cells were collected, stained with appropriate antibodies (BM cells), resuspended in 45 µl of staining buffer, and analyzed by flow cytometry for 40 s (LSRII; BD).
μCT and histomorphometry.
Femurs were removed from soft tissue and stored in 70% EtOH at 4°C before μCT quantitation. Trabecular morphometry of the distal metaphysis was scanned using a μCT-35 instrument (Scanco). For histomorphometry, undecalcified femurs embedded in methylmethacrylate were sectioned and stained with 2% toluidine blue for light microscopy. For measurements of bone formation rate, mice were intraperitoneally injected with 20 mg/kg calcein in a 2% sodium bicarbonate solution and labeled for 7 d and at least 1 d before sacrifice. Sections were examined in a blinded fashion to measure static and dynamic histomorphometric parameters using the OsteoMeasure software program.
Serum CTX measurements.
Serum CTXs were measured using a commercially available EIA (Ratlaps EIA; Immunodiagnostic Inc.).
Two-photon intravital imaging of mouse calvarial BM.
Mice were anesthetized and the mouse calvarium was surgically prepared as described previously (Pereira et al., 2009a). Animals were immobilized on a custom-built stage. Deep-tissue images were collected using a BX61WI fluorescence microscope (Olympus) with a 20×, 0.95 NA water immersion objective (Olympus), and dedicated single-beam TriM laser-scanning microscope (LaVision Biotec), controlled by Imspector software (Abberior Instruments GmbH). The light source was a Chameleon Vision II Ti:Sapphire laser (Coherent) with pulse precompensation. For four-dimensional analysis of cell migration, stacks of 16–20 optical sections with 3-µm z spacing were acquired every 30 s for 30 min with the laser tuned to a wavelength of 895 nm. Each xy plane spanned 400 µm in each dimension. Videos were analyzed using Imaris, and cell tracks were automatically generated and then verified manually. Cell velocities and mean squared displacements were calculated using software programmed in MATLAB (The Math Works, Inc.)
Preparation of whole-mount tissues and frozen sections of long bones.
Femurs and tibias were removed of soft tissue, fixed in 4% PFA, and cryoprotected in 30% sucrose at 4°C. Bones were then placed in Tissue-Tek optimum cutting temperature compound (Sakura) and immediately frozen in EtOH and dry ice and stored at −80°C. Cryostat sections 7 µm in thickness were transferred to CJ1X adhesive–coated slides with the Cryojane Tape-Transfer system (Instrumedics) according to the manufacturer’s protocol. For whole-mount immunostaining and three-dimensional histology by two-photon microscopy, ∼250 µm of femur bone and marrow tissue was excised longitudinally using a cryostat to fully expose BM and bone for imaging.
Three-dimensional histology by two-photon microscopy.
In whole long bone tissues the metaphysis (primary spongiosa) and isolated trabecular bones in the metaphysis region were scanned with femtosecond titanium/sapphire laser pulses at 900-nm wavelength. The total imaging volume was 500 × 500 × 100 µm. At least 10 three-dimensional images were analyzed per femur using Imaris software. OCs were visualized by TRAPRed fluorescence intensity and anatomical position adjacent to bone detected by second harmonic excitation. Assessment of OC volumes was performed using Imaris software. In brief, the “surfaces” module was used to calculate OC volume based on TRAPRed fluorescence intensity. For measuring the distances between GFP+ adoptively transferred cells from the bone surfaces, the “Measurement Points” module was used. In brief, measurement distances were calculated from two points: the edge of GFP+ cells closest to the bone surface and the edge of the bone surface as detected by second harmonics. If the distance of GFP+ cells to the bone surface was greater than the distance of the cell to any edge of the 500 × 500–µm field of view, we determined this measurement invalid because of the possibility of bone being present outside the field of view.
Immunohistochemistry and immunofluorescent microscopy.
For TRAP staining, bone sections were rehydrated in H2O for 5 min, and TRAP staining solution (Kamiya) was applied for 5 min at 37°C according to the manufacturer’s protocol. The reaction was stopped with H2O. For Von Kossa staining of calcium, bone sections were rehydrated in H2O, 5% silver nitrate in H2O (Alpha Aesar) was applied for 15 min, and the reaction was stopped by 5% sodium thiosulfate (Sigma-Aldrich) in H2O. Fluorescent TRAP staining was performed using conventional TRAP histochemistry procedures in combination with ELF97 endogenous phosphatase detection reagent (Invitrogen), as described previously (Filgueira, 2004). Images were obtained using an Axio Observer inverted microscope (Carl Zeiss) using 40× objective.
Immunostaining and confocal imaging of whole mount tissues.
Long bone whole mount tissues were washed in PBS, for 10 min, three times. Tissues were blocked and permeabilized with 20% normal goat serum and 0.5% Triton-X in PBS for 4–6 h and then incubated in primary antibodies for 2 d at 4°C, followed by secondary antibody incubation for 1–2 d at 4°C with repeated washes in between. The primary antibody was rabbit anti-GFP (Invitrogen). Finally, tissues were incubated with fluorochrome-conjugated antibodies for 6–8 h, with DAPI added for the last hour to reveal nuclei. Images were acquired on an SP8 confocal microscope (Leica) using 20× objective. The total imaging volume was 580 × 580 × 75 µm. Images were analyzed using Imaris software.
BM chimeras.
BM chimeras were performed as previously described (Beck et al., 2014). In brief, ∼2.0 × 106 total BM cells from donor mice were transferred into adult recipient mice that had been exposed to two rounds of 6.35 Gy separated by 3 h. Chimeras were analyzed at least 6 wk after reconstitution.
Ovariectomy.
Mice were ovariectomized or sham operated at 16–20 wk of age under general anesthesia and kept on warming pads during prep, surgery, and recovery. 4 wk after surgery, successful ovariectomy was confirmed by measuring uterus weight.
RNA isolation and quantitative real-time PCR.
Total RNA was isolated from cultured murine BMDMs, OCs, and OBs using the RNeasy kit (QIAGEN). Quantitative real-time PCR was performed with the SensiFAST SYBR Lo-ROX kit (Bioline) and the Stratagene Mx3000P QPCR System cycler (SA Biosciences). Hprt mRNA levels were used as a control. PCR primer sequences are as follows: Tnfsf11 forward, 5′-TCACCATCAATGCTGCCAG-3′; Tnfsf11 reverse, 5′-AGGTAATAGAAGCCATCTTGG-3′; Acp5 forward, 5′-GCTGGAAACCATGATCACCT-3′; Acp5 reverse, 5′-TGAAGCGCAAACGGTAGTAA-3′; Bgalp forward, 5′-GGGCAATAAGGTAGTGAACAG-3′; Bgalp reverse, 5′-GCAGCACAGGTCCTAAATAGT-3′; Runx2 forward, 5′-TTACCTACACCCCGCCAGTC-3′; Runx2 reverse, 5′-TGCTGGTCTGGAAGGGTCC-3′; Tm7sf4 forward, 5′-GGAGGAACCTAAGCGGAACT-3′; Tm7sf4 reverse, 5′-CTAGGGCTTCGTGGAAACAC-3′; Cd47 forward, 5′-GGCCTTCAACACTGACCAAC-3′; Cd47 reverse, 5′-TCATTTGGAGAAAACCACGA-3′; Tcirg1 forward, 5′-GGACGCTACAGGGAAGTTA-3′; Tcirg1 reverse, 5′-CACATCGCCAAACATCACA-3′; Nfatc1 forward, 5′-CCTCGAACCCTATCGAGTGT-3′; Nfatct reverse, 5′-CCGATGACTGGGTAGCTGTC-3′; Rank forward, 5′-GTGCTGCTCGTTCCACTG-3′; Rank reverse, 5′-CCGTCCGAGATGCTCATAAT-3′; Ch25h forward, 5′-TGCTACAACGGTTCGGAGC-3′; Ch25h reverse, 5′-AGAAGCCCACGTAAGTGATGAT-3′; Cyp7b1 forward, 5′-AGCTGCTTACTGATGACGACC-3′; Cyp7b1 reverse, 5′-AGTGAGCCACAGAATGCAAAT-3′; Ebi2 forward, 5′-ACAACGGAGGTCCTAGCCA-3′; Ebi2 reverse, 5′-GCTGTGGTGGGCATAGAGA-3′; Irf7 forward, 5′-CACGGTCTTGCTCCTGGC-3′; Irf7 reverse, 5′-GCTTGGATCTACTGTGGGC-3′; Hprt forward, 5′-AGGTTGCAAGCTTGCTGGT-3′; and Hprt reverse, 5′-TGAAGTACTGATTATAGTCAAGGGCA-3′.
Online supplemental material.
Video 1 shows the distribution of Ebi2GFP/+ cells and MPC-derived cells in the BM acquired by two-photon microscopy. Video 2 shows the distribution of OCs in the BM of Ebi2GFP/+ TRAPRed mice acquired by two-photon microscopy. Video 3 shows time-lapse microscopy imaging of Ebi2+/+ and Ebi2−/− BMDM motility in vitro (3 h). Video 4 shows time-lapse microscopy imaging of Ebi2+/+ and Ebi2−/− OC motility in vitro (3 h). Video 5 shows intravital imaging of OCP migration and fusion with a TRAP+ OC in vivo (30 min). Video 6 shows the measurement of OC volume in three dimensions using two-photon microscopy. Video 7 shows intravital imaging of EBI2 signaling–deficient and –sufficient monocytes and OCPs in the calvaria (30 min). Table S1 displays dynamic histomorphometry of Ebi2+/+ and Ebi2−/− mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150088/DC1.
Supplementary Material
Acknowledgments
We are grateful to Drs. Karl Insogna and Karen Rosenberg for invaluable discussions and for critical review of the manuscript. We are in debt to Prof. Lorenzo (University of Connecticut Health Center, Farmington, CT) for providing bone disks and for helping setup up bone pit assays. We thank Dr. David W. Russell for sharing Ch25h−/− mice. We would like to thank Dr. Diane Krause, Shangqin Guo, and members of the Krause laboratory for assistance with Vivaview time-lapse microscopy. We thank Vivian Lim and Ana C. Gomes for sharing MPC RNA. We would like to thank the Yale Core Center for Musculoskeletal Disorders for technical assistance with μCT and histomorphometric analyses.
E. Nevius is the recipient of a postdoctoral fellowship from the National Institutes of Health (NIH; 1F32AR063574-01A1). This work was partially supported by the NIH (R56AI098996-01 and RO1AI113040), by pilot awards from Yale Core Center for Musculoskeletal Disorders, and by Careers in Immunology Fellowship from The American Association of Immunologists.
The authors declare no competing financial interests.
Author contributions: E. Nevius designed experiments, performed experiments, and wrote the paper. F. Pinho, M. Dhodapkar, H. Jin, K. Nadrah, and M.C. Horowitz performed experiments. J. Kikuta and M. Ishii generated TRAPRed reporter mice. J.P. Pereira designed and performed experiments, supervised experiments, and wrote the paper.
Footnotes
Abbreviations used:
- 7α,25-OHC
- 7α,25-dihydroxycholesterol
- cMoP
- common monocyte progenitor
- CTX
- carboxy-terminal collagen cross-link
- GPCR
- Gαi protein–coupled receptor
- MDP
- macrophage and dendritic cell precursor
- MPC
- mesenchymal progenitor cell
- μCT
- microcomputed tomography
- OB
- osteoblast
- OC
- osteoclast
- OCP
- OC precursor
- RANKL
- ligand for receptor activator of nuclear factor kappa binding
- TRAP
- tartrate-resistant acid phosphatase
References
- Bauman D.R., Bitmansour A.D., McDonald J.G., Thompson B.M., Liang G., and Russell D.W.. 2009. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc. Natl. Acad. Sci. USA. 106:16764–16769. 10.1073/pnas.0909142106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T.C., Gomes A.C., Cyster J.G., and Pereira J.P.. 2014. CXCR4 and a cell-extrinsic mechanism control immature B lymphocyte egress from bone marrow. J. Exp. Med. 211:2567–2581. 10.1084/jem.20140457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder N.B., Niederreiter B., Hoffmann O., Stange R., Pap T., Stulnig T.M., Mack M., Erben R.G., Smolen J.S., and Redlich K.. 2009. Estrogen-dependent and C-C chemokine receptor-2-dependent pathways determine osteoclast behavior in osteoporosis. Nat. Med. 15:417–424. 10.1038/nm.1945 [DOI] [PubMed] [Google Scholar]
- Burnett S.H., Kershen E.J., Zhang J., Zeng L., Straley S.C., Kaplan A.M., and Cohen D.A.. 2004. Conditional macrophage ablation in transgenic mice expressing a Fas-based suicide gene. J. Leukoc. Biol. 75:612–623. 10.1189/jlb.0903442 [DOI] [PubMed] [Google Scholar]
- Charles J.F., Hsu L.Y., Niemi E.C., Weiss A., Aliprantis A.O., and Nakamura M.C.. 2012. Inflammatory arthritis increases mouse osteoclast precursors with myeloid suppressor function. J. Clin. Invest. 122:4592–4605. 10.1172/JCI60920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R.E. 2001. Metastatic bone disease: clinical features, pathophysiology and treatment strategies. Cancer Treat. Rev. 27:165–176. 10.1053/ctrv.2000.0210 [DOI] [PubMed] [Google Scholar]
- Croke M., Ross F.P., Korhonen M., Williams D.A., Zou W., and Teitelbaum S.L.. 2011. Rac deletion in osteoclasts causes severe osteopetrosis. J. Cell Sci. 124:3811–3821. 10.1242/jcs.086280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E.A., Deems R.A., Harkewicz R., Quehenberger O., Brown H.A., Milne S.B., Myers D.S., Glass C.K., Hardiman G., Reichart D., et al. 2010. A mouse macrophage lipidome. J. Biol. Chem. 285:39976–39985. 10.1074/jbc.M110.182915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diczfalusy U., Olofsson K.E., Carlsson A.M., Gong M., Golenbock D.T., Rooyackers O., Fläring U., and Björkbacka H.. 2009. Marked upregulation of cholesterol 25-hydroxylase expression by lipopolysaccharide. J. Lipid Res. 50:2258–2264. 10.1194/jlr.M900107-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., and Morrison S.J.. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 495:231–235. 10.1038/nature11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filgueira L. 2004. Fluorescence-based staining for tartrate-resistant acidic phosphatase (TRAP) in osteoclasts combined with other fluorescent dyes and protocols. J. Histochem. Cytochem. 52:411–414. 10.1177/002215540405200312 [DOI] [PubMed] [Google Scholar]
- Gatto D., Paus D., Basten A., Mackay C.R., and Brink R.. 2009. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity. 31:259–269. 10.1016/j.immuni.2009.06.016 [DOI] [PubMed] [Google Scholar]
- Gatto D., Wood K., Caminschi I., Murphy-Durland D., Schofield P., Christ D., Karupiah G., and Brink R.. 2013. The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat. Immunol. 14:446–453. 10.1038/ni.2555 [DOI] [PubMed] [Google Scholar]
- Geissmann F., Manz M.G., Jung S., Sieweke M.H., Merad M., and Ley K.. 2010. Development of monocytes, macrophages, and dendritic cells. Science. 327:656–661. 10.1126/science.1178331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser W.A., Baumann A.P., Petras S.F., Harwood H.J. Jr., Devalaraja R., Renkiewicz R., Baragi V., Thompson D.D., and Paraklar V.M.. 2003. Regulation of osteoclast differentiation by statins. J. Musculoskelet. Neuronal Interact. 3:53–62. [PubMed] [Google Scholar]
- Grassi F., Piacentini A., Cristino S., Toneguzzi S., Cavallo C., Facchini A., and Lisignoli G.. 2003. Human osteoclasts express different CXC chemokines depending on cell culture substrate: molecular and immunocytochemical evidence of high levels of CXCL10 and CXCL12. Histochem. Cell Biol. 120:391–400. 10.1007/s00418-003-0587-3 [DOI] [PubMed] [Google Scholar]
- Hannedouche S., Zhang J., Yi T., Shen W., Nguyen D., Pereira J.P., Guerini D., Baumgarten B.U., Roggo S., Wen B., et al. 2011. Oxysterols direct immune cell migration via EBI2. Nature. 475:524–527. 10.1038/nature10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinig M., Petretto E., Wallace C., Bottolo L., Rotival M., Lu H., Li Y., Sarwar R., Langley S.R., Bauerfeind A., et al. Cardiogenics Consortium. 2010. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 467:460–464. 10.1038/nature09386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettinger J., Richards D.M., Hansson J., Barra M.M., Joschko A.C., Krijgsveld J., and Feuerer M.. 2013. Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 14:821–830. 10.1038/ni.2638 [DOI] [PubMed] [Google Scholar]
- Hoshino A., Iimura T., Ueha S., Hanada S., Maruoka Y., Mayahara M., Suzuki K., Imai T., Ito M., Manome Y., et al. 2010. Deficiency of chemokine receptor CCR1 causes osteopenia due to impaired functions of osteoclasts and osteoblasts. J. Biol. Chem. 285:28826–28837. 10.1074/jbc.M109.099424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Ueha S., Hanada S., Imai T., Ito M., Yamamoto K., Matsushima K., Yamaguchi A., and Iimura T.. 2013. Roles of chemokine receptor CX3CR1 in maintaining murine bone homeostasis through the regulation of both osteoblasts and osteoclasts. J. Cell Sci. 126:1032–1045. 10.1242/jcs.113910 [DOI] [PubMed] [Google Scholar]
- Hurley M.M., Lee S.K., Raisz L.G., Bernecker P., and Lorenzo J.. 1998. Basic fibroblast growth factor induces osteoclast formation in murine bone marrow cultures. Bone. 22:309–316. 10.1016/S8756-3282(97)00292-5 [DOI] [PubMed] [Google Scholar]
- Ishii M., Egen J.G., Klauschen F., Meier-Schellersheim M., Saeki Y., Vacher J., Proia R.L., and Germain R.N.. 2009. Sphingosine-1-phosphate mobilizes osteoclast precursors and regulates bone homeostasis. Nature. 458:524–528. 10.1038/nature07713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M., Kikuta J., Shimazu Y., Meier-Schellersheim M., and Germain R.N.. 2010. Chemorepulsion by blood S1P regulates osteoclast precursor mobilization and bone remodeling in vivo. J. Exp. Med. 207:2793–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome-Galarza C.E., Lee S.K., Lorenzo J.A., and Aguila H.L.. 2013. Identification, characterization, and isolation of a common progenitor for osteoclasts, macrophages, and dendritic cells from murine bone marrow and periphery. J. Bone Miner. Res. 28:1203–1213. 10.1002/jbmr.1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquelin S., Licata F., Dorgham K., Hermand P., Poupel L., Guyon E., Deterre P., Hume D.A., Combadière C., and Boissonnas A.. 2013. CX3CR1 reduces Ly6Chigh-monocyte motility within and release from the bone marrow after chemotherapy in mice. Blood. 122:674–683. 10.1182/blood-2013-01-480749 [DOI] [PubMed] [Google Scholar]
- Jansen I.D., Vermeer J.A., Bloemen V., Stap J., and Everts V.. 2012. Osteoclast fusion and fission. Calcif. Tissue Int. 90:515–522. 10.1007/s00223-012-9600-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J.D., Park-Min K.H., Shen Z., Fajardo R.J., Goldring S.R., McHugh K.P., and Ivashkiv L.B.. 2009. Inhibition of RANK expression and osteoclastogenesis by TLRs and IFN-γ in human osteoclast precursors. J. Immunol. 183:7223–7233. 10.4049/jimmunol.0900072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly L.M., Pereira J.P., Yi T., Xu Y., and Cyster J.G.. 2011. EBI2 guides serial movements of activated B cells and ligand activity is detectable in lymphoid and nonlymphoid tissues. J. Immunol. 187:3026–3032. 10.4049/jimmunol.1101262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta J., Wada Y., Kowada T., Wang Z., Sun-Wada G.H., Nishiyama I., Mizukami S., Maiya N., Yasuda H., Kumanogoh A., et al. 2013. Dynamic visualization of RANKL and Th17-mediated osteoclast function. J. Clin. Invest. 123:866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Magno C.L., Day C.J., and Morrison N.A.. 2006. Induction of chemokines and chemokine receptors CCR2b and CCR4 in authentic human osteoclasts differentiated with RANKL and osteoclast like cells differentiated by MCP-1 and RANTES. J. Cell. Biochem. 97:512–518. 10.1002/jcb.20649 [DOI] [PubMed] [Google Scholar]
- Koizumi K., Saitoh Y., Minami T., Takeno N., Tsuneyama K., Miyahara T., Nakayama T., Sakurai H., Takano Y., Nishimura M., et al. 2009. Role of CX3CL1/fractalkine in osteoclast differentiation and bone resorption. J. Immunol. 183:7825–7831. 10.4049/jimmunol.0803627 [DOI] [PubMed] [Google Scholar]
- Kotani M., Kikuta J., Klauschen F., Chino T., Kobayashi Y., Yasuda H., Tamai K., Miyawaki A., Kanagawa O., Tomura M., and Ishii M.. 2013. Systemic circulation and bone recruitment of osteoclast precursors tracked by using fluorescent imaging techniques. J. Immunol. 190:605–612. 10.4049/jimmunol.1201345 [DOI] [PubMed] [Google Scholar]
- Lean J.M., Murphy C., Fuller K., and Chambers T.J.. 2002. CCL9/MIP-1γ and its receptor CCR1 are the major chemokine ligand/receptor species expressed by osteoclasts. J. Cell. Biochem. 87:386–393. 10.1002/jcb.10319 [DOI] [PubMed] [Google Scholar]
- Li X., Qin L., Bergenstock M., Bevelock L.M., Novack D.V., and Partridge N.C.. 2007. Parathyroid hormone stimulates osteoblastic expression of MCP-1 to recruit and increase the fusion of pre/osteoclasts. J. Biol. Chem. 282:33098–33106. 10.1074/jbc.M611781200 [DOI] [PubMed] [Google Scholar]
- Liu C., Yang X.V., Wu J., Kuei C., Mani N.S., Zhang L., Yu J., Sutton S.W., Qin N., Banie H., et al. 2011. Oxysterols direct B-cell migration through EBI2. Nature. 475:519–523. 10.1038/nature10226 [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma’ayan A., Enikolopov G.N., and Frenette P.S.. 2010. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 466:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., Ninomiya K., Sonoda K.H., Miyauchi Y., Hoshi H., Iwasaki R., Miyamoto H., Yoshida S., Sato Y., Morioka H., et al. 2009. MCP-1 expressed by osteoclasts stimulates osteoclastogenesis in an autocrine/paracrine manner. Biochem. Biophys. Res. Commun. 383:373–377. 10.1016/j.bbrc.2009.04.020 [DOI] [PubMed] [Google Scholar]
- Mundy G.R., Varani J., Orr W., Gondek M.D., and Ward P.A.. 1978. Resorbing bone is chemotactic for monocytes. Nature. 275:132–135. 10.1038/275132a0 [DOI] [PubMed] [Google Scholar]
- Muto A., Mizoguchi T., Udagawa N., Ito S., Kawahara I., Abiko Y., Arai A., Harada S., Kobayashi Y., Nakamichi Y., et al. 2011. Lineage-committed osteoclast precursors circulate in blood and settle down into bone. J. Bone Miner. Res. 26:2978–2990. 10.1002/jbmr.490 [DOI] [PubMed] [Google Scholar]
- Nakashima T., Hayashi M., Fukunaga T., Kurata K., Oh-Hora M., Feng J.Q., Bonewald L.F., Kodama T., Wutz A., Wagner E.F., et al. 2011. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 17:1231–1234. 10.1038/nm.2452 [DOI] [PubMed] [Google Scholar]
- Oba Y., Lee J.W., Ehrlich L.A., Chung H.Y., Jelinek D.F., Callander N.S., Horuk R., Choi S.J., and Roodman G.D.. 2005. MIP-1α utilizes both CCR1 and CCR5 to induce osteoclast formation and increase adhesion of myeloma cells to marrow stromal cells. Exp. Hematol. 33:272–278. 10.1016/j.exphem.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Okada T., and Cyster J.G.. 2007. CC chemokine receptor 7 contributes to Gi-dependent T cell motility in the lymph node. J. Immunol. 178:2973–2978. 10.4049/jimmunol.178.5.2973 [DOI] [PubMed] [Google Scholar]
- Park K., and Scott A.L.. 2010. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 88:1081–1087. 10.1189/jlb.0610318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlos N.J., and Ng P.Y.. 2012. “Fusion and fission” unveils remarkable insights into osteoclast plasticity. Calcif. Tissue Int. 91:157–158. 10.1007/s00223-012-9620-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J.P., An J., Xu Y., Huang Y., and Cyster J.G.. 2009a. Cannabinoid receptor 2 mediates the retention of immature B cells in bone marrow sinusoids. Nat. Immunol. 10:403–411. 10.1038/ni.1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J.P., Kelly L.M., Xu Y., and Cyster J.G.. 2009b. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 460:1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J.P., Kelly L.M., and Cyster J.G.. 2010a. Finding the right niche: B-cell migration in the early phases of T-dependent antibody responses. Int. Immunol. 22:413–419. 10.1093/intimm/dxq047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J.P., Xu Y., and Cyster J.G.. 2010b. A role for S1P and S1P1 in immature-B cell egress from mouse bone marrow. PLoS ONE. 5:e9277 10.1371/journal.pone.0009277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit I., Szyper-Kravitz M., Nagler A., Lahav M., Peled A., Habler L., Ponomaryov T., Taichman R.S., Arenzana-Seisdedos F., Fujii N., et al. 2002. G-CSF induces stem cell mobilization by decreasing bone marrow SDF-1 and up-regulating CXCR4. Nat. Immunol. 3:687–694. 10.1038/ni813 [DOI] [PubMed] [Google Scholar]
- Ponomaryov T., Peled A., Petit I., Taichman R.S., Habler L., Sandbank J., Arenzana-Seisdedos F., Magerus A., Caruz A., Fujii N., et al. 2000. Induction of the chemokine stromal-derived factor-1 following DNA damage improves human stem cell function. J. Clin. Invest. 106:1331–1339. 10.1172/JCI10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D.W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72:137–174. 10.1146/annurev.biochem.72.121801.161712 [DOI] [PubMed] [Google Scholar]
- Saitoh Y., Koizumi K., Sakurai H., Minami T., and Saiki I.. 2007. RANKL-induced down-regulation of CX3CR1 via PI3K/Akt signaling pathway suppresses Fractalkine/CX3CL1-induced cellular responses in RAW264.7 cells. Biochem. Biophys. Res. Commun. 364:417–422. 10.1016/j.bbrc.2007.09.137 [DOI] [PubMed] [Google Scholar]
- Sakai H., Chen Y., Itokawa T., Yu K.P., Zhu M.L., and Insogna K.. 2006. Activated c-Fms recruits Vav and Rac during CSF-1-induced cytoskeletal remodeling and spreading in osteoclasts. Bone. 39:1290–1301. 10.1016/j.bone.2006.06.012 [DOI] [PubMed] [Google Scholar]
- Semerad C.L., Christopher M.J., Liu F., Short B., Simmons P.J., Winkler I., Levesque J.P., Chappel J., Ross F.P., and Link D.C.. 2005. G-CSF potently inhibits osteoblast activity and CXCL12 mRNA expression in the bone marrow. Blood. 106:3020–3027. 10.1182/blood-2004-01-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Jia T., Mendez-Ferrer S., Hohl T.M., Serbina N.V., Lipuma L., Leiner I., Li M.O., Frenette P.S., and Pamer E.G.. 2011. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 34:590–601. 10.1016/j.immuni.2011.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., and Nagasawa T.. 2006. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 25:977–988. 10.1016/j.immuni.2006.10.016 [DOI] [PubMed] [Google Scholar]
- Teitelbaum S.L. 2000. Bone resorption by osteoclasts. Science. 289:1504–1508. 10.1126/science.289.5484.1504 [DOI] [PubMed] [Google Scholar]
- Walzer T., Chiossone L., Chaix J., Calver A., Carozzo C., Garrigue-Antar L., Jacques Y., Baratin M., Tomasello E., and Vivier E.. 2007. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat. Immunol. 8:1337–1344. 10.1038/ni1523 [DOI] [PubMed] [Google Scholar]
- Wang Y., Lebowitz D., Sun C., Thang H., Grynpas M.D., and Glogauer M.. 2008. Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. J. Bone Miner. Res. 23:260–270. 10.1359/jbmr.071013 [DOI] [PubMed] [Google Scholar]
- Worbs T., Mempel T.R., Bölter J., von Andrian U.H., and Förster R.. 2007. CCR7 ligands stimulate the intranodal motility of T lymphocytes in vivo. J. Exp. Med. 204:489–495. 10.1084/jem.20061706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright L.M., Maloney W., Yu X., Kindle L., Collin-Osdoby P., and Osdoby P.. 2005. Stromal cell-derived factor-1 binding to its chemokine receptor CXCR4 on precursor cells promotes the chemotactic recruitment, development and survival of human osteoclasts. Bone. 36:840–853. 10.1016/j.bone.2005.01.021 [DOI] [PubMed] [Google Scholar]
- Yang M., Mailhot G., MacKay C.A., Mason-Savas A., Aubin J., and Odgren P.R.. 2006. Chemokine and chemokine receptor expression during colony stimulating factor-1–induced osteoclast differentiation in the toothless osteopetrotic rat: a key role for CCL9 (MIP-1γ) in osteoclastogenesis in vivo and in vitro. Blood. 107:2262–2270. 10.1182/blood-2005-08-3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T., and Cyster J.G.. 2013. EBI2-mediated bridging channel positioning supports splenic dendritic cell homeostasis and particulate antigen capture. eLife. 2:e00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T., Wang X., Kelly L.M., An J., Xu Y., Sailer A.W., Gustafsson J.A., Russell D.W., and Cyster J.G.. 2012. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 37:535–548. 10.1016/j.immuni.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Huang Y., Collin-Osdoby P., and Osdoby P.. 2003. Stromal cell-derived factor-1 (SDF-1) recruits osteoclast precursors by inducing chemotaxis, matrix metalloproteinase-9 (MMP-9) activity, and collagen transmigration. J. Bone Miner. Res. 18:1404–1418. 10.1359/jbmr.2003.18.8.1404 [DOI] [PubMed] [Google Scholar]
- Yu X., Huang Y., Collin-Osdoby P., and Osdoby P.. 2004. CCR1 chemokines promote the chemotactic recruitment, RANKL development, and motility of osteoclasts and are induced by inflammatory cytokines in osteoblasts. J. Bone Miner. Res. 19:2065–2077. 10.1359/jbmr.040910 [DOI] [PubMed] [Google Scholar]
- Zhou B.O., Yue R., Murphy M.M., Peyer J.G., and Morrison S.J.. 2014. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 15:154–168. 10.1016/j.stem.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.