Hu et al. show that the chemokine receptor CCR4 is involved in thymocyte medullary entry, interactions with dendritic cells, and negative selection. In the absence of CCR4, central tolerance is not established, promoting autoimmunity.
Abstract
Autoimmunity results from a breakdown in central or peripheral tolerance. To establish central tolerance, developing T cells must enter the thymic medulla, where they scan antigen-presenting cells (APCs) displaying a diverse array of autoantigens. If a thymocyte is activated by a self-antigen, the cell undergoes either deletion or diversion into the regulatory T cell (T reg) lineage, thus maintaining self-tolerance. Mechanisms promoting thymocyte medullary entry and interactions with APCs are incompletely understood. CCR4 is poised to contribute to central tolerance due to its expression by post-positive selection thymocytes, and expression of its ligands by medullary thymic dendritic cells (DCs). Here, we use two-photon time-lapse microscopy to demonstrate that CCR4 promotes medullary entry of the earliest post-positive selection thymocytes, as well as efficient interactions between medullary thymocytes and DCs. In keeping with the contribution of thymic DCs to central tolerance, CCR4 is involved in regulating negative selection of polyclonal and T cell receptor (TCR) transgenic thymocytes. In the absence of CCR4, autoreactive T cells accumulate in secondary lymphoid organs and autoimmunity ensues. These studies reveal a previously unappreciated role for CCR4 in the establishment of central tolerance.
As T cells develop, they migrate within distinct thymic microenvironments, where they interact with stromal cells that provide signals critical for thymocyte survival, proliferation, differentiation, and selection (Love and Bhandoola, 2011; Hu et al., 2015). Immature thymocytes are restricted to the thymic cortex, where they interact primarily with cortical thymic epithelial cells (cTECs) that provide differentiation and survival cues (Shah and Zúñiga-Pflücker, 2014). More mature CD4+CD8+ double-positive (DP) thymocytes rely on signaling through TCRαβ antigen receptors for further differentiation. Failure to signal through the TCR at this stage results in cell death, whereas moderate signaling allows cells to pass the positive selection checkpoint, resulting in survival and differentiation to the CD4+ single-positive (CD4SP) or CD8+ single-positive (CD8SP) lineages (Klein et al., 2014). These post-positive selection thymocytes migrate into the thymic medulla to undergo maturation and selection before egress as naive T cells to secondary lymphoid organs (Takahama, 2006; Ehrlich et al., 2009; Love and Bhandoola, 2011; Ross et al., 2014).
The thymic medulla is a specialized microenvironment for the establishment of T cell tolerance. Diverse tissue-restricted antigens (TRAs), proteins that are otherwise expressed only in peripheral tissues, are displayed by medullary APCs to delete or tolerize autoreactive thymocytes (Klein et al., 2014). Two main classes of medullary APCs have been implicated in TRA presentation: MHCIIhiCD80hi medullary thymic epithelial cells (mTEChi) and DCs. mTEChi cells express a wide range of TRAs due to expression of the chromatin modulator AIRE, which promotes transcription at epigenetically silenced loci (Anderson et al., 2002; Metzger and Anderson, 2011; Sansom et al., 2014; Brennecke et al., 2015; Meredith et al., 2015). mTEChi cells can directly present TRAs to thymocytes to induce negative selection (i.e., apoptosis) or T reg cell differentiation (Aschenbrenner et al., 2007; Hinterberger et al., 2010; Klein et al., 2014). In addition, thymic DCs can acquire TRAs from mTEChi cells for presentation to thymocytes (Koble and Kyewski, 2009). DCs also acquire autoantigens from blood or peripheral tissues to tolerize thymocytes to these autoantigens (Bonasio et al., 2006; Baba et al., 2009; Atibalentja et al., 2011). A recent report confirms that both mTEChi cells and DCs contribute to thymocyte negative selection and T reg cell generation, while demonstrating that Sirpα− DCs are mainly responsible for presentation of TRAs acquired from mTEChi cells (Perry et al., 2014). Thus, to circumvent autoimmunity, thymocytes are required to interact efficiently with multiple classes of medullary APCs (Anderson et al., 2002; Bonasio et al., 2006; Proietto et al., 2008; Hinterberger et al., 2010).
SP thymocytes must migrate into the medulla to encounter APCs that induce central tolerance. Chemokine receptors have been widely implicated in promoting migration and localization of lymphocytes in primary and secondary lymphoid (Petrie and Zúñiga-Pflücker, 2007; Love and Bhandoola, 2011; Zlotnik and Yoshie, 2012; Hu et al., 2015). The chemokine receptor CCR7, which is up-regulated following positive selection, governs chemotaxis of SP thymocytes toward the medulla and accumulation therein (Ueno et al., 2004; Ehrlich et al., 2009). CCR7 deficiency impairs SP medullary entry, leading to defective negative selection against TRAs and ensuing autoimmune disease (Kurobe et al., 2006; Nitta et al., 2009). Our previous studies demonstrated that other G protein-coupled receptors (GPCRs) must also contribute to medullary entry, and thus likely to the induction of central tolerance (Ehrlich et al., 2009).
The chemokine receptor CCR4 is a candidate GPCR that could contribute to medullary entry and central tolerance. In the periphery, CCR4 is predominantly expressed by Th2 cells, T reg cells, and skin-homing T cells. CCR4 has been implicated in Th2-mediated allergic disorders, such as asthma and atopic dermatitis, and in mature T cell malignancies (Yoshie and Matsushima, 2014). CCR4 also promotes interactions of activated T cells with DCs in secondary lymphoid tissues (Tang and Cyster, 1999). In the thymus, CCR4 is expressed on post-positive selection thymocytes (Campbell et al., 1999), and its ligands CCL17 and CCL22 are expressed by myeloid cells in the medulla (Bleul and Boehm, 2000). CCR4 induces chemotaxis of CD4SP thymocytes in vitro (Campbell et al., 1999; Bleul and Boehm, 2000). Collectively, these studies implicate a role for CCR4 in thymocyte medullary entry and tolerance induction. However, the contribution of CCR4 to these processes has not been confirmed.
In this study, we examined the contribution of CCR4 to the induction of thymocyte central tolerance. We demonstrate that post-positive selection DP and CD4SP thymocytes express CCR4, while CCR4 ligands are expressed predominantly by Sirpα+ DCs in the thymus, suggesting CCR4 could promote thymocyte medullary entry and interactions with DCs. Indeed, CCR4 deficiency impairs medullary accumulation of post-positive selection thymocytes, and diminishes the efficiency of interactions with medullary DCs. Polyclonal and monoclonal CCR4-deficient thymocytes are impaired in their ability to undergo negative selection to endogenous autoantigens. We find that CCR4 is required for efficient negative selection of TCR transgenic thymocytes responding to low concentrations of cognate antigen, and CCR4 shifts the balance of clonal deletion versus T reg cell generation toward negative selection. Consistent with a defect in central tolerance, CCR4-deficient mice have an increase in autoreactive naive CD4 T cells in secondary lymphoid organs, and they develop lymphocytic infiltrates in lacrimal glands and anti-nuclear autoantibodies, resembling the late onset autoimmune disorder seen in patients with Sjögren’s syndrome (Yamamoto, 2003). Collectively, our findings demonstrate a critical role for CCR4 in the induction of T cell central tolerance.
RESULTS
Expression patterns and chemotactic activity of CCR4 and its ligands are consistent with a role in thymocyte chemotaxis toward the medulla
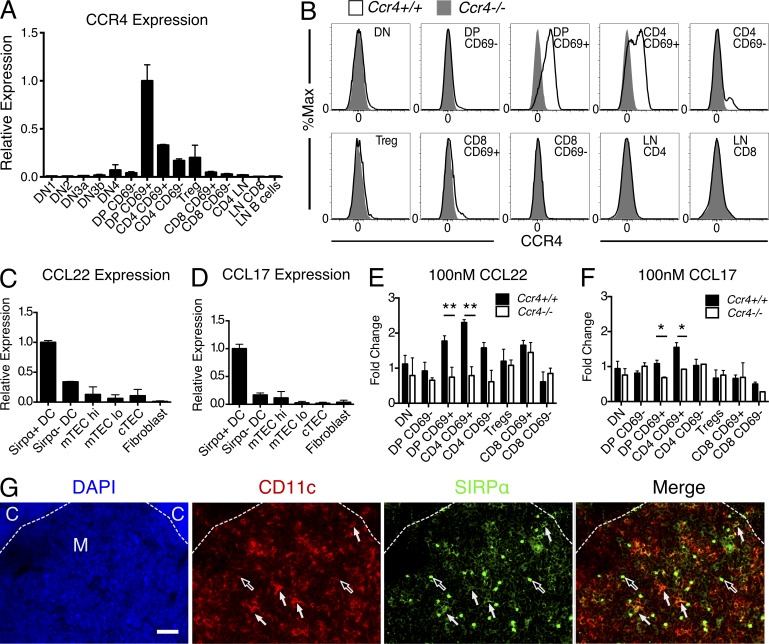
We recently developed a gene expression database for thymocyte and stromal cell subsets, enabling us to identify receptor–ligand pairs that could contribute to thymocyte–stromal cell crosstalk (Ki et al., 2014). By searching for chemokine receptors that might be involved in medullary entry, we found that Ccr4 was expressed by post-positive selection DP and CD4SP thymocytes, whereas its ligands were expressed by thymic DCs (not depicted). RT-PCR and flow cytometric analyses confirmed CCR4 expression in DP CD69+ and CD4SP CD69+ thymocytes, with subsequent down-regulation at the mature CD4SP CD69− stage (Fig. 1, A and B), consistent with previous studies (Cowan et al., 2014). RT-PCR analyses of FACS-purified thymic stromal subsets revealed that the CCR4 ligands CCL17 and CCL22 were expressed predominantly by Sirpα+ DCs (Fig. 1, C and D). Consistent with the aforementioned expression patterns, DP CD69+ and CD4SP CD69+ thymocytes underwent chemotaxis toward CCL22 and CCL17 in a CCR4-dependent manner (Fig. 1, E and F). CCL17 was less potent than CCL22, likely due to its lower affinity for CCR4 (Imai et al., 1998). Previous studies demonstrated that CCL17 and CCL22 are expressed mainly in the thymic medulla (Bleul and Boehm, 2000; Alferink et al., 2003). However, Sirpα+ DCs, which express the highest levels of CCR4 ligands, have been identified in perivascular regions in the thymic cortex (Baba et al., 2009). Immunostaining of thymic cryosections for CD11c and Sirpα+ revealed abundant Sirpα+ DCs in medullary regions (Fig. 1 G), reconciling the medullary localization of CCL17 and CCL22 messages with expression by Sirpα+ DCs. Collectively, these results suggest that CCR4 could promote medullary entry of post-positive selection thymocytes and/or interactions with DCs.
Figure 1.
CCR4 is expressed on post-positive selection thymocytes, whereas CCR4 ligands are expressed by thymic DCs. (A) Ccr4 mRNA expression levels in FACS-purified thymocyte and lymphocyte subsets, normalized to expression in DP CD69+ cells, as determined by qRT-PCR. Means + SEM for one of three representative experiments with three technical replicates each. (B) Flow cytometric profiles of cell surface CCR4 levels on thymocytes and lymph node T cell subsets. Representative of three mice per group. (C and D) mRNA expression levels of the CCR4 ligands Ccl22 and Ccl17 in FACS-purified thymic stromal subsets, normalized to expression in Sirpα+ DCs, as determined by qRT-PCR. Means + SEM for one of two representative experiments with two technical replicates each. (E and F) Fold change in the number of cells of the indicated subsets migrating toward 100 nM CCL22 (E) or 100 nM CCL17 (F) relative to media alone in Transwell chemotaxis assays. Means + SEM for one of three representative experiments with three technical replicates each. (G) Immunostaining of CD11c (red), Sirpα (green), and DAPI (blue) in thymic cryosections from C57Bl6/J mice. The boundary between cortex (C) and medulla (M) is indicated with a dashed line. White arrows indicate Sirpα+ DCs; black arrows indicate globular Sirpα+ CD11c− cells, presumably macrophages. Bar, 50 µm. Representative of two mice analyzed in two separate experiments. *, P < 0.05; **, P < 0.01 (unpaired Student’s t test).
CCR4 deficiency does not grossly impair thymic architecture or T cell development
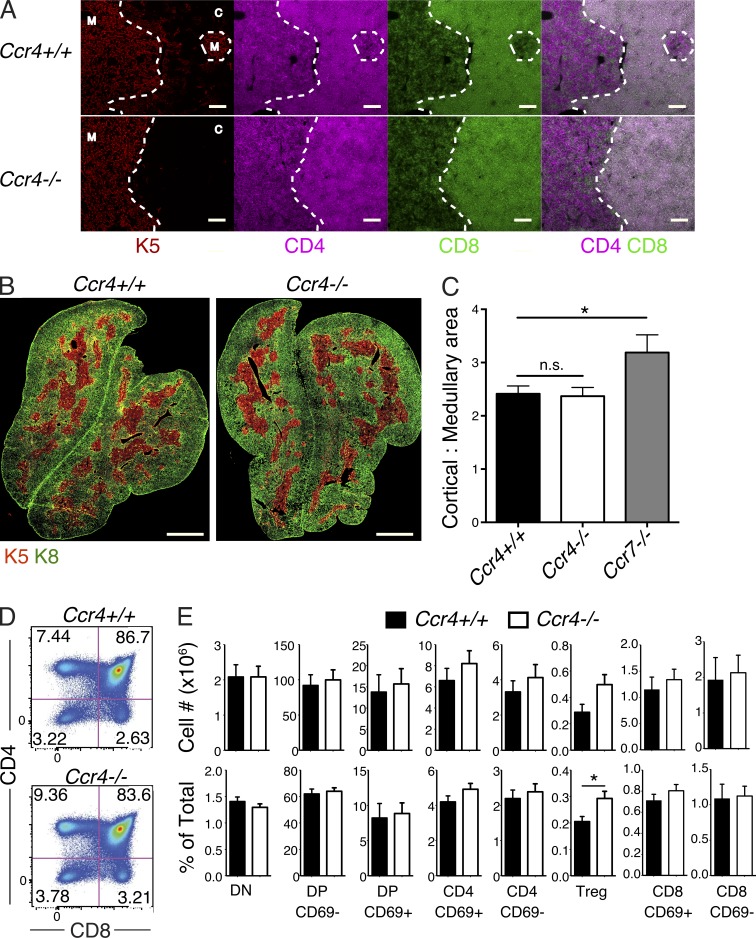
To determine whether CCR4 impacts thymocyte differentiation and selection, we first analyzed the steady-state organization of CCR4-deficient (Ccr4−/−) thymi. CCR7 deficiency impairs the ability of SP thymocytes to accumulate in the medulla (Ueno et al., 2004; Ehrlich et al., 2009). However, localization of Ccr4−/− DP and SP thymocytes to the cortex and medulla, respectively, was not grossly impaired (Fig. 2 A). Upon medullary entry, CD4SP cells induce mTEC proliferation and maturation (Nitta et al., 2011). Thus, in Ccr7−/− thymi, mTEC maturation is impaired, resulting in small medullary regions, and an increase in the total area of the cortex versus medulla (Ueno et al., 2004; Fig. 2 C). Cortical and medullary organization was grossly normal in Ccr4−/− thymi (Fig. 2, B and C). Thus, thymic architecture and thymocyte localization are largely unimpaired in the absence of CCR4.
Figure 2.
Thymic architecture and thymocyte cellularity are largely unaltered in Ccr4−/− mice. (A) Immunostaining of CD4 (purple), CD8 (green), and mTEC-specific keratin 5 (K5; red) in thymic cryosections from Ccr4+/+ versus Ccr4−/− mice. Bar, 100 µm. Representative of two mice in each group. (B) Representative immunostaining of K5 (red) and keratin 8 (K8; green), marking medullary and cortical epithelium, respectively, in Ccr4+/+ and Ccr4−/− thymic cryosections. Bar, 1 mm. (C) The ratio of cortical to medullary areas in Ccr4+/+, Ccr4−/−, and Ccr7−/− thymic cryosections was calculated from images as in B. Mean + SEM are compiled from analysis of 5 Ccr4+/+ mice, 6 Ccr4−/− mice, and 3 Ccr7−/− mice. (D) Representative flow cytometric analysis of CD4 versus CD8 profiles from Ccr4+/+ and Ccr4−/− thymi. (E) The absolute cell numbers (above) and percentages (below) of thymocyte subsets in Ccr4+/+ and Ccr4−/− thymi were calculated from flow cytometric analysis, as in D. Means + SEM compiled from six independent experiments with a total of 11 Ccr4+/+ and 16 Ccr4−/− mice. *, P < 0.05 (unpaired Student’s t test).
We next determined whether CCR4 deficiency impacted thymocyte differentiation. Neither the absolute numbers nor percentages of most thymocyte subsets, including the CCR4-expressing DP CD69+ and CD4SP CD69+ subsets, were significantly altered in Ccr4−/− thymi (Fig. 2, D and E). However, we observed a significant increase in the percentage of T reg cells in Ccr4−/− thymi (Fig. 2 E). We also observed a consistent, though not statistically significant, increase in the number of CD4SP and T reg cells in Ccr4−/− thymi.
CCR4 is required for efficient negative selection of polyclonal thymocytes
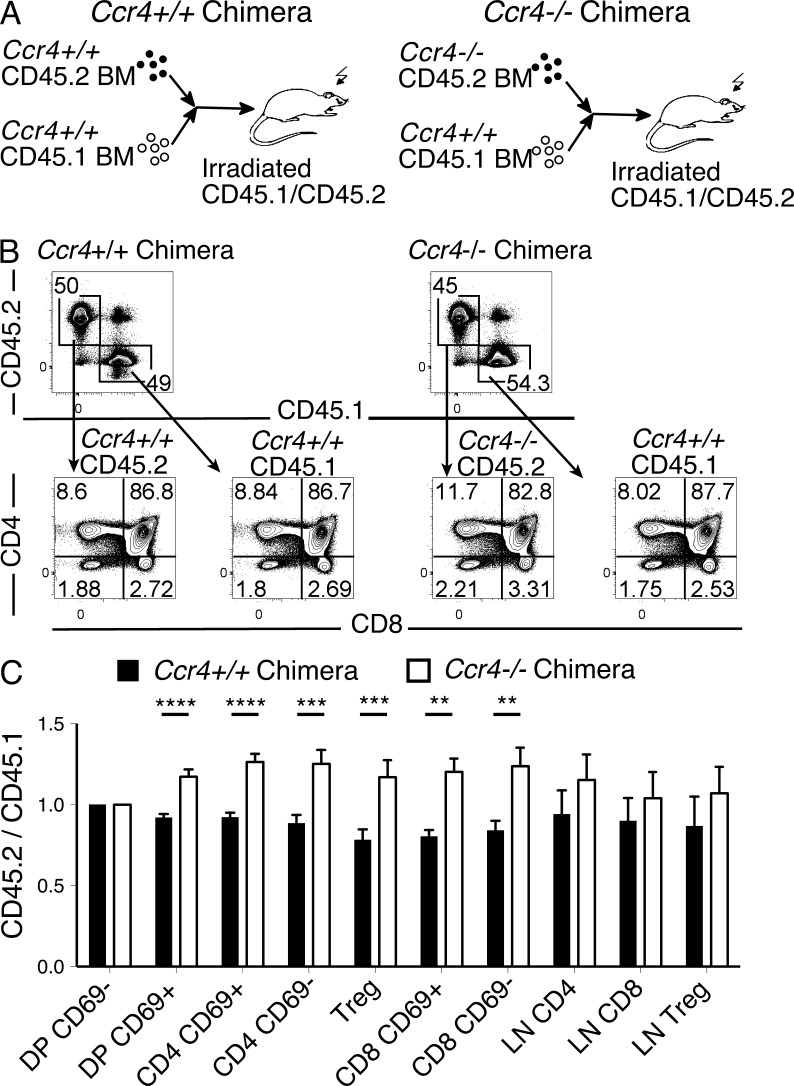
Given the trend toward increased cellularity of CD4SP and T reg cell subsets in Ccr4−/− thymi, we generated mixed bone marrow chimeras to determine if the effects of CCR4 deficiency might be exacerbated in a competitive environment. Lethally irradiated CD45.1/CD45.2 recipients were reconstituted with lineage-depleted bone marrow consisting of Ccr4+/+ CD45.1 cells mixed at a 1:1 ratio with either Ccr4+/+ CD45.2 cells (Ccr4+/+ chimeras) or Ccr4−/− CD45.2 cells (Ccr4−/− chimeras; Fig. 3 A). Thymic chimerism was analyzed 6 wk after transplantation, and the ratio of CD45.2 to CD45.1 cells for each thymocyte subset was quantified and normalized for chimerism to the DP CD69− subset, in which CCR4 is not yet expressed. Ccr4−/− thymocytes were overrepresented starting at the DP CD69+ stage and in all downstream thymocyte subsets (Fig. 3, B and C), consistent with the hypothesis that CCR4 is required for efficient negative selection.
Figure 3.
Competitive bone marrow chimeras indicate CCR4 is required for negative selection. (A) Experimental schematic for the generation of competitive bone marrow chimeras. Thymic and lymph node chimerism was analyzed by flow cytometry 6 wk after transplantation. (B) Representative CD45.1 versus CD45.2 and CD4 versus CD8 flow cytometric plots from the indicated recipient thymi. (C) The ratios of CD45.2/CD45.1 cellularity for each specified thymocyte and lymph node subset were calculated and normalized to the DP CD69− population. Data are compiled from four experiments with a total of 11 mice per group. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (unpaired Student’s t test).
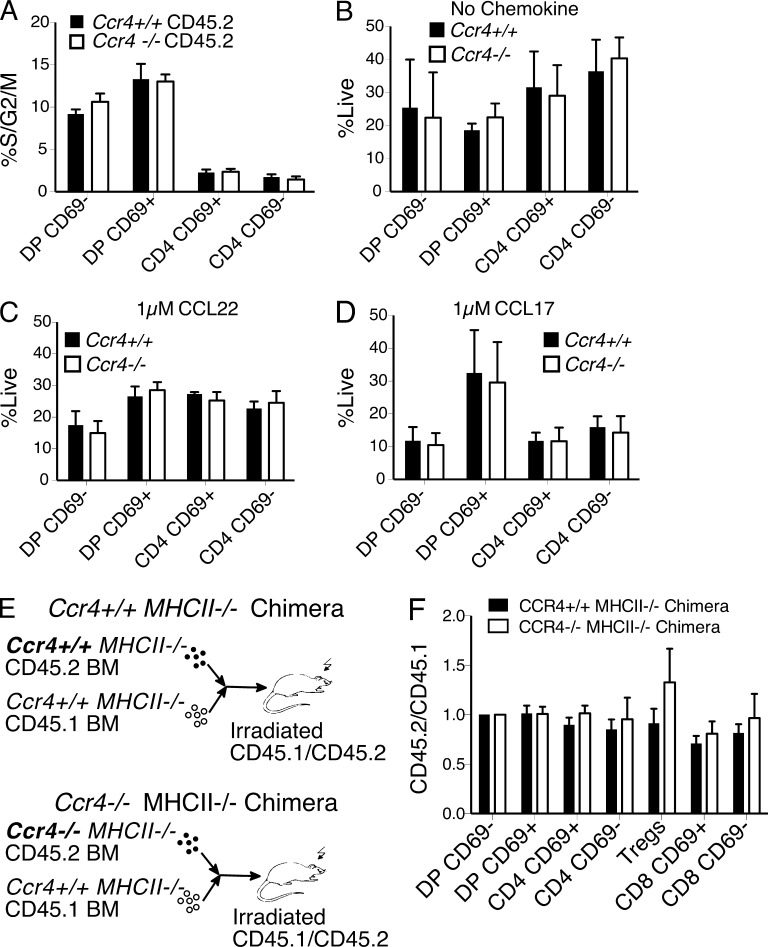
We performed additional experiments to evaluate whether the increased cellularity of Ccr4−/− post-positive selection thymocytes in bone marrow chimeras was a result of impaired negative selection. An alternative explanation is that CCR4 signaling could impede cell cycle progression, such that proliferation of Ccr4−/− thymocyte subsets would be increased, resulting in cellular expansion. To address this possibility, we analyzed the DNA content of Ccr4+/+ versus Ccr4−/− thymocytes in competitive chimeras. CCR4 deficiency did not increase the percentage of cells in S/G2/M at any stage of T cell development (Fig. 4 A), demonstrating that the increase in Ccr4−/− post-positive selection thymocytes was not caused by enhanced proliferation. Alternatively, CCR4 signaling could directly induce cellular apoptosis, such that Ccr4−/− thymocytes would have a survival advantage over WT (C57BL/6J) cells. To test this, we cultured Ccr4+/+ and Ccr4−/− thymocytes in vitro in the presence or absence of CCR4 ligands and quantified viability by flow cytometry. Survival of Ccr4+/+ and Ccr4−/− cells was comparable under all conditions (Fig. 4, B–D). Next, we evaluated mixed bone marrow chimeras in which the hematopoietic compartment was MHC-II–deficient (Fig. 4 E). We reasoned that if increased cellularity of Ccr4−/− thymocytes were caused by a defect in negative selection in response to antigens presented by DCs, which express CCR4 ligands, ablating the capacity of DCs to present antigens would abrogate this increase. As expected, the CD4SP compartment was expanded in all of these chimeras, reflecting the essential contribution of hematopoietic APCs to negative selection (unpublished data; Proietto et al., 2008; Yamano et al., 2015). Notably, Ccr4−/− thymocytes were not overrepresented relative to Ccr4+/+ thymocytes at any stage (Fig. 4 F). Collectively, these data demonstrate that CCR4 is required for efficient negative selection of polyclonal thymocytes to autoantigens presented by hematopoietic APCs.
Figure 4.
Overrepresentation of CCR4-deficient post-positive selection thymocytes in mixed bone marrow chimeras requires antigen presentation by hematopoietic cells and does not reflect cell-intrinsic changes in proliferation or apoptosis. (A) The percentage of thymocytes in cell cycle (S/G2/M) was calculated by flow cytometric analysis of DNA content in the indicated CD45.2+ cell subsets from mixed bone marrow chimeric recipients analyzed in Fig. 3. Means + SEM compiled from three independent experiments with a total of nine mice per group. (B–D) Ccr4+/+ and Ccr4−/− thymocytes were incubated in the presence or absence of 1 µM CCL22 or CCL17, as indicated. The percentage of viable cells relative to the total number of viable cells input for each subset was quantified after 24 h. Mean + SEM from three independent experiments with three technical replicates per experiment. (E) Experimental schematic for the generation of MHCII−/− competitive mixed bone marrow chimeras. Thymic chimerism was analyzed by flow cytometry 6 wk after transplantation. (F) The ratio of CD45.2/CD45.1 cellularity was calculated and normalized for each thymocyte subset as in Fig. 3. Data are compiled from two experiments with a total of six mice per group. No significant differences were observed based on unpaired Student’s t tests in all panels.
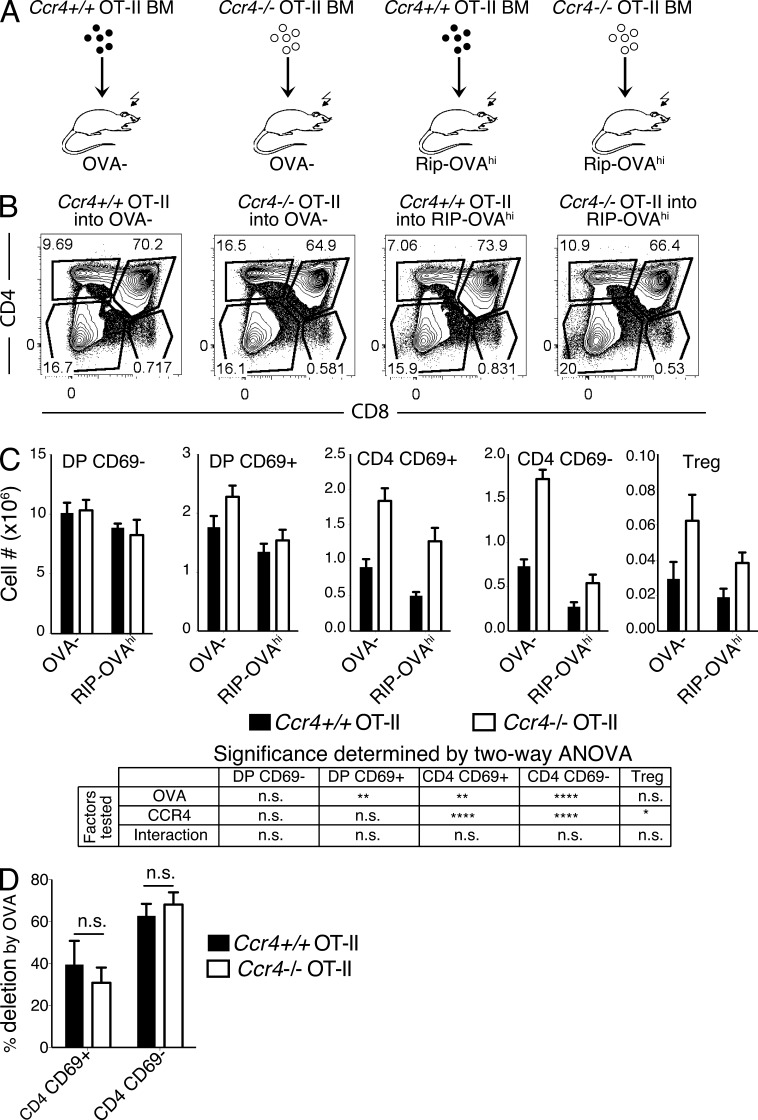
CCR4 deficiency impairs deletion of OT-II thymocytes to an endogenous antigen and alters the TCR repertoire of polyclonal thymocytes
We also evaluated whether CCR4 is required for negative selection of OT-II TCR transgenic thymocytes (Barnden et al., 1998) in response to a model TRA. RIP-OVAhi transgenic mice express a secreted form of OVA in mTECs (Kurts et al., 1998), and negative selection of OT-II thymocytes to this cognate TRA is dependent on AIRE and DCs (Hubert et al., 2011). T cell–depleted bone marrow from Ccr4+/+ OT-II or Ccr4−/− OT-II donors was transferred into lethally irradiated WT (OVA−) or RIP-OVAhi recipients (Fig. 5 A), and thymocyte cellularity was assessed after 6 wk. Negative selection of post-positive selection Ccr4+/+ OT-II thymocytes occurred in response to the RIP-OVAhi TRA, as expected (Fig. 5, B and C). CCR4 deficiency resulted in increased cellularity of OT-II CD4SP cells in RIP-OVAhi recipients, seemingly consistent with a defect in negative selection (Fig. 5 C). Surprisingly, however, CCR4 deficiency also resulted in increased OT-II CD4SP cellularity in OVA− recipients, which do not express the OVA TRA (Fig. 5 C). Moreover, CCR4 deficiency did not significantly alter the percentage of CD4SP OT-II thymocytes deleted in the presence of OVA (Fig. 5 D), indicating that CCR4 is not required for negative selection to this mTEC-expressed TRA. To further evaluate whether CCR4 deficiency and/or the presence of OVA significantly impacted OT-II cellularity at each stage of T cell development, we used a two-way ANOVA (Fig. 5 C). As expected, cellularity of prepositive selection DP CD69− cells was not affected by OVA, CCR4, or the interaction of these two factors. OVA induced significant deletion of DP CD69+ and subsequent CD4SP subsets, indicating that negative selection against this TRA first occurs in post-positive selection DP cells. CCR4 deficiency significantly increased the cellularity of CD4SP cells. However, there was no significant interaction between OVA and CCR4, confirming that CCR4 was not required for negative selection of OT-II CD4SP against the RIP-OVAhi TRA. Similar results were obtained with RIP-mOVA transgenic mice, in which the TRA is a membrane-bound form of OVA (Kurts et al., 1996; Hubert et al., 2011; unpublished data). T reg cellularity was significantly increased in the absence of CCR4 (Fig. 5 C), consistent with the observed increase in polyclonal Ccr4−/− T reg cells (Fig. 2 E and Fig. 3 C).
Figure 5.
CCR4 is required for negative selection of OT-II thymocytes to an endogenous antigen, but not to the model TRA RIP-OVAhi. (A) Experimental schematic for transplantation of Ccr4+/+ or Ccr4−/− OT-II bone marrow into OVA- or RIP-OVAhi recipient mice. Thymi were analyzed by flow cytometry 6 wk after transplantation. (B) Representative CD4 versus CD8 flow cytometry plots from the indicated chimeric mice. (C) Cellularity of indicated thymocyte subsets from recipient mice. Means + SEM from three independent experiments, with nine mice total per group. Two-way ANOVA was used to determine if cellularity at the indicated stages was significantly affected by CCR4, OVA, or the interaction of these two factors. (D) Plots display the percentage of thymocytes deleted by the OVA TRA, calculated as the percent decrease in cellularity between OVA− and RIP-OVAhi recipients for the indicated subsets and genotypes from data in C. No significance was observed based on unpaired Student’s t tests.
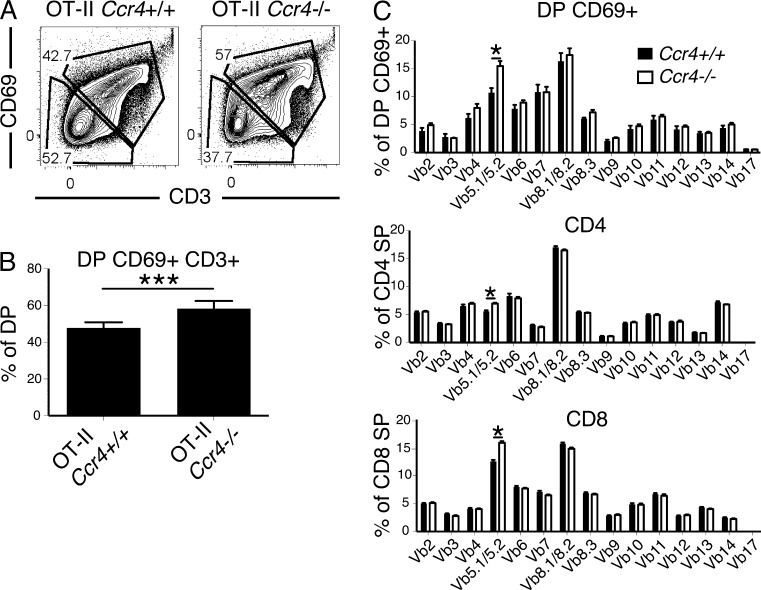
The finding that CCR4 deficiency resulted in increased cellularity of OT-II CD4SP cells in WT recipients suggests that CCR4 is required for clonal deletion of OT-II thymocytes to an endogenous autoantigen. While initially surprising, previous reports indicate that OT-II thymocytes are subject to negative selection against endogenous antigens presented by DCs on the C57BL/6 background (Barnden et al., 1998; Bouillet et al., 2002; Hubert et al., 2011). Comparison of OT-II Ccr4+/+ and OT-II Ccr4−/− thymocytes revealed that the frequency of CD69+ cells within the DP compartment increased in CCR4-deficient mice (Fig. 6, A and B), providing further evidence that CCR4 is required for negative selection of post-positive selection OT-II DP cells to an endogenous autoantigen.
Figure 6.
CCR4 deficiency impairs deletion of OT-II thymocytes to an endogenous antigen and alters the TCR repertoire of polyclonal thymocytes. (A) Representative flow cytometry plots of CD3 versus CD69 within the DP thymocyte compartment of Ccr4+/+ OT-II and Ccr4−/− OT-II mice. (B) Quantification of the percentage of CD69+CD3+ cells within the DP compartment of the indicated strains, calculated from data as in A. Means + SEM compiled from two independent experiments with a total of six mice per group. (C) Comparison of TCRVβ usage in the indicated thymocyte subsets of Ccr4+/+ versus Ccr4−/− mice. Means + SEM compiled from three independent experiments with a total of 10 mice per group. *, P < 0.05; ***, P < 0.001 (unpaired Student’s t test).
Previous studies suggested that because the OT-II TCRβ chain consists of Vβ5.2, it is subject to low-level negative selection against the endogenous retroviral Mtv-9 superantigen, which is likely displayed inefficiently on I-Ab in C57BL/6 mice (Woodland et al., 1990; Barnden et al., 1998). If CCR4 were required for negative selection to superantigens, which are preferentially displayed on DCs (Speiser et al., 1989; Jenkinson et al., 1992), then the TCR Vβ repertoire of polyclonal thymocytes should also be altered in Ccr4−/− mice. Indeed, the frequency of Vβ5.1/Vβ5.2 expressing post-positive selection thymocytes was significantly higher in Ccr4−/− mice (Fig. 6 C), consistent with defective negative selection of Ccr4−/− polyclonal thymocytes to the endogenous Mtv-9 superantigen.
CCR4 deficiency impairs negative selection of OT-II thymocytes to low concentrations of OVA and biases the cell fate decision toward T reg cell generation, away from deletion in response to rare autoantigens
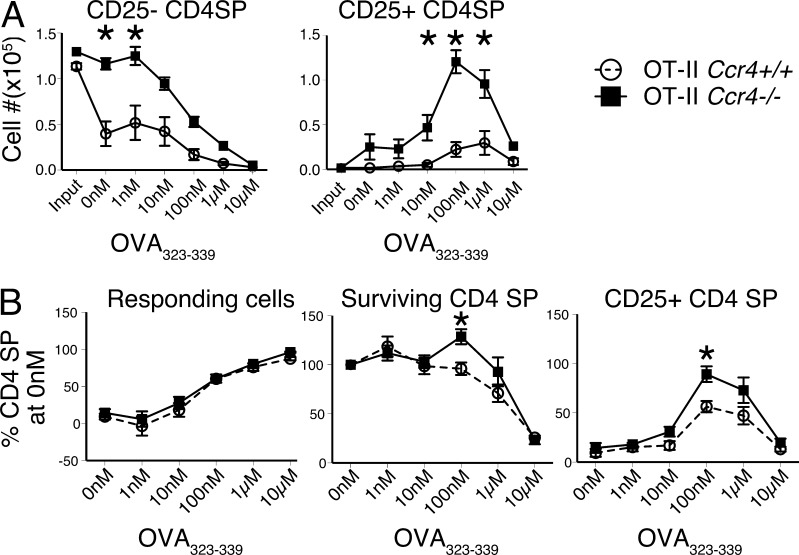
Efficient negative selection of Ccr4−/− OT-II thymocytes in the presence of the RIP-OVAhi TRA could indicate that CCR4 is not required for deletion to antigens expressed by mTECs and/or is not required for deletion against high affinity/abundant autoantigens. To test whether CCR4 modulates the efficiency of negative selection to rare cognate antigens, we introduced TCR transgenic thymocytes along with a range of concentrations of a negatively selecting peptide into live thymic slices, and assayed thymocyte cellularity after 24 h (Dzhagalov et al., 2013). WT thymic slices were pulsed with graded concentrations of the OT-II agonist peptide OVA323-339, and bulk CD45 congenic Ccr4+/+ OT-II and Ccr4−/− OT-II thymocytes were incubated with the slices. After 24 h, cellularity of CD4SP OT-II cells within the slices was compared with the initial input cellularity, normalizing for slice entry. Strikingly, without any peptide addition, Ccr4+/+ OT-II thymocytes underwent deletion to an endogenous antigen in WT thymic slices, while Ccr4−/− OT-II cells did not (Fig. 7 A). Furthermore, high concentrations of OVA323-339 induced equivalent deletion of OT-II CD4SP thymocytes regardless of CCR4 expression (Fig. 7 A). Thus, negative selection of OT-II thymocytes in thymic slices recapitulates two key observations in vivo: (1) CCR4 is required for deletion of OT-II thymocytes to an endogenous autoantigen; and (2) negative selection of OT-II thymocytes to abundant OVA is CCR4 independent. Also, at intermediate OVA323-339 concentrations, Ccr4−/− OT-II thymocytes gave rise to a higher number of CD25+ CD4SP T reg cell progenitors (Fig. 7 A), consistent with the increase in T reg cells in Ccr4−/− mice and chimeras.
Figure 7.
CCR4 is required for efficient negative selection and shifts the cell fate decision to negative selection from T reg cell generation in response to rare autoantigens. (A) Ccr4+/+ OT-II and Ccr4−/− OT-II thymocytes were incubated for 24 h on thymic slices in the presence of the indicated concentration of OVA323-339 peptide, after which the normalized cellularity of the indicated subsets was quantified. The quantity of thymocytes initially loaded onto the slices was calculated as Input. (B) The percentage of OT-II CD4SP thymocytes (left) responding (undergoing negative selection or up-regulating CD25), (middle) surviving, or (right) up-regulating CD25 in response to the indicated concentration of OVA323-339 relative to 0nM peptide was calculated from data in A. Means ± SEM compiled from three independent experiments with three technical replicates per experiment. *, P < 0.05 (unpaired Student’s t test).
In the presence of OVA323-339, a reduction in OT-II cellularity in thymic slices reflects OVA-mediated deletion for both Ccr4+/+ and Ccr4−/− cells, in addition to endogenous antigen-mediated deletion for Ccr4+/+ OT-II thymocytes; thus, the impact of CCR4 deficiency on OVA-mediated negative selection is not revealed by absolute cells numbers. To address whether CCR4 deficiency specifically alters OVA-mediated deletion, we quantified the percentage of CD4SP cells that were either negatively selected or differentiated into T reg cells at each OVA323-339 concentration relative to the cellularity of CD4SP cells of the same CCR4 genotype in slices without peptide (0 nM). For all concentrations of OVA, the percentage of cells that responded to antigen by undergoing either deletion or T reg cell differentiation was equivalent (Fig. 7 B, left). However, a higher percentage of CCR4-deficient OT-II CD4SP cells survived at 100 nM OVA323-339, indicating less clonal deletion had occurred at this low peptide concentration (Fig. 7 B, center). Also at 100 nM OVA323-339, a higher percentage of CD4SP differentiated into CD25+ CD4SP T reg cell precursors (Fig. 7 B, right). Together, these data demonstrate that in the context of rare/low avidity autoantigens, CCR4 is required for efficient negative selection. In lieu of undergoing apoptosis to rare high affinity autoantigens, Ccr4−/− thymocytes divert to a T reg cell lineage. Thus, CCR4 modulates the threshold of autoantigen levels required for efficient negative selection versus diversion to T reg cell fate.
CCR4 is required for medullary accumulation of post-positive selection DP and CD4SP thymocytes
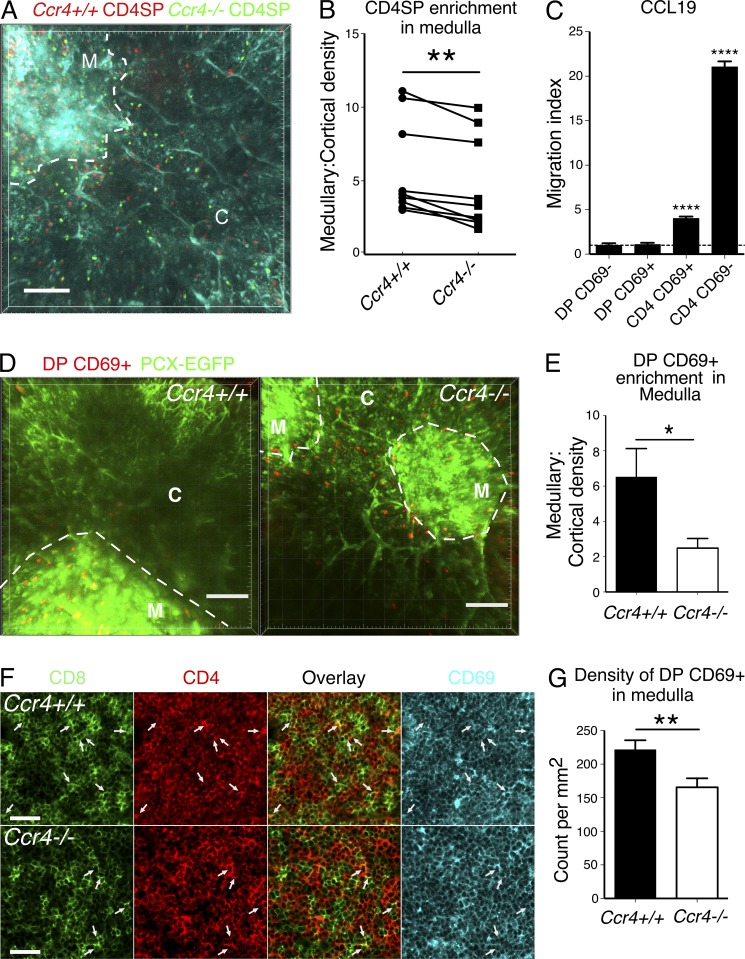
We next sought to identify mechanisms by which CCR4 deficiency impacts negative selection. Because CCR4 ligands are expressed by medullary DCs (Fig. 1, C and D), a gradient of CCR4 ligands emanating from the medulla could promote efficient CD4SP medullary entry and accumulation. To test this, we first isolated Ccr4−/− and Ccr4+/+ CD4SP cells, differentially labeled them with fluorescent dyes, and allowed them to migrate into live thymic slices. Analysis of cellular densities in cortical and medullary regions revealed that Ccr4−/− CD4SP thymocytes accumulated less efficiently in the medulla than Ccr4+/+ cells (Fig. 8, A and B; and Video 1). Swapping dyes did not impact the results. Although statistically significant, the impact of CCR4 deficiency was not as severe as the previously documented impact of CCR7 deficiency on CD4SP medullary accumulation. Ccr7−/− thymocytes distributed almost uniformly across the cortex and medulla (Ehrlich et al., 2009), whereas Ccr4−/− CD4SP cells had a mean medullary: cortical density ratio of 4.6 relative to the WT mean of 5.7.
Figure 8.
CCR4 is required for medullary accumulation of post-positive selection DP and CD4SP thymocytes. (A) A maximum intensity projection of Ccr4+/+ (red) and Ccr4−/− (green) CD4SP thymocytes in live pCX-EGFP (blue) thymic slices. The boundary between cortex (C) and medulla (M) is indicated with a dashed line Video 1 shows corresponding two-photon imaging data. Bar, 100 µm. (B) Densities of Ccr4+/+ and Ccr4−/− CD4SP thymocytes in medulla and cortex (cells/µm3) were quantified from data as in A, and the ratio of medullary to cortical density was plotted for each dataset. Data represent three independent experiments with three time-lapse movies per experiment. **, P < 0.01 (paired Student’s t test). (C) Migration of WT thymocytes toward 100 nM CCL19 was assayed using in vitro Transwell chemotaxis assays. The migration index is the ratio of thymocytes migrating to the bottom chamber in response to CCL19 versus media alone. The horizontal line at 1 indicates no chemotaxis. Means + SEM compiled from three independent experiments, with three technical replicates per experiment. (D) Maximum intensity projections of Ccr4+/+ or Ccr4−/− DP CD69+ thymocytes migrating in pCX-EGFP (green) thymic slices. The boundary between cortex (C) and medulla (M) is outlined with a dashed line. Videos 2 and 3 show corresponding two-photon imaging data. Bar, 100 µm. (E) The ratios of medullary/cortical density for Ccr4+/+ and Ccr4−/− DP CD69+ cells were calculated from data, as in D. Means + SEM from three independent experiments with three time-lapse movies acquired for each group per experiment. (F) Representative confocal images of CD8 (green), CD4 (red), and CD69 (blue) on immunostained cryosections from Ccr4+/+ and Ccr4−/− thymi. White arrows indicate CD4+CD8+ DP thymocytes. Bar, 40 µm. (G) Nine medullary regions from three Ccr4+/+ and three Ccr4−/− thymi immunostained as in F were analyzed to calculate the mean number of DP CD69+ thymocytes per square millimeter. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (unpaired Student’s t test unless otherwise indicated). See also Fig. S1.
Because CCR4 is first expressed at the DP CD69+ stage (Fig. 1, A and B) while CCR7 is not up-regulated until the single positive stage (Suzuki et al., 1999; Cowan et al., 2014), CCR4 might be most important for guiding DP CD69+ cells into the medulla. Consistent with this, DP CD69+ cells migrate toward CCR4 ligands, but not CCR7 ligands in vitro, whereas CD4SP CD69+ cells undergo chemotaxis toward both CCR4 and CCR7 ligands (Fig. 1, E and F; and Fig. 8 C). To directly test the role of CCR4 in promoting medullary entry of post-positive selection DP cells, we FACS sorted DP CD69+ cells from Ccr4+/+ and Ccr4−/− thymi and analyzed their localization after entry into thymic slices. Strikingly, whereas Ccr4+/+ DP CD69+ cells were present mainly in the medulla, accumulation of Ccr4−/− DP CD69+ in the medulla was significantly reduced (Fig. 8, D and E; and Videos 2 and 3). Confocal imaging of immunostained Ccr4+/+ and Ccr4−/− thymic cryosections also revealed that CCR4 deficiency significantly diminished the density of medullary DP CD69+ cells (Fig. 8, F and G). We objectively scored colocalization of CD4 and CD8 signals to identify medullary DP thymocytes, which were all CD69+. Because SP cells are packed at high density in a steady-state medulla, immunostaining likely identifies more false-positive medullary DP cells than live imaging of FACS-purified DP thymocytes, accounting for the smaller fold change observed by immunostaining relative to live imaging. Together, these data demonstrate that post-positive selection DP cells enter the medulla and that CCR4 is essential for their efficient medullary accumulation.
CCR4 is required for efficient interactions between post-positive selection thymocytes and thymic DCs
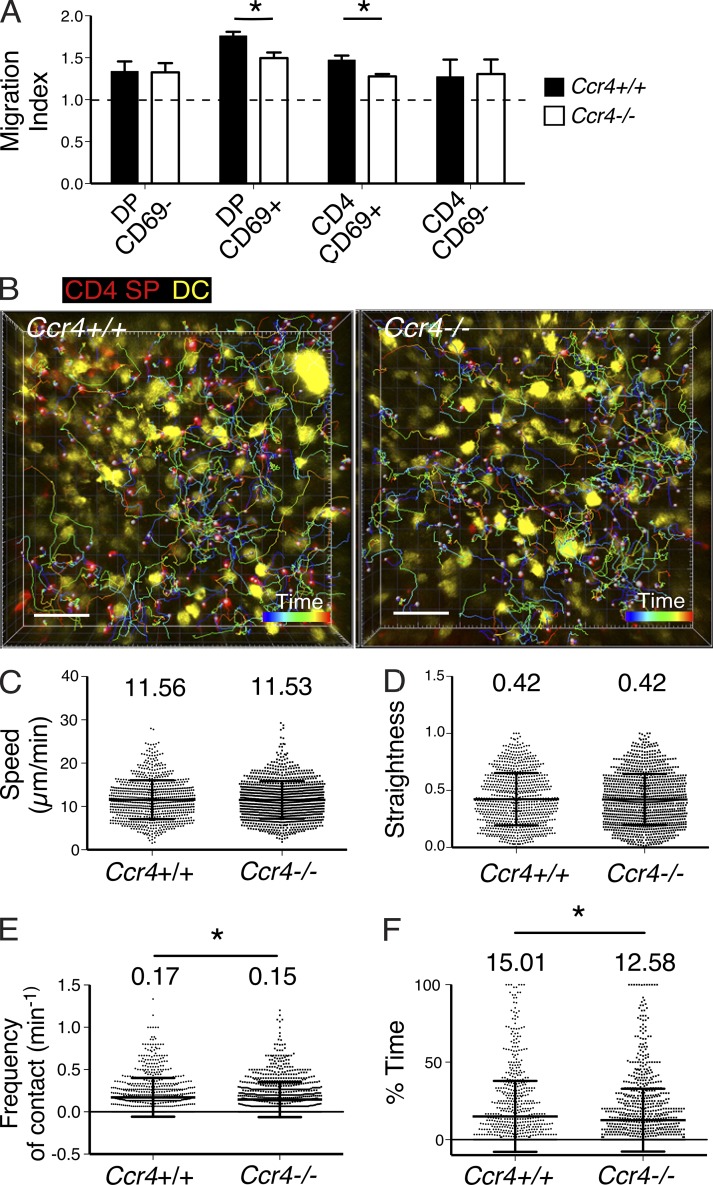
We next addressed the possibility that CCR4 deficiency could reduce the efficiency of interactions between thymocytes and medullary DCs, one of the thymic APCs implicated in negative selection (Klein et al., 2014; Perry et al., 2014). We first tested if CCR4 was required for chemotaxis of thymocytes toward supernatant from thymic DCs in vitro. DP CD69+ and CD4SP CD69+ subsets migrated toward thymic DC supernatant, and CCR4 deficiency significantly reduced chemotaxis of these subsets. CCR4 independent migration of all DP and CD4SP cells was also observed, indicating that other DC-derived factors may impact thymocyte migration (Fig. 9 A). These results demonstrate that CCR4 promotes migration of DP CD69+ and CD4SP CD69+ cells toward ligands produced by thymic DCs.
Figure 9.
CCR4 is required for efficient interactions between post-positive selection thymocytes and thymic DCs. (A) The migration index of Ccr4+/+ and Ccr4−/− thymocytes toward supernatant from thymic DC cultures was assayed using in vitro Transwell chemotaxis assays, as in Fig. 8 C. Means + SEM were compiled from three independent experiments, with three technical replicates per experiment. A line at 1 indicates no chemotaxis. *, P < 0.05 (unpaired Student’s t test). (B) A maximum intensity projection of Ccr4+/+ or Ccr4−/− CD4SP cells (red) migrating among DCs (yellow) in a live CD11c-EYFP thymic slice. Color-encoded time tracks depict the location of individual CD4SP cells throughout the 15-min time course. Bar, 50 µm. Videos 4 and 5 show corresponding two-photon microscopy time-lapse imaging data. Mean cell velocities (C), migratory straightness (D), frequency of thymocyte/DC contacts (E), and percent of time in which CD4SP cells contacted DCs (F) were quantified from two-photon time-lapse imaging data as in B. Data are compiled from five independent experiments, with a total of five mice per group. Each dot represents a single tracked cell, and bar and whiskers indicate means ± SD. Mean values are listed above each plot. (C–F) *, P < 0.05 (Mann-Whitney test). n = 832, Ccr4+/+ thymocytes; n = 1,119, Ccr4−/− thymocytes.
To directly test whether CCR4 promotes efficient interactions between CD4SP cells and medullary DCs in the thymic environment, we used two-photon time-lapse microscopy to image CD4SP thymocytes migrating in CD11c-EYFP live thymic slices (Fig. 9 B and Videos 4 and 5). We found that the velocity and straightness of CD4SP cells in the medulla was not impacted by CCR4 deficiency (Fig. 9, C and D). However, Ccr4−/− CD4SP thymocytes made less frequent contacts with DCs, and spent significantly less of their time in contact with DCs (Fig. 9, E and F), demonstrating that CCR4 promotes efficient interactions between CD4SP thymocytes and medullary DCs, which are likely critical for negative selection to lower avidity autoantigens.
CCR4-deficient mice are prone to autoimmune disease
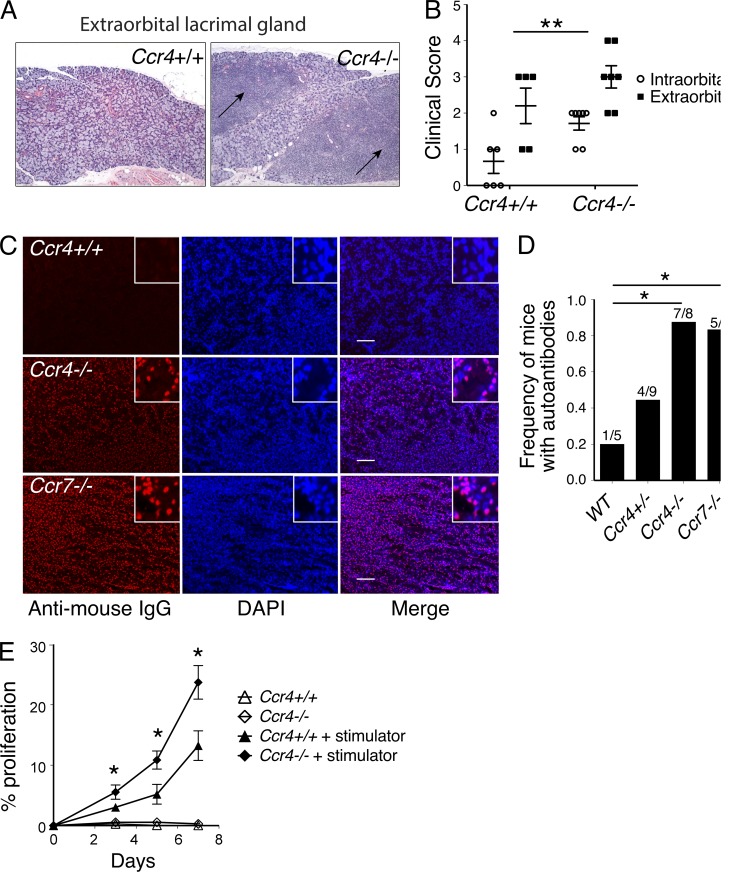
If Ccr4−/− thymocytes do not undergo efficient clonal deletion, autoreactive T cells should accumulate in the periphery, potentially leading to autoimmune disease. Thus, we analyzed Ccr4−/− versus Ccr4+/+ mice at 9–13 mo of age for signs of autoimmunity. CCR4 deficiency resulted in increased severity of lymphocytic infiltrates in lacrimal glands (Fig. 10, A and B). Antinuclear autoantibodies were also detected in the serum of 7 out of 8 Ccr4−/− mice, similar to the frequency of antinuclear antibodies in the autoimmune Ccr7−/− strain (Fig. 10, C and D; Kurobe et al., 2006). Previous studies demonstrated that CCR4 is required for T reg cell trafficking in the periphery (Yuan et al., 2007). Therefore, autoimmunity observed in Ccr4−/− mice could result from impaired T reg cell trafficking or from an increase in autoreactive T cells as a result of defective negative selection. Impaired negative selection would result in an increased frequency of autoreactive naive CD4+ T cells in secondary lymphoid organs, which we tested with syngeneic mixed lymphocyte reactions. Compared with Ccr4+/+ T cells, an increased frequency of Ccr4−/− naive CD4+ T cells underwent proliferation to WT splenic stimulators (Fig. 8 E), indicating that the pool of autoreactive naive CD4+ T cells is expanded in the periphery of CCR4-deficient mice. Together, these data indicate that defective negative selection of CD4SP thymocytes likely contributes to autoimmunity in Ccr4−/− mice.
Figure 10.
Ccr4−/− mice are prone to autoimmune disease. (A) Representative hematoxylin and eosin stains of extraorbital lacrimal gland sections from Ccr4−/− and Ccr4+/+ mice at 8–13 mo of age. Arrows indicate lymphocytic infiltrates. (B) Severity of lymphocytic infiltrates in intraorbital and extraorbital lacrimal glands of 8–13-mo-old Ccr4+/+ or Ccr4−/− mice, with seven mice analyzed per group. Two-way ANOVA was used to determine if CCR4 or the type of lacrimal gland impacted severity; **, P < 0.01. The significance of CCR4 sufficiency is displayed. (C) Representative images of Rag2−/− kidney sections immunostained with serum from 8-13-mo-old WT, Ccr4−/−, or Ccr7−/− mice. Autoantibodies were detected with anti–mouse IgG (red) and nuclei with DAPI (blue). Bars, 100 µm. Digitally magnified regions are shown in insets to facilitate visualization of colocalization of autoantibodies and nuclei. (D) Proportion of aged mice (8–13 mo) containing serum autoantibodies was quantified, as determined by immunostaining as in C. *, P < 0.05 (Fisher’s exact test). (E) Naive splenic T cells were isolated from Ccr4+/+ or Ccr4−/− mice, labeled with CMFDA, and incubated in the presence or absence of irradiated syngeneic splenic stimulator cells. At the indicated time points, the percentage of cells that had undergone at least one cell division was quantified. Mean ± SEM are representative of three experiments with five technical replicates per group. *, P < 0.05 (Student’s t test with Bonferroni correction).
DISCUSSION
Post-positive selection thymocytes migrate into the medulla, where they encounter APCs displaying autoantigens that tolerize autoreactive cells through clonal deletion or T reg cell induction (Klein et al., 2014). The mechanisms that promote efficient medullary entry and interactions with APCs are incompletely understood. Here, we demonstrate that CCR4 induces post-positive selection thymocytes to accumulate in the medulla and to interact with medullary DCs, rendering CCR4 critical to the induction of central tolerance. Ccr4−/− thymocytes do not undergo efficient clonal deletion to low avidity autoantigens, resulting in accumulation of autoreactive naive T cells in secondary lymphoid organs. Ccr4−/− mice display autoimmune symptoms resembling Sjögren’s syndrome, a late-onset human autoimmune disorder characterized by antinuclear antibodies and autoimmunity against lacrimal glands (Yamamoto, 2003). Thus, identification of mechanisms promoting SP medullary entry and interactions with APCs may illuminate novel etiologies of human autoimmune disorders.
Prior analyses of Ccr7−/− mice established that chemokine receptors governing medullary entry promote central tolerance. Because they cannot efficiently accumulate in the medulla (Ueno et al., 2004; Ehrlich et al., 2009), Ccr7−/− thymocytes do not undergo negative selection to TRAs, and autoimmunity ensues (Kurobe et al., 2006; Nitta et al., 2009). We previously determined that other GPCRs must contribute to medullary entry, and thus likely to central tolerance (Ehrlich et al., 2009). Given expression of CCR4 by post-positive selection thymocytes, and its cognate ligands by medullary DCs, CCR4 is poised to contribute to medullary entry and central tolerance induction. Indeed, we find that CCR4 is critical for accumulation of DP CD69+ thymocytes in the medulla and contributes to medullary accumulation of CD4SP cells as well. Because DP CD69+ thymocytes express CCR4, but not CCR7, whereas CD4SP cells express both CCR4 and CCR7 (Cowan et al., 2014), it is not surprising that CCR4 deficiency has a more dramatic impact on medullary localization of DP CD69+ cells. As CD4SP cells mature, up-regulation of CCR7 promotes chemotaxis toward and accumulation within the medulla. Immunostaining reveals a steady-state accumulation of DP cells in the cortex and SP cells in the medulla, so initially we did not expect to observe medullary entry of DP CD69+ cells. However, a recent study comparing the timing of medullary entry after positive selection with down-regulation of CD4 or CD8 also concluded that post-positive selection DP cells enter the medulla (Ross et al., 2014). Together, these findings demonstrate that post-positive selection DP cells efficiently accumulate within the medulla in a CCR4-dependent manner, whereas CCR7 is critical for medullary accumulation of later SP cells.
We find that CCR4 is required for efficient interactions between CD4SP cells and medullary DCs, and our data suggest that such interactions are critical for sampling rare or low-affinity autoantigens for tolerance induction. When OT-II thymocytes encounter abundant or high-affinity antigens, such as the RIP-OVAhi TRA in vivo or high concentrations of OVA peptide in thymic slices, CCR4 is dispensable for negative selection. In these cases, thymocytes likely encounter sufficient peptide/MHC to mediate deletion either by encountering DCs via random migration or by interacting with mTECs, perhaps via CCR7 signaling. mTECs express OVA in the TRA models, and likely present sufficient exogenous peptide for deletion in thymic slices when the peptide is provided at high concentrations (Klein et al., 2001). However, in thymic slices, exogenously added peptides would be most efficiently presented by thymic DCs (Klein et al., 2001), which produce CCR4 ligands, suggesting CCR4 could impact deletion in this model, particularly at low-peptide concentrations. Consistent with this hypothesis, CCR4 deficiency impaired negative selection of OT-II thymocytes to low concentrations of OVA peptide in thymic slices. CCR4 deficiency also impaired negative selection of polyclonal thymocytes and altered their TCR repertoire, suggesting efficient interactions with DCs are also critical for deletion of polyclonal thymocytes to rare or low-affinity antigens. The concept that chemokine guidance is particularly important for encountering rare antigens is supported by computational simulations (Vroomans et al., 2012). In addition, CCR7 can directly promote tethering of peripheral T cells to APCs (Friedman et al., 2006), and integrin-mediated adhesion of thymocytes to APCs can be impacted by chemokine receptor signaling (Ueda et al., 2012; Choi et al., 2014), suggesting a mechanism by which chemokine receptors could enhance the sensitivity of thymocytes to rare antigens. As DCs present low levels of antigens acquired in the periphery or from mTECs, and mTEChi cells express low levels of TRAs for presentation to medullary thymocytes, the ability of thymocytes to efficiently interact with medullary APCs, thus promoting negative selection to rare antigens, is likely critical for many, if not most, natural thymic self-antigens.
CCR4 and CCR7 ligands are preferentially expressed by DCs and mTECs, respectively (Ueno et al., 2002; Ki et al., 2014). This raises the interesting possibility that CCR4 and CCR7 cooperate to promote respective interactions with DCs and mTEChi cells, ensuring thymocytes thoroughly sample multiple medullary APC subsets for tolerance induction. Recent data demonstrate that negative selection by thymic B cells is critical for tolerance induction to B cell-specific antigens (Yamano et al., 2015); similarly, CCR4-mediated negative selection could be particularly critical for tolerance of T cells against the proteome of activated DCs, which they will encounter during priming in secondary lymph nodes. This is consistent with enhanced proliferation of Ccr4−/− T cells in syngeneic MLRs, in which naive T cells respond to autoantigens presented by splenic APCs, including DCs.
Although, we demonstrate that Sirpα+ DCs express the highest levels of CCR4 ligands and are present in the medulla, a previous study found Sirpα+ DCs in perivascular regions in the cortex (Baba et al., 2009). Furthermore, thymocytes undergoing cortical negative selection accumulate near and require DCs (McCaughtry et al., 2008). Thus, in addition to promoting medullary entry of DP CD69+ cells and interactions with DCs in the medulla, CCR4 may also promote interactions with cortical Sirpα+ DCs that contribute to negative selection. Given that CCR4 is up-regulated before CCR7, post-positive selection thymocytes may first become tolerized to autoantigens on DCs before interacting with autoantigens on mTECs.
The increased cellularity of Ccr4−/− versus Ccr4+/+ OT-II CD4SP cells in the absence of OVA in vivo and in WT thymic slices, suggests that endogenous antigens can delete OT-II thymocytes. Previous studies demonstrated that the absence of MHCII on bone-marrow–derived cells resulted in increased OT-II thymocyte cellularity, indicating that thymic DCs present endogenous antigens that delete OT-II cells (Hubert et al., 2011). Furthermore, the endogenous superantigen Mtv-9 can delete Vβ5 containing TCRs, including OT-II TCRs (Woodland et al., 1990; Barnden et al., 1998), in keeping with reduced cellularity of OT-II thymi (Bouillet et al., 2002). In conjunction with the finding that Vβ5 usage increases in polyclonal Ccr4−/− thymocytes, these data are consistent with a model in which CCR4 is critical for thymocyte negative selection against superantigens presented by DCs; other autoantigens expressed by DCs for which CCR4 is required for tolerance induction remain to be identified.
CCR4 and CCR7 both contribute to T cell trafficking in periphery. CCR7 is required for entry of naive T cells into lymph nodes (Förster et al., 2008), whereas CCR4 promotes migration of activated Th2 cells to the skin or lung (Kunkel et al., 2002; Mikhak et al., 2013; Yoshie and Matsushima, 2014). Interestingly, both CCR4- and CCR7-deficient mice are prone to autoimmunity, despite impaired peripheral T cell trafficking. This should be considered when developing CCR4 antagonists as antiinflammatory drugs, since long-term CCR4 inhibition may impair thymic central tolerance and potentially enhance autoimmunity.
Defective negative selection in Ccr4−/− mice was accompanied by increased cellularity of T reg cells, as has been observed in other models, such as Ccr7−/− (Nitta et al., 2009; Cowan et al., 2014), C2TA knockdown (Hinterberger et al., 2010), and TCRζ mutant mice (Hwang et al., 2012). In these models, TCR signaling is reduced, but not abolished, either by disrupting interactions between thymocytes and APCs (Ccr7−/− and Ccr4−/−), down-regulating antigen presentation (C2TA knockdown), or reducing TCR signaling (TCR ζ mutation). Increased T reg cell differentiation with concomitant defects in negative selection is consistent with the avidity model of thymocyte selection, in which thymocytes that receive the highest avidity TCR signals undergo apoptosis, while those receiving more moderate signals can differentiate into T reg cells (Lee et al., 2012; Klein et al., 2014). Thus, by modulating the ability of thymocytes to engage APCs and thus efficiently scan for autoantigens, particularly low avidity antigens, chemokine receptor deficiencies likely skew some thymocytes that would normally undergo deletion toward a T reg cell fate.
Although a recent study did not reveal defects in negative selection in Ccr4−/− mice, an increase in thymic T reg cellularity was reported (Cowan et al., 2014), consistent with our findings. Also, competitive bone marrow chimera data from the previous study revealed a slight, though nonsignificant, increase in Ccr4−/− CD4SP cellularity. The discrepancy between the studies may result from differences in analysis: our chimera data were normalized for chimerism at the DP CD69− stage, whereas the previous findings were not normalized. Interestingly, the previous study did reveal increased numbers of SP thymocytes in the medullas of Ccr4−/− versus Ccr4+/+ thymi, consistent with defective negative selection in the Ccr4−/− group.
There is still much to be learned about the mechanisms by which SP thymocytes enter the medulla and encounter APCs therein to induce central tolerance. Although CCR4 promotes interactions between CD4SP cells and thymic DCs, a comparable chemokine receptor that guides interactions of CD8SP thymocytes with DCs has not been identified. In addition, the chemokine receptor promoting interactions between SP cells and mTEChi cells has not been identified, although CCR7 is a likely candidate. Furthermore, we do not know if different chemokine receptors guide stromal interactions of cells destined to a T reg cell versus clonal deletion fate. Elucidation of cellular interactions and molecular cues critical for efficient negative selection of autoreactive cells should enable the development of rational interventions for autoimmune disorders.
MATERIALS AND METHODS
Mice.
C57BL/6J (CD45.2), B6.SJL-Ptprca PepCb (CD45.1), B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II), C57BL/6-Tg(Ins2-TFRC/OVA)296Wehi/WehiJ (Rip-mOVA), B6(Cg)-Rag2tm1.1Cgn/J (Rag2−/−), B6.129P2(C)-Ccr7tm1Rfor/J (Ccr7−/−), B6.Cg-Tg(Itgax-Venus)1Mnz/J (CD11c-EYFP), and B6.129S2-H2dlAb1-Ea/J (MHC-II−/−) mice were obtained from The Jackson Laboratory. Rip-OVAhi (Kurts et al., 1998), Ccr4−/− (Chvatchko et al., 2000), and pCX-EGFP (Wright et al., 2001) strains were generously provided by W.R. Heath (University of Melbourne, Melbourne, Australia), A.D. Luster (Massachusetts General Hospital, Boston, MA), and Irving L. Weissman (Stanford University, Stanford, CA), respectively. Ccr4−/− OT-II, Ccr4−/−MHC-II−/−, CD45.1/CD45.2, and CD45.1 OT-II mice were bred in-house. Experiments were performed using mice 4–8 wk of age, unless otherwise noted. All strains were bred and maintained under specific pathogen–free conditions in the University of Texas at Austin animal facility. Mouse maintenance and experimental procedures were performed with approval from the University of Texas at Austin’s Institutional Animal Care and Use Committee.
Antibodies.
For flow cytometry and FACS purification of thymocyte subsets and thymic stroma, the following fluorochrome or biotin-conjugated antibodies (from eBioscience or BioLegend unless indicated) were used: anti-CD3 (145-2C11), -CD4 (RM4-5), -CD8 (53–6.7), -CD69 (H1.2F3), -CD25 (PC61.5), -CD45.1 (A20), -CD45.2 (104), -Vβ5 (MR9-4), -Foxp3 (FJK-16s), -EpCAM (G8.8), -TER-119 (TER-119), -CD11c (N418), -CD31 (390), -Sirpα (P84), -B220 (RA3-6B2), -I-A/I-E (M5/114.15.2), -CD80 (16-10A1), -CD45 (30-F11; BD), -Gr1 (RB6-8C5), -CD11b (M1/70), -NK1.1 (PK136), -cKit (2B8), and Ly-51 (6C3).
For immunofluorescent analyses, the following antibodies were used: anti-keratin 5 (polyclonal; Covance), -pan-cytokeratin (C-11; Sigma-Aldrich), -CD8-Alexa Fluor 488 (53–6.7; eBioscience), -CD4-APC (RM4-5; eBioscience), -CD4 (GK1.5; Bio X cell), -CD69-Biotin (H1.2F3; BioLegend), -CD11c-Biotin (N418; BioLegend), -Sirpα (P84; BioLegend), donkey-anti–rabbit IgG DyLight 549 (poly-clonal; Jackson ImmunoResearch Laboratories), and Donkey-anti–rat IgG DyLight 488 (polyclonal; Jackson ImmunoResearch Laboratories), Streptavidin Alexa Fluor 488 (Life Technologies), and Streptavidin Alexa 647 (Life Technologies).
Flow cytometric analyses.
Single-cell suspensions of thymocytes were obtained by manually dissociating thymi and passing the suspension through a 40-µM cell strainer (Thermo Fisher Scientific). 107 thymocytes were stained with fluorochrome-conjugated or biotinylated antibodies for 20 min in the dark at 4°C in FACS wash (FW; PBS+ 4% bovine calf serum [BCS]; GemCell). Cells were washed once in FW before detection of biotinylated antibodies with Streptavidin Qdot 605 (Life Technologies). 1 µg/ml Propidium iodide (PI; Enzo) was added to identify dead cells, and the suspension was analyzed with a LSRFortessa cell analyzer (BD). FlowJo ver.9.5.3 (Tree Star) was used for analysis of flow cytometry data. For FOXP3 staining, thymocytes were first immunostained with antibodies against cell surface markers, and then fixed, permeabilized, and stained with anti-FOXP3 using the FOXP3/Transcription Factor Staining Buffer Set (eBioscience) following the manufacturer’s recommendations. To analyze DNA content, cells were stained with antibodies against surface markers, and then permeabilized with 70% ethanol and stained with 1 µg/ml PI in PBS.
FACS purification of thymocyte and thymic stromal cell subsets.
To isolate DP and SP subsets for RT-PCR analysis, thymocytes from a 1-mo-old C57BL/6J mouse were immunostained with anti-CD3, CD4, CD8, CD69, Gr1, CD11b, NK1.1, CD25, and PI. 106 cells from each indicated thymocyte subset were sorted to >95% purity on a FACSAria II (BD). To isolate DN thymocyte subsets, thymocytes from 6 C57BL/6J mice were pooled and DP and CD8 SP thymocytes were depleted by immunostaining with anti-CD8 antibody, followed by magnetic depletion with anti–rat IgG DynaBeads (Life Technologies) according to the manufacturer’s guidelines. The remaining cells were stained with fluorescently conjugated antibodies to CD4, CD8, CD25, CD44, and cKit. 5 × 104 DN1-4 cells were sorted to >95% purity on a FACSAria II (BD).
Thymic stromal cell subsets were sorted as previously described (Ki et al., 2014). In brief, thymi from 1-mo-old male C57BL/6J mice were digested with Collagenase D (Roche), followed by Collagenase/Dispase (Roche) in the presence of DNase I, as described previously (Gray et al., 2008). Cells were immunostained with FITC-conjugated Ulex europaeus agglutinin I (UEA-1; Vector Laboratories) and the following fluorochrome-conjugated antibodies: EpCAM, TER-119, CD11c, CD31, Sirpα, B220, I-A/I-E, CD80, CD45, and Ly-51. Stromal subsets were FACS purified by double sorting to >95% purity on a FACSAria II (BD).
cDNA preparation and quantitative PCR.
106 FACS-purified thymocytes of each subset were pelleted by centrifugation and resuspended in 1 ml of TRIzol. RNA was purified according to manufacturer’s recommendations, and cDNA was generated using the SuperScript III RT kit, according to manufacturer’s recommendations. qRT-PCR of thymocyte cDNA was performed using the SYBR Green Real-Time PCR Master Mix on a ViiA 7 Real-Time PCR System. All reagents and instruments were manufactured by Life Technologies. The following primers were used: CCR4 forward 5′-GGAAGGTATCAAGGCATTTGGG-3′, CCR4 reverse 5′-GTACACGTCCGTCATGGACTT-3′, CCL22 forward 5′-AGGTCCCTATGGTGCCAATGT-3′, CCL22 reverse 5′-CGGCAGGATTTTGAGGTCCA-3′, CCL17 forward 5′-GACGACAGAAGGGTACGGC-3′, and CCL17 reverse 5′-GCATCTGAAGTGACCTCATGGTA-3′. cDNA preparation and qRT-PCR from thymic stromal cell subsets was performed as previously described (Ki et al., 2014). In brief, for each FACS-sorted stromal cell subset, cDNA was synthesized from 300 ng RNA using qScriptTM cDNA SuperMix (Quanta). cDNAs were diluted and preamplified with 2X TaqMan preamp Master Mix (Life Technologies). qPCRs were performed on cDNA distributed on 48.48 Dynamic Arrays (Fluidigm).
DC isolation.
For in vitro chemotaxis assays toward DC supernatants, 10 thymi were pooled from C57BL/6J mice, and were dissociated with Collagenase/Dispase as per the thymic stromal cell isolation procedure above. DCs were isolated by incubating the resultant cell suspension with biotinylated anti-CD11c, followed by streptavidin MicroBeads (Miltenyi Biotec), and CD11c+ cells were enriched on a MACS column (Miltenyi Biotec) following manufacturer’s instructions. Purities were determined by flow cytometry to be >90%.
Chemotaxis assays.
Thymocyte chemotaxis assays using CCR4 ligands were performed as previously described (Campbell et al., 1999). In brief, 5 × 105 cells were added to the top chamber of 5 µm pore, polycarbonate 24-well tissue culture inserts (Costar) in 100 µl medium, with 600 µl of RPMI 1640 + 10% FBS + the specified concentration of chemokine in the bottom chamber. All assays were conducted at 37°C in 5% CO2 for 2 h. Recombinant mouse CCL22, CCL17, or CCL19 (R&D Systems) were added as indicated. 104 15 µm polystyrene beads (Polyscience) were added to the bottom chambers after the 2 h incubation to enable quantitation of the cells that migrated into the bottom chamber. Cells and beads from the bottom chambers, or 5 × 105 cells from the input cell suspension for the assay were immunostained stained with fluorochrome or biotin-conjugated antibodies to CD3, CD4, CD8, CD25, and CD69. Samples were analyzed on an LSRFortessa flow cytometer (BD). Percent migration was determined as the percentage of input cells of a given subset that were present in the bottom chamber of the chemotaxis assay. The fold increase in migration was calculated as the ratio of cells in the bottom well in the presence of the indicated concentration of chemokine versus media alone.
Chemotaxis assays in which thymocytes migrated toward DC supernatant were performed as previously described, with a few modifications (Proietto et al., 2008). In brief, 5 × 105 DCs were incubated at 37°C in 8% CO2 for 3 h in 600 µl complete RPMI (RPMI-1640 medium [Gibco] supplemented with 1× GlutaMAX [2 mM l-alanyl-l-glutamine], 1× Penicillin [100 U/ml]-S Streptomycin [100 µg/ml]-Glutamine [300 µg/ml; Gibco], 1 mM Sodium Pyruvate, 1× MEM NEAA, and 50 µM 2-mercaptoethanol [all from Gibco], and 10% FBS [Hyclone]). Supernatant from the DC cultures was then placed into the bottom chamber of Transwell plates. 100 µl complete RPMI containing 5 × 105 thymocytes was place in the top well, and cells were allowed to migrate for 6 h at 37°C in 8% CO2. Cells migrating to the bottom of the Transwell were analyzed to determine the percentage of each input subset that migrated toward the DC supernatant.
Immunofluorescent analyses of thymic cryosections and detection of autoantibodies.
7-µm thymic cryosections were fixed in acetone (−20°C for 20 min), washed in PBS, and stained with the indicated combinations of the following primary antibodies for 1 h at room temperature in humidified chambers in FW: anti-keratin 5, pan-keratin, CD11c-biotin, CD8-Alexa Fluor 488, CD69-biotin, Sirpα, CD4, and CD4-APC. Donkey-anti–rabbit IgG DyLight 594, donkey-anti–rat IgG DyLight 488, donkey-anti–rat IgG DyLight 594, streptavidin Alexa Fluor 594, or streptavidin Alexa Fluor 647 (Life Technologies) were used as secondary detection reagents. 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies) was used to detect nuclei.
To detect autoantibodies in mouse serum, 7-µm cryosections were prepared from Rag2−/− kidneys (C57BL/6J background). Cryosections were fixed in acetone as described, and incubated with undiluted mouse serum from the indicated mice overnight at 4°C. After a wash in PBS + 0.5% Tween-20 (Thermo Fisher Scientific), the sections were incubated with the secondary reagent donkey-anti–mouse IgG DyLight594, to detect murine autoantibodies, followed by a DAPI nuclear counterstain.
Immunostained sections were mounted in ProLong Antifade (Life Technologies) and were observed under an Axio Imager A1 microscope with a 10× objective, NA 0.45 or 20×, 0.8 (Carl Zeiss). Images were acquired using AxioVision 4.4.2.0 (Carl Zeiss). For the medullary DP CD69+ analysis, immunostained sections were imaged using an Olympus FV1000 confocal microscope with a 40× objective, NA 1.3 (Olympus). Images were acquired using FV10-ASW3.1 (Olympus). Photoshop (Adobe) was used to overlay the single color TIFFs. All comparable images were taken at a fixed exposure for each channel, and identical image processing was performed uniformly across the entire image to increase the “Exposure” for optimal visualization. The tiled thymic sections were imaged with Eclipse 80i microscope (Nikon) and were stitched together using the NIS-Elements 3.0 software (Nikon). The K-mean clustering function in the ImageJ (National Institutes of Health) package Toolkit was used to quantify the Cortical and Medullary area in the tiled images by segmenting the image into three regions, cortex, medulla and blank regions, followed by quantitation of the area in each region. To quantify the number of medullary DP CD69+ cells, a constant stringent threshold was applied to CD4 and CD8 images, and thresholded images were converted into binary masks using in ImageJ. The two binary images were multiplied using the image calculator in Fiji, such that only CD4+CD8+ pixels had the value of 1, and the Analyze Particle algorithm in Fiji was used to identify groups of CD8+ CD4+ double-positive pixels. These groups were then manually inspected to verify and count CD69+ DP cells; manual verification was necessary because sometimes a single region consisted of multiple adjacent cell membranes.
Negative selection assays in thymic slices.
C57BL/6J thymi were embedded in 4% low-melt agarose and 400-µm thick live slices were generated using a VT 1000S Microtome (Leica), as previously described (Ehrlich et al., 2009). Slices were incubated on a 0.4-µm cell culture insert (Millipore) in a 35-mm tissue culture dish (Thermo Fisher Scientific) filled with 1 ml complete RPMI containing 0–10 µM OVA323–339, as indicated. 106 total thymocytes from CD45.1 Ccr4+/+ OT-II, CD45.2 Ccr4−/− OT-II, and CD45.1 mice each were stained with CMF2HC Cell tracker blue (Life Technologies), according to the manufacturer’s instructions, and were incubated at 37°C, 5% CO2 together on a thymic slice. After 24 h, the slices were gently washed in PBS + 2% BSA and then manually disrupted to obtain single-cell suspensions. To quantify the number of thymocytes within each subset input into the slices, an aliquot of the thymocyte mixture was maintained at 4°C for 24 h, and was subsequently analyzed by flow cytometry in parallel with cells cultured in slices. Thymocytes were immunostained for CD3, CD4, CD8, CD69-biotin, CD25, Vβ5, CD45.1, and CD45.2, followed by Streptavidin Qdot 605. Samples were analyzed on an LSRFortessa flow cytometer (BD). Ccr4+/+ CD25− CD4SP OT-II (CD4+, CD25−, CD3+, Vβ5+, CMF2HC+, and CD8−), and Ccr4+/+ CD25+ CD4SP OT-II (CD4+, CD25+, CD3+, Vβ5+, CMF2HC+, and CD8−) subsets were identified within the CD45.1+ population. The corresponding Ccr4−/− OT-II subsets were identified within the CD45.2+ population. Cell numbers of the identified subsets were normalized to the cell number of CD45.1 CD4SP (CD45.1+ Vβ5−, CMF2HC+, CD4+ CD8−) cells in the slice to normalize for variability in thymic slice entry. To calculate the number and percentage of cells responding to OVA, we assume that a CD4+ CD25− cell makes one of three decisions: survive, undergo apoptosis (negative selection), or differentiate into a CD25+CD4SP T reg cell precursor. The number of CD4SP cells undergoing negative selection at the indicated concentration of OVA323–339 was calculated as the number of CD4SP OT-II cells in slices with no peptide minus the number of CD4SP OT-II cells in slices loaded with that concentration of OVA323–339. The number of surviving cells was the number of CD4SP OT-II cells in slices at each concentration of peptide. The number of responding cells was calculated as the number of CD4SP OT-II cells deleted by a given concentration of OVA323–339 plus the number of CD25+ CD4SP OT-II cells at that peptide concentration. To calculate the percentage of CD4SP cells responding, surviving, or diverting to the T reg cell lineage at a given peptide concentration, the number of cells undergoing each of these three fates at each OVA323–339 concentration, was divided by the number of CD4SP OT-II cells in slices without OVA323–339, and converted to a percentage.
Syngeneic mixed lymphocyte reactions.
Splenocytes were harvested from Ccr4−/− and Ccr4+/+ mice. Red blood cells were lysed by incubation in red blood cell lysis buffer (0.155 M NH4Cl, 10 mM KHCO3, and 0.1 mM Na2EDTA) for 2 min on ice and naive T cells were isolated by incubating the remaining cells for 30 min at 4°C with the following lineage antibodies: anti-Mac1, Gr1, Ter119, CD8, CD25, and B220. The cells were then magnetically depleted using anti–rat IgG DynaBeads (Life Technologies) for 10 min at room temperature. Magnetic depletion resulted in 90% purity of naive CD4 T cells. Naive T cells were then stained with 5 µM CMFDA (Life Technologies), according to manufacturer’s recommendations. Splenocytes from an additional C57BL/6J mice were irradiated at 3,000 cGy using a 43855 Series x-ray irradiator (Faxitron). 3 × 105 naive T cells were cultured with 106 irradiated splenocytes in 96-well plates. 3 × 105 naive T cells were cultured alone as a control. Cells were analyzed after 3, 5, and 7 d by flow cytometry to determine the percentage of cells that had undergone at least one cell division based on CMFDA dilution.
Mouse necropsy.
Full necropsies of aged mice (8–13 mo) were performed by the Histology and Tissue Processing core at the University of Texas MD Anderson Cancer Center Science Park (Smithville, TX), and clinical scores of lymphocytic infiltrates in numerous organs were assessed by a veterinary pathologist.
Two-photon imaging.
To analyze CD4SP interactions with DCs, CD4SP cells were isolated from Ccr4−/− OT-II or Ccr4+/+ OT-II mice by depleting DP, CD8SP, CD25+ DN and other hematopoietic lineages using anti-Mac1, -Gr1, -Ter119, -CD8, and -CD25, followed by anti–rat IgG DynaBeads (Life Technologies) and magnetic depletion, as previously described (Ehrlich et al., 2009). Isolated cells were stained with CMTPX CellTracker Red (Life Technologies) and incubated on 400 µm live thymic slices. Thymic slices were generated from CD11c-EYFP thymi using a VT 1000S Microtome (Leica), as previously described (Ehrlich et al., 2009). After incubation for 2 h at 37°C 5% CO2, images of thymic slices were acquired every 15 s, through a depth of 40 µm, at 5-µm intervals using an Ultima IV microscope with 20× objective, NA 0.5 and PraireView software (Prairie). The sample was illuminated with a MaiTai titanium/sapphire laser (Spectra Physics) tuned to 840 nm. Emitted light was passed through 535/50 and 607/45 bandpass filters (Chroma) to separate PMTs for detection of EYFP and CMTPX fluorescence, respectively.
Imaris v7.5 (Bitplane) was used to evaluate cell migration parameters and the distances between cell surfaces. Velocities were computed as path length divided by time observed. Track straightness was computed as cell displacement divided by path length. Thymocytes were scored as contacting DCs when the CD4SP cells were within 3 µm of the surface of a DC. The frequency of contacts was computed as the number of contacts for a given cell track divided by tracking time. The percent of time contacting DCs was calculated as the time a CD4SP spent contacting DCs divided by total tracking time.
To analyze the accumulation of CD4SP cells in the medulla, CD4SP cells were isolated from Ccr4−/− or Ccr4+/+ mice as previously described, and were stained with CellTracker CMPTX Red (Life Technologies) or CellTracker Blue CMF2HC (Life Technologies), respectively. The CellTracker for Ccr4+/+ and Ccr4−/− CD4SP were switched between independent repeats to eliminate color effects. A 1:1 mixture of 106 cells was incubated on 400-µm-thick live thymic slices from pCX-EGFP mice. After incubation for 2 h at 37°C 5% CO2, slices were imaged as above, except the samples were illuminated simultaneously with two MaiTai lasers one tuned to 740 nm, and the other to 900 nm, to excite Cell Tracker Blue and EGFP/CMTPX, respectively. Emitted light was passed through 480/40, 535/50, and 607/45 bandpass filters (Chroma) to separate PMTs for detection of CMF2HC, EGFP, and CMTPX fluorescence, respectively. Imaris was used to identify migrating cells in the medulla or cortex, as well as to calculate the volume of medullary and cortical regions, based on the EGFP channel, so as to calculate the density of cells in each region. The fold enrichment of cells in the medulla relative to the cortex was calculated by dividing the density of cells in the medulla by the cell density in the cortex.
To analyze medullary accumulation of DP CD69+ cells, 106 Ccr4+/+ and Ccr4−/− DP CD69+ cells (CD4+, CD8+, CD3+, and CD69+) were sorted to >95% purity (Fig. S1 A) using fluorophore conjugated anti-CD3, -CD4, -CD8, -CD69, and FACSAria II cell sorter (BD). The sorted cells were stained with CMTPX, loaded on CD11c-EYFP or pCX-EGFP thymic slices, imaged by two-photon microscopy, and analyzed for medullary and cortical densities as above.
In vitro cultures with CCR4 ligands.
106 Ccr4−/− or Ccr4+/+ thymocytes were cultured in 200 µl of complete RPMI +/− 1,000 nM of rmCCL22 or rmCCL17 (R&D Systems). After 24 h, cells were stained with the following antibodies: CD3, CD4, CD8, CD69-biotin, CD25, Annexin V-PE, and PI. The number of PI-Annexin V− cells in each thymocyte subset was quantified. The percentage of live cells in each subset is calculated by dividing the number of live cells after culture by the number of live cells in the input.
Generation and analysis of bone marrow chimeras.
For competitive bone marrow chimeras, bone marrow was extracted from femurs of CD45.1 Ccr4+/+ (C57BL/6J) and CD45.2 Ccr4−/− mice. T cells were magnetically depleted using 25 µg/ml anti-CD3, followed by incubation with sheep anti-rat IgG DynaBeads (Life Technologies) at a 4:1 cell to bead ratio. CD45.1/CD45.2 mice were lethally irradiated in two split doses of 450 rad, separated by 3 h. 5 × 106 CD45.1 Ccr4+/+ and 5 × 106 CD45.2 Ccr4−/− bone marrow cells were mixed and injected into each recipient mouse following irradiation. MHCII−/−Ccr4+/+: MHCII−/−Ccr4−/− competitive chimeras were generated as above, and BM cells were isolated from CD45.1 MHCII−/− Ccr4+/+, CD45.2 MHCII−/− Ccr4+/+, and CD45.2 MHCII−/− Ccr4−/− mice for these experiments. Chimeric mice were sacrificed 6 wk after reconstitution. T cells and thymocytes from chimeric recipients were stained with fluorophore conjugated anti-CD3, CD4, CD8, CD69-biotin, CD25, CD45.1, and CD45.2, followed by a streptavidin Qdot 605 (Life Technologies). Following fixation and permeabilization, the cells were stained with anti-FOXP3 using the FOXP3/Transcription Factor Staining Buffer Set (eBioscience) and were analyzed on an LSRFortessa cell analyzer (BD). To analyze DNA content, cells were stained with the surface antibodies as above, and were then permeabilized with 70% ethanol and stained with PI. FlowJo ver.9.5.3 (Tree Star) was used to analyze flow cytometry data.
For analysis of OT-II T cell development and selection in bone marrow chimeras, bone marrow cells from Ccr4+/+ OT-II and Ccr4−/− OT-II mice were prepared and T cell depleted as above. Lethally irradiated C57BL/6J, RIP-OVAhi, or RIP-mOVA recipient mice were reconstituted with 107 Ccr4+/+ OT-II or Ccr4−/− OT-II bone marrow cells. Chimeric mice were sacrificed 6 wk after reconstitution and analyzed by flow cytometry as above, except anti-Vα2 and -Vβ5 were included to detect OT-II thymocytes.
Vβ profiling.
Thymocytes from Ccr4+/+ and Ccr4−/− mice were stained with anti–CD4-Brilliant Violet 510, CD8-Pacific Blue, CD4-PE-Cy7, CD25-Alexa Fluor 700, CD69-Biotin followed by Streptavidin Qdot 605 stain. The stained cells from each mouse were then divided equally into 15 aliquots, each of which was stained with an Alexa Fluor 488–conjugated antibody against a different Vβ chains using the anti–mouse TCR Vβ Screening Panel (BD). The samples were then stained with PI and analyzed by flow cytometry.
Statistical analyses.
D’Agostino and Pearson omnibus normality tests were used to test the normality of the data in all experiments. To test the effect of OVA and CCR4 on clonal deletion in OT-II competitive chimera experiments, two-way ANOVA was performed using OVA and CCR4 as the two factors. Statistical significance of lymphocytic infiltration in WT versus Ccr4−/− lacrimal glands was calculated using two-way ANOVA with CCR4 and the type of lacrimal gland (extraorbital or intraorbital) as the two factors, and the significance level for CCR4 was displayed. To test for significance of differences in migratory parameters or DC contact between Ccr4−/− and Ccr4+/+ SP cells, a nonparametric Mann-Whitney test was used because the distribution of data were nonnormal. To test if CCR4 affected medullary accumulation of CD4SP cells, a two-tailed paired student’s t test was performed because Ccr4+/+ and Ccr4−/− CD4SP thymocytes were analyzed together on the same thymic slices. Unpaired student’s t tests were used to calculate p-values for the remaining experiments. In all cases, the F test was used to test the equal variance assumption between samples, and the appropriate t test, with or without the assumption of equal variance, was then used accordingly. When more than one statistical test was performed simultaneously, the Holm–Bonferroni multiple testing adjustment was used. All statistical analysis was performed using Prism (GraphPad).
Online supplemental material.
Fig. S1 shows the purity of DP CD69+ cells before and after FACS sorting for imaging experiments. Video 1 shows the two-photon time-lapse video microscopy of Ccr4+/+ (red) and Ccr4−/− (green) CD4SP thymocytes migrating in a live thymic slice from a pCX-EGFP (blue) transgenic mouse. Videos 2 and 3 show two-photon time-lapse video microscopy of Ccr4+/+ (Video 2) or Ccr4−/− (Video 3) DP CD69+ thymocytes (red) migrating in a pCX-EGFP live thymic slice (green). Videos 4 and 5 show two-photon time-lapse video microscopy of Ccr4+/+ (Video 4) or Ccr4−/− (Video 5) CD4SP cells (red) migrating among medullary DCs (yellow) in a live CD11c-EYFP thymic slice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150178/DC1.
Supplementary Material
Acknowledgments
We thank Andrew Luster, William Heath, and Irving Weissman for mice, Stephen Trent and Kyle Miller for use of their microscopes, Donna Kusewitt for analysis of autoimmune infiltrates, Andy Tran for thymic slice preparation, Sanghee Ki for analysis of CCR4 ligand expression, and Hilary Selden for technical assistance.
This work was supported by the Cancer Prevention and Research Institute of Texas (R1003; to L.I.R. Ehrlich) and the National Institutes of Health/NIAID (R01AI104870; to L.I.R. Ehrlich).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CD4SP
- CD4+ single-positive thymocyte
- CD8SP
- CD8+ single-positive thymocyte
- cTEC
- cortical thymic epithelial cell
- DP
- double-positive thymocyte
- GPCR
- G protein–coupled receptor
- mTEChi
- MHC-IIhiCD80hi medullary thymic epithelial cell
- PI
- propidium iodide
- T reg cell
- regulatory T cell
- TRA
- tissue-restricted antigen
References
- Alferink J., Lieberam I., Reindl W., Behrens A., Weiss S., Hüser N., Gerauer K., Ross R., Reske-Kunz A.B., Ahmad-Nejad P., et al. 2003. Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. J. Exp. Med. 197:585–599. 10.1084/jem.20021859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.S., Venanzi E.S., Klein L., Chen Z., Berzins S.P., Turley S.J., von Boehmer H., Bronson R., Dierich A., Benoist C., and Mathis D.. 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science. 298:1395–1401. 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- Aschenbrenner K., D’Cruz L.M., Vollmann E.H., Hinterberger M., Emmerich J., Swee L.K., Rolink A., and Klein L.. 2007. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8:351–358. 10.1038/ni1444 [DOI] [PubMed] [Google Scholar]
- Atibalentja D.F., Murphy K.M., and Unanue E.R.. 2011. Functional redundancy between thymic CD8α+ and Sirpα+ conventional dendritic cells in presentation of blood-derived lysozyme by MHC class II proteins. J. Immunol. 186:1421–1431. 10.4049/jimmunol.1002587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Nakamoto Y., and Mukaida N.. 2009. Crucial contribution of thymic Sirp alpha+ conventional dendritic cells to central tolerance against blood-borne antigens in a CCR2-dependent manner. J. Immunol. 183:3053–3063. 10.4049/jimmunol.0900438 [DOI] [PubMed] [Google Scholar]
- Barnden M.J., Allison J., Heath W.R., and Carbone F.R.. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40. 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- Bleul C.C., and Boehm T.. 2000. Chemokines define distinct microenvironments in the developing thymus. Eur. J. Immunol. 30:3371–3379. [DOI] [PubMed] [Google Scholar]
- Bonasio R., Scimone M.L., Schaerli P., Grabie N., Lichtman A.H., and von Andrian U.H.. 2006. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat. Immunol. 7:1092–1100. 10.1038/ni1385 [DOI] [PubMed] [Google Scholar]
- Bouillet P., Purton J.F., Godfrey D.I., Zhang L.-C., Coultas L., Puthalakath H., Pellegrini M., Cory S., Adams J.M., and Strasser A.. 2002. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 415:922–926. 10.1038/415922a [DOI] [PubMed] [Google Scholar]
- Brennecke P., Reyes A., Pinto S., Rattay K., Nguyen M., Küchler R., Huber W., Kyewski B., and Steinmetz L.M.. 2015. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat. Immunol. 16:933–941. 10.1038/ni.3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.J., Pan J., and Butcher E.C.. 1999. Cutting edge: developmental switches in chemokine responses during T cell maturation. J. Immunol. 163:2353–2357. [PubMed] [Google Scholar]
- Choi Y.I., Duke-Cohan J.S., Chen W., Liu B., Rossy J., Tabarin T., Ju L., Gui J., Gaus K., Zhu C., and Reinherz E.L.. 2014. Dynamic control of β1 integrin adhesion by the plexinD1-sema3E axis. Proc. Natl. Acad. Sci. USA. 111:379–384. 10.1073/pnas.1314209111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chvatchko Y., Hoogewerf A.J., Meyer A., Alouani S., Juillard P., Buser R., Conquet F., Proudfoot A.E., Wells T.N., and Power C.A.. 2000. A key role for CC chemokine receptor 4 in lipopolysaccharide-induced endotoxic shock. J. Exp. Med. 191:1755–1764. 10.1084/jem.191.10.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan J.E., McCarthy N.I., Parnell S.M., White A.J., Bacon A., Serge A., Irla M., Lane P.J.L., Jenkinson E.J., Jenkinson W.E., and Anderson G.. 2014. Differential requirement for CCR4 and CCR7 during the development of innate and adaptive αβT cells in the adult thymus. J. Immunol. 193:1204–1212. 10.4049/jimmunol.1400993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhagalov I.L., Chen K.G., Herzmark P., and Robey E.A.. 2013. Elimination of self-reactive T cells in the thymus: a timeline for negative selection. PLoS Biol. 11:e1001566 10.1371/journal.pbio.1001566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich L.I.R., Oh D.Y., Weissman I.L., and Lewis R.S.. 2009. Differential contribution of chemotaxis and substrate restriction to segregation of immature and mature thymocytes. Immunity. 31:986–998. 10.1016/j.immuni.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster R., Davalos-Misslitz A.C., and Rot A.. 2008. CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 8:362–371. 10.1038/nri2297 [DOI] [PubMed] [Google Scholar]
- Friedman R.S., Jacobelli J., and Krummel M.F.. 2006. Surface-bound chemokines capture and prime T cells for synapse formation. Nat. Immunol. 7:1101–1108. 10.1038/ni1384 [DOI] [PubMed] [Google Scholar]
- Gray D.H.D., Fletcher A.L., Hammett M., Seach N., Ueno T., Young L.F., Barbuto J., Boyd R.L., and Chidgey A.P.. 2008. Unbiased analysis, enrichment and purification of thymic stromal cells. J. Immunol. Methods. 329:56–66. 10.1016/j.jim.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Hinterberger M., Aichinger M., Prazeres da Costa O., Voehringer D., Hoffmann R., and Klein L.. 2010. Autonomous role of medullary thymic epithelial cells in central CD4(+) T cell tolerance. Nat. Immunol. 11:512–519. 10.1038/ni.1874 [DOI] [PubMed] [Google Scholar]
- Hu Z., Lancaster J.N., and Ehrlich L.I.R.. 2015. The contribution of chemokines and migration to the induction of central tolerance in the thymus. Front. Immunol. 6:398 10.3389/fimmu.2015.00398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert F.-X., Kinkel S.A., Davey G.M., Phipson B., Mueller S.N., Liston A., Proietto A.I., Cannon P.Z., Forehan S., Smyth G.K., et al. 2011. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood. 118:2462–2472. 10.1182/blood-2010-06-286393 [DOI] [PubMed] [Google Scholar]
- Hwang S., Song K.-D., Lesourne R., Lee J., Pinkhasov J., Li L., El-Khoury D., and Love P.E.. 2012. Reduced TCR signaling potential impairs negative selection but does not result in autoimmune disease. J. Exp. Med. 209:1781–1795. 10.1084/jem.20120058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Chantry D., Raport C.J., Wood C.L., Nishimura M., Godiska R., Yoshie O., and Gray P.W.. 1998. Macrophage-derived chemokine is a functional ligand for the CC chemokine receptor 4. J. Biol. Chem. 273:1764–1768. 10.1074/jbc.273.3.1764 [DOI] [PubMed] [Google Scholar]
- Jenkinson E.J., Anderson G., and Owen J.J.. 1992. Studies on T cell maturation on defined thymic stromal cell populations in vitro. J. Exp. Med. 176:845–853. 10.1084/jem.176.3.845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ki S., Park D., Selden H.J., Seita J., Chung H., Kim J., Iyer V.R., and Ehrlich L.I.R.. 2014. Global transcriptional profiling reveals distinct functions of thymic stromal subsets and age-related changes during thymic involution. Cell Reports. 9:402–415. 10.1016/j.celrep.2014.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L., Roettinger B., and Kyewski B.. 2001. Sampling of complementing self-antigen pools by thymic stromal cells maximizes the scope of central T cell tolerance. Eur. J. Immunol. 31:2476–2486. [DOI] [PubMed] [Google Scholar]
- Klein L., Kyewski B., Allen P.M., and Hogquist K.A.. 2014. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14:377–391. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koble C., and Kyewski B.. 2009. The thymic medulla: a unique microenvironment for intercellular self-antigen transfer. J. Exp. Med. 206:1505–1513. 10.1084/jem.20082449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel E.J., Boisvert J., Murphy K., Vierra M.A., Genovese M.C., Wardlaw A.J., Greenberg H.B., Hodge M.R., Wu L., Butcher E.C., and Campbell J.J.. 2002. Expression of the chemokine receptors CCR4, CCR5, and CXCR3 by human tissue-infiltrating lymphocytes. Am. J. Pathol. 160:347–355. 10.1016/S0002-9440(10)64378-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurobe H., Liu C., Ueno T., Saito F., Ohigashi I., Seach N., Arakaki R., Hayashi Y., Kitagawa T., Lipp M., et al. 2006. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 24:165–177. 10.1016/j.immuni.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Kurts C., Heath W.R., Carbone F.R., Allison J., Miller J.F., and Kosaka H.. 1996. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184:923–930. 10.1084/jem.184.3.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurts C., Miller J.F.A.P., Subramaniam R.M., Carbone F.R., and Heath W.R.. 1998. Major histocompatibility complex class I-restricted cross-presentation is biased towards high dose antigens and those released during cellular destruction. J. Exp. Med. 188:409–414. 10.1084/jem.188.2.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-M., Bautista J.L., Scott-Browne J., Mohan J.F., and Hsieh C.-S.. 2012. A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity. 37:475–486. 10.1016/j.immuni.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love P.E., and Bhandoola A.. 2011. Signal integration and crosstalk during thymocyte migration and emigration. Nat. Rev. Immunol. 11:469–477. 10.1038/nri2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughtry T.M., Baldwin T.A., Wilken M.S., and Hogquist K.A.. 2008. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J. Exp. Med. 205:2575–2584. 10.1084/jem.20080866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith M., Zemmour D., Mathis D., and Benoist C.. 2015. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat. Immunol. 16:942–949. 10.1038/ni.3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T.C., and Anderson M.S.. 2011. Control of central and peripheral tolerance by Aire. Immunol. Rev. 241:89–103. 10.1111/j.1600-065X.2011.01008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhak Z., Strassner J.P., and Luster A.D.. 2013. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. J. Exp. Med. 210:1855–1869. 10.1084/jem.20130091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T., Nitta S., Lei Y., Lipp M., and Takahama Y.. 2009. CCR7-mediated migration of developing thymocytes to the medulla is essential for negative selection to tissue-restricted antigens. Proc. Natl. Acad. Sci. USA. 106:17129–17133. 10.1073/pnas.0906956106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T., Ohigashi I., Nakagawa Y., and Takahama Y.. 2011. Cytokine crosstalk for thymic medulla formation. Curr. Opin. Immunol. 23:190–197. 10.1016/j.coi.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Perry J.S.A., Lio C.-W.J., Kau A.L., Nutsch K., Yang Z., Gordon J.I., Murphy K.M., and Hsieh C.-S.. 2014. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. 41:414–426. 10.1016/j.immuni.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie H.T., and Zúñiga-Pflücker J.C.. 2007. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 25:649–679. 10.1146/annurev.immunol.23.021704.115715 [DOI] [PubMed] [Google Scholar]
- Proietto A.I., van Dommelen S., Zhou P., Rizzitelli A., D’Amico A., Steptoe R.J., Naik S.H., Lahoud M.H., Liu Y., Zheng P., et al. 2008. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc. Natl. Acad. Sci. USA. 105:19869–19874. 10.1073/pnas.0810268105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J.O., Melichar H.J., Au-Yeung B.B., Herzmark P., Weiss A., and Robey E.A.. 2014. Distinct phases in the positive selection of CD8+ T cells distinguished by intrathymic migration and T-cell receptor signaling patterns. Proc. Natl. Acad. Sci. USA. 111:E2550–E2558. 10.1073/pnas.1408482111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom S.N., Shikama-Dorn N., Zhanybekova S., Nusspaumer G., Macaulay I.C., Deadman M.E., Heger A., Ponting C.P., and Holländer G.A.. 2014. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24:1918–1931. 10.1101/gr.171645.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D.K., and Zúñiga-Pflücker J.C.. 2014. An overview of the intrathymic intricacies of T cell development. J. Immunol. 192:4017–4023. 10.4049/jimmunol.1302259 [DOI] [PubMed] [Google Scholar]
- Speiser D.E., Lees R.K., Hengartner H., Zinkernagel R.M., and MacDonald H.R.. 1989. Positive and negative selection of T cell receptor V beta domains controlled by distinct cell populations in the thymus. J. Exp. Med. 170:2165–2170. 10.1084/jem.170.6.2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G., Sawa H., Kobayashi Y., Nakata Y., Nakagawa Ki., Uzawa A., Sakiyama H., Kakinuma S., Iwabuchi K., and Nagashima K.. 1999. Pertussis toxin-sensitive signal controls the trafficking of thymocytes across the corticomedullary junction in the thymus. J. Immunol. 162:5981–5985. [PubMed] [Google Scholar]
- Takahama Y. 2006. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 6:127–135. 10.1038/nri1781 [DOI] [PubMed] [Google Scholar]
- Tang H.L., and Cyster J.G.. 1999. Chemokine Up-regulation and activated T cell attraction by maturing dendritic cells. Science. 284:819–822. 10.1126/science.284.5415.819 [DOI] [PubMed] [Google Scholar]
- Ueda Y., Katagiri K., Tomiyama T., Yasuda K., Habiro K., Katakai T., Ikehara S., Matsumoto M., and Kinashi T.. 2012. Mst1 regulates integrin-dependent thymocyte trafficking and antigen recognition in the thymus. Nat. Commun. 3:1098 10.1038/ncomms2105 [DOI] [PubMed] [Google Scholar]
- Ueno T., Hara K., Willis M.S., Malin M.A., Höpken U.E., Gray D.H., Matsushima K., Lipp M., Springer T.A., Boyd R.L., et al. 2002. Role for CCR7 ligands in the emigration of newly generated T lymphocytes from the neonatal thymus. Immunity. 16:205–218. 10.1016/S1074-7613(02)00267-4 [DOI] [PubMed] [Google Scholar]
- Ueno T., Saito F., Gray D.H.D., Kuse S., Hieshima K., Nakano H., Kakiuchi T., Lipp M., Boyd R.L., and Takahama Y.. 2004. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 200:493–505. 10.1084/jem.20040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroomans R.M., Marée A.F., de Boer R.J., and Beltman J.B.. 2012. Chemotactic migration of T cells towards dendritic cells promotes the detection of rare antigens. PLOS Comput. Biol. 8:e1002763 10.1371/journal.pcbi.1002763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland D., Happ M.P., Bill J., and Palmer E.. 1990. Requirement for cotolerogenic gene products in the clonal deletion of I-E reactive T cells. Science. 247:964–967. 10.1126/science.1968289 [DOI] [PubMed] [Google Scholar]
- Wright D.E., Cheshier S.H., Wagers A.J., Randall T.D., Christensen J.L., and Weissman I.L.. 2001. Cyclophosphamide/granulocyte colony-stimulating factor causes selective mobilization of bone marrow hematopoietic stem cells into the blood after M phase of the cell cycle. Blood. 97:2278–2285. 10.1182/blood.V97.8.2278 [DOI] [PubMed] [Google Scholar]
- Yamamoto K. 2003. Pathogenesis of Sjögren’s syndrome. Autoimmun. Rev. 2:13–18. 10.1016/S1568-9972(02)00121-0 [DOI] [PubMed] [Google Scholar]
- Yamano T., Nedjic J., Hinterberger M., Steinert M., Koser S., Pinto S., Gerdes N., Lutgens E., Ishimaru N., Busslinger M., et al. 2015. Thymic B Cells Are Licensed to Present Self Antigens for Central T Cell Tolerance Induction. Immunity. 42:1048–1061. 10.1016/j.immuni.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Yoshie O., and Matsushima K.. 2014. CCR4 and its ligands: from bench to bedside. Int. Immunol. 4:1–10. 10.1093/intimm/dxu079 [DOI] [PubMed] [Google Scholar]
- Yuan Q., Bromley S.K., Means T.K., Jones K.J., Hayashi F., Bhan A.K., and Luster A.D.. 2007. CCR4-dependent regulatory T cell function in inflammatory bowel disease. J. Exp. Med. 204:1327–1334. 10.1084/jem.20062076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik A., and Yoshie O.. 2012. The chemokine superfamily revisited. Immunity. 36:705–716. 10.1016/j.immuni.2012.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.