Abstract
E3 ubiquitin ligases have been implicated in the ubiquitination and proteasome-mediated degradation of several key regulators of cell cycle. Owing to their pleotropic behavior, E3 ubiquitin ligases are tightly regulated both at transcriptional and post-translational levels. The E3 ubiquitin ligase TRUSS (tumor necrosis factor receptor-associated ubiquitous scaffolding and signaling protein) which negatively regulates c-Myc, are found down-regulated in most human cancer cell lines. However, the mechanism of regulation of intracellular levels of TRUSS remains elusive. Here we show that TRUSS is expressed majorly during the G1 phase of cell cycle and its level starts to decline with the expression of S-phase specific E3 ligase Skp2. Enforced expression of Skp2 led to a marked increase in the ubiquitination of TRUSS after its phosphorylation by GSK3β and followed by rapid proteolytic degradation. Our co-immunoprecipitation studies suggested a direct interaction between Skp2 and TRUSS through the LRR motif of Skp2. Interestingly, the human tumor samples that exhibited elevated expression of Skp2, showed relatively poor expression of TRUSS. Further, enforced expression of HBx, the oncoprotein of Hepatitis B virus which is known to stabilize c-Myc and enhance its oncogenic potential, led to the intracellular accumulation of TRUSS as well as c-Myc. Apparently, HBx also interacted with TRUSS which negatively impacted the TRUSS-c-Myc and TRUSS-Skp2 interactions leading to stabilization of TRUSS. Thus, the present study suggests that TRUSS is a novel substrate of E3 ligase Skp2 and that disruption of TRUSS-Skp2 interaction by viral oncoproteins could lead to pathophysiological sequelae.
Keywords: cell cycle, c-Myc, E3 ubiquitin ligase, HBx, Skp2, TRUSS, ubiquitination
Abbreviations
- ANCT
adjacent non-cancerous tissues
- CRL4
Culin ring finger ligase-4
- DCAFs
DDB1-Cul4 associated factors
- CHX
cycloheximide
- DCAFs
DDB1 and Cullin1-associated factors
- DDB1
damage-specific DNA-binding protein 1
- GSK3β
glycogen synthase kinase β
- HBx
hepatitis B virus X protein
- IB
immuno-blotting
- IHC
immunohistochemistry
- IP
immune-precipitation
- LRR
leucine-rich repeat
- RT-PCR
Reverse transcriptase-PCR
- RT-qPCR
real time quantitative PCR
- SCF
Skp1-cullin1-F box protein
- Skp2
S-phase kinase-associated protein-2
- TRUSS
Tumor necrosis factor receptor-associated ubiquitous scaffolding and signaling protein
- UPS
ubiquitin proteasome system.
Introduction
The ubiquitin-proteasome system (UPS) serves as a pivotal regulator of cellular homeostasis via destruction of regulatory proteins within the cell. It is essential for the regulation of cell cycle, transcription, DNA damage repair and even apoptosis.1 It consists of 3 different enzymes E1 (Ubiquitin activating enzyme), E2 (Ubiquitin-conjugation enzyme) and E3 (ubiquitin ligase) working in a chain to transfer the ubiquitin moiety to the target proteins followed by their degradation by 26S proteasome.2 Cullin-based E3 ubiquitin ligases are the largest class of UPS with a central role in cell cycle progression and includes Skp1-cullin1-F box protein (SCF), Elongin B/C-cullin 2-VHL protein and Cullin 4-based E3 ligase.3 SCF is the most studied class of E3 ligases having multiple F-box proteins as a substrate recognition component including Skp2, Fbw7 and βTrCP. Both Skp2 and Fbw7 functions as a positive regulator of cell cycle progression by regulating the intracellular levels of c-Myc, cyclin D1, cyclin A, cyclin E, p57, BRCA2, ORC1, E2F, c-Jun, and Notch. Therefore, it is not surprising to note that the Skp2 and Fbw7 levels are found deregulated in many cancers.4,5 Cullin4, which is conserved from yeast to human uses damaged DNA binding protein 1 (DDB1) as a linker to interact with substrate recognition subunit called DDB1 and cullin1-associated factors (DCAFs), and regulates a wide array of cellular process including cell proliferation, cell survival, DNA repair and genomic integrity.6 More than 2 dozen proteins are reported to be degraded by CRL4s E3 ubiquitin ligases including xeroderma pigmentosum group C proteins (XPC) and damaged DNA binding protein 2 (DDB2) that are involved in DNA damage repair, and histone H2A, H3 and H4 that are essential for chromatin assembly. Recent studies on CRL4s have suggested over 50 different substrate recognition components or DCAFs.7-11 Tumor necrosis factor receptor-associated ubiquitous scaffolding and signaling protein is one of the DCAFs known to regulate c-Myc are well known to be overexpressed in several hematological and solid tumors. Though, deregulated expression of TRUSS has been observed in many cancers,12 very little is known about its other cellular targets and functions. Alterations, such as mutations in the myc gene, its overexpression, translocation, gene amplification or even enhanced protein stability contribute to Myc-induced oncogenesis.13,14
CRLs are targeted by several viruses, which exploit the ubiquitination pathway in order to evade the innate cellular antiviral response and help viral propagation. Viruses belonging to paramyxovirus family such as simian virus 5, human parainfluenza virus type 2 and mumps virus use DDB1-Cul4 to degrade signal transducer and activator of transcription protein which otherwise would mount an antiviral interferon response.15-17 The HBx protein of Hepatitis B virus (HBV) is also known to bind DDB1 possibly to facilitate viral replication by extending the S phase of cell cycle.18,19 Further, HBx is shown to stabilize c-Myc by disrupting the SCFSkp2 complex and interfering with its ubiquitination.20
Although both TRUSS and Skp2 regulate the intracellular levels of c-Myc, it is not clear if both regulators work in synergy or act sequentially during different phases of the cell cycle. Thus, the selective disruption of SCFSkp2 complex by HBx in order to regulate Myc functions, prompted us to study the cellular regulation of TRUSS activity under the HBx microenvironment. In the present study, we show that intracellular TRUSS levels are negatively regulated by Skp2 and that TRUSS levels are stabilized in the presence of viral HBx or upon down-regulation of Skp2.
Results
Skp2 interacts with TRUSS
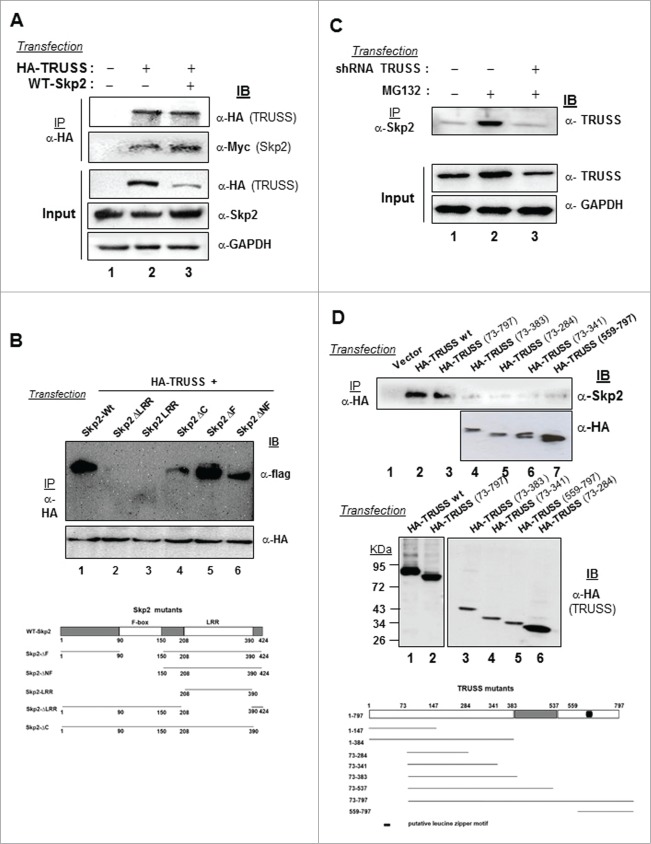
TRUSS has been identified as a Myc regulator whose levels are downregulated in cancerous cells.12 Considering the importance of c-Myc in cell cycle regulation, we analyzed the expression of TRUSS in the synchronized population of human hepatoma Huh-7 cells (Fig. S1A). We observed that TRUSS protein is expressed during first 6 h of release and then declined as cells progressed to S phase coinciding with the appearance of S phase kinase protein 2 (Skp2) albeit its mRNA levels remains unchanged during the same period (Fig. S1B, C). These results suggested 2 things: (a) TRUSS protein level gets down-regulated at the onset of S phase independent of its gene expression, and (b) TRUSS could be a substrate of Skp2. To investigate the regulation of TRUSS by Skp2, we first investigated the interaction between 2 proteins. Our co-immunoprecipitation studies suggested that TRUSS bound to Skp2 in a cellular milieu as HA-tagged recombinant TRUSS could pull down Skp2 (Fig. 1A). The TRUSS-Skp2 interaction was, however, disrupted in the presence of ΔLRR-Skp2 but not ΔF-Skp2 suggesting the involvement of LRR motif in substrate binding (Fig. 1B).The specificity of this interaction was further evident from the abrogation of Skp2 binding to endogenous TRUSS in the presence of TRUSS-specific shRNA (Fig. 1C; Fig. S2). Further our mutational studies with TRUSS deletion mutants suggested that the entire length of TRUSS except for its N terminal 72 amino acids, was required for Skp2 binding (Fig. 1D). Together these results substantiated that Skp2 specifically interacts with TRUSS in a cellular milieu.
Figure 1.
Interaction of TRUSS with Skp2. Huh-7 cells were transfected with the expression vectors for HA-TRUSS and WT-Skp2 (2.5 μg each) (A) or HA TRUSS (1μg) along with WT-Skp2 and ΔF-Skp2, ΔNF-Skp2, ΔLRR-Skp2, ΔC-Skp2 and LRR-Skp2 (2 μg each) (B), or with 2.5 μg of shRNA TRUSS (C) or with 2.5 μg each of different deletion mutants of TRUSS (D). The cell lysates were immunoprecipitated with indicated antibodies followed by immuno-blotting for Skp2, FLAG, TRUSS and GAPDH.
Skp2 mediates degradation of TRUSS by promoting its polyubiquitination
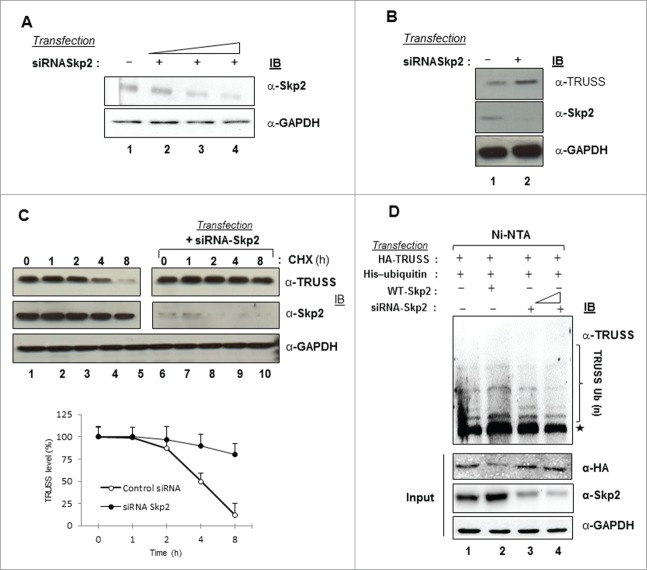
The levels of E3 ubiquitin ligases are considered to be proteolytically regulated by each other during the cell cycle.21 Since the TRUSS level declined as cells moved from the G1 to S phase, we studied the regulation of TRUSS in the presence of Skp2, a key E3 ubiquitin ligase involved in the S phase progression. The intracellular level of TRUSS was measured after co-expressing either Skp2 or its deletion mutants ΔC-Skp2 and ΔF-Skp2 (shown in Fig. 1D). Enforced expression of Skp2 led to a dramatic decline in the levels of TRUSS protein. Interestingly, the TRUSS level was restored in the presence of dominant negative mutant ΔF-Skp2 but not by ΔC-Skp2 suggesting the involvement of F box region of Skp2 in TRUSS degradation (Fig. 2A). Further, increased expression of Skp2 was associated with a progressive decline in the TRUSS levels (Fig. 2B). As expected, the endogenous TRUSS was also destabilized following Skp2 overexpression but was stabilized in the presence of ΔF-Skp2 (Fig. 2C). Interestingly, no significant change in the TRUSS mRNA level was observed under these conditions (Fig. S3) suggesting a post-transcriptional regulation of TRUSS protein. This conjecture was confirmed by enforced expression of CDH1 which is a known regulator of Skp2.21 There was a marked increase in the stability of TRUSS in the presence of CDH1 probably due to the degradation of Skp2 (Fig. S4). As TRUSS appeared to be a substrate of Skp2, we next checked the half-life of TRUSS in the presence of Skp2. Cells transfected with TRUSS and/or Skp2 expression vectors were subjected to cycloheximide (CHX) treatment to inhibit de novo protein biosynthesis. As shown in Figure 2D, enforced expression of Skp2 resulted in a considerable decrease in the steady-state levels of TRUSS protein in the presence of CHX due to its destabilization by Skp2. Further, metabolic labeling of cells in presence of 35S labeled amino acids followed by pulse chase and densitometric analyses of autoradiographs suggested a considerable decrease in the steady state levels of TRUSS in the presence of Skp2 (Fig. 2E). While the TRUSS-transfected cells showed only a marginal (˜25%) decline in TRUSS level in first 2h, and nearly 50% decrease by 4h. In contrast, cells co-expressing TRUSS and Skp2 showed a rapid decline (˜60% decrease within first 2h) in TRUSS level. To investigate the role of ubiquitin-proteasome system (UPS) in this process, TRUSS protein was immunoprecipitated in the presence of MG132 (a proteasome inhibitor). As shown in Figure 2F, there was a marked increase in the polyubiquitination of TRUSS in the presence of Skp2 (lane 3). As expected, the ΔF-Skp2 mutant that did not destabilize TRUSS also did not promote the polyubiquitination of TRUSS (lane 4). Together, these results suggested that TRUSS is a substrate of Skp2 and that its intracellular levels are regulated via polyubiquitination.
Figure 2.
Regulation of TRUSS expression by Skp2. Huh-7 cells were transfected with expression vectors of HA-TRUSS (1 μg) along with 1.5 μg each of WT–Skp2, Flag-ΔF-Skp2 or ΔC Skp2 (A) or with increasing concentrations in combination with (0.5, 1 and 2 μg) of either Skp2 and Flag ΔF-Skp2 (1 μg) (B) or with WT–Skp2 and Flag-ΔF-Skp2 (C). Cell lysates were western-blotted for recombinant TRUSS, Skp2, c-Myc. Huh-7 cells were transiently transfected either with 2.5 μg WT-TRUSS alone or along with WT-Skp2 (D). After 48 h, cells were incubated with cycloheximide for indicated time points and the cell lysates were western-blotted for TRUSS, Skp2 and GAPDH. Huh-7 cells were metabolically labeled with [35S] methionine/cysteine mix, pulse chased for indicated time periods flowed by the immunoprecipitation of cell lysates with anti-HA antibody resolved by SDS/PAGE and auto-radiographed (E). The total cell lysates were also western blotting for TRUSS and Skp2. GAPDH was used as internal control. (F) For ubiquitination assay, HEK293 cells were transfected with wild-type HA-TRUSS (1 μg), along with 2 μg each of His-Ub, WT-Skp2 and ΔF-Skp2 as indicated. The ubiquitinated proteins were pulled down under denaturing conditions using Ni-NTA agarose beads and were analyzed via protein gel blotting. The asterisk indicates a band that may correspond to TRUSS nonspecifically bound to the Ni-NTA beads.
Downregulation of Skp2 stabilizes TRUSS by inhibiting its ubiquitination
To further confirm the role of Skp2 in the regulation of intracellular TRUSS levels, we downregulated endogenous Skp2 using specific small interfering RNAs (siRNA) (Fig. 3A). Knocking down of Skp2 expression resulted in the stabilization of TRUSS protein (Fig. 3B) an overall increase (P < 0.001) in the steady-state level of TRUSS in the presence of CHX (Fig. 3C). Also, there was a marked decline in the polyubiquitination of TRUSS in the presence of Skp2 siRNA (Fig. 3D, compare lane 3 to 4). Together these results suggested that Skp2 is a regulator of polyubiquitination and degradation of TRUSS.
Figure 3.
Skp2 depletion promotes TRUSS accumulation. Huh-7 cells were transfected with different concentrations of Skp2 siRNA (A) (B). Cell lysates were western-blotted for TRUSS, Skp2, and GAPDH. Huh-7 cells were transiently transfected with Skp2 siRNA (C). After 48 h, cells were incubated with cycloheximide for indicated time points and the cell lysates were western-blotted for TRUSS, Skp2 and GAPDH. Data are shown as mean ± SD of 3 independent observations (P < 0.001). For ubiquitination assay, HEK293 cells were transfected with wild-type HA TRUSS (1 μg), in combination with 2 μg of His-Ub, WT-Skp2 (1 μg) and Skp2 siRNA {20nM (lane 3) and 50 nM (lane 4)} (D). The ubiquitinated proteins were pulled down under denaturing conditions using Ni-NTA agarose beads and were analyzed via western blotting. The asterisk indicates a band that may correspond to TRUSS nonspecifically bound to the Ni-NTA beads.
Elevated expression of Skp2 correlates with low expression of TRUSS in human tumors
As we observed that overexpression of Skp2 down-regulated TRUSS levels and an earlier report suggested low levels of TRUSS in a number of human cancer cell lines,12 we next investigated the pattern of TRUSS and Skp2 protein expression in different tumor tissues. Serial sections of different human tumors (one each of HCC, pancreatic adenocarcinoma, colonic adenocarcinoma and breast invasive ductal carcinoma) and their adjacent non-cancerous tissues (ANCT) prepared from the paraffin blocks, were analyzed by immunohistochemistry (IHC) using specific antibodies. Our IHC results revealed that Skp2 was expressed at high levels in all the tumor samples analyzed while a diffused expression of TRUSS was observed in these tumor samples (Fig. 4). In contrast, the ANCT samples, with the exception of pancreatic tissue, showed relatively low expression of both TRUSS and Skp2 (Fig. S5). Together these results suggested that poor expression of TRUSS in tumor tissues is possibly due to its in vivo regulation by elevated levels of Skp2.
Figure 4.
TRUSS and Skp2 protein expression in different human tumor tissues. Serial tissue sections of HCC, pancreas, colon and breast tumor tissues were analyzed for the expression of TRUSS and Skp2 using specific antibodies. Incubation of samples with IgG was used as negative control. The hematoxylin and eosin (H-E)-stained sections of different tumor tissues are also shown. All images, original magnification, × 100.
Phosphorylation of TRUSS by GSK3β is essential for its recognition by Skp2
The intracellular levels and activity of many regulatory proteins are determined by their site-specific phosphorylation by specific protein kinases.22 To find whether phosphorylation of TRUSS was required for its Skp2-mediated degradation and to identify the kinase(s) involved in the process, we performed a bioinformatical analysis of the primary sequence of TRUSS. Our analysis predicted at least 9 putative phosphorylation sites for GSK3β within TRUSS sequence (Fig. 5A). To establish the involvement of GSK3β in TRUSS phosphorylation, we treated the Skp2-expressing cells with increasing concentrations (5, 10 and 20μM) of GSK3β inhibitor and measured TRUSS levels. Our immunoblot analysis showed a marked increase in TRUSS stability in the presence of GSK3β inhibitor despite Skp2 overexpression (Fig. 5B). Moreover, our pulse-chase analysis revealed that the GSK3β inhibitor delayed the turn-over of TRUSS (Fig. 5C). We next investigated the possible role of phosphorylation in the ubiquitination of TRUSS. Our ubiquitination assay revealed that cells treated with different doses (5, 10 and 20μM) of GSK3β inhibitor showed a progressive inhibition in the ubiquitination of TRUSS (Fig. 5D). Together these data suggested that GSK3β-mediated phosphorylation of TRUSS is essential for its Skp2-dependent ubiquitination and proteasomal degradation.
Figure 5.
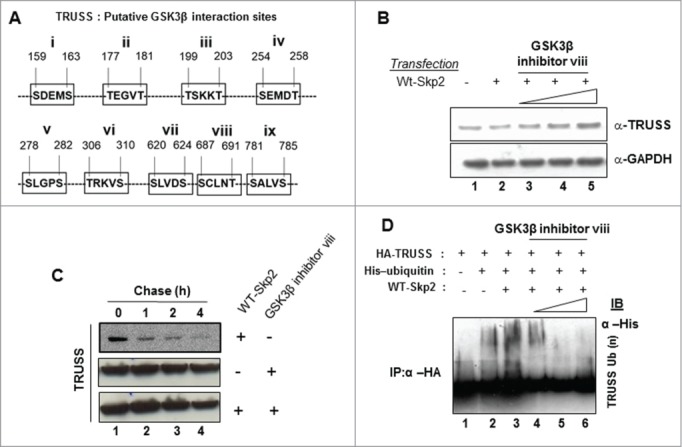
GSK3β facilitates TRUSS degradation. Huh-7 cells were transfected with expression vectors of WT-Skp2 with increasing concentrations of GSK3β inhibitor viii (5 μM, 10 μM and 20 μM) (B). Cell lysates were western-blotted for TRUSS and GAPDH. Huh-7 cells were transiently transfected either with 2.5 μg WT-Sp2 alone or along with GSK3β inhibitor viii (20 μM) (C). After 48 h, cells were incubated with cycloheximide for indicated time points and the cell lysates were western-blotted for TRUSS, and GAPDH. Cells were transfected with wild-type HA TRUSS (1 μg), in combination with 2 μg His-Ub, WT-Skp2 (1 μg) and increasing doses of GSK3β inhibitor viii (5 μM, 10 μM and 20 μM) (D). The cell lysates were harvested after MG132 treatment and immunoprecipitated with anti-HA antibody followed by immunoblotting for His.
Viral oncoprotein HBx interacts with TRUSS and enhances its intracellular stability
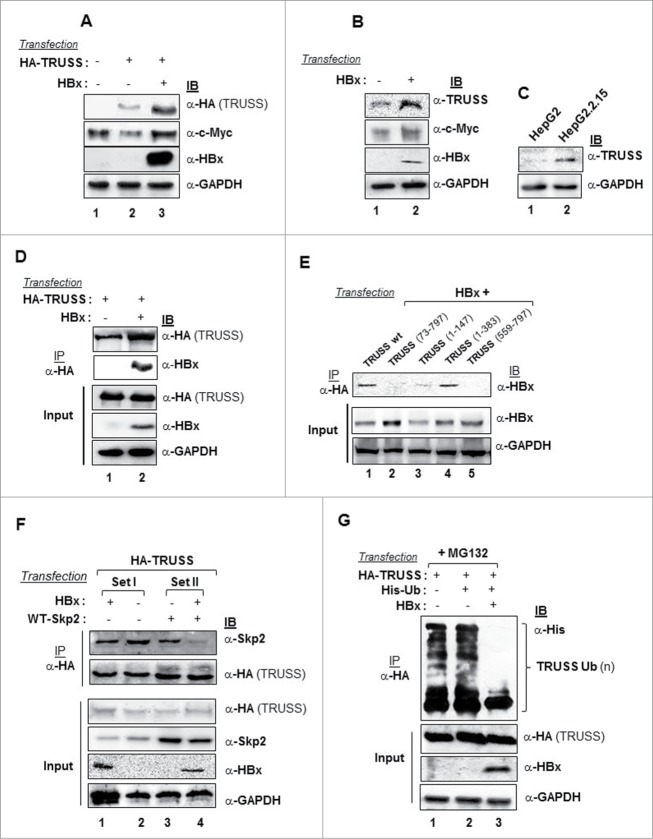
Accumulation of Myc protein has been implicated in the development of hepatocellular carcinoma,14 while TRUSS levels are generally downregulated in most human cancer cell lines.12 Therefore, we measured the levels of TRUSS in hepatoma cells following enforced expression of HBx. There was a marked increase in the level of recombinant HA-TRUSS protein as well as c-Myc in the presence of HBx (Fig. 6A). Further, the levels of endogenous TRUSS also increased, albeit marginally, in the presence of HBx (Fig. 6B). These findings could be corroborated in HepG2.2.15 cells that carry chromosomally integrated sequences of the HBV genome and express the HBx gene. HepG2.2.15 cells showed the accumulation of native TRUSS as compared to HepG2 cells (Fig. 6C). As TRUSS protein got stabilized in the presence of HBx, we next examined the possible interaction between TRUSS and HBx. Our Co-immunoprecipitation studies suggested an interaction between HBx and TRUSS in a cellular milieu (Fig. 6D). Further, confocal microscopy revealed the co-localization of HBx and TRUSS in the perinuclear region of cells (Fig. S6). To fine map the HBx interacting domain of TRUSS, we used the deletion mutants of TRUSS described in the previous section. As shown in Figure 6E, the N-terminal (1–72) and (148–383) domains of TRUSS were essential for its interaction as deletions of both N-terminal (1–72) amino acids (lane 2), and C-terminal (148–797) amino acids (lane 3) compromised its binding with HBx but remained intact in presence of N-terminal (1–383) amino acids (lane 4). Notably, the Skp2-TRUSS interaction also seemed to be compromised in the presence of HBx (Fig. 6F, compare lane 1 and 2) as well as in overexpressed Skp2 conditions (Fig. 6F, compare lane 3 and 4) and thus, be a reason for TRUSS stabilization. Further, the interaction between HBx and TRUSS led to a dramatic reduction in the ubiquitination of TRUSS (Fig. 6G, compare lanes 2 and 3) and enhancing its stability and accumulation. Apparently, HBx stabilizes TRUSS by compromising the Skp2-TRUSS interaction and preventing its ubiquitination.
Figure 6.
Stabilization of TRUSS in the presence of viral HBx. Huh-7 cells were co-transfected with the HBx expression vector X0 and HA-TRUSS (1.5 µg each) (A) or X0 alone (B) and the cell lysates were protein gel blotted for TRUSS, c-Myc and HBx. (C) Cell lysates of un-transfected HepG2 and HepG2.2.15 cells were western-blotted for endogenous TRUSS. Huh-7 cells were transfected with X0 (2.5 µg) along with 2.5 µg each of either wild-type HA-TRUSS (D) or TRUSS deletion mutants (E), and the cell lysates were immunoprecipitated with anti HA antibody followed by immuno-blotting for HBx or recombinant TRUSS. Huh-7 cells were transfected with expression vectors HA-TRUSS (1 µg) along with the expression vectors of HBx (2 µg) (set I) or 2 µg each of HBx and Skp2 (Set II) (F), the cell lysates were immunoprecipitated with anti HA antibody followed by immuno-blotting for HBx. (G) For ubiquitination assay, cells were transfected with HA-TRUSS in combination with Ub and or X0 and the cell lysates were harvested after MG132 treatment and immunoprecipitated with anti-HA antibody for the recombinant TRUSS followed by immuno-blotting with anti-His antibody.
HBx disrupts Skp2-c-Myc and TRUSS-c-Myc interactions leading to c-Myc accumulation
Earlier we have shown that HBx can disrupt the SCFSkp2 complex leading to the accumulation of its target protein c-Myc.20 Although, TRUSS has a role in Myc degradation,12 it was surprising to note that Myc level did not decrease and rather increased, despite elevated levels of TRUSS under the HBx microenvironment (Fig. 6A). Therefore, we examined the status of TRUSS/Myc and Skp2/Myc interactions in the presence of HBx. We found that the interaction of Skp2 with c-Myc was abrogated in the presence of HBx (Fig. 7A). Further, the interaction between TRUSS and c-Myc was also compromised in presence of HBx, just as Skp2-c-Myc interaction (Fig. 7B). Together, these results suggested a competitive binding of HBx with Skp2 and TRUSS leading to interference with the functions of CRL4TRUSS complex.
Figure 7.
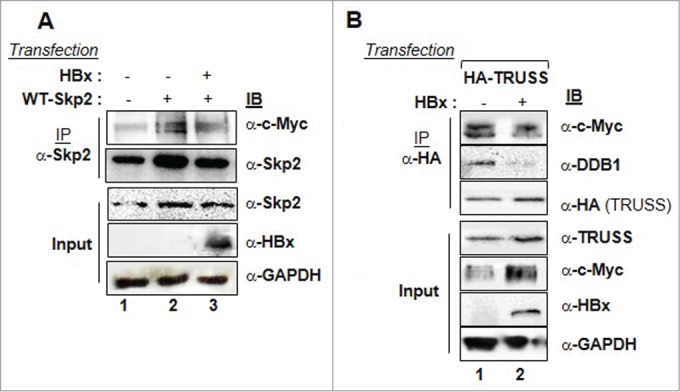
Destabilization of the CRL4TRUSS complex and accumulation of c-Myc in the presence of HBx. Huh-7 cells were transfected with WT-Skp2 (1 µg) alone or along with the expression vectors of HBx (2 µg) (A) or HA-TRUSS alone or along with expression vector of HBx (B). The cell lysates were immunoprecipitated with anti-Skp2 (A) or anti-HA antibodies (B) followed by their immunoblotting with indicated antibodies. GAPDH was used as internal control in all experiments.
Discussion
Regulated protein degradation is essential for maintaining homeostasis in the cell. The E3 ubiquitin ligases reportedly play a central role in this process as their deregulation often results in diseased states including cancers.23 Skp2, which is recognized as the key regulator of c-Myc in the cell,24 also functions as a cell cycle regulator by controlling the levels of cyclin-dependent kinase inhibitors such as p27, p130, p57 and p21,25-28 and replication control protein such as hORC1 and Cdt1,29-30 Cyclin E31 and E2F132 has been reported differently expressed in several cancers. More recently, Skp2 has been shown to exhibit homeostatic regulation of tumor suppressor DAB2IP and suppression of JARID1B demethylase activity in prostate carcimona.33,34 Likewise cullin1 based E3 ubiquitin ligases, cullin4-based E3 ubiquitin ligases also play an important role in cell proliferation, DNA damage repair7 and ubiquitylation of histone10 via different DCAFs like Cul4-DDB1-Det1-Cop1 regulate intracellular c-jun level.35
The present study showed that TRUSS expression peaked during the G1 phase of cell cycle and as the cells progress from G1 to S phase TRUSS level gets down-regulated coinciding with Skp2 expression. TRUSS is a regulator of c-Myc but very little is known about other biological functions of TRUSS and its regulation. Here we have shown that Skp2 post-translationally degraded TRUSS protein without affecting its mRNA level. The regulation of TRUSS by Skp2 involved a direct interaction between 2 proteins and required the LRR motif of Skp2 and nearly full length TRUSS except the N-terminal 1–72 amino acids (Fig. 1A-D). The specificity of TRUSS regulation by Skp2 was evident from the restoration of its level in the presence of dominant negative Skp2 (Fig. 2A-C). Further, a sharp decrease in the half-life of TRUSS in the CHX treated cells and TRUSS labeled with 35S under Skp2 microenvironment corroborated with our findings that Skp2 indeed was a regulator of TRUSS (Fig. 2D, E). Further, enhanced polyubiquitination and degradation of TRUSS following Skp2 overexpression (Fig. 2F) and accumulation of TRUSS protein or increase in its half-life in a Skp2-depleted environment confirmed the regulatory role of Skp2 in maintaining intracellular TRUSS levels (Fig. 3). We also present compelling evidence supporting that Skp2-mediated TRUSS degradation is facilitated by GSK3β. We found that inhibition in GSK3β activity inhibited the polyubiquitination of TRUSS and confirmed that phosphorylation of TRUSS by GSK3β was necessary for its degradation (Fig. 5).
The regulation of TRUSS levels by Skp2 observed in cell culture experiments were adequately substantiated by our in vivo studies. Immunohistochemical analysis of 4 different human cancer samples (liver, pancreas, colon and breast) showed enhanced expression of Skp2 (Fig. 4) as compared to adjacent non-cancerous tissues (ANCT) (Fig. S5). Interestingly, serial sections of the same tumors showed poor expression of TRUSS (Fig. 4) indicating its possible regulation by Skp2 in vivo.
Viral oncoproteins are known to target the ubiquitin-proteasomal pathways to support their replication and propagation.7 Viruses not only regulate the host-encoded E3 ligases but also encode their own Ub ligases in order to exploit the cellular homeostasis.36-40 The viral HBx oncoprotein which is implicated in hepatocellular carcinoma (HCC),41 is also known to regulate the proteasomal functions.42 HBx can destabilize the SCFSkp2 complex resulting in increased intracellular stability of c-Myc,20 and its improved co-operation with c-Myc in ribosomal biogenesis and cell transformation.43 Since TRUSS is a regulator of Myc protein, analysis of TRUSS levels indicated its accumulation in HepG2.2.15 cells that constitutively express the HBx oncoprotein (Fig. 6C). Akin to this observation, a high TRUSS level was also seen in the HBx-transfected hepatoma cells (Fig. 6A, B). Interestingly, HBx reduces the polyubiquitination of TRUSS which leads to the stabilization of TRUSS (Fig. 6G). The stabilization of TRUSS also involved a direct interaction between TRUSS and HBx (Fig. 6D) and involved the N-terminal 1–72 amino acids of TRUSS (Fig. 6E). Thus, it is quite possible that HBx binding to TRUSS could mask the domain required for Skp2 interaction. While the interaction of Skp2 with TRUSS results was followed by ubiquitination and proteasomal degradation TRUSS, HBx seemed to interfere with TRUSS degradation by disrupting the TRUSS-Skp2 interaction (Fig. 6F).
Since Myc is a target of TRUSS, the elevated TRUSS level under HBx microenvironment is expected to destabilize c-Myc. To our surprise, we noted that Myc levels also remained higher under these conditions (Fig. 6A, B). The elevated Myc levels possibly be due to a compromise in the interaction between Skp2/c-Myc and TRUSS/c-Myc and destabilization of the CRL4TRUSS complex in the presence of HBx (Fig. 7). The mechanism by which HBx can override the Skp2 functions has been reported earlier.20 Herein, we established the mechanism of HBx-mediated destabilization of CRL4TRUSS complex. Our domain mapping studies suggested that amino acid regions 1–72 of TRUSS are crucial for its interaction with HBx (Fig. 6E). Note that the same N-terminal 1–72 amino acids of TRUSS are critical for recruiting DDB1-Cul4 complex and mediating Myc degradation.12 Thus, the HBx binding with TRUSS is likely to mask and prevent DDB1-Cul4 interaction. As a result the CRL4TRUSS complex gets destabilized resulting in the accumulation of c-Myc levels.
Thus, the present study suggests that Skp2 is a major regulator of TRUSS activity during the G1/S transition. The degradation of TRUSS during G1/S transition may be a mechanism to stabilize certain positive regulators of cell cycle. Thus, TRUSS turnover by Skp2 could be an inbuilt mechanism in the regulation of cell cycle. Further, it involves a direct interaction between Skp2 and TRUSS followed by ubiquitination and degradation of TRUSS. The viral HBx seems to destabilize both SCFSkp2 and CRL4TRUSS complexes to get control over their targets including c-Myc (Fig. 8). A deregulated c-Myc level might be leading to cell transformation and creating a ground for other players to trigger hepatocarcinogenesis.
Figure 8.
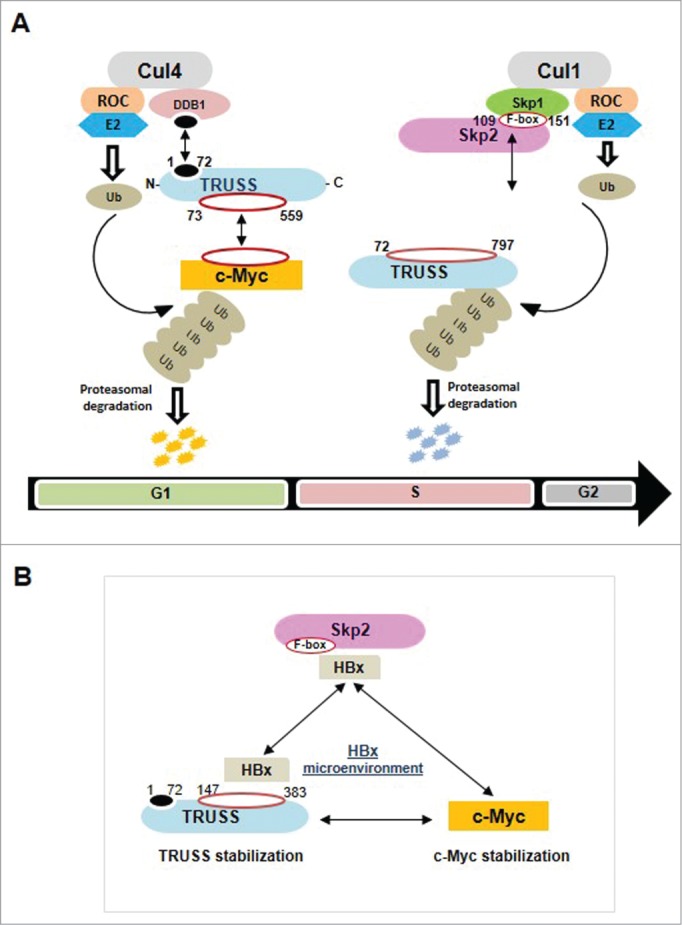
Schematic representation of the regulatory network of E3 ubiquitin ligases TRUSS and Skp2. (A) Regulation of c-Myc and TRUSS by Skp2 and c-Myc by TRUSS. (B) Stabilization of TRUSS and c-Myc in the presence of HBx oncoprotein of hepatitis B virus.
Materials and Methods
DNA recombinants
The expression vector for wild-type HBx (X0) has been described previously.44 The HA-tagged wild type and mutant TRUSS expression vectors were kindly provided by Michael D Cole.12 Details of expression vectors for full length Skp2 with N-terminal Myc tag (pcDNA3 MT-Skp2), F box deleted Skp2 with FLAG tag (pcDNA-FLAG/ΔF Skp2), full length Ubiquitin with His tag (His-Ub) and C-terminal truncated mutant of Skp2 (ΔC-Skp2) can be found elsewhere.20 Expression vector details for FLAG-Skp2, FLAG-Skp2-ΔF, FLAG-Skp2-ΔLRR, FLAG-Skp2-LRR, FLAG-Skp2-ΔNF and HA-DDB1 are available upon request. Expression vectors for wild-type CDH1 (HA-CDH1) and myc tag ubiquitin (pUI-myc-Ub) were kind gifts from Vishwa M. Dixit and Michael M. Lai.45,46
Antibodies and chemical inhibitors
Antibodies for c-Myc, Skp2, Ubiquitin, CDH1, CDC6, DDB1, HA-tag, HIS-Tag, and GAPDH were purchased from Santa Cruz Biotechnologies (California, USA). TRUSS antibody was purchased from Abcam (Cambridge, USA) and Proteintech (USA). Development of a monoclonal antibody against HBx has been described earlier.47 Protein-A, protein-G sepharose and protease inhibitor cocktail were procured from Amersham Biosciences, UK. The proteasome inhibitor MG132 (carbobenzoxy-L-leucycl-L-leucy-L-leucinal; 20 μM) was purchased from Calbiochem (Germany). Cycloheximide (30 μg/ml) was purchased from Sigma-Aldrich. The GSK3β inhibitor viii (AR-A014418; N-(4-Methoxybenzyl)-N′-(5-nitro-1,3-thiazol-2-yl)urea) was purchased from Santa Cruz Biotechnologies (California, USA).
Cell culture, DNA and siRNA transfections
The human hepatoma Huh-7 cells, Human embryonic kindey HEK293, hepatoblastoma HepG2 and HepG2.2.15 cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies, USA) supplemented with 10% fetal bovine serum at 37°C in a humidified atmosphere with 5% CO2. Cells in 60 mm (0.6 × 106 cells) or 100 mm (2 × 106 cells) culture dishes were transfected respectively with a total of 2.5 and 5.0 μg of individual or combination of plasmids using Lipofectamine (Life Technologies, USA) according to the manufacturer's instructions. For expression/interaction studies, cells were harvested 48 h post-transfection while for pulse–chase experiments, cells were harvested at the indicated time intervals 30 h post-transfection. For Skp2 knock-down studies Skp2 siRNA (Silencer select Skp2 siRNA) and negative control siRNA (Silencer select Negative control siRNA) were transfected in Huh-7 cells using Lipofectamine RNAiMAX according to the manufacturer's protocol. Cells were then harvested after 48 hours and used for further experiments. Sense and Antisense Skp2 siRNA oligos sequences are available upon request.
RNA isolation and its quantitation
Total RNA was isolated from cells using TRIzol reagent as per the supplier's instructions (Life Technologies, USA). Reverse transcriptase PCR (RT-PCR) was performed with M-MuLV reverse transcriptase (Fermentas, Thermo Fisher Scientific, USA) according to the manufacturer's guidelines. The real-time quantitative PCR (RT-qPCR) was done using specific primers for TRUSS, forward (TRUSS-F) 5′-AGCAGCAGCTCGCTAATTTC-3′ and reverse (TRUSS-R) 5′-AAAGCCAGGAATGCTGAGAA-3′ and GAPDH, forward (GAPDH-F) 5′- CGACCACTTTGTCAAGCTCA-3′ and reverse (GAPDH-R) 5′- AGGGGTCTACATGGCAACTG-3′ as described previously.48
Cell synchronization and flow cytometry
Huh-7 cells were synchronized by serum starvation for 72 h and stimulated by replacing complete medium containing 10% fetal bovine serum. The cell-cycle kinetics was done by growing cells in the absence or presence of serum using a FACS Calibur flow cytometer (Becton Dickinson, USA) after staining with propidium iodide. Cell-cycle analysis was performed using FlowJo software (Tree Star, Ashland, USA).
Immuno-precipitation and immuno-blotting
Cell lysates were prepared in buffer A [50 mM Tris/HCl (pH 7.6), 120 mM NaCl and 0.5% (v/v) Nonidet P40]. Protein estimation was carried out in the cleared cell lysates using Bradford's reagent. Equal amount of the extracts were incubated with different primary antibodies (1 μg) for overnight at 4°C. Protein A–Sepharose beads (Amersham Biosciences, UK) were added to each sample (10%, v/v) and incubated further for 3 h at 4°C. After five washes with buffer A, samples were recovered in 30 μl of 3x SDS gel-loading buffer, resolved by SDS/PAGE (10% gel) and electro-transferred on to a nitrocellulose membrane. Protein bands were visualized by enhanced chemiluminescence detection system (Cell Signaling Technology, USA). For immuno-blotting (IB), equal amounts of protein samples were electrophoretically separated, transferred onto nitrocellulose membrane, and visualized as above. To determine the half-life of TRUSS, Huh-7 cells were transfected with TRUSS expression vector and treated with 30 μg/ml cycloheximide for indicated time points and the cell lysates were immunoblotted with anti-HA antibody.
Pulse-chase analysis
To determine the half-life of TRUSS, Huh-7 cells were transfected with TRUSS expression vector with or without Skp2 or transfected with Skp2 siRNA for 48 h and treated with 30 μg/ml cycloheximide for indicated time points and the cell lysates were immunoblotted with anti-HA and anti-TRUSS antibody. Further to find the turn-over of TRUSS protein, Huh-7 cells transfected with TRUSS expression construct for 30h, and metabolically labeled in the presence of 110 µCi of [35S] cysteine/methionine mix (NEN Life Science Product) for 2h, followed by incubation with an excess of unlabeled methionine and cysteine for different time periods. After washing with ice-cold PBS, cell lysates were prepared in buffer A, immunoprecipitated using anti HA (TRUSS) antibody, resolved by SDS PAGE (8% gel) and autoradiographed.
Immunofluorescence staining
For immunofluorescence staining, cells were seeded on coverslips in 12-well culture plates, transfected as described above for 36 h then starved for 72 hours and harvested at indicated time points. After a PBS wash, cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Tween-20 for 20 min at room temperature. After rehydration in PBS for 30 min, cells were incubated for 1 h at room temperature with appropriate antibodies in PBS containing 5% BSA. After a quick wash, cells were incubated further for 1 h either with anti-mouse IgG coupled to Alexa Fluor 488 dye and/or with anti-rabbit IgG coupled to Alexa Fluor 594 dye in PBS with 5% BSA at a 1:2000 dilution. After a PBS wash, the coverslips were mounted in anti-fade reagent containing DAPI (Invitrogen); the fluorescence images were acquired at ×60 using a Nikon A1R confocal microscope and analyzed with the help of NIS-Elements software (Nikon).
In-vivo ubiquitination assay
The ubiquitination assay was done following 2 different protocols: (1) Huh-7 cells were transfected with the indicated plasmids for 48 h were treated with 30 µM MG132 for 1 h before harvest and lysed by the denatured buffer (50 mM Tris-Cl (pH 8.0), 5 mM DTT, and 1% SDS). The lysates were sonicated, and supernatants were recovered after centrifugation at 13,000 rpm for 10 min. HA-TRUSS was immunoprecipitated by anti-HA antibodies. Ubiquitinated TRUSS protein in the immunoprecipitate was detected by SDS-PAGE and immunoblotting with anti-ubiquitin and anti-His antibodies. (2) HEK293 cells were transfected with the indicated expression plasmids as above and treated with 30 µM MG132 for 2 h. Cell pellets were lysed in buffer I (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl [pH 8.0], 10 mM ß- mercaptoethanol) and incubated with Ni-NTA beads at room temperature for 6 h. Beads were sequentially washed once with buffer I, buffer II (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl [pH 8.0], 10 mM ß-mercaptoethanol), and buffer III (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl [pH 6.3], 10 mM ß-mercaptoethanol). Proteins were eluted from beads in buffer IV (200 mM imidazole, 0.15 M Tris-HCl [pH 6.7], 30% glycerol, 0.72 M ß-mercaptoethanol, and 5% SDS) for 30 minutes at room temperature. Eluted proteins were analyzed by Western blotting with anti-TRUSS antibody.
Patient's Tissue Specimens and Ethics Statement
A total of 4 tumor tissues including one each of HCC, pancreas, breast and colon carcinoma along with 4 matched available adjacent non-cancerous tissue (ANCT) specimens were used from the paraffin blocks archived in the All India Institute of Medical Sciences hospital. Necessary ethical clearance for this study was obtained from the Institutional Human Ethical Committee of the All India Institute of Medical Sciences, New Delhi (Ref. No. IESC/T-20/04.01.2013).
Immunohistochemical Analysis
Thin tissue sections (5 µm) were first stained with hematoxylin and eosin (H&E) followed by analysis for the expression of TRUSS and Skp2 proteins by immunohistochemistry (IHC) using rat polyclonal anti-TRUSS and rat polyclonal anti-Skp2 antibodies or control IgG. IHC was performed on positively charged glass slides coated with 10% poly 1-lysine (Sigma-Aldrich) for the detection of Skp2 and TRUSS proteins. After de-paraffinization, the sections were boiled in a water bath (Widsons Scientific Works, New Delhi, India) for 15 min in 10 mM citrate buffer, pH 6.0. After cooling to room temperature, blocking for endogenous peroxidase activity was carried out by treating the sections with 5% H2O2 in methanol for 30 min, followed by treatment with commercially available Ultra Block V solution (LabVision Corporation, Thermo Scientific, CA, USA) for 5 min. The slides were then incubated overnight at 4°C with anti-TRUSS and anti-Skp2 antibodies at 1:100 and 1:50 dilution respectively. The HRP polymer linked secondary antibody-based detection kit (UltraVision LP Large Volume Detection System HRP Polymer; LabVision Corporation) was used for the detection. The slides were then incubated with primary antibody enhancer for 30 min followed by HRP polymer for 30 min and developed using diaminobenzidine as a chromogen and images captured at 100x magnification using a Nikon Eclipse 80i microscope.
Funding
This work was supported by grant no. BT/PR12658/BRB/10/712/2009 of the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India (to VK). Azfar Jamal worked as a Junior Research Fellow of DBT while Manickavinayaham Swarnalatha has been a Research Associate of DBT in the above project. Vijay Kumar was a recipient of the JC Bose National Fellowship from the Department of Science and Technology, Government of India.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to the following scientists for kindly providing us the expression vectors for genes: Dr Michael D Cole (Dartmouth Medical School, New Hampshire, USA) for HA-tagged wild type and mutant TRUSS recombinants; Dr Michael M Lai (University of Napoli, Italy) for myc tagged ubiquitin (pUI-myc-Ub). Technical assistance by R Kumar and T Choeden is gratefully acknowledged.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol 2009; 49:73-96; PMID:18834306; http://dx.doi.org/ 10.1146/annurev.pharmtox.051208.165340 [DOI] [PubMed] [Google Scholar]
- 2.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 2006; 6:369-81; PMID:16633365; http://dx.doi.org/ 10.1038/nrc1881 [DOI] [PubMed] [Google Scholar]
- 3.Kitagawa K, Kotake Y, Kitagawa M. Ubiquitin-mediated control of oncogene and tumor suppressor gene products. Cancer Sci 2009; 100:1374-81; PMID:19459846; http://dx.doi.org/ 10.1111/j.1349-7006.2009.01196.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer 2008; 8:438-49; PMID:18500245; http://dx.doi.org/ 10.1038/nrc2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 2008; 8:83-93; PMID:18094723; http://dx.doi.org/ 10.1038/nrc2290 [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell 2007; 26:775-80; PMID:17588513 [DOI] [PubMed] [Google Scholar]
- 7.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci 2009; 34:562-70; PMID:19818632; http://dx.doi.org/ 10.1016/j.tibs.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al.. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 2005; 121:387-00; PMID:15882621; http://dx.doi.org/ 10.1016/j.cell.2005.02.035 [DOI] [PubMed] [Google Scholar]
- 9.Nag A, Bondar T, Shiv S, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol 2001; 21:6738-47; PMID:11564859; http://dx.doi.org/ 10.1128/MCB.21.20.6738-6747.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 2006; 22:383-94; PMID:16678110; http://dx.doi.org/ 10.1016/j.molcel.2006.03.035 [DOI] [PubMed] [Google Scholar]
- 11.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS. The DDB1–CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci U SA 2006; 103:2588-93; PMID:16473935; http://dx.doi.org/ 10.1073/pnas.0511160103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev 2010; 24:1236-41; PMID:20551172; http://dx.doi.org/ 10.1101/gad.1920310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene 1999; 18:3004-16; PMID:10378696; http://dx.doi.org/ 10.1038/sj.onc.1202746 [DOI] [PubMed] [Google Scholar]
- 14.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol 2006; 16:318-30; PMID:16934487; http://dx.doi.org/ 10.1016/j.semcancer.2006.07.015 [DOI] [PubMed] [Google Scholar]
- 15.Andrejeva J, Poole E, Young DF, Goodbourn S, Randall RE. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J Virol 2002; 76:11379-86; PMID:12388698; http://dx.doi.org/ 10.1128/JVI.76.22.11379-11386.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulane CM, Horvath CM. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 2002; 304:160-66; PMID:12504558; http://dx.doi.org/ 10.1006/viro.2002.1773 [DOI] [PubMed] [Google Scholar]
- 17.Ulane CM, Rodriguez JJ, Parisien JP, Horvath CM. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J Virol 2003; 77:6385-93; PMID:12743296; http://dx.doi.org/ 10.1128/JVI.77.11.6385-6393.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leupin O, Bontron S, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol 2005; 79:4238-45; PMID:15767425; http://dx.doi.org/ 10.1128/JVI.79.7.4238-4245.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leupin O, Bontron S, Strubin M. Hepatitis B virus X protein and simian virus 5 V protein exhibit similar UV-DDB1 binding properties to mediate distinct activities. J Virol 2003; 77:6274-83; PMID:12743284; http://dx.doi.org/ 10.1128/JVI.77.11.6274-6283.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalra N, Kumar V. The X protein of hepatitis B virus binds to the F box protein Skp2 and inhibits the ubiquitination and proteasomal degradation of c-Myc. FEBS Lett 2006; 580:431-36; PMID:16376880; http://dx.doi.org/ 10.1016/j.febslet.2005.12.034 [DOI] [PubMed] [Google Scholar]
- 21.Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF (Skp2-Cks1) ubiquitin ligase by the APC/C (Cdh1) ubiquitin ligase. Nature 2004; 11:190-93; PMID:15014502; http://dx.doi.org/ 10.1038/nature02330 [DOI] [PubMed] [Google Scholar]
- 22.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 2004; 23:2116-25; PMID:15103331; http://dx.doi.org/ 10.1038/sj.emboj.7600217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004; 5:739-51; PMID:15340381; http://dx.doi.org/ 10.1038/nrm1471 [DOI] [PubMed] [Google Scholar]
- 24.von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI et al.. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 2003; 11:1189-00; PMID:12769844; http://dx.doi.org/ 10.1016/S1097-2765(03)00193-X [DOI] [PubMed] [Google Scholar]
- 25.Carrano AC, Eytan E, Hershko A, Pagano M. Skp2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1999; 1:193-99; PMID:10559916; http://dx.doi.org/ 10.1038/12013 [DOI] [PubMed] [Google Scholar]
- 26.Tedesco D, Lukas J, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev 2002; 16:2946-57; PMID:12435635; http://dx.doi.org/ 10.1101/gad.1011202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem 2003; 278:25752-57; PMID:12730199; http://dx.doi.org/ 10.1074/jbc.M301774200 [DOI] [PubMed] [Google Scholar]
- 28.Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Nakayama KI. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci U SA 2003; 100:10231-36; PMID:12925736; http://dx.doi.org/ 10.1073/pnas.1831009100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell 2002; 9:481-91; PMID:11931757; http://dx.doi.org/ 10.1016/S1097-2765(02)00467-7 [DOI] [PubMed] [Google Scholar]
- 30.Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Bio Chem 2003; 278:30854-58; PMID:12840033; http://dx.doi.org/ 10.1074/jbc.C300251200 [DOI] [PubMed] [Google Scholar]
- 31.Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Hatakeyama S. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J 2000; 19:2069-81; PMID:10790373; http://dx.doi.org/ 10.1093/emboj/19.9.2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol 1999; 1:14-19; PMID:10559858; http://dx.doi.org/ 10.1038/8984 [DOI] [PubMed] [Google Scholar]
- 33.Tsai YS, Lai CL, Lai CH, Chang KH, Wu K, Tseng SF, Fazli L, Gleave M, Xiao G, Gandee L, et al.. The role of homeostatic regulation between tumor suppressor DAB2IP and oncogenic Skp2 in prostate cancer growth. Oncotarget 2014; 5:6425-36; PMID:25115390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu W, Liu S, Li B, Xie Y, Adhiambo C, Yang Q, Ballard BR, Nakayama KI, Matusik RJ, Chen Z. SKP2 inactivation suppresses prostate tumorigenesis by mediating JARID1B ubiquitination. Oncotarget 2015; 6:771-88; PMID:25596733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wertz IE, O'Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. Human de-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 2004; 303:1371-74; PMID:14739464; http://dx.doi.org/ 10.1126/science.1093549 [DOI] [PubMed] [Google Scholar]
- 36.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993; 75:495-05; PMID:8221889; http://dx.doi.org/ 10.1016/0092-8674(93)90384-3 [DOI] [PubMed] [Google Scholar]
- 37.Harty RN, Brown ME, McGettigan JP, Wang G, Jayakar HR, Huibregtse JM, Whitt MA, Schnell MJ. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J Virol 2011; 75:10623-29; PMID:11602704; http://dx.doi.org/ 10.1128/JVI.75.22.10623-10629.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winberg G, Matskova L, Chen F, Plant P, Rotin D, Pawson T. Latent Membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol Cell Biol 2002; 20:8526-35; PMID:11046148; http://dx.doi.org/ 10.1128/MCB.20.22.8526-8535.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coscoy L, Sanchez DJ, Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol 2001; 155:1265-73; PMID:11756476; http://dx.doi.org/ 10.1083/jcb.200111010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bour S, Perrin C, Akari H, Strebel K. The human immunodeficiency virus type 1 Vpu protein inhibits NF-kappa B activation by interfering with beta TrCP-mediated degradation of Ikappa B. J Biol Chem 2001; 276:15920-28; PMID:11278695; http://dx.doi.org/ 10.1074/jbc.M010533200 [DOI] [PubMed] [Google Scholar]
- 41.Kumar V, Sarkar DP. Hepatitis B virus X protein (HBx): structure-function relationship and role in viral pathogenesis in: Handbook of Experimental Pharmacology. Triezenberg SJ, Kaufman J, Gossen M, Eds., Germany: Springer-Verlag; 2004, pp.377-407. [Google Scholar]
- 42.Sirma H, Weil R, Rosmorduc O, Urban S, Israel A, Brechot C. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene 1998; 16: 2051-63; PMID:9572486; http://dx.doi.org/ 10.1038/sj.onc.1201737 [DOI] [PubMed] [Google Scholar]
- 43.Shukla SK, Kumar V. HBx and c-myc cooperate in the up-regulation of ribosome biogenesis and in cellular transformation. FEBS J 2012; 279:3859-71; PMID:22889122; http://dx.doi.org/ 10.1111/j.1742-4658.2012.08745.x [DOI] [PubMed] [Google Scholar]
- 44.Kumar V, Jayasuryan N, Kumar R. A truncated mutant (residues 58–140) of the hepatitis B virus X protein retains transactivation function. Proc Natl Acad Sci U S A 1996; 93:5647-52; PMID:8643631; http://dx.doi.org/ 10.1073/pnas.93.11.5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, Kirkpatrick DS, Jackson PK, Fang G, Dixit VM. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell 2011; 20:511-23; PMID:21596315; http://dx.doi.org/ 10.1016/j.molcel.2011.03.027 [DOI] [PubMed] [Google Scholar]
- 46.Liao TL, Wu CY, Su WC, Jeng KS, Lai MM. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J 2010; 29:3879-90; PMID:20924359; http://dx.doi.org/ 10.1038/emboj.2010.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar V, Jayasuryan N, Reddi H, Sahal D, Panda SK. A monoclonal antibody against the X protein of hepatitis B virus: fine mapping of its epitope and application in a quantitative ELISA of the X protein in sera of hepatitis B patients. Hybridoma 1998; 17:157-64; PMID:9627056; http://dx.doi.org/ 10.1089/hyb.1998.17.157 [DOI] [PubMed] [Google Scholar]
- 48.Janbandhu VC, Singh AK, Mukherji A, Kumar V. p65 negatively regulate transcription of the cyclin E gene. J Biol Chem 2010; 285:17453-64; PMID:20385564; http://dx.doi.org/ 10.1074/jbc.M109.058974 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.