Viny et al. show that depletion of the cohesin subunit Smc3 affects HSC function through disruption of chromatin structure and that cohesin haploinsufficiency cooperates with Flt3-ITD to induce transformation of stem/progenitor cells and leukemia development.
Abstract
Cohesin complex members have recently been identified as putative tumor suppressors in hematologic and epithelial malignancies. The cohesin complex guides chromosome segregation; however, cohesin mutant leukemias do not show genomic instability. We hypothesized that reduced cohesin function alters chromatin structure and disrupts cis-regulatory architecture of hematopoietic progenitors. We investigated the consequences of Smc3 deletion in normal and malignant hematopoiesis. Biallelic Smc3 loss induced bone marrow aplasia with premature sister chromatid separation and revealed an absolute requirement for cohesin in hematopoietic stem cell (HSC) function. In contrast, Smc3 haploinsufficiency increased self-renewal in vitro and in vivo, including competitive transplantation. Smc3 haploinsufficiency reduced coordinated transcriptional output, including reduced expression of transcription factors and other genes associated with lineage commitment. Smc3 haploinsufficiency cooperated with Flt3-ITD to induce acute leukemia in vivo, with potentiated Stat5 signaling and altered nucleolar topology. These data establish a dose dependency for cohesin in regulating chromatin structure and HSC function.
Normal hematopoiesis is orchestrated by master regulatory transcription factors that are expressed in specific cell types at discrete differentiation stages (Novershtern et al., 2011; Lara-Astiaso et al., 2014). Tight coordination of these networks maintains the balance between hematopoietic stem/progenitor cell (HSPC) self-renewal and lineage commitment. In contrast, acute leukemias are characterized by increased self-renewal and impaired differentiation, often in the setting of mutations in genes with a known or postulated role in regulating transcriptional output (Dawson et al., 2012). Recent cancer genome sequencing studies have identified loss of function mutations in cohesin complex factors among patients with solid tumors (Solomon et al., 2011; Balbás-Martínez et al., 2013) and with myeloid malignancies (Jan et al., 2012; Welch et al., 2012; Cancer Genome Atlas Research Network, 2013; Kon et al., 2013; Thol et al., 2014; Thota et al., 2014), suggesting a role for cohesin as a tumor suppressor.
Cohesin is a multiprotein ring-like complex known to regulate sister chromatid alignment during mitosis; thus, it has been suggested that cohesin mutations will induce chromosomal instability (Solomon et al., 2011). However, cohesin mutations are not associated with aneuploidy, suggesting an alternate pathophysiologic mechanism (Thota et al., 2014). Recent research has suggested a role for cohesin in regulation of gene expression by stabilizing interactions between promoters and distal cis-regulatory elements (or enhancers). This research suggests cohesin functions as an insulator factor, protecting promoters from distal enhancers, thus establishing boundaries around actively transcribed chromatin domains (Bell et al., 1999; Kagey et al., 2010; Merkenschlager and Odom, 2013). These activities are often in direct physical association with the DNA-binding factor CTCF (CCCTC-binding factor; Parelho et al., 2008; Rubio et al., 2008; Stedman et al., 2008; Wendt et al., 2008), although cohesin can also form promoter–enhancer and enhancer–enhancer loops in a CTCF-independent manner (Dowen et al., 2014). The formation of specific chromatin architecture through cohesin-mediated looping mediates the recruitment and activity of RNA polymerase II, facilitating transcriptional activation (Schaaf et al., 2013).
Genomic data strongly suggest cohesin complex members function as tumor suppressors, but the underlying mechanism by which these mutations disrupt hematopoiesis and promote transformation has not been delineated. Of note, myeloid malignancies such as myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML) with cohesin mutations are characterized by monoallelic mutations in any one of the cohesin complex members and never by complete loss of a cohesin complex member or by multiple heterozygous mutations. These data suggest a dose response for cohesin function in hematopoiesis, with heterozygous loss of a single cohesin complex member sufficient to contribute to leukemic transformation. We therefore sought to investigate the impact of complete loss of a specific cohesin subunit, Smc3, compared with Smc3 haploinsufficiency on HSPC function in vivo.
RESULTS AND DISCUSSION
Development of a conditional Smc3 allele
To delineate the role of Smc3 in hematopoietic function, we generated a conditional allele targeting Smc3 in vivo. We used embryonic stem cells in which two LoxP sites were inserted flanking exon 4, a critical coil-coil domain, as well as Frt sites surrounding a neomycin cassette in an upstream intron (Fig. 1 A). After Frt-mediated excision of the neo cassette, mice were then crossed to the inducible cre-recombinase, Mx1. Smc3 deletion was achieved after treatment with the IFN-α–stimulating polyinosinic:polycytidylic acid (PIpC).
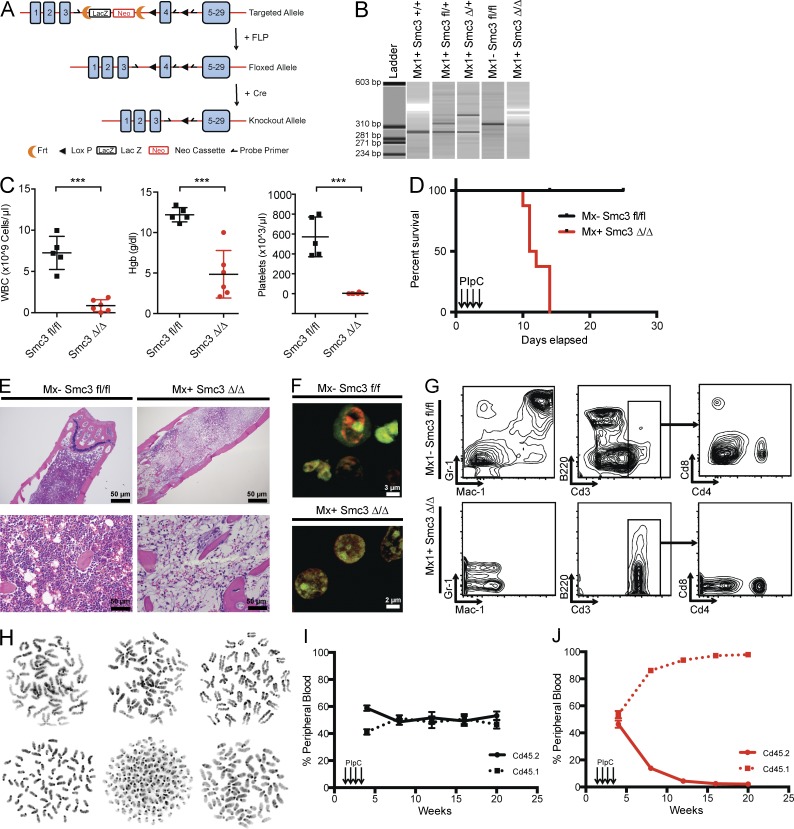
Figure 1.
Smc3 is required for HSC function. (A) Schematic depiction of the targeted Smc3 allele. Exon 4 is targeted and flanked by LoxP sites upon Frt-mediated deletion of the Neo cassette. (B) Qiaxel gel electrophoresis image of PCR genotyping of the Smc3 WT allele (287 bp), Smc3 floxed allele (313 bp), and Cre recombination allele (349 bp). The genotype is shown for Smc3+/+, Smc3fl/+, Smc3Δ/+, Smc3fl/fl, and Smc3Δ/Δ. (C) Peripheral blood WBCs, hemoglobin (Hgb), and platelets after postnatal deletion of Smc3. Counts are compared with age-matched controls injected with PIpC (n = 6 for each genotype). (D) Kaplan-Meier curve of primary mice after postnatal Smc3 deletion (n = 10 for each genotype). (E) Histological (H&E) analysis of Mx1-Cre Smc3Δ/Δ and Cre-negative Smc3fl/fl control BM. (F) Nucleolar stain of Mx1-Cre Smc3Δ/Δ BM reveals fragmented and supernumerary nucleoli. ACK-lysed BM was stained with TOTAL-NUCLEAR-ID fluorescent reagents, allowing simultaneous staining of both the nucleoli (green) and total nucleus (red). (G) Flow cytometric enumeration of B220+, Cd11b+/Gr1+, Cd3+, and Cd4/8 ratio of cells in the peripheral blood of Mx1-Cre Smc3fl/fl mice and cre-negative controls. (H) Metaphase karyotyping upon exposure to colcemid (45 min): Representative metaphases with partial or complete premature sister chromatid separation resulting in tetraploidy/polyploidy in 44/200 cells (22%). (I and J) Competitive transplantation of 500K donor-derived Cd45.1 whole BM was injected by tail vein after lethal irradiation with either 500K Cd45.2 whole BM from Mx− Smc3fl/fl or 500K Mx+ Smc3Δ/Δ (n = 5 mice each genotype). Mice were treated with PIpC at week 2, and chimerism was measured by percentage of Cd45.2 in the peripheral blood every 4 wk. Error bars represent ± SD (C, I, and J); ***, P < 0.001 (Mann–Whitney U test).
Smc3 is required for hematopoietic stem cell (HSC) function and for normal hematopoiesis
We characterized the effects of homozygous Smc3 loss by crossing mice bearing floxed alleles to Mx1-cre mice. Mice were treated with PIpC at 4 wk of age, resulting in complete recombination in Mx1-cre Smc3fl/fl mice (Fig. 1 B). We observed severe pancytopenia with decreased WBC (P < 0.001), decreased hemoglobin (P < 0.001), and decreased platelet count (P < 0.001) compared with littermate cre-negative controls (Fig. 1 C). Moreover, Smc3 deletion in the hematopoietic compartment (Smc3Δ/Δ) resulted in 100% lethality with a median survival of 11.5 d from the start of PIpC (P < 0.001; Fig. 1 D). At time of death, we observed evidence of cerebral hemorrhage in coronal brain sections and local tissue necrosis in spleen and liver tissue. Pathological analysis of femurs from Smc3 KO mice revealed marked hypocellularity, and nucleolar staining of aspirates taken on day 5 after PIpC revealed increased numbers of discrete nucleoli (green) per cell with the TOTAL-NUCLEAR-ID red/green nucleolar/nuclear immunofluorescent staining kit on confocal microscopy (Fig. 1, E and F). Flow cytometry of peripheral blood at euthanasia revealed an absence of myeloid elements in Smc3Δ/Δ mice, with T lymphocytes (Mac1− Gr1− Cd3+ B220−) as the only remaining circulating WBCs (Fig. 1 G). These remaining cells did not show recombined floxed alleles and expressed Smc3 protein (not depicted). Chromosome morphology was difficult to assess, and we observed premature sister chromatid separation and tetraploidy/polyploidy in 44/200 (22%) of scored metaphases in Smc3 KO mice (Fig. 1 H) compared with normal metaphase cytogenetics and sister chromatid separation in cre-negative littermate controls (not depicted). To assess the effects of Smc3 deletion on HSC function in vivo, 500,000 BM cells from 6-wk-old Cd45.2 Mx1-cre Smc3fl/fl mice or cre-negative Smc3fl/fl littermate controls were transplanted with an equal number of 6-wk-old Cd45.1 competitor BM cells into lethally irradiated Cd45.1 recipient mice. Mice were treated with PIpC after engraftment. After PIpC-mediated deletion, Smc3 KO cells showed complete loss of HSC function, as KO cells were completely outcompeted by WT BM in vivo (Fig. 1, I and J).
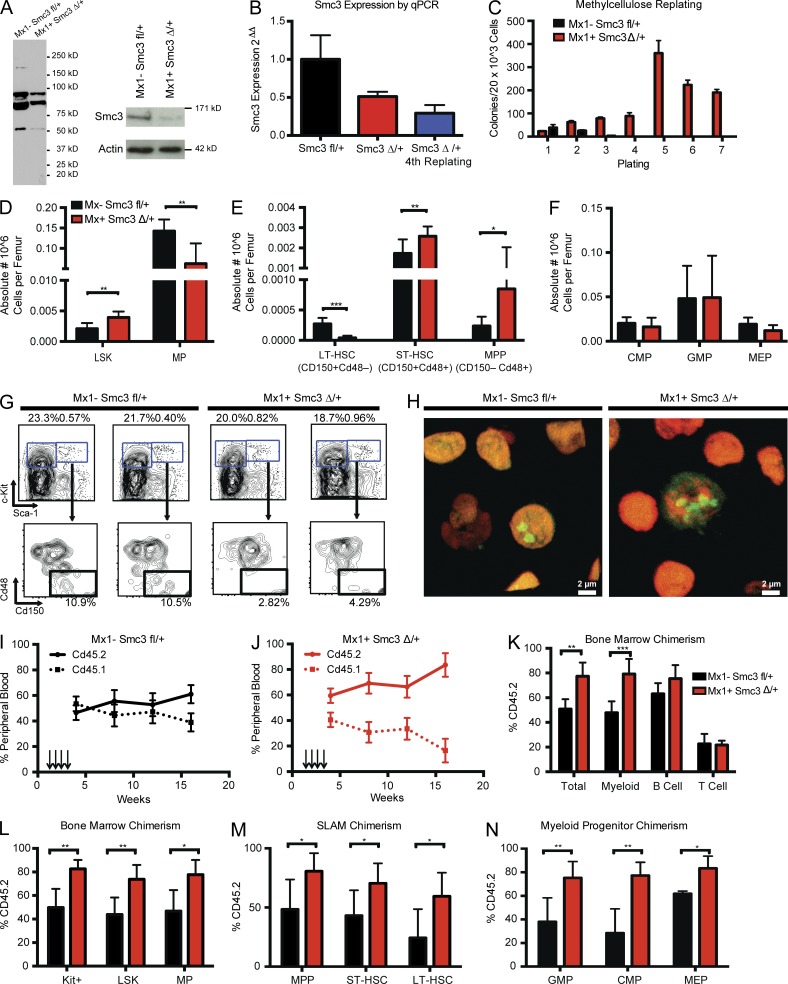
Impact of Smc3 haploinsufficiency on hematopoiesis in vitro and in vivo
Mx1-cre mice bearing one floxed allele were treated with PIpC at 4 wk of age, resulting in one recombined allele and one WT allele (Fig. 1 B). We observed reduced Smc3 protein expression and we observed a 50% reduction in Smc3 mRNA expression (Fig. 2, A and B). In methylcellulose plating assays (rmIL-3, rmSCF, rh-EPO, and eh-IL6), Smc3Δ/+ cells showed increased serial replating capacity, whereas cells derived from Smc3fl/+ mice failed to replate after the third plating. Cells taken from the fourth plating continued to express a single Smc3 allele (Fig. 2, B and C). 4 wk after PIpC treatment, we observed an increase in the absolute number of immunophenotypically defined HSPCs in Smc3Δ/+ mice, with a significant increase in absolute number of LSK (Lin− Sca-1+ c-Kit+; P = 0.005) but a decrease in myeloid progenitor (MP) cells (Lin− Sca-1− c-Kit+; P = 0.003; Fig. 2, D and G). Within the LSK compartment, we observed an increase in short-term HSCs (ST-HSCs; Cd150+ Cd48+ Lin− Sca-1+ c-Kit+; P = 0.008), a significant decrease in the frequency of long-term HSCs (LT-HSCs; Cd150+ Cd48− Lin− Sca-1+ c-Kit+; P < 0.001) and a significant increase in the absolute number of multipotent progenitor (MPP) populations (Cd150− Cd48+ Lin− Sca-1+ c-Kit+; P = 0.003; Fig. 2 E). No differences were seen within the MP populations (Fig. 2 F). Nucleolar staining revealed increased numbers of discrete nucleoli on confocal microscopy in Smc3Δ/+ BM, whereas cre-negative Smc3fl/+ BM had singlet or doublet nucleoli exclusively (Fig. 2 H).
Figure 2.
Smc3 haploinsufficiency increases stem cell self-renewal. (A) Mx1-cre–mediated deletion of Smc3 confirmed by Western blot of BM. (B) qRT-PCR showing relative expression level of Smc3 in BM and persistence of the decreased expression in the cells that replate in methylcellulose. (C) Serial plating and colony counts in methylcellulose for Smc3fl/+ or Smc3Δ/+ BM. Whole BM was plated in triplicate at 20K cells/well. (D–F) Flow cytometric enumeration of BM c-Kit+, LSK (Lin− Sca1+ Kit+), and MP (Lin− Sca1− Kit+) cells (D), LT-HSCs (LSK CD150+ CD48−), ST-HSCs (LSK, Cd150+, Cd48+), and MPP cells (LSK CD150− CD48+; E), and percentage of common myeloid progenitors (CMPs; lineage−, c-Kit+, Sca-1−, FcγR−, CD34+), GMPs (lineage− c-Kit+ Sca-1−, FcγR+ CD34+), and MEPs (lineage− c-Kit+ Sca-1− FcγR− CD34−; F) in Smc3fl/+ and Smc3Δ/+ mice (n = 4–6 mice per genotype). Mice were taken down at 4 wk after PIpC injection. Excision was confirmed at takedown. Data are expressed as frequency of live cells per femur. (G) Representative FACS analysis of BM stem cell populations of Smc3fl/+ and Smc3Δ/+ mice. (H) Nucleolar staining of Smc3fl/+ and Smc3Δ/+ BM reveals occasional cells with supernumerary nucleoli in Smc3Δ/+ cells. (I and J) Competitive transplantation of 500K donor-derived Cd45.1 whole BM was injected by tail vein after lethal irradiation with either 500K Cd45.2 whole BM from Smc3fl/+ or 500K Smc3Δ/+. Chimerism was measure by percentage of Cd45.2 in the peripheral blood every 4 wk. Mice were sacrificed at 20 wk. (K–N) Flow cytometric chimerism of unsorted peripheral blood, Mac1+Gr1+ myeloid cells, B220+ B cells, and Cd3+ T cells (K), BM c-Kit+, LSK (Lin− Sca1+ Kit+), and MP (Lin− Sca1− Kit+) cells (L), LT-HSCs (LSK CD150+ CD48−), ST-HSCs (LSK, Cd150+, Cd48+), and MPP cells (LSK CD150− CD48+; M), and percentage of CMPs (lineage−, c-Kit+, Sca-1−, FcγR−, CD34+), GMPs (lineage− c-Kit+ Sca-1−, FcγR+ CD34+), and MEPs (lineage− c-Kit+ Sca-1− FcγR− CD34−; N) in Smc3fl/+ and Smc3Δ/+ mice (n = 5 mice per genotype). Error bars represent ± SD (B–F and I–N); *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Mann–Whitney U test).
Smc3 haploinsufficiency increases self-renewal in vivo
To assess the effects of Smc3 haploinsufficiency in vivo, 500,000 BM cells from 6-wk-old Cd45.2 Mx1-cre Smc3fl/+ mice or cre-negative Smc3fl/+ littermate controls were transplanted in competition with an equal number of 6-wk-old Cd45.1 competitor BM cells into lethally irradiated Cd45.1 recipient mice. Mice were treated with PIpC after engraftment 2 wk later. In contrast to the competitive disadvantage seen with Smc3 KO BM in transplant assays, Smc3 haploinsufficient BM outcompeted WT BM, consistent with increased self-renewal in vivo (Fig. 2, I and J). We observed increased chimerism in total BM (P = 0.001) and in the myeloid (P = 0.002) compartment with a trend toward increased chimerism in B lymphocytes (P = 0.08) but not in T lymphocytes (P = 0.81; Fig. 2 K). The increase in self-renewal observed in the competitive transplant assays was seen in all HSPC compartments (Fig. 2, L–N).
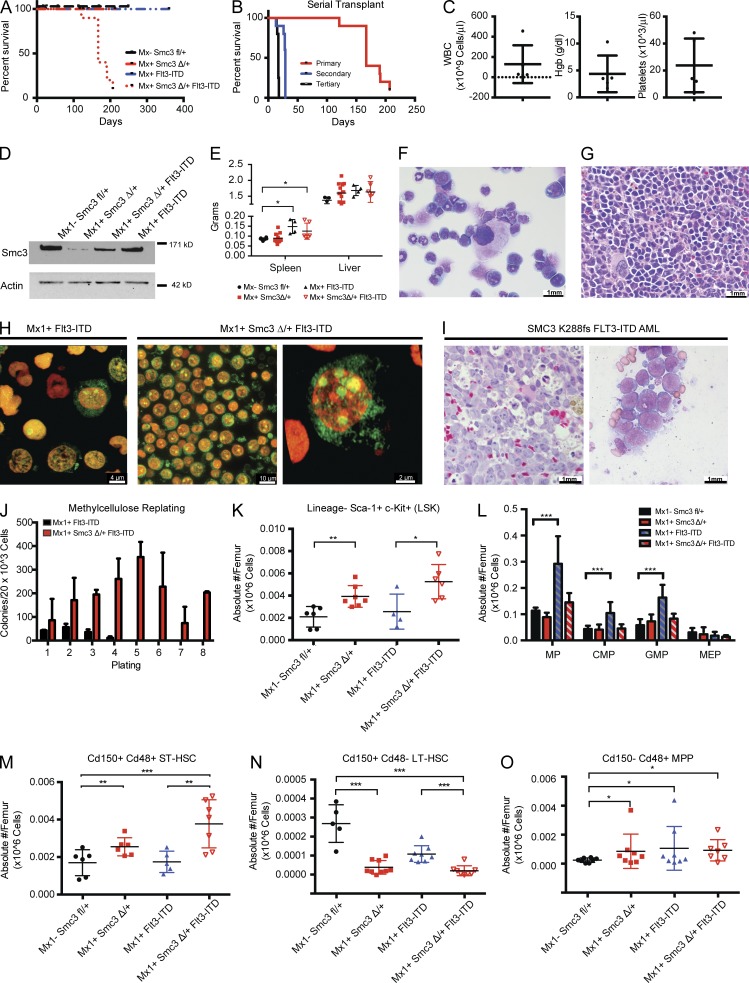
Heterozygous Smc3 loss cooperates with Flt3-ITD to induce AML
AML patients with cohesin mutations often have concurrent FLT3-ITD mutations (Cancer Genome Atlas Research Network, 2013). We therefore investigated the phenotype of mice with concurrent Smc3 haploinsufficiency and expression of the Flt3ITD allele from the endogenous locus, compared with mice with Flt3ITD alone (Lee et al., 2007). Whole BM from primary Mx1-cre Smc3fl/+, cre-negative Smc3fl/+ littermate control, Mx1-cre Flt3ITD, and Mx1-cre Smc3fl/+ Flt3ITD was injected into lethally irradiated recipients along with support control marrow. Mice were PIpC injected after 2 wk and followed longitudinally. All Mx1-cre Smc3Δ/+ Flt3ITD died of acute leukemia (median survival 167 d; P < 0.001), whereas there were no deaths in the Mx1-cre Smc3Δ/+, cre-negative Smc3fl/+, or Mx1-cre Flt3ITD cohorts (P = 0.05; Fig. 3 A). Serial transplantation of whole BM from leukemic Mx1-cre Smc3Δ/+ Flt3ITD mice resulted in further anticipation of the leukemic disease with shortened disease latency (Fig. 3 B). All recipients developed AML hallmarked by leukocytosis, anemia, and thrombocytopenia (Fig. 3 C). Protein lysates from all four genotypes revealed decreased Smc3 expression in Smc3 haploinsufficient and Mx1-cre Smc3Δ/+ Flt3ITD BM without loss of the remaining Smc3 allele or complete loss of Smc3 protein expression (Fig. 3 D). We noted splenomegaly in both Flt3ITD and Smc3fl/+ Flt3ITD mice (P = 0.03; Fig. 3 E). Cytospins revealed that Mx1-cre Smc3Δ/+ Flt3ITD mice were characterized by expansion of blasts in the blood and BM, with several discrete nucleoli in each blast. Blasts were evident throughout the marrow and spleen with marked dysmyelopoiesis, increased nucleoli, and dysplastic, hypolobated megakaryocytes (Fig. 3, F and G). We observed splenomegaly and a leukemoid appearance of the bones consistent with the increased number of stem and progenitor cells, and cytogenetic analysis revealed normal metaphase cytogenetics in all mice (not depicted). Although Mx1-cre Flt3ITD stem and progenitor cells had singlet or doublet nucleoli, nearly every cell from the BM of Mx1-cre Smc3Δ/+ Flt3ITD had three or more nucleoli (Fig. 3 G and Videos 1 and 2). Consistent with the morphological alterations observed in our mouse model, pathological analysis of a patient sample with concurrent SMC3 R288fs and FLT3-ITD mutations revealed an increase in nucleolar frequency in the blast population (Fig. 3 I). Mx1-cre Smc3Δ/+ Flt3ITD BM cells showed increased serial replating, similar to the phenotype observed with Smc3 haploinsufficiency but with higher numbers of colonies per plating (Fig. 3 J).
Figure 3.
Smc3 haploinsufficiency cooperated with Flt3-ITD to induce AML. (A) Kaplan-Meier survival curve of primary transplant recipients of 106 total BM cells from Smc3fl/+ (n = 6), Smc3Δ/+ (n = 5), Flt3ITD (n = 5), and Smc3Δ/+ Flt3ITD mice (n = 5). (B) Kaplan-Meier survival curve of secondary and tertiary transplantation of BM from Smc3Δ/+ Flt3ITD mice (n = 5 each). (C) Peripheral blood counts of secondary transplanted mice with BM from Smc3fl/+ Flt3-ITD primary mice at the time of death (n = 5). (D) Mx1-cre–mediated deletion of one Smc3 allele reduces Smc3 expression in Smc3Δ/+ and Smc3Δ/+ Flt3ITD mice compared with age-matched controls (Smc3fl/+ and Flt3ITD). (E) Liver and spleen weights for primary mice (n = 4–8 for each genotype). (F and G) H&E stain of BM cytospin (F) and spleen (G). (H) Nucleolar stain of Flt3ITD and Smc3Δ/+ Flt3ITD BM. ACK-lysed BM was stained with TOTAL-NUCLEAR-ID fluorescent reagents, allowing simultaneous staining of both the nucleoli (green) and total nucleus (red). (I) H&E stain of MDS patient with refractory anemia with excess blasts-1 (RAEB-1) with molecular genetics identifying SMC3 K288fs and FLT3-ITD. Further experiments also identified IDH2 R140Q. Light microscopy at 100 magnification reveals multiple nucleoli in the blast population. (J) Serial plating and colony counts in methylcellulose for Smc3Δ/+ Flt3ITD and Flt3ITD BM. Whole BM was plated in triplicate at 20K cells/well. (K) Flow cytometric enumeration of BM LSK (Lin− Sca1+ c-Kit+). (L–O) enumeration of MPs (Lin− Sca1− c-Kit+), CMPs (lineage− c-Kit+ Sca-1− FcγR− CD34+), GMPs (lineage− c-Kit+ Sca-1− FcγR+ CD34+), and MEPs (lineage− c-Kit+ Sca-1− FcγR− CD34−; L) and flow cytometric enumeration of ST-HSCs (LSK Cd150+ Cd48+; M), LT-HSCs (LSK CD150+ CD48−; N), and MPP cells (LSK CD150− CD48+; O; n = 5–8 mice per genotype). Data are expressed as frequency of live cells per femur. Error bars represent ± SD (C, E, and J–O); *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Mann–Whitney U test).
Alterations in the stem/progenitor compartment induced by concomitant Smc3 heterozygous loss and expression of Flt3-ITD
We next assessed the effects of Smc3 loss in concert with Flt3-ITD in the HSC and progenitor populations. We observed a significant increase in the absolute number of LSK cell (Lin− Sca-1+ c-Kit+) Mx1-cre Smc3Δ/+ Flt3ITD mice compared with control (P = 0.003) and compared with Mx1-cre Flt3ITD (P = 0.008; Fig. 3 K). Mx1-cre Flt3ITD mice were characterized by an increase in MPs (P = 0.018) and granulocyte-macrophage progenitors (GMPs; lineage− c-Kit+ Sca-1−, FcγR+ CD34+; P = 0.05) compared with control, as previously reported (Lee et al., 2007; Shih et al., 2015). In contrast, concomitant Smc3 haploinsufficiency in the setting of the Flt3ITD significantly reduced the number of GMPs (P = 0.04; Fig. 3 L). The overall increase in the LSK population seen in Mx1-cre Smc3Δ/+ Flt3ITD mice was driven mostly by an overall increase in the absolute number of ST-HSCs (P = 0.005 and 0.008 compared with Smc3 and Flt3 single mutations, respectively; Fig. 3 M), with a statistically significant decrease in LT-HSCs compared with both cre-negative Smc3fl/+ and Mx1-cre Flt3ITD (P = 0.002 and 0.001, respectively; Fig. 3 N) and an increase in MPPs (Lin− Sca-1+ c-Kit+ Cd48+ Cd150−; P = 0.047; Fig. 3 O). These data suggest that concurrent Flt3-ITD mutation and Smc3 heterozygosity alter the stem/progenitor milieu.
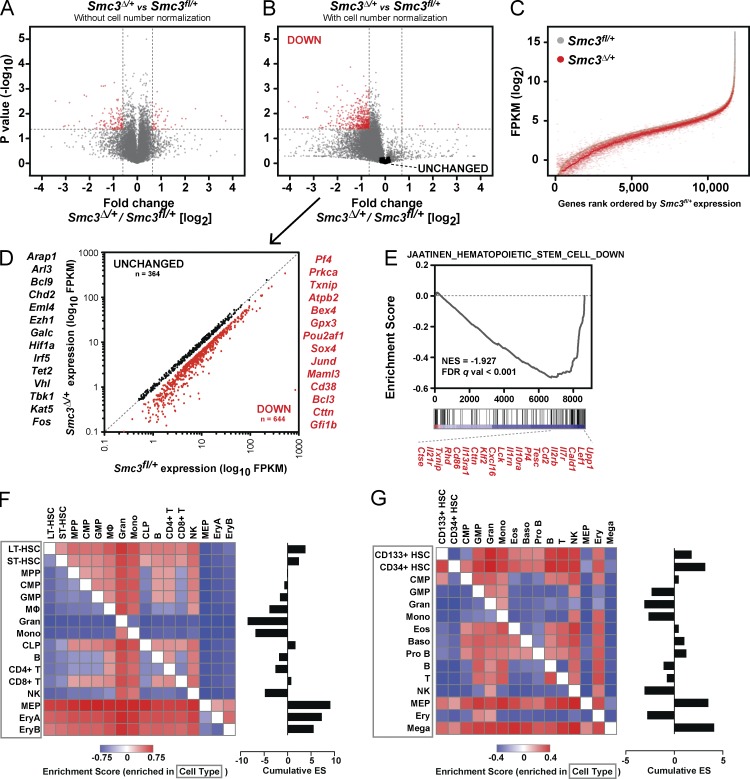
Smc3 haploinsufficiency results in a distinct reduction of mRNA transcriptional output
We performed RNA sequencing of poly-A selected transcripts (mRNA-seq) to measure global gene expression of c-Kit+ hematopoietic progenitors 4 wk after conditional deletion of a single Smc3 allele. Samples were harvested from individual mice, with a high correlation between samples of the same genotype. We hypothesized that Smc3 haploinsufficiency would result in global and locus-specific effects on cis-regulatory elements influencing coordinated gene expression programs. We used standardized spiked-in synthetic RNAs to normalize RNA expression to the input cell number to allow us to discern the impact of Smc3 haploinsufficiency on global patterns of gene expression (Lovén et al., 2013). When RNA expression changes were measured without input cell number normalization, modest effects on gene expression were observed with only a small proportion of genes significantly up- or down-regulated (Fig. 4 A). However, when the data were normalized to input cell number, we observed a significant decrease of a large subset of transcripts in Smc3Δ/+ hematopoietic cells (Fig. 4, B and C), consistent with a global reduction in transcription. Whereas the magnitude of global transcriptional dampening observed with Smc3 haploinsufficiency was modest throughout the transcriptome (Fig. 4 C), we identified a subset of genes with disproportionate dysregulation of gene expression. We found a subset of genes to be significantly down-regulated (n = 644), whereas another subset of genes were relatively unchanged between genotypes (n = 364; Fig. 4 D and Table S1). We found that among the unchanged gene set were known regulators of stem cell maintenance, including Hif1a and Tet2 (Ko et al., 2011; Kim et al., 2014). In contrast, we observed a marked decrease in the expression of genes indicative of lineage-committed cells, including the lymphoid-specific transcription factors Pou2af1 and Bcl3 and the myeloerythroid transcription factor Gfi1b. These data suggest that cohesin haploinsufficiency alters the transcriptional balance between stem cell self-renewal and lineage commitment.
Figure 4.
Smc3 haploinsufficiency perturbs mRNA transcriptional output. RNA-seq using synthetic RNA spike-in controls reveals RNA transcript levels are globally decreased in Smc3Δ/+ c-Kit+ cells when compared with Smc3fl/+ WT. (A and B) The widespread decrease in overall mRNA levels is obscured when spike-in control reads are not used (A) but apparent with cell number normalization (B); fold change values represent the mean of duplicate samples. (C) These effects are apparent across gene expression levels; gray line represents gene expression of Smc3fl/+ cells rank ordered by expression level; red dotted line represents LOESS curve through Smc3Δ/+ matched values. (D) Scatterplot of genes identified as unchanged and down-regulated with Smc3 haploinsufficiency with exemplary genes in each group shown. Gene lists generated as defined in Materials and methods; full gene lists annotated in Table S1. (E) GSEA using a human hematopoietic progenitor gene set, with several enriched down-regulated genes highlighted. (F and G) Pairwise GSEA of mouse (F) and human (G) hematopoietic cells using the down-regulated gene set. Cumulative enrichment scores represent the sum of enrichment scores for each cell type when compared across all other cell types.
To comprehensively explore transcriptional programs influenced by Smc3 loss in an unbiased manner, we performed gene set enrichment analysis (GSEA) on the Smc3fl/+ and Smc3Δ/+ RNA sequencing data (Subramanian et al., 2005). Smc3 haploinsufficiency enriched for genes that are differentially expressed in human cord blood stem/progenitor cells (Fig. 4 E; Jaatinen et al., 2006). To further investigate the nature of the down-regulated gene set, we performed GSEA using this gene set searching for enrichment in a pairwise fashion with gene expression datasets from a compendium of mouse and human hematopoietic progenitor and lineage cells (Novershtern et al., 2011; Lara-Astiaso et al., 2014). We found that the down-regulated gene set was enriched not only in HSCs, but also in lineage-committed progenitor cells and differentiated hematopoietic cells (Fig. 4, F and G). Enrichment was especially apparent in megakaryocyte-erythrocyte progenitors (MEPs) and differentiated mouse erythrocytes (Ery) and also in the common lymphoid progenitor (CLP) population. However, these genes are specifically not enriched in myeloid cell types (granulocytes, monocytes, and macrophages). These data reveal that many of the genes most disrupted by cohesin haploinsufficiency are regulatory factors required for lineage commitment, mechanistically corroborating the increase in self-renewal and impaired differentiation with cohesin haploinsufficiency
Cohesin haploinsufficiency disrupts expression of genes with distinct chromatin organization
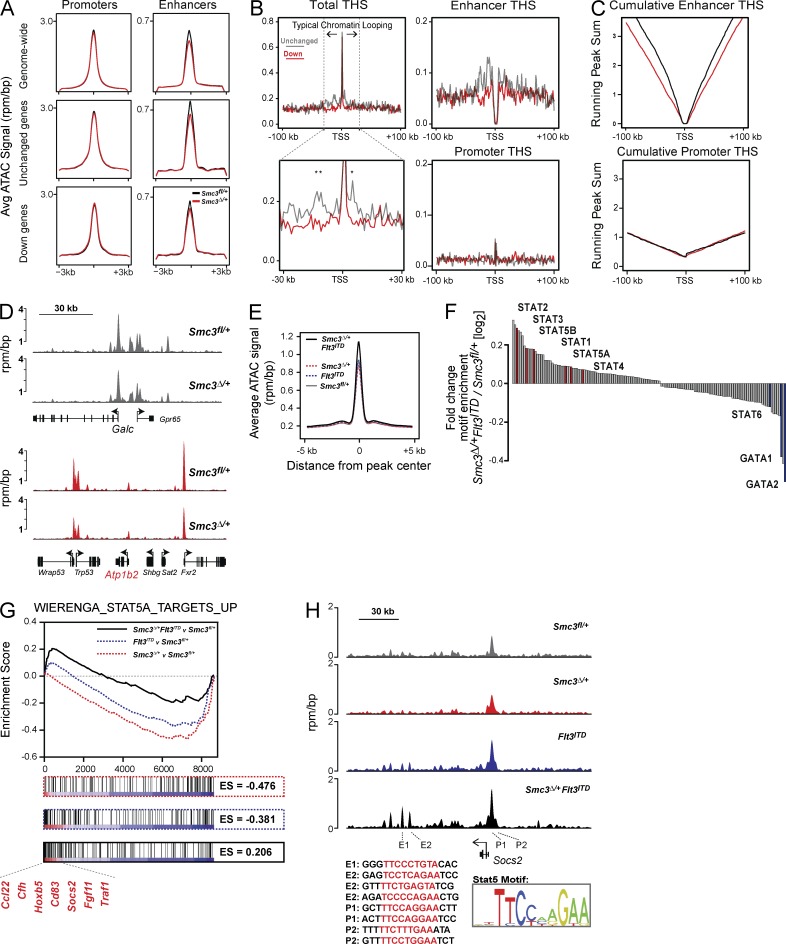
We next performed assays of transposase-accessible chromatin sequencing (ATAC-seq) on Smc3fl/+ and Smc3Δ/+ cells. This allowed us to assess chromatin accessibility at transposase hypersensitive sites (THSs) across the Smc3Δ/+ genome compared with WT cells (Fig. 5 A). When THSs were analyzed based on specific genomic features (promoters and enhancers), we only observed a modest decrease in chromatin accessibility at enhancer regions. We then compared the local chromatin structure of down-regulated genes compared with unchanged genes. Surprisingly, chromatin structure at these genes remained largely unchanged, revealing that reduced Smc3 levels did not dramatically change chromatin accessibility, even at gene loci that displayed significant decreases in mRNA levels. We observed at THSs outside of promoter regions, indicative of transcription factor–bound enhancer regulatory elements, that overall chromatin accessibility was lower at down-regulated gene loci compared with unchanged genes. To further explore chromatin characteristics that may indicate an increased susceptibility to reduced Smc3 levels, we analyzed the density of chromatin accessibility immediately surrounding transcriptional start sites (TSSs) at the unchanged and down-regulated gene loci. We observed significantly less THS density surrounding TSSs of down-regulated genes when compared with unchanged genes (Fig. 5 B, left). This difference in ATAC-seq signal is observable within ∼30 kb in the 5′ direction and ∼10 kb in the 3′ direction of the TSS. This size domain generally corresponds to the size of typical local chromatin looping interactions that facilitate transcription factor–mediated transactivation, as observed by high-throughput chromatin conformation capture experiments in a variety of cell types (Jin et al., 2013). These data establish a model whereby promoters that are associated with fewer local cis-regulatory elements regulate genes most affected by cohesin complex depletion.
Figure 5.
Chromatin accessibility profiles of Smc3 haploinsufficiency. (A) Global chromatin accessibility assessed by ATAC-seq (top row). Overall THS density was analyzed genome wide and found in promoter regions (±1 kb of annotated RefSeq TSS) and enhancer sites (>1 kb from annotated TSS). All plots were generated using a LOESS curve through the mean of duplicate ATAC-seq measurements. Analysis was also performed on THSs of unchanged and down-regulated genes (middle and bottom rows). THSs included in analysis were within 30 kb of annotated TSSs of either unchanged or down-regulated genes. (B) ATAC-seq peak density was quantified within a 200-bp window of gene TSSs in 1-kb bins (top left). A highlighted 60-kb region immediately surrounding the TSSs is shown in the bottom left panel (* represents individual bins with P < 0.005, Student’s t test). The value for each bin is the mean of replicate samples with a fitted LOESS curve. Enhancer-only THS density is shown in the top right panel, whereas promoter-only peaks are shown in the bottom right panel. (C) Cumulative THS enhancer and promoter peaks counting outwards from TSSs. (D) Chromatin accessibility of the unchanged gene locus Galc and the down-regulated gene locus Atp1b2. (E) Genome-wide chromatin accessibility at THSs as measured by ATAC-seq in each genotype. Plotted is the mean ATAC-seq signal at all THSs genome wide. (F) Transcription factor motif site enrichment in ATAC-seq peaks gained and lost in Smc3Δ/+ Flt3-ITD cells compared with Smc3fl/+. (G) GSEA using RNA-seq data from each genotype compared with Smc3fl/+ controls and a human HSC STAT5 target gene set. (H) Chromatin accessibility of the Stat5 target gene Socs2 showing potentiation of Stat5 signaling at specific regulatory elements. Shown are ATAC-seq profiles of the entire locus for each genotype. Stat5 motifs are found within each labeled regulatory element (sequence highlighted in red).
We next analyzed THS density of either enhancer- or promoter-specific sites. We did not observe significant THS density of other local promoter sites at either unchanged or down-regulated genes, indicating that the THS density differences we observe are not a result of a high density of active genes (Fig. 5 B, bottom right). In contrast, we found that THS density was specifically depleted at enhancer sites predominantly immediately upstream of the down-regulated gene set (Fig. 5 B, top right). In addition to decreased local THS enhancer density at down-regulated genes, we observed lower absolute numbers of enhancer, but not promoter, THSs within at 200-kb window of TSSs (Fig. 5 C). These analyses show that a particular subset of lineage commitment genes in HSCs are particularly reliant on normal expression levels of cohesin subunits and that their chromatin structure locally around the TSS has fewer enhancer elements. In contrast, genes with stable expression in the setting of Smc3 attenuation have a greater THS density surrounding their promoter regions, suggesting Smc3 haploinsufficiency does not disrupt the more complex promoter/enhancer interactions in these transcriptional complexes. For example, the Galc locus remains stable with Smc3 haploinsufficiency and contains a large region of chromatin accessibility immediately upstream of its active promoter. In contrast, down-regulated genes such as Atp1b2 have lower levels of chromatin accessibility at their locus, with fewer local enhancer THS sites (Fig. 5 D).
Combined Smc3 haploinsufficiency and Flt3-ITD results in potentiation of Stat5-driven transcription
To investigate how Smc3 haploinsufficiency may cooperate with the known myeloid leukemia oncogene Flt3-ITD to promote leukemia in mice, we analyzed the gene expression and chromatin accessibility profiles of c-Kit+ hematopoietic cells from mice harboring Flt3ITD alone and in combination with Smc3 monoallelic deletion. We found that THSs in Smc3Δ/+ Flt3ITD cells generally were more accessible than in cells of the other genotypes (Fig. 5 E). This suggests that the combination of Smc3Δ/+ and Flt3ITD in hematopoietic cells potentiates alterations in chromatin factor association through either gain or loss of functional cis-regulatory elements. To understand which TFs may be driving the observed increase in genome-wide chromatin accessibility, we analyzed 146 transcription factor recognition motifs within the THSs differentially observed in Smc3Δ/+ Flt3ITD and WT cells (Fig. 5 F). Notably, the THSs that are lost in the Smc3Δ/+ Flt3ITD cells are enriched in both Gata1- and Gata2-binding sites, whereas THSs that are gained in Smc3Δ/+ Flt3ITD cells are enriched in Stat family transcription factor–binding sites, including Stat5. As Flt3-ITD alters gene expression through the STAT transcription factors, we reason that increased accessibility at THS cis-regulatory elements and the alterations in gene expression seen in cells with combined Smc3 haploinsufficiency and Flt3ITD may be in a large part driven by potentiated Stat signaling at chromatin. We next investigated the impact of combined Smc3 haploinsufficiency and Flt3ITD on a STAT5 target gene expression signature derived from expression of activated STAT5A in human CD34+ HSCs/progenitors (Wierenga et al., 2008). We observed enrichment of the STAT5A signature in the Smc3Δ/+ Flt3ITD cells compared with Smc3Δ/+, Flt3ITD, and WT cells, suggesting the divergent mutations cooperate to potentiate oncogenic Stat5 signaling in HSCs (Fig. 5 G). Among the Stat5 target genes most differentially activated in our dataset is Socs2, a suppressor of cytokine signaling found to be up-regulated in AML patients, a known target gene of the STAT5 transcription factor, and a marker of poor prognosis (Laszlo et al., 2014). We observed increased chromatin accessibility at enhancer THSs at the Socs2 locus in both Flt3ITD mutant hematopoietic cells and, at a greater magnitude, in Smc3Δ/+ Flt3ITD cells (Fig. 5 H). These data suggest that the lower Smc3 level in these cells potentiates Stat5 chromatin association at the locus, perhaps by establishing a more permissive chromatin architecture, thus increasing accessibility for Stat5 binding. Using the TCGA dataset for de novo AML (Cancer Genome Atlas Research Network, 2013), we assessed gene expression changes in non-APL cohesin mutant AML compared with FLT3-ITD, cohesin WT AML patients. Similar to our mouse results, cohesin mutant AML had increased SOCS2 expression (Log2 fold change 2.2, false discovery rate < 3.64 × 10−6).
Gene discovery studies have identified somatic, loss of function mutations in most major subunits of the cohesin complex, including STAG2, SMC1A, SMC3, and RAD21, in patients with myeloid malignancies such as MDS and AML (Jan et al., 2012; Welch et al., 2012; Cancer Genome Atlas Research Network, 2013; Kon et al., 2013; Thol et al., 2014; Thota et al., 2014). These mutations are almost always monoallelic and mutually exclusive, suggesting that proper cohesin function in HSCs requires a specific dosage, which is altered in myeloid transformation. Here we demonstrate a dose-dependent requirement for Smc3 in hematopoiesis and in stem cell function. Whereas biallelic loss of Smc3 abrogates HSC function, heterozygous loss of Smc3 increases HSC function and alters the balance toward self-renewal at the expense of lineage commitment. These data underscore the critical role for cohesin in regulating cell specification during pivotal developmental transitions in hematopoietic cells. In addition, our data are consistent with the clinical observation that cohesin mutations are not associated with chromosomal instability, but rather alter the chromatin state and transcriptional output of hematopoietic cells.
Smc3 haploinsufficiency results in an expansion of stem/progenitor cells in the BM coupled with dysmegakaryopoiesis, which are seen in chronic myeloid malignancies such as MDS and in preleukemic clonal hematopoiesis (Welch et al., 2012; Jaiswal et al., 2014). Moreover, Smc3 haploinsufficient cells showed a capacity for serial replating and competitive advantage in BM transplant assays, consistent with an increase in self-renewal. Consistent with these data, we observed marked alterations in the transcriptome of Smc3 haploinsufficient stem/progenitor cells. The majority of differentially expressed genes were down-regulated, including marked reduction in the expression of genes involved in lineage priming such as Pou2af1 and Gfi1b. The specific loss of lineage factors, normally expressed at constitutive levels in early stem/progenitor cells, likely constrains the capability of these cells to commit to downstream lineages.
The majority of genes that showed altered expression in Smc3 haploinsufficient cells show decreased RNA expression, in concordance with the notion that the cohesin complex plays an important role in maintaining active transcription complexes. Importantly, the genes that were not altered with cohesin attenuation shared chromatin characteristics that suggest locally complex transcriptional regulatory domains are relatively resistant to reduced Smc3 expression. This is in contrast to other recent studies where near-complete disruption of cohesin subunit expression more symmetrically alters gene expression, including increased expression of many target genes (Ing-Simmons et al., 2015). Our data suggest that this level of Smc3 expression is sufficient to maintain most transcription regulatory elements to a degree required for HSC proliferation and self-renewal, while reducing the expression of genes required for lineage determination and differentiation. The genes that maintain stem cell identity are often characterized by highly complex regulatory elements with several promoter–enhancer and enhancer–enhancer contacts in the form of so-called “super-enhancers” (Whyte et al., 2013). Although these complex regulatory elements may be sensitive to disruption of cohesin complexes, in normal hematopoiesis cohesin may be titrated at a higher efficiency to these elements by DNA-binding transcription factors like CTCF than more “typical” enhancers at less complexly regulated loci. In contrast, the factors that govern lineage determination may have less complex transcriptional regulation in the stem cell state, which is lost with reduced cohesin expression.
The observation that cohesin mutations can co-occur with tyrosine kinase mutations in AML led us to perform experiments that showed that Smc3 monoallelic deletion cooperates with Flt3-ITD to induce AML in vivo. We have recently shown cooperativity between a chromatin-associated mutant allele (Tet2) and Flt3ITD in inducing AML in a mouse model (Shih et al., 2015). Notably, similar effects on gene expression were observed in our current, cohesin mutant AML model, including loss of Gata1 and Gata2 regulation and up-regulation of Socs2. These data suggest cohesin mutations can confer a chromatin structure permissive to oncogenic signaling from growth factor–independent and somatically altered oncogenes. Mechanistically, combined Smc3 and Flt3 mutations resulted in potentiation of Stat5-mediated transcriptional output. Although Flt3-ITD has been shown to activate Stat5 nuclear localization and transactivation activity (Hayakawa et al., 2000; Mizuki et al., 2000; Zhang et al., 2000), the combination with Smc3 haploinsufficiency increased the expression of Stat5 targets by altering chromatin structures at Stat5-binding motifs. This potentiation may be a result of relaxed chromatin architecture, which allows for increased Stat5 chromatin association in the setting of mutant-driven kinase activation.
Interestingly, we observed a preponderance of disrupted multifocal nucleoli present in Smc3Δ/+ and, to a greater extent, in Smc3Δ/+ Flt3ITD hematopoietic progenitors. This phenotype resembles the nucleolar alterations seen in Roberts syndrome, a known cohesinopathy. There, mutation in the human ESCO2 gene inactivates the SMC3 acetyltransferase required to induce cohesiveness of the cohesin ring complex (Harris et al., 2014). Roberts syndrome nuclei display strikingly similar gross nucleolar architecture, with disrupted ribosomal RNA expression and ribosome biogenesis. Thus, we speculate that leukemogenesis facilitated by reduction in functional cohesin complexes may also arise as a result of disrupted protein synthesis pathways in addition to perturbed gene expression. Most provocatively, our data showing that complete loss of cohesin complex abrogates hematopoiesis suggest a potential therapeutic opportunity in disrupting cohesin function in Smc3 haploinsufficient malignancies. Subsequent studies are required to delineate whether malignancies that feature cohesin haploinsufficiency as a mechanism of transformation are sensitive to further attenuation of cohesin function and whether this can be leveraged to therapeutic benefit.
MATERIALS AND METHODS
Animals.
All animals were housed at Memorial Sloan Kettering Cancer Center. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at Memorial Sloan Kettering Cancer Center.
Generation of Smc3-deficient mice.
The Smc3 allele was deleted by targeting exon 4 in a construct obtained from the EUCOMM consortium (Smc3tm1a(EUCOMM)Wtsi). Two LoxP sites flanking exon 4 and an Frt-flanked neomycin selection cassette were inserted in the upstream intron (Fig. 1 A). Positive embryonic stem clones were expanded and injected into blastocysts. The generated mice (Smc3fl/fl) were initially crossed to a germline Flp-deleter (The Jackson Laboratory), to eliminate the neomycin cassette, and subsequently to the IFN-α–inducible Mx1-cre (The Jackson Laboratory; Kühn et al., 1995; Lakso et al., 1996). Mice were backcrossed for six generations to C57BL/6 mice. Smc3fl/fl, Smc3fl/+, and Smc3+/+ littermate mice were genotyped by PCR with primers Smc3-FloxF (5′-TGTTGCCTCCCTGTGTTCTCAGGC-3′), Smc3-CommonF (5′-TCTTCGTCCAGAGCAGCGATTGGC-3′), and Smc3-CommonR (5′-TGAGCCATCATGTGGATGCT-3′) using the following parameters: 95°C for 4 min, followed by 35 cycles of 95°C for 45 s, 56°C for 45 s, and 72°C for 1 min, and then 72°C for 5 min. The WT allele was detected as a band at 287 bp, whereas the floxed allele was detected as a band of 313 bp. Excision after Cre recombination was confirmed by PCR with primers to detect a band at 349 bp.
In vivo experiments.
Mx1-cre Smc3fl/fl conditional and Cre− Smc3fl/fl control mice received four intraperitoneal injections of PIpC every other day at a dose of 20 mg/kg of body weight starting at 2 wk after birth. Mice were analyzed between 1 and 60 wk of age. BM, spleen, and peripheral blood were analyzed by flow cytometry. Formalin-fixed paraffin-embedded tissue sections were stained with hematoxylin and eosin (H&E). Peripheral blood was smeared on a slide and stained using the Wright-Giemsa staining method. Tissue sections and blood smears were evaluated by a hematopathologist (A. Chiu). Deletion of the Smc3 allele and transcript was measured by genomic PCR and Western blot analysis.
BM transplantation.
Freshly dissected femurs and tibias were isolated from Smc3fl/fl CD45.2+ or Mx1-cre+ Smc3fl/fl CD45.2+ mice. BM was flushed with a 3-cc insulin syringe into PBS supplemented with 3% FBS. The BM was spun at 0.5 g by centrifugation at 4°C, and RBCs were lysed in ACK bicarbonate lysis buffer for 5 min. After centrifugation, cells were resuspended in PBS plus 3% FBS, passed through a cell strainer, and counted. Finally, 0.5 × 106 total BM cells from Smc3fl/fl CD45.2+ or Mx1-cre+ Smc3fl/fl CD45.2+ mice were mixed with 0.5 × 106 WT CD45.1+ support BM and transplanted via tail vein injection into lethally irradiated (two times 450 cGy) CD45.1+ host mice. Chimerism was measured by FACS in peripheral blood at 2 wk after transplant (week 0, pre-PIpC). Chimerism was followed via FACS in the peripheral blood every 4 wk (week 0, 4, 6, 8, 12, 16, and 20 after PIpC injection). Additionally, for each bleeding, whole blood cell counts were measured on a blood analyzer, and peripheral blood smears were scored. Chimerism in the BM, spleen, and thymus was evaluated at 20 wk via animal sacrifice and subsequent FACS analysis. The above procedure was also repeated with Smc3fl/+ CD45.2+, Mx1-cre+ Smc3fl/+ CD45.2+ mice for competitive transplantation of mice with monoallelic loss of Smc3. For noncompetitive serial transplantation experiments, 106 total BM cells of Smc3fl/+ Flt3ITD CD45.2+ mice were injected into lethally irradiated (two times 450 cGy) CD45.1+ host mice. Recipient mice were then followed until moribund.
In vitro colony-forming assays.
BM of Smc3fl/+ and littermate Mx1-cre Smc3Δ/+ mice was seeded at a density of 20,000 cells/replicate into cytokine-supplemented methylcellulose medium (MethoCult M3434; STEMCELL Technologies). Colonies propagated in culture were scored at day 7. Representative colonies were isolated from the plate for cytospins and flow cytometry. Remaining cells were resuspended and counted, and a portion was taken for replating (20,000 cells/replicate) for a total of seven platings. Cytospins were performed by resuspending in warm PBS and spinning onto the slides at 350 g for 5 min. Slides were air-dried and stained using the Giemsa-Wright method.
Antibodies, FACS, and Western blot analysis.
Antibody staining and FACS analysis were performed as previously described (Abdel-Wahab et al., 2013). BM or spleen mononuclear cells were stained with a lineage cocktail comprised of antibodies targeting CD4, CD8, B220, NK1.1, Gr-1, CD11b, Ter119, and IL-7Rα. Cells were also stained with antibodies against c-Kit, Sca-1, FcγRII/III, and CD34. Cell populations were analyzed using a FACS-LSRII (BD) and sorted with a FACSAria instrument (BD). All antibodies were purchased from BD or eBioscience. We used the following antibodies: c-Kit (2B8), Sca-1 (D7), Mac-1/CD11b (M1/70), Gr-1 (RB6-8C5), NK1.1 (PK136), Ter-119, IL7-Rα (A7R34), CD34 (RAM34), FcγRII/III (2.4G2), CD4 (RM4-5), CD4 (H129.19), CD8 (53-6.7), CD45.1 (A20), CD45.2 (104), CD150 (9D1), and CD48 (HM48-1). The following antibodies were used for Western blot analysis: Smc3 (Bethyl Laboratories, Inc.) and Actin (EMD Millipore).
Nucleolar detection analyses.
Mouse BM was washed with PBS, and 1.5 × 106 cells were centrifuged at 400 g for 5 min and then stained with 50 µl of dual detection reagent TOTAL-NUCLEAR-ID (Enzo Life Sciences). Cells were incubated for 30 min at 37°C and then washed with 200 µl of 1× buffer solution. Cells were centrifuged at 400 g for 5 min and resuspended in 200 µl of 1× buffer solution and seeded into an 8-well chamber slide (Lab-Tek). Z-stack images were taken using a TCS SP8 confocal laser-scanning microscope (Leica) equipped with a 63× objective. Images were deconvolved and processed using Imaris 3D software (Bitplane AG).
Histological analyses.
Mice were sacrificed and autopsied, and then dissected tissue samples were fixed for 24 h in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4 µm and stained with H&E. Images were acquired using an Axio Observer A1 microscope (Carl Zeiss).
Peripheral blood analysis.
Blood was collected by retroorbital bleeding using heparinized microhematocrit capillary tubes (Thermo Fisher Scientific). Automated peripheral blood counts were obtained using a ProCyte Dx (IDEXX Laboratories) according to standard manufacturer’s instruction. Differential blood counts were realized on blood smears stained using Wright-Giemsa staining and visualized using an Axio Observer A1 microscope.
Cytogenetic analysis and metaphase karyotyping.
BM aspirates after documented excision or day 5 after PIpC (for Smc3Δ/Δ) were injected into T25 flasks containing 5 ml RPMI 10% FCS supplemented with 2 mM l-glutamine. Harvested cells were cultured with 25 µl colcemid (10 mg/ml; Gibco) for 45 min and 4 h, respectively, resuspended in 0.075 mol/l KCl for 10 min at 37°C, and fixed in methanol/acetic acid (3:1). Metaphases were scored and counted. Chromosome analysis was performed on a minimum of 20 DAPI-banded metaphases, and all metaphases were fully karyotyped.
RNA-seq and quantitative real-time PCR (qRT-PCR) analysis.
For qRT-PCR experiments, all samples were prepared in biological triplicate. Whole BM was positively selected for Cd117 (c-Kit) using antibodies conjugated to magnetic beads and separated over a column (Cd117 microbeads; Miltenyi Biotec). Total RNA was isolated using TRIzol (Invitrogen), and cDNA was synthesized using the SuperScript First-Strand kit (Invitrogen). Quantitative PCR was performed using SYBR green iMaster and a LightCycler 480 (Roche). For mRNA-seq analysis, samples were prepared and analyzed in biological duplicate (r2 = 0.9724, Smc3fl/+ sample 1 vs. 2; 0.9809, Smc3Δ/+ sample 1 vs. 2; 0.9760, Flt3ITD sample 1 vs. 2; 0.9401, Smc3Δ/+ Flt3ITD sample 1 vs. 2). We used ERCC RNA Spike-In Mix (Life Technologies) for cell number normalization. Fastq files were aligned to mm9 using HiStat with default parameters. Transcript abundances were calculated using the cuffquant module of Cufflinks. FPKM values were calculated and normalized using Cuffnorm. We considered genes that had a P < 0.05 and log2 fold change value ≤0.58 to be significantly down-regulated between genotypes. To determine “unchanged” genes between genotypes, we considered genes with <25% variance between replicates and log2 fold changes <0.2. GSEA was performed using default parameters with the following exceptions: -nperm 1000, -rnd_seed 149, -set_max 1500, -norm meandiv. RNA-seq data are deposited in NCBI’s GEO database, accessible through accession no. GSE73218.
ATAC-seq.
Chromatin accessibility assays using the bacterial Tn5 transposase were performed as described previously (Buenrostro et al., 2013) with minor modifications. For ATAC-seq analysis, samples were prepared and analyzed in biological duplicate. 50 × 103 cells were lysed and incubated with transposition reaction mix for 60 min at 37°C. Samples were PCR amplified and sequenced on a NextSeq 500 (Ilumina). Fastq files were aligned to mm9 with Bowtie, and peaks were called using Zinba. ATAC peak density near unchanged versus down-regulated gene TSSs was determined by extending the TSS region by 100 kb in the 5′ and 3′ direction. The 200-kb window was binned into 1-kb regions, and overlapping ATAC peaks in each window were counted. Regions were further subdivided into regions overlapping enhancers or promoters. The mean ATAC peak density in each window across each gene set was plotted as a function of distance from the TSS. A LOESS fit line was added to capture overall trends. Density plots were determined by binning 3- or 5-kb windows centered at ATAC peaks into 200 50-bp bins. Signal density at each region was calculated, and the mean density in that region was plotted in units of reads per million per base pair (rpm/bp). Transcription factor motif analysis was performed using the FIMO database of the MEME suite. Transcription factor motif densities were calculated for pairwise comparisons of unique ATAC peaks for motifs with annotated position weight matrices (PWMs; Matys et al., 2006). To generate expected motif background frequencies, ATAC-enriched regions were, respectively, scrambled to maintain dinucleotide frequencies akin to the actual genomic sequences. Motifs were considered present in these regions if they had >1.5-fold change between observed frequency ratios in unique ATAC-enriched regions and a p-value <0.05 for each pairwise comparison. The statistical significance of differences of motif occurrences between genotypes was assessed using Fisher’s exact test. ATAC-seq data are deposited in NCBI’s GEO database, accessible through accession no. GSE73218.
Online supplemental material.
Video 1 shows Smc3fl/+ BM nucleolar 3D reconstruction. Video 2 shows Smc3Δ/+ Flt3-ITD BM nucleolar 3D reconstruction. Table S1, included as a separate Excel file, shows the Smc3Δ/+ stable and unstable gene list. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151317/DC1.
Supplementary Material
Acknowledgments
We would like to thank Dr. Katia Manova and the staff of the Molecular Cytology Core Facility for their assistance in imaging and processing of microscopy data and the Memorial Sloan Kettering Center Support Grant (P30 CA008748).
This work was supported by a grant from the Pershing Square Sohn Prize for Cancer Research (to R.L. Levine); a Damon Runyon Cancer Research Foundation Postdoctoral Fellowship Award, an American Society of Hematology Research Training Award for Fellows, an NCI Training Grant, Clinical Scholars Biomedical Research Training Program (T32 CA009512), and philanthropic support from the Dana Foundation and Lymphoma Foundation (to A.D. Viny); and a Research Fellow Award from the Leukemia and Lymphoma Society and an NCI Pathway to Independence Award (K99CA190861; to C.J. Ott).
The authors declare no competing financial interests.
Author contributions: Conceptualization, A.D. Viny, C.J. Ott, J.E. Bradner, and R.L. Levine. Methodology, A.D. Viny, C.J. Ott, B. Spitzer, M. Rivas, C. Meydan, J. Reyes, A. Chiu, Y. Romin, V. Boyko, A. Melnick, J.E. Bradner, and R.L. Levine. Investigation, A.D. Viny, C.J. Ott, E. Papalexi, D. Yelin, K. Shank, J.E. Bradner, and R.L. Levine. Writing the original draft, A.D. Viny and C.J. Ott. Writing: review, and editing, J.E. Bradner and R.L. Levine. Funding acquisition, J.E. Bradner and R.L. Levine. Resources, S. Thota, J.P. Maciejewski, A. Melnick, J.E. Bradner, and R.L. Levine. Supervision, J.E. Bradner and R.L. Levine.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- GMP
- granulocyte-macrophage progenitor
- GSEA
- gene set enrichment analysis
- HSC
- hematopoietic stem cell
- HSPC
- hematopoietic stem/progenitor cell
- LT-HSC
- long-term HSC
- MDS
- myelodysplastic syndrome
- MEP
- megakaryocyte-erythrocyte progenitor
- MP
- myeloid progenitor
- MPP
- multipotent progenitor
- PIpC
- polyinosinic:polycytidylic acid
- qRT-PCR
- quantitative real-time PCR
- ST-HSC
- short-term HSC
- THS
- transposase hypersensitive site
- TSS
- transcriptional start site
References
- Abdel-Wahab O., Gao J., Adli M., Dey A., Trimarchi T., Chung Y.R., Kuscu C., Hricik T., Ndiaye-Lobry D., Lafave L.M., et al. 2013. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 210:2641–2659. 10.1084/jem.20131141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbás-Martínez C., Sagrera A., Carrillo-de-Santa-Pau E., Earl J., Márquez M., Vazquez M., Lapi E., Castro-Giner F., Beltran S., Bayés M., et al. 2013. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat. Genet. 45:1464–1469. 10.1038/ng.2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A.C., West A.G., and Felsenfeld G.. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 98:387–396. 10.1016/S0092-8674(00)81967-4 [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., and Greenleaf W.J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10:1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.A., Kouzarides T., and Huntly B.J.. 2012. Targeting epigenetic readers in cancer. N. Engl. J. Med. 367:647–657. 10.1056/NEJMra1112635 [DOI] [PubMed] [Google Scholar]
- Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schuijers J., Lee T.I., Zhao K., and Young R.A.. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 159:374–387. 10.1016/j.cell.2014.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris B., Bose T., Lee K.K., Wang F., Lu S., Ross R.T., Zhang Y., French S.L., Beyer A.L., Slaughter B.D., et al. 2014. Cohesion promotes nucleolar structure and function. Mol. Biol. Cell. 25:337–346. 10.1091/mbc.E13-07-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa F., Towatari M., Kiyoi H., Tanimoto M., Kitamura T., Saito H., and Naoe T.. 2000. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 19:624–631. 10.1038/sj.onc.1203354 [DOI] [PubMed] [Google Scholar]
- Ing-Simmons E., Seitan V.C., Faure A.J., Flicek P., Carroll T., Dekker J., Fisher A.G., Lenhard B., and Merkenschlager M.. 2015. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 25:504–513. 10.1101/gr.184986.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaatinen T., Hemmoranta H., Hautaniemi S., Niemi J., Nicorici D., Laine J., Yli-Harja O., and Partanen J.. 2006. Global gene expression profile of human cord blood–derived CD133+ cells. Stem Cells. 24:631–641. 10.1634/stemcells.2005-0185 [DOI] [PubMed] [Google Scholar]
- Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371:2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M., Snyder T.M., Corces-Zimmerman M.R., Vyas P., Weissman I.L., Quake S.R., and Majeti R.. 2012. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 4:149ra118 10.1126/scitranslmed.3004315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., and Ren B.. 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 503:290–294. 10.1038/nature12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S., et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 467:430–435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Klarmann K.D., and Keller J.R.. 2014. Gfi-1 regulates the erythroid transcription factor network through Id2 repression in murine hematopoietic progenitor cells. Blood. 124:1586–1596. 10.1182/blood-2014-02-556522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Bandukwala H.S., An J., Lamperti E.D., Thompson E.C., Hastie R., Tsangaratou A., Rajewsky K., Koralov S.B., and Rao A.. 2011. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. USA. 108:14566–14571. 10.1073/pnas.1112317108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A., Shih L.Y., Minamino M., Sanada M., Shiraishi Y., Nagata Y., Yoshida K., Okuno Y., Bando M., Nakato R., et al. 2013. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 45:1232–1237. 10.1038/ng.2731 [DOI] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., and Rajewsky K.. 1995. Inducible gene targeting in mice. Science. 269:1427–1429. 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., and Westphal H.. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA. 93:5860–5865. 10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S., et al. 2014. Chromatin state dynamics during blood formation. Science. 345:943–949. 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo G.S., Ries R.E., Gudgeon C.J., Harrington K.H., Alonzo T.A., Gerbing R.B., Raimondi S.C., Hirsch B.A., Gamis A.S., Meshinchi S., and Walter R.B.. 2014. High expression of suppressor of cytokine signaling-2 predicts poor outcome in pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. Leuk. Lymphoma. 55:2817–2821. 10.3109/10428194.2014.893305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Tothova Z., Levine R.L., Anderson K., Buza-Vidas N., Cullen D.E., McDowell E.P., Adelsperger J., Fröhling S., Huntly B.J., et al. 2007. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell. 12:367–380. 10.1016/j.ccr.2007.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., and Young R.A.. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153:320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., et al. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34:D108–D110. 10.1093/nar/gkj143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., and Odom D.T.. 2013. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 152:1285–1297. 10.1016/j.cell.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Mizuki M., Fenski R., Halfter H., Matsumura I., Schmidt R., Müller C., Grüning W., Kratz-Albers K., Serve S., Steur C., et al. 2000. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 96:3907–3914. [PubMed] [Google Scholar]
- Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T., et al. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144:296–309. 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 132:422–433. 10.1016/j.cell.2008.01.011 [DOI] [PubMed] [Google Scholar]
- Rubio E.D., Reiss D.J., Welcsh P.L., Disteche C.M., Filippova G.N., Baliga N.S., Aebersold R., Ranish J.A., and Krumm A.. 2008. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. USA. 105:8309–8314. 10.1073/pnas.0801273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf C.A., Kwak H., Koenig A., Misulovin Z., Gohara D.W., Watson A., Zhou Y., Lis J.T., and Dorsett D.. 2013. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 9:e1003382 10.1371/journal.pgen.1003382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A.H., Jiang Y., Meydan C., Shank K., Pandey S., Barreyro L., Antony-Debre I., Viale A., Socci N., Sun Y., et al. 2015. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 27:502–515. 10.1016/j.ccell.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon D.A., Kim T., Diaz-Martinez L.A., Fair J., Elkahloun A.G., Harris B.T., Toretsky J.A., Rosenberg S.A., Shukla N., Ladanyi M., et al. 2011. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 333:1039–1043. 10.1126/science.1203619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W., Kang H., Lin S., Kissil J.L., Bartolomei M.S., and Lieberman P.M.. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27:654–666. 10.1038/emboj.2008.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F., Bollin R., Gehlhaar M., Walter C., Dugas M., Suchanek K.J., Kirchner A., Huang L., Chaturvedi A., Wichmann M., et al. 2014. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 123:914–920. 10.1182/blood-2013-07-518746 [DOI] [PubMed] [Google Scholar]
- Thota S., Viny A.D., Makishima H., Spitzer B., Radivoyevitch T., Przychodzen B., Sekeres M.A., Levine R.L., and Maciejewski J.P.. 2014. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 124:1790–1798. 10.1182/blood-2014-04-567057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J.S., Ley T.J., Link D.C., Miller C.A., Larson D.E., Koboldt D.C., Wartman L.D., Lamprecht T.L., Liu F., Xia J., et al. 2012. The origin and evolution of mutations in acute myeloid leukemia. Cell. 150:264–278. 10.1016/j.cell.2012.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 451:796–801. 10.1038/nature06634 [DOI] [PubMed] [Google Scholar]
- Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., and Young R.A.. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153:307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga A.T., Vellenga E., and Schuringa J.J.. 2008. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol. Cell. Biol. 28:6668–6680. 10.1128/MCB.01025-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Fukuda S., Lee Y., Hangoc G., Cooper S., Spolski R., Leonard W.J., and Broxmeyer H.E.. 2000. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J. Exp. Med. 192:719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.