Sullivan et al. showed that pharmacological suppression of BET proteins by a highly selective brain-permeable inhibitor, results in the suppression of genes that have been linked to autism spectrum disorders in humans. BET inhibitor treatment of young mice led to the induction of an autism-like syndrome characterized by alterations in social behaviors. These findings suggest a key role of the BET-controlled gene networks in the development of ASD.
Abstract
Studies investigating the causes of autism spectrum disorder (ASD) point to genetic, as well as epigenetic, mechanisms of the disease. Identification of epigenetic processes that contribute to ASD development and progression is of major importance and may lead to the development of novel therapeutic strategies. Here, we identify the bromodomain and extraterminal domain–containing proteins (BETs) as epigenetic regulators of genes involved in ASD-like behaviors in mice. We found that the pharmacological suppression of BET proteins in the brain of young mice, by the novel, highly specific, brain-permeable inhibitor I-BET858 leads to selective suppression of neuronal gene expression followed by the development of an autism-like syndrome. Many of the I-BET858–affected genes have been linked to ASD in humans, thus suggesting the key role of the BET-controlled gene network in the disorder. Our studies suggest that environmental factors controlling BET proteins or their target genes may contribute to the epigenetic mechanism of ASD.
The search for the cause of autism spectrum disorder (ASD) revealed the disease’s associations with numerous gene mutations. Many of the ASD candidate genes encode proteins that control neuronal network formation and function (Chen et al., 2015). A significant fraction of ASD-associated genes, however, encode rather ubiquitous regulators of gene expression (De Rubeis et al., 2014). The latter finding underscores the possibility of an epigenetic etiology of ASD, where aberrant control of gene expression rather than gene mutations can lead to abnormal neuronal development and function.
One of the major challenges for testing the role of abnormal gene regulation in brain function is the lack of experimental models where changes in gene expression can be achieved in a controlled fashion and without the generalized negative impact on neuronal development, survival, and function. This obstacle could be potentially overcome by using brain-permeable compounds that regulate transcription in a selective and time-dependent fashion. Pharmacological suppression of well-defined transcriptional processes would allow one to determine the impact of acute or chronic transcriptional deregulation on animal behavior at any developmental time point and would also enable the identification of genes affected by the temporal impairment of these processes.
Our earlier studies describe the pharmacological modulation of transcription by inhibitors of the bromodomain and extraterminal domain–containing proteins (BETs; Nicodeme et al., 2010). In humans and mice, somatic cells, including neurons, express three independent BET proteins: Brd2, Brd3, and Brd4 (Sanchez et al., 2014). All three BET proteins contain two N-terminal bromodomains (BD1 and BD2) that recognize acetylated lysines within the N-terminal domain of histone H4, as well as other lysine-acetylated proteins (Filippakopoulos and Knapp, 2012). The binding of the BET bromodomain to acetylated histone H4 lysines initiates a chain of biochemical and molecular events that leads to the formation of elongation-competent transcriptional complexes containing RNA Polymerase II phosphorylated at serine 2 (Brès et al., 2008). Pharmacological inhibitors of BET proteins, such as I-BET (Nicodeme et al., 2010) or JQ1 (Filippakopoulos et al., 2010), bind with high specificity to the acetyl-lysine–binding pocket of the tandem bromodomains of all members of the BET family. The inhibitor binding prevents BET protein association with acetylated histone H4 and affects gene transcription (Nicodeme et al., 2010; Barbieri et al., 2013). However, despite the widespread BET binding to numerous genes, the impact of the BET inhibitors on gene expression in various cell types is surprisingly limited (Nicodeme et al., 2010; Dawson et al., 2011; Delmore et al., 2011). The mechanism of the selective impact of BET inhibitors on gene expression is not well understood. Our earlier studies showed that susceptibility to I-BET in activated macrophages correlates with the gene’s dependence on SWI/SNF-mediated chromatin remodeling (Nicodeme et al., 2010). Other studies have suggested a link between the susceptibility to BET inhibitors and BET-association with specific enhancer clusters (super-enhancers; Lovén et al., 2013).
To address the impact of BET suppression on neuronal gene expression and brain function, we developed a novel, brain-permeable I-BET (I-BET858). Here, we provide evidence for the selective impact of BET protein functions on neuronal gene expression in vitro and in vivo. We show that I-BET858 treatment preferentially suppresses genes associated with neuronal differentiation and synaptic function and has no effect on neuronal housekeeping or early response genes. The suppressive effects of I-BET858 correlate directly with gene length. A significant number of I-BET858–suppressed genes, including genes of extended length, are potential ASD candidate genes in humans (Basu et al., 2009; King et al., 2013). Furthermore, suppression of specific neuronal genes by I-BET858 in young mice leads to the development of an autism-like syndrome. Our findings describe for the first time a pharmacologically induced model of ASD and point to the selective I-BET858 suppressed genes as potential key contributors to ASD in mice.
RESULTS AND DISCUSSION
Identification of a novel brain-permeable BET inhibitor
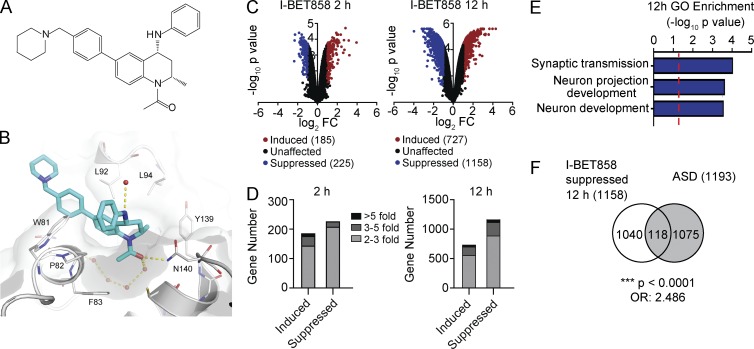
Brain permeability is a crucial factor in assessing the impact of BET inhibitors on neuronal gene expression and associated behaviors. To investigate the effects of BET inhibition within the central nervous system, we identified a novel BET inhibitor compound with higher brain permeability than the original I-BET151 and I-BET762. Through in silico assessment of ∼1,000 potential brain-penetrant BET inhibitor compounds, we identified the novel tetrahydroquinoline class (THQ; Gosmini et al., 2014) compound I-BET858 (Fig. 1 A). I-BET858 displayed an appropriate profile for progression into in vivo rodent models based on its calculated physicochemical properties, pharmacokinetics, BET inhibitory activity in cell-based assays, and enhanced brain penetrance (Fig. S1, A and B). The binding mode of I-BET858 to BRD4-BD1 was determined by x-ray crystallography (Fig. 1 B and Fig. S1, C and D). The curvature of the THQ template complements that of the BET proteins and results in high affinity of I-BET858 for the BET subfamily with >1,000-fold selectivity over 34 other bromodomain-containing proteins (Fig. S2).
Figure 1.
Effect of the brain-permeable inhibitor of BET proteins (I-BET858) on gene expression in primary neurons in vitro. (A) Chemical structure of I-BET858. (B) X-ray structure of I-BET858 (blue) bound to the acetylated lysine (Kac) recognition pocket of BRD4-BD1 (red spheres, water network; yellow dashed lines, hydrogen bonds). The WPF shelf (W81, P82, F83), as well as the asparagine N140 essential for acetylated lysine (Kac) binding, are labeled accordingly. (C and D) Gene expression in primary cortical neurons treated with I-BET858 (1 µM) for 2 and 12 h was analyzed using microarray analysis (n = 3/group, experiment was performed in duplicate). (C) Volcano plot shows the genes that are significantly suppressed (P < 0.05; >2-fold; blue) or induced (red) after I-BET858 treatment. (D) Bar graph shows the number and fold gene expression changes. (E) Gene Ontology (GO) enrichment of I-BET858–induced (top) or suppressed (bottom) genes (−log10 p-value; red line, P = 0.05). (F) Venn diagram shows the overlap between I-BET858–suppressed genes and ASD-associated genes. Statistics: χ2 test, ***, P < 0.0001; OR, 2.486.
Selective impairment of neuronal gene expression by pharmacological BET inhibition in vitro
The impact of I-BET858 on neuronal gene expression was initially assessed in vitro. Primary cortical neurons isolated from E18 mouse brain were grown in an in vitro culture system for 7 d, followed by treatment with I-BET858 at a concentration of 1 µM. As in other cell types treated with I-BET or JQ1 (Nicodeme et al., 2010; Dawson et al., 2011; Delmore et al., 2011), the treatment of primary cultured neurons with I-BET858 leads to a time-dependent up- and down-regulation of numerous genes (Fig. 1, C and D). Opposite to gene induction, the down-regulation of gene expression by BET inhibitors has been shown to correlate directly with BET-association at the gene loci before the BET inhibitor treatment and loss of BET-association following exposure to the inhibitors (Nicodeme et al., 2010). Therefore, in our studies we focus exclusively on genes that are suppressed by I-BET858.
Treatment of primary cortical neurons with I-BET858 results in time-dependent changes in gene expression, characterized by the moderate (2.75-fold, on average) down-regulation of 225 and 1,158 genes at 2 and 12 h after I-BET858 treatment, respectively (Fig. 1, C and D, and Table S1). Notably, I-BET858 preferentially affects genes controlling synaptic transmission, neuronal development, and morphogenesis (Fig. 1 E). Moreover, ∼10% of the suppressed genes (118/1,158; P < 0.0001; OR, 2.486) are previously identified ASD risk genes (combined lists from Basu et al. [2009] and King et al. [2013]) in humans (Fig. 1 F and Table S1). Several of the ASD candidate genes that show strong suppression in response to I-BET858, such as Met, Gabra1, Cntn6, Pde4b, Npas2, Cdh10, and Foxp1, have been shown to play an important role in neuronal development and function in circuits relevant to core behavioral domains of ASD (Zhou et al., 1997; Qiu et al., 2014; Bacon et al., 2015; Ozburn et al., 2015). The brain-specific deficiency of Foxp1-expression was recently shown to impair neuronal development, causing autistic-like behaviors in mice (Bacon et al., 2015).
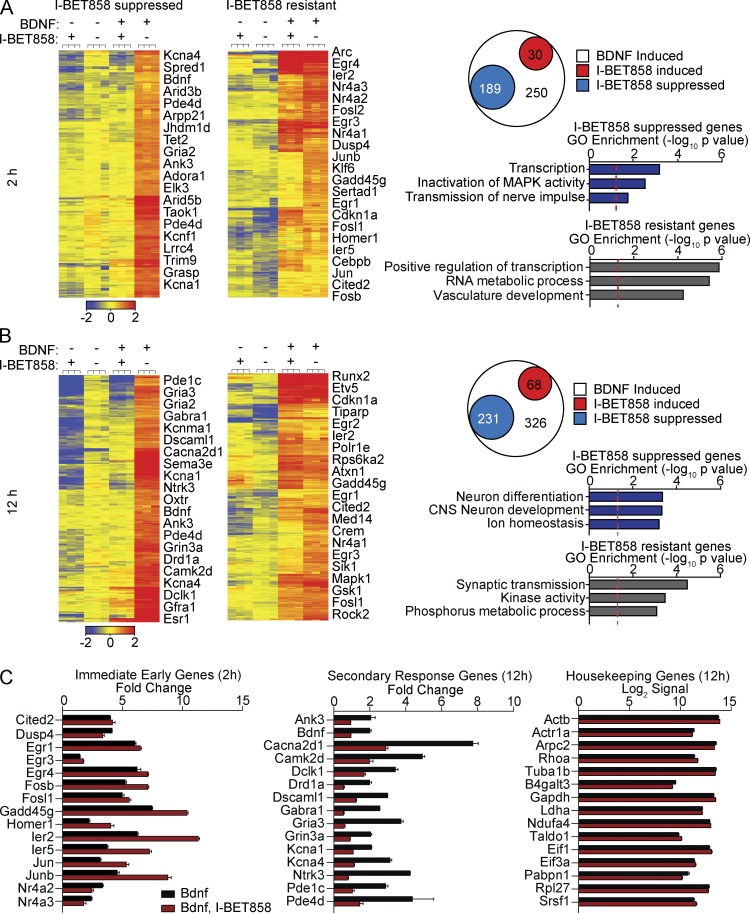
The ability of I-BET858 to modify gene expression in neurons was further demonstrated by the profound yet selective effect of I-BET858 on the expression of genes induced by the brain-derived neurotrophic factor (BDNF; Fig. 2, A and B). The BDNF-controlled genes play a crucial role during brain development and are essential for normal neuronal function and survival (Park and Poo, 2013). Treatment of primary cortical neurons with BDNF results in the induction of numerous genes that regulate neuronal maturation, axogenesis, synapse formation and synaptic transmission in neurons (Table S2). The BDNF-induced transcriptional changes follow a well-defined temporal pattern with the induction of early response genes, such as Egr1-4, Fos, FosB, Ier2-5, Jun, JunB, and Nr4a2/3, within 20 min after BDNF stimulation. The expression of these immediate-early genes (IEGs) is followed by the induction of secondary/late response genes that control the specific BDNF-induced changes in neuronal morphology and function (Alder et al., 2003; Calella et al., 2007).
Figure 2.
I-BET858 effect on BDNF-inducible gene expression in vitro. Heat maps show normalized expression values using microarray analysis for all BDNF-inducible and I-BET–suppressed (left, more than twofold) or I-BET–resistant (right) genes in cortical neurons at 2 h (A) and 12 h (B) after treatment (n = 3/group; experiment was performed in duplicate). Selected genes are indicated. Circle plot shows the number of BDNF-inducible genes that are unchanged (white), significantly suppressed (blue), or induced (red) more than twofold after I-BET858 treatment. Bar graphs display selected GO enrichments (−log10 p-value; red line, P = 0.05). (C) I-BET858 effect on the expression of selected immediate-early (left) or secondary (middle) BDNF-inducible genes or housekeeping genes (right) is shown.
We found that treatment with I-BET858 at 1 µM resulted in a highly selective and time-dependent suppression of gene expression in BDNF-stimulated cortical neurons (Fig. 2, A and B). I-BET858 has no impact on housekeeping gene expression but specifically affects the expression of a selected group of BDNF-induced neuronal genes at 2 and 12 h after BDNF treatment (Fig. 2, A–C; and Table S2). Many BDNF-inducible and I-BET858–suppressed genes are regulators of important neuronal processes, including neuronal transmission via ion channels (such as Cacna2d1, Kcnma1, and Kcna1/4), neurotransmitter receptor signaling (Drd1a, Gabra1, Gria1-3, Grin3a, and Pde1c/4d), and dendrite and axon development (Ank3, Bdnf, Camk2d, Dscaml1, Dclk1, Ntrk3, and Sema3a; Fig. 2, A–C).
In contrast, the expression of IEGs, such as Arc, Dusp1/4, Egr1-4, Fosb, Homer1, Ier2/5, Junb, Nr4a1/2/3, and Srf in response to BDNF treatment after 2 and 12 h was not affected or even slightly increased by I-BET858 (Fig. 2, A–C). These data suggest that I-BET858 treatment in neurons does not disrupt BDNF-induced neuronal signal responses or subsequent IEG induction, but specifically represses the transcription of a selected group of secondary/late response genes in response to BDNF treatment (Fig. 2, A–C).
Selective impairment of neuronal gene expression by pharmacological BET inhibition in vivo
The development of the brain-permeable I-BET858 enables us to address for the first time the impact of pharmacological suppression of BET proteins on gene expression in neurons in vivo. We found that I-BET858 enters the mouse brain shortly after an i.p. injection (Fig. S1 B). A single acute i.p. administration of I-BET858 at 30 mg/kg allows I-BET858 detection in the brain at a concentration of ≥1 µM lasting for several hours after the injection (Fig. S1 B). The tissue concentration of 1 µM matches the concentration of I-BET required for the therapeutic impact during systemic inflammation (Nicodeme et al., 2010).
In our initial studies, we addressed the impact of acute and chronic I-BET858 administration on gene expression in the striatum. We chose the striatum because of (a) its relatively homogenous neuronal population with 90% of striatal neurons being D1 or D2 receptor-expressing medium spiny neurons, (b) the well-known pattern of medium spiny neuron gene expression (Heiman et al., 2008), (c) the well-established role of the striatum in animal behavior, and (d) the contribution of impaired striatal function to neuronal disorders, including ASD (Di Martino et al., 2011; Peça et al., 2011; Rothwell et al., 2014).
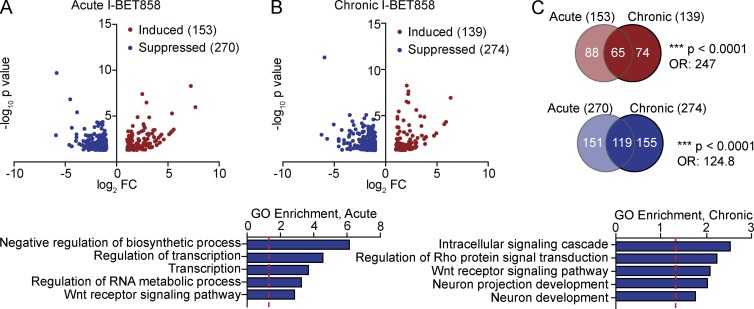
Acute I-BET858 treatment results in the suppression of 270 genes 4 h after a single i.p. injection (Fig. 3 A and Table S3). The chronic administration of I-BET858 for 2 wk does not affect the total number, but rather the pattern of affected genes (Fig. 3, B and C; and Table S3). Approximately 45% of genes suppressed by acute I-BET858 treatment remain down-regulated 12 h after chronic daily I-BET858 injections, whereas 155 genes are suppressed by I-BET858 only after long-term treatment (Fig. 3 C; P < 0.0001; OR, 124.8). A significant fraction of the in vivo I-BET858–suppressed genes (69/270 after acute; 51/274 after chronic; P < 0.0001; OR, 7.701 and 5.042) overlap with the I-BET–repressed genes in primary neurons in vitro (Table S3). Most importantly, acute and chronic I-BET858 treatment results in significant down-regulation of 34 genes that have been previously implicated in ASD (P = 0.0025; OR, 1.755). Several of the I-BET858–suppressed ASD-associated genes include potent transcriptional regulators, such as the histone lysine demethylase Phf2, the Set-domain binding protein Setbp1, and the transcription factors Foxp1, Nr4a2, Zbtb20, and Tcf3 (Table S3). The latter finding points to BET proteins as transcriptional regulators of gene-controlling chromatin circuits that can broaden the impact of BET inhibition on gene expression in neurons. This may, in part, explain the distinct patterns of genes only altered with chronic dosing (Fig. 3 C). Similar to our in vitro studies, in vivo administration of I-BET858 has no impact on housekeeping gene expression (not depicted) but preferentially suppresses genes involved in neuronal development, and dendrite and synapse formation and function (Fig. 3 B).
Figure 3.
Selective effect of I-BET858 on gene expression in vivo. Volcano plots show genes that are significantly suppressed (blue) or induced (red) more than twofold after acute (A) or chronic (B) I-BET858 treatment in the mouse striatum using RNA sequencing analysis (n = 2). Bar graphs display selected GO enrichments (bottom; −log10 p-value; red line, P = 0.05). (C) Venn diagrams show the overlap of genes after acute or chronic I-BET858 treatment. Statistics: χ2 test, ***, P < 0.0001; OR, 247 and 124.8.
I-BET858–mediated gene suppression correlates with gene length
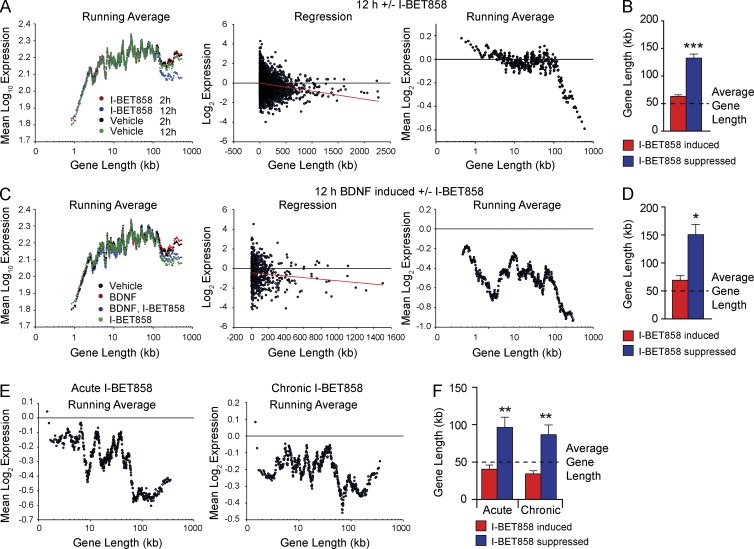
Earlier studies revealed a link between gene length and human ASD risk genes (King et al., 2013; Gabel et al., 2015). Remarkably, many of the genes suppressed by I-BET858 are on average significantly longer than the unaffected genes (Fig. 4). Treatment with I-BET858 for 12 h, but not for 2 h, of in vitro–cultured, nonstimulated neurons suppresses preferentially extra-long genes >100 kb with a correlation between gene length and reduced expression (Fig. 4 A; Pearson’s r = −0.147877; P < 0.0001). The average gene length of I-BET858–suppressed genes is ∼130 kb and exceeds the average length of genes (∼50 kb) expressed in these neurons by ∼2.5-fold (Fig. 4 B). Notably, 20% of the I-BET858–suppressed genes (194/1,158) are extremely long genes that range in length from 200–2,000 kb (Table S4). In accordance with previous data pointing to a possible correlation between extra-long gene length and human ASD risk genes (King et al., 2013), we found that 42 of the 100 longest I-BET858–repressed genes are potential ASD candidate genes (P < 0.0001; OR, 15.44; Table S4).
Figure 4.
I-BET858–mediated gene suppression correlates with gene length in vitro and in vivo. (A) Distribution of gene expression levels versus gene length in primary neurons after 2 h (red) or 12 h (blue) I-BET858 or vehicle treatment (2 h, black; 12 h, green) using microarray analysis (n = 3). Linear regression of log2 gene expression fold changes (middle) and running averages (right) are shown (Pearson’s r = −0.147877, red line; P < 0.0001). (B) Bar graph shows average gene length of I-BET858–induced (red) or –suppressed (blue) genes after 12 h using microarray analysis (n = 3); dashed line, average neuronal gene length. (C) Distribution of gene expression versus gene length in primary neurons after 12-h treatment with BNDF (red), vehicle (black), I-BET858 (green), or BDNF + I-BET858 (blue) using microarray analysis (n = 3) is shown. Linear regression (middle) and running averages (right) of log2 expression fold changes of 12-h BDNF induced genes ± I-BET are shown (Pearson’s r = −0.119946, red line; P = 0.004). (D) Bar graph shows average gene length of I-BET858–induced (red) and –suppressed (blue) BDNF-inducible genes after 12 h. (E) Distribution of gene expression versus gene length using RNA-seq analysis of mouse striatum (n = 2) after acute (left) or chronic (right) I-BET858 treatment are shown. (F) Bar graph shows average gene length of I-BET858–induced (red) and –suppressed (blue) genes after acute and chronic I-BET858 treatment, respectively. Statistics: two-tailed unpaired Student’s t test, error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
A similar correlation between gene length and susceptibility to I-BET858–mediated transcriptional suppression is observed in BDNF-treated neurons (Fig. 4, C and D). BDNF treatment leads to a slight increase in the expression of long genes (>100 kb) that are also strongly and specifically suppressed by I-BET858 treatment (Fig. 4 C; Pearson’s r = −0.119946; P = 0.004). The gene length of I-BET858 suppressed genes is approximately threefold higher than the average length of genes expressed in these neurons (Fig. 4 D, Table S4).
The strong correlation between gene length and I-BET858 susceptibility is also observed in vivo (Fig. 4 E). Similar to in vitro–cultured neurons, I-BET858 affected gene transcripts in the mouse brain in vivo are on average approximately twofold longer than those induced or unaffected by I-BET858 (Fig. 4 F and Table S4). Furthermore, 18 (acute) and 13 (chronic) of the 100 longest I-BET858–repressed genes in vivo are ASD candidate genes (P < 0.0001; OR, 4.428; P < 0.001; OR, 3.001). These genes include the transcriptional regulators Foxp1, Npas2, Phf2, Setbp1, Tcf3, and Zbtb20, and the neuronal signaling and adhesion proteins Antxr1, Cdh10, Cdh11, Grm1, Lamb1, Nrg1, and Slitrk5, many of which are similarly controlled by I-BET858 in cultured neurons in vitro. The preferential impact of I-BET858 on long genes in neurons may reflect the known role of BET proteins in regulation of transcriptional elongation (Zhou et al., 2012; Patel et al., 2013). It is possible that the successful generation of a nascent transcript for long genes >100 kb is more susceptible to perturbation of elongation efficiency as compared with shorter genes.
I-BET858 treatment induces autism-like behavior in mice
The selective impact of BET protein inhibition on neuronal gene expression and the preferential I-BET858–dependent inhibition of long genes suggested that treatment with I-BET may lead to the development of ASD-like behaviors in mice. The key behavioral symptoms of ASD in humans include abnormal social interactions, communication deficits, anxiety and repetitive behaviors. A combination of alterations in social behaviors, anxiety, and repetitive behaviors are therefore frequently used to diagnose autism-like phenotypes in mice (Peça et al., 2011; Ellegood and Crawley, 2015).
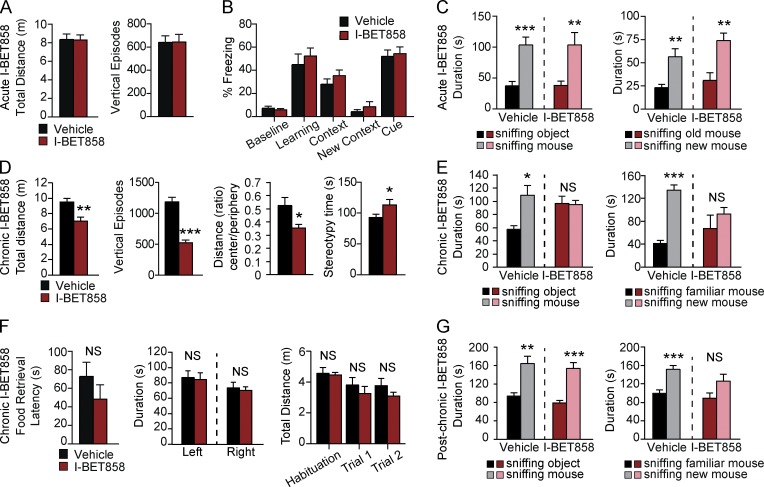
The impact of acute I-BET858 on mouse behavior was determined by assessing basal motor activity, anxiety-like behaviors, social interaction, and memory (Fig. 5). We did not observe any signs of distress, anxiety, or other behavioral or motor abnormalities in mice of various ages acutely treated with I-BET858 (Fig. 5 A and not depicted). Acute I-BET858 treatment also did not affect memory formation in mice. A single injection of I-BET858, after training mice in a fear-conditioning paradigm, has no effect on mouse memory of the foot-shock–associated context or auditory cue 24 h later, as compared with vehicle-injected controls (Fig. 5 B).
Figure 5.
I-BET858 treatment induces an autism-like syndrome in mice. (A–C) Effects of acute I-BET858 treatment on motor activity and exploration in the open field (A; n = 8 mice per group), memory (B; n = 10), and social interaction (C; n = 8) in mice are shown. (D and E) Effects of chronic I-BET858 treatment on motor activity, anxiety, and stereotypic behavior in the open field (D; n = 10), social behavior (E; n = 10), hidden food retrieval (F; n = 5), left versus right bias and total distance moved during the social interaction paradigm (n = 10) in mice are shown. (G) Social behavior in mice 5 mo after chronic I-BET858 treatment is shown (n = 5). Statistics: Student’s t test, error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
Contrary to acute BET inhibition, chronic daily I-BET858 administration starting at 4 wk of age led to the development of behavioral abnormalities consistent with an autism-like syndrome (Fig. 5, D and E). After 2 wk of chronic I-BET858 treatment, 6-wk-old mice displayed reduced explorative motor activity and heightened anxiety-like behavior in the open field (Fig. 5 D). I-BET858–treated mice also spend slightly more time engaging in stereotypic behaviors as compared with their vehicle-treated controls (Fig. 5 D). Using the three-chamber social approach task that is commonly used to reveal ASD-like social deficits in mice (Ellegood and Crawley, 2015), we found that chronic (Fig. 5 E), but not acute (Fig. 5 C), 6-wk-old I-BET858–treated mice displayed greatly reduced sociability. When tested for their preference to explore a novel mouse versus an inanimate novel object, the chronic I-BET858–treated mice spent equal amounts of time sniffing the mouse and the object, unlike vehicle-treated littermates that prefer to explore the novel mouse (Fig. 5 E). In addition to the reduced sociability, chronic I-BET858–treated mice showed a significant reduction in their preference for social novelty. Preference for social novelty requires functional social recognition and is defined as the test mouse spending more time exploring/sniffing a novel mouse than a familiar mouse. Unlike vehicle-injected mice, chronic I-BET858–treated mice displayed no significant preference in exploring a novel mouse over a familiar mouse (Fig. 5 E). Importantly, the observed effects on social preference in the I-BET858–treated mice are not due to defective olfaction or reduced overall explorative activity in the three-chamber assay; I-BET858-treated mice displayed no differences in hidden food retrieval, no left versus right bias differences, and no reduction in velocity, total distance moved, or time spent sniffing/exploring during the social interaction paradigm (Fig. 5 F and not depicted) as compared with control, vehicle-injected mice. Notably, mice that were chronically treated with I-BET858 between 4–6 wk of age showed a partial reversal of the social behavioral deficits with age (Fig. 5 G).
In summary, our findings reveal a key role of BET proteins in the regulation of selected genes that control normal neuronal development and function. The significance of BET proteins in the regulation of mouse behavior is underscored by the development of an ASD-like syndrome in mice treated with brain permeable I-BET858. The data also suggest that environmental factors that have the ability to affect BET protein function or BET target gene expression may contribute to ASD. The pharmacological induction of ASD-like symptoms in mice provides a valuable tool for the identification of genes that may play a pivotal role in the etiology of the disease or for the development of novel drugs targeting ASD. Moreover, our studies suggest that the pharmacological control of BET proteins or BET-dependent gene expression regulation could be used not only for modeling of autism-like behaviors, but possibly for the correction of aberrantly expressed genes in ASD and other neurodevelopmental disorders.
MATERIALS AND METHODS
Animals
4-wk-old male C57BL/6 mice were purchased from The Jackson Laboratory. Mice were housed at five animals per cage on a 12-h light/dark cycle (lights on from 0700 to 1900 h) at constant temperature (23°C) with ad libitum access to food and water. All studies were conducted in accordance with the GSK Policy on the Care, Welfare, and Treatment of Laboratory Animals and were reviewed by the Institutional Animal Care and Use Committee at GSK and the IACUC at Icahn School of Medicine at Mount Sinai where the work was performed.
BET inhibitors
Inhibitor characterization.
Fluorescence resonance energy transfer (FRET), surface plasmon resonance (SPR), and lipopolysaccharide-stimulated IL-6 cytokine production from human peripheral blood mononuclear cells (PBMCs) or whole blood (WB) to compare potencies of I-BET858, I-BET762, and I-BET151 was performed as previously described (Dawson et al., 2011).
In vivo pharmacokinetics.
I-BET858 penetration in mouse brain tissue was determined 4 h after a single I-BET858 i.p. injection of 30 mg/kg (n = 3 mice), and after continuous i.v. infusion to steady state at 0.35 mg/kg in rat. Pharmacokinetics were determined in mouse after i.p. injection at 3 and 30 mg/kg (n = 3 mice) and in rat after 1-h continuous i.v. infusion at 1 or 5 mg/kg p.o. administration (n = 3 rats). For time course measurements, mouse whole trunk blood and brain tissues were collected at 2, 16, and 26 h after a single injection of 30 mg/kg I-BET858 (n = 4–10/time point).
BRD4-BD1/ligand cocrystallization and x-ray structure determination.
BRD4-BD1 protein was produced for crystallographic studies as previously described (Nicodeme et al., 2010). BRD4-BD1 ligand complex was cocrystallized with at least 3:1 excess of compound at 9 mg/ml in 120 nl + 120 nl sitting drops using a 96-well MRC plate with a well solution of 0.1 M MIB (pH 4.0) 25% PEG1500 at 20°C and cryoprotected by adding 20% ethylene glycol before flash freezing in liquid nitrogen. Data from a single crystal was collected at the European Synchotron Radiation Facilities (Grenoble) and processed to 2.01 Å using XDS and AIMLESS within AUTOPROC (Vonrhein et al., 2011). Manual model building was performed using COOT (Emsley and Cowtan, 2004) and refined using REFMAC within the CCP4 suite of software (Project, 1994). The final crystal structures were deposited in the Protein Data Bank under the accession code 5acy.
Bromodomain profiling.
Bromodomain profiling was provided by DiscoveRx Corp. on the basis of BROMOscan. This screen accounted for the determination of the Kd between I-BET858 and each of the 34 DNA-tagged bromodomains included in the assay, by binding competition against a reference immobilized ligand (DiscoveRx; Theodoulou et al., 2015).
Primary cortical neuron culture
Embryonic day 18 (E18) timed-pregnant female mice were anesthetized with CO2 and sacrificed by cervical dislocation. In a dissection hood, 24–26 embryos per experiment were collected through an incision of the mother’s abdomen, taken out of the amniotic sacs, and decapitated in ice-cold Hank’s Balanced Salt Solution (HBSS). Using fine scissors and forceps, brains were rapidly dissected and the cortex cleared from meninges and isolated under a dissection microscope. Cortices were collected in ice-cold HBSS and kept on ice until all embryos had been dissected. In a tissue culture hood, HBSS was removed and the cortex tissue was digested by 0.25% Trypsin-EDTA for 12 min at 37°C, followed by DNase1 treatment for 10 min at 37°C. The tissue was dissociated by serial trituration with a 25-ml serological pipette, followed by trituration with 10 and 5 ml serological pipettes. Cell suspension was washed once with DMEM medium, supplemented with 10% FBS and 1% penicillin/streptomycin, and passed through a 40 µM cell strainer before being counted on a hemocytometer. Single cells were seeded on poly-d-lysine (0.1 mg ml−1)–coated wells at a density of 106 cells per well on a 12-well plate. Cells were grown in neurobasal medium, complemented with B27 supplement, N2 supplement, and 0.5 mM l-glutamine and maintained at 37°C in 5% CO2 for 1 wk. Cultures were treated with DMSO (0.2%), I-BET858 (1 µM, 0.2% DMSO), BDNF (50 ng/ml), or BDNF + I-BET858 for 2 and 12 h on day 7 in vitro. Cultures were washed with PBS on ice and processed for RNA isolation using TRIzol/Chloroform extraction.
In vivo I-BET858 treatment
I-BET858 solutions used for in vivo studies (3 mg/ml) were prepared in 10% Kleptose buffer with 5% DMSO (pH 6.5) and maintained at 4°C. I-BET858 was made fresh daily; buffers were made fresh each week. 30 mg/kg I-BET858 or vehicle (Kleptose buffer, pH 6.5) was delivered by i.p injection. For acute treatment, 6-wk-old mice received a single I-BET858 injection. For chronic I-BET858 treatment, 4-wk-old mice received a total of 14 i.p. injections (1/d) over 2 wk. Gene expression and behavioral changes were assessed in 6-wk-old mice at 4 h (acute, n = 8–10 for behavior, n = 2 for gene expression analysis) or 12 h (chronic, n = 10 for behavior, n = 2 for gene expression analysis) after the (final) I-BET858 injection, respectively.
RNA isolation
For in vitro gene expression analysis, neuronal culture plates were washed once with PBS, and TRIzol was added directly to cultures to isolate RNA (n = 3 per group). For in vivo gene expression analysis, mice were anesthetized with CO2 and decapitated. The striata from two control and acute or chronic I-BET–treated mice were rapidly dissected on ice and frozen in liquid nitrogen. RNA extraction from frozen samples was performed using TRIzol/Chloroform according to manufacturer’s instructions (Invitrogen). RNA was precipitated overnight at −80°C in isopropanol with 0.15 M sodium acetate, washed twice with 70% ethanol, air-dried, and resuspended in RNase-free water. RNA samples were purified using RNeasy Micro columns (QIAGEN) with on-column DNase treatment as specified by manufacturer. RNA integrity was assayed using nanodrop and RNA Pico chip on Bioanalyzer 2100 (Agilent) for quality RIN > 9.
Microarray analysis
Total RNA samples were prepared for microarray analysis as described previously (Schaefer et al., 2009). In brief, total RNA was converted to cDNA using the Superscript GeneChip Expression 3′-Amplification Reagents Two-Cycle cDNA Synthesis kit (Affymetrix) and the GeneChip T7-Oligo(dT) Primer. Affymetrix Mouse Genome 430 2.0 arrays were used in all experiments. Mouse Genome 430 2.0 arrays were scanned using the GeneChip Scanner 3000 (Affymetrix) and globally scaled to 150 using the Affymetrix GeneChip Operating Software (GCOS v1.4). Three biological replicates were performed for each experiment. GeneChip CEL files were imported into GeneSpring GX 13.0 (Agilent Technologies), processed with the GC-RMA algorithm, and expression values on each chip were normalized to that chip’s 50th percentile. Statistical analysis was performed to determine which genes are differentially expressed in BDNF-, I-BET858-, or BDNF + I-BET858–treated neurons as compared with the DMSO controls. Genes were filtered for a raw expression level of >20 and a fold change of >2, followed by moderate t test with a p-value cutoff of 0.05. P values were adjusted using the Benjamini-Hochberg correction. Gene expression changes are shown using Volcano plots where the corrected P value (-log10) is plotted versus fold-change (log2). Heatmaps were created by hierarchical gene clustering on entities using the Euclidean distance metric and the Ward’s linkage rule using GeneSpring GX 13.0.
RNA sequencing
Double-stranded cDNA was generated from 1–5 ng purified RNA using Ovation V2 kit (NuGEN) following manufacturer’s instructions. 500 ng of cDNA per sample were sonicated to obtain fragments of 200 bp using Covaris-S2 system (duty cycle,10%; intensity, 5.0; Bursts per second, 200; duration, 120 s; mode, frequency sweeping; power, 23 W; temperature, 5.5–6°C; Covaris Inc.). These fragments were then used to produce sequencing libraries with the TruSeq DNA Sample kit (Illumina). The quality of the libraries was ensured using the 2200 TapeStation (Agilent). Duplexed libraries were sequenced on HiSeq 2000, typically yielding on average 60 million, 100-bp-long single-end reads per sample (Illumina). All samples were mapped at a rate of 79–80%. After filtering out adaptor and low-quality reads, reads were mapped using TopHat (version 2.0.8) to the mm9 mouse genome. The Cufflinks/Cuffdiff suite was used to estimate gene-level expression values as fragments per kilobase of exon model per million mapped fragments (FPKM). Differentially expressed autosomal genes between control and acute I-BET858 or chronic I-BET858 libraries were determined using a p-value <0.05 and fold change >2. Values with a FPKM <0.5 were excluded.
Gene list statistics
Overlaps between gene sets were tested for statistical significance using the χ2 test by using GraphPad Prism 5.01. The total number of expressed genes in neurons as measured by microarray (25,788) was used to calculate the χ2 test for all lists generated by microarray. For RNA sequencing data, the total number of protein-coding genes in the mouse genome according to Mouse Genome Informatics (24,979) was used for the χ2 test. The ASD candidate gene list (n = 1193) was obtained from the SFARI homepage (Basu et al., 2009) and supplemented with the list from (King et al. (2013; Table S1).
Pathway analysis
Bioinformatic network and pathway analyses of I-BET858–suppressed genes have been performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6. Representative biological pathways from the top 25 enriched categories are shown. Pathway enrichment was calculated as −log10 p-value. The p-value cut-off (0.05) for significance is indicated by red dashed line.
Gene length analysis
Gene symbols were annotated with their gene start and end (in bp) using the mm9 database. Gene length was then calculated from these values. Replicate symbols were filtered out. To determine the relationship between fold-change (e.g., I-BET858 vs. DMSO) and gene length, the log2 fold-change was plotted versus length for all genes expressed in neurons. Pearson correlation coefficients and P values were calculated from the resulting scatter plot using linear regression. Genes were binned by length and a sliding window was used to calculate the average of the log2 fold changes for the genes within each length window (in vitro: window, 200 bp, step, 40 bp; in vivo: window, 800 bp, step, 20 bp). For the BDNF-induced, I-BET858–suppressed gene list, different parameters were necessary due to the significantly smaller list size (625 genes; window, 80 bp, step, 1 bp). To assess changes in gene expression levels in regards to length, raw microarray values were transformed using log10 and plotted versus length for each condition. A running average was calculated using window size of 400 bp and a step size of 20 bp. The code used to generate expression versus length plots can be found at https://github.com/GeneExpressionScripts/ibet858_expression_plots.
Behavioral analysis
All behavioral tests were performed on 6-wk-old adult C57BL/6 males (n = 8–10; The Jackson Laboratory) and were performed between 0700 and 1900 h. For all behavioral experiments, experimenters were blinded to the treatment of the animals. GraphPad Prism version 5.01 for Windows (Graph-Pad Software) was used for statistical analysis of the data. Samples corresponding to data points that are more than two standard deviations from the sample mean were excluded from analyses. All procedures were conducted in strict accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the IACUC at Icahn School of Medicine at Mount Sinai.
Locomotion and exploration.
Locomotion, exploratory, and thigmotaxis/anxiety behavior was measured using the open field analysis as previously described (Schaefer et al., 2009). Mice were assessed for 60 min and data were collected in 5-min intervals (n = 10). Locomotor activity was assessed by total distance traveled (m), anxiety-like behavior was defined by counts of rearing (number of vertical episodes in counts), and the ratio of the distance moved in the center versus the periphery (thigmotaxis), and stereotypical behavior (time spent). Mice were tested in the open field 4 h after acute (n = 8) and 12 h after chronic (n = 10) I-BET858 or vehicle treatment.
Memory and learning.
Memory and learning in mice acutely treated with I-BET858 or vehicle (n = 10) was analyzed using a standard fear conditioning paradigm (Med Associates) as described previously (Schaefer et al., 2009). In brief, a mouse was placed in the test chamber (house lights on) and allowed to explore freely for 2 min. A white noise (80 dB) was then presented for 30 s, coterminating with a mild foot shock (2 s, 0.5 mA). 2 min later, the same sequence of auditory cue-shock pairing was repeated. The mouse was removed from the chamber 30 s later and returned to its home cage. Freezing behavior was continuously recorded during the time spent in the test chamber. Mice were injected with either I-BET858 (30 mg/kg) or vehicle 10 min after the training. 24 h later, the mouse was placed back into the test chamber for 5 min and freezing behavior was recorded (context test). 2 h later, the mouse was tested for freezing in response to the auditory cue. Environmental and contextual cues were changed for the auditory cue test. The auditory cue test was divided into two phases: 3 min in the absence of the auditory cue and 3 min of the auditory cue. The time freezing during each test was converted to a percentage of freezing value.
Social interaction paradigm.
Social preference and social memory was performed as previous described (Ellegood and Crawley, 2015) using a Plexiglas chamber divided into three compartments. The two edge compartments contain an empty wire cup. Mice were habituated to the testing room for at least 1 h before the experiment. Stimulus mice are 6–8-wk-old C67Bl/6 male mice that were housed in separate areas of the animal facility and had no prior contact with the test mice. Stimulus mice were habituated to the wire cup before testing. For the sociability test, the test mouse is introduced to the middle chamber and allowed to freely move and habituate to all three compartments for 10 min. Then, the mouse was restricted to the middle chamber using the dividers, while a novel object (Lego) was placed under the wire cup in one chamber and an unfamiliar mouse in the other. The test mouse was then allowed to investigate the whole apparatus for 10 min. After, the mouse was again restricted to the middle chamber while the object was replaced by a second, unfamiliar mouse. The test mouse was allowed 10 min to investigate. Data were acquired using the Ethovision system (Noldus) to automatically track motion while manual scoring was used to quantify time spent sniffing the stimuli. Counterbalancing was used to control for potential left-right preferences.
Olfaction.
Olfaction was tested by exposing mice (n = 5) to a small amount of palatable food (Cinnamon Toast Crunch cereal; General Mills) once per day for 2 d. Mice were deprived of food overnight before the test. A clean cage was filled with roughly 3 inches of fresh bedding, and the stimulus food was buried in the bedding until it was not visible. Mice were then placed in the cage one at a time and allowed to freely explore. The latency to localize and retrieve the food was measured. All mice retrieved the food within 2 min. Bedding was mixed in between trials and tested mice were placed in a new holding cage until all cage-mates had been tested. After this, all mice were returned to their original cage and ad libitum food access was restored.
Accession nos.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO; Edgar et al., 2002) and are accessible through GEO SuperSeries accession no. GSE72149.
Online supplemental material
In Fig. S1, the characterization of the novel brain-permeable inhibitor of BET proteins (I-BET858) is shown. Fig. S2 shows the affinity analysis of I-BET858 for bromodomain-containing proteins. Table S1 contains the effects of I-BET858 treatment on gene expression in primary neurons in vitro (2 and 12 h). Table S2 lists the effects of I-BET858 on BDNF-inducible gene expression in primary neurons in vitro (2 and 12 h). Table S3 displays the effects of acute and chronic I-BET858 on striatal gene expression in vivo. Table S4 shows the gene lengths analysis of I-BET858 suppressed genes in vitro and in vivo. Online supplemental material are available at http://www.jem.org/cgi/content/full/jem.20151271/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Jacob Varley for his assistance with gene length analyses and Joe Scarpa for his help with the statistical analyses.
This work was supported by the National Institutes of Health (NIH) Director New Innovator Award DP2 MH100012-01 (A. Schaefer), 1R01GM112811-01 (A. Tarakhovsky), NIH Neuroscience Training grant T32MH096678 (J. Sullivan), CURE Challenge Award (A. Schaefer), the Emerald Foundation Inc. (A. Tarakhovsky), the Seaver Autism Center (A. Schaefer), and NARSAD Young Investigator Award #22802 (M. von Schimmelmann).
J. Gray, C.-w. Chung, N. Garton, N. Smithers, H. Lewis, and R.K. Prinja are employees and shareholders of GlaxoSmithKline. The authors declare no additional competing financial interests.
Footnotes
Abbreviations used:
- ASD
- autism spectrum disorder
- BDNF
- brain-derived neurotrophic factor
- BET
- bromodomain and extraterminal domain–containing proteins
- H4
- histone 4
- IEGs
- immediate early genes
- THQ
- tetrahydroquinoline
References
- Alder J., Thakker-Varia S., Bangasser D.A., Kuroiwa M., Plummer M.R., Shors T.J., and Black I.B.. 2003. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J. Neurosci. 23:10800–10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon C., Schneider M., Le Magueresse C., Froehlich H., Sticht C., Gluch C., Monyer H., and Rappold G.A.. 2015. Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behaviour. Mol. Psychiatry. 20:632–639. 10.1038/mp.2014.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I., Cannizzaro E., and Dawson M.A.. 2013. Bromodomains as therapeutic targets in cancer. Brief Funct Genomics. 12:219–230. 10.1093/bfgp/elt007 [DOI] [PubMed] [Google Scholar]
- Basu S.N., Kollu R., and Banerjee-Basu S.. 2009. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 37:D832–D836. 10.1093/nar/gkn835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V., Yoh S.M., and Jones K.A.. 2008. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 20:334–340. 10.1016/j.ceb.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calella A.M., Nerlov C., Lopez R.G., Sciarretta C., von Bohlen und Halbach O., Bereshchenko O., Minichiello L.. 2007. Neurotrophin/Trk receptor signaling mediates C/EBPalpha, -beta and NeuroD recruitment to immediate-early gene promoters in neuronal cells and requires C/EBPs to induce immediate-early gene transcription. Neural Dev. 2:4 10.1186/1749-8104-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.A., Peñagarikano O., Belgard T.G., Swarup V., and Geschwind D.H.. 2015. The emerging picture of autism spectrum disorder: genetics and pathology. Annu. Rev. Pathol. 10:111–144. 10.1146/annurev-pathol-012414-040405 [DOI] [PubMed] [Google Scholar]
- Dawson M.A., Prinjha R.K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W.I., Robson S.C., Chung C.W., Hopf C., Savitski M.M., et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 478:529–533. 10.1038/nature10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., et al. UK10K Consortium. 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 515:209–215. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 146:904–917. 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A., Kelly C., Grzadzinski R., Zuo X.N., Mennes M., Mairena M.A., Lord C., Castellanos F.X., and Milham M.P.. 2011. Aberrant striatal functional connectivity in children with autism. Biol. Psychiatry. 69:847–856. 10.1016/j.biopsych.2010.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., and Lash A.E.. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood J., and Crawley J.N.. 2015. Behavioral and Neuroanatomical Phenotypes in Mouse Models of Autism. Neurotherapeutics. 12:521–533. 10.1007/s13311-015-0360-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P., and Knapp S.. 2012. The bromodomain interaction module. FEBS Lett. 586:2692–2704. 10.1016/j.febslet.2012.04.045 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., et al. 2010. Selective inhibition of BET bromodomains. Nature. 468:1067–1073. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel H.W., Kinde B., Stroud H., Gilbert C.S., Harmin D.A., Kastan N.R., Hemberg M., Ebert D.H., and Greenberg M.E.. 2015. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 522:89–93. 10.1038/nature14319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmini R., Nguyen V.L., Toum J., Simon C., Brusq J.M., Krysa G., Mirguet O., Riou-Eymard A.M., Boursier E.V., Trottet L., et al. 2014. The discovery of I-BET726 (GSK1324726A), a potent tetrahydroquinoline ApoA1 up-regulator and selective BET bromodomain inhibitor. J. Med. Chem. 57:8111–8131. 10.1021/jm5010539 [DOI] [PubMed] [Google Scholar]
- Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E., Suárez-Fariñas M., Schwarz C., Stephan D.A., Surmeier D.J., et al. 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 135:738–748. 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I.F., Yandava C.N., Mabb A.M., Hsiao J.S., Huang H.S., Pearson B.L., Calabrese J.M., Starmer J., Parker J.S., Magnuson T., et al. 2013. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 501:58–62. 10.1038/nature12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., and Young R.A.. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153:320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E., Jeffrey K.L., Schaefer U., Beinke S., Dewell S., Chung C.W., Chandwani R., Marazzi I., Wilson P., Coste H., et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature. 468:1119–1123. 10.1038/nature09589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozburn A.R., Falcon E., Twaddle A., Nugent A.L., Gillman A.G., Spencer S.M., Arey R.N., Mukherjee S., Lyons-Weiler J., Self D.W., and McClung C.A.. 2015. Direct regulation of diurnal Drd3 expression and cocaine reward by NPAS2. Biol. Psychiatry. 77:425–433. 10.1016/j.biopsych.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., and Poo M.M.. 2013. Neurotrophin regulation of neural circuit development and function. Nat. Rev. Neurosci. 14:7–23. 10.1038/nrn3379 [DOI] [PubMed] [Google Scholar]
- Patel M.C., Debrosse M., Smith M., Dey A., Huynh W., Sarai N., Heightman T.D., Tamura T., and Ozato K.. 2013. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol. Cell. Biol. 33:2497–2507. 10.1128/MCB.01180-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peça J., Feliciano C., Ting J.T., Wang W., Wells M.F., Venkatraman T.N., Lascola C.D., Fu Z., and Feng G.. 2011. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 472:437–442. 10.1038/nature09965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Project C.C. Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- Qiu S., Lu Z., and Levitt P.. 2014. MET receptor tyrosine kinase controls dendritic complexity, spine morphogenesis, and glutamatergic synapse maturation in the hippocampus. J. Neurosci. 34:16166–16179. 10.1523/JNEUROSCI.2580-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell P.E., Fuccillo M.V., Maxeiner S., Hayton S.J., Gokce O., Lim B.K., Fowler S.C., Malenka R.C., and Südhof T.C.. 2014. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 158:198–212. 10.1016/j.cell.2014.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R., Meslamani J., and Zhou M.M.. 2014. The bromodomain: from epigenome reader to druggable target. Biochim. Biophys. Acta. 1839:676–685. 10.1016/j.bbagrm.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A., Sampath S.C., Intrator A., Min A., Gertler T.S., Surmeier D.J., Tarakhovsky A., and Greengard P.. 2009. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 64:678–691. 10.1016/j.neuron.2009.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou N.H., Bamborough P., Bannister A.J., Becher I., Bit R.A., Che K.H., Chung C.W., Dittmann A., Drewes G., Drewry D.H., et al. 2015. Discovery of I-BRD9, a selective cell active chemical probe for bromodomain containing protein 9 inhibition. J. Med. Chem. 10.1021/acs.jmedchem.5b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., and Bricogne G.. 2011. Data processing and analysis with the autoPROC toolbox. Acta. Crystallogr. D. Biol. Crystallogr. 67:293-302. 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Li T., and Price D.H.. 2012. RNA polymerase II elongation control. Annu. Rev. Biochem. 81:119–143. 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.D., Barnard M., Tian H., Li X., Ring H.Z., Francke U., Shelton J., Richardson J., Russell D.W., and McKnight S.L.. 1997. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. USA. 94:713–718. 10.1073/pnas.94.2.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.