Abstract
Protein phosphatase 2A (PP2A) is the major serine-threonine phosphatase that regulates a number of cell signaling pathways. PP2A activity is controlled partially through protein degradation; however, the underlying mechanism is not fully understood. Here we show that PP2A/C, a catalytic subunit of PP2A, is degraded by the Cullin3 (Cul3) ligase-mediated ubiquitin-proteasome pathway. In response to death receptor signaling by tumor-necrosis factor-related apoptosis-inducing ligand (TRAIL), PP2A/C, caspase-8 and Cul3, a subunit of the cullin family of E3 ligases, are recruited into the death-inducing signaling complex (DISC) where the Cul3 ligase targets PP2A/C for ubiquitination and subsequent degradation. Functionally, knockdown of PP2A/C expression by siRNA or pharmacological inhibition of PP2A activity increases TRAIL-induced apoptosis. In cancer cells that have developed acquired TRAIL resistance, PP2A phosphatase activity is increased, and PP2A/C protein is resistant to TRAIL-induced degradation. Thus, this work identifies a new mechanism by which PP2A/C is regulated by Cul3 ligase-mediated degradation in response to death receptor signaling and suggests that inhibition of PP2A/C degradation may contribute to resistance of cancer cells to death receptor-induced apoptosis.
Keywords: apoptosis resistance, Cullin3, PP2A, TRAIL, ubiquitination
Introduction
Protein phosphorylation is one of the important posttranslational modifications that play a critical role in regulating cell signaling. It is regulated by functionally opposing kinases and phosphatases. In eukaryotic cells, kinases predominately phosphorylate proteins on serine, threonine and tyrosine residues. Conversely, phosphatases act in opposition to kinases to dephosphorylate these residues. Protein phosphatase 2A (PP2A) is the major serine-threonine phosphatase that regulates a number of cell signaling pathways.1 PP2A is a trimeric holoenzyme that consists of a catalytic subunit (C subunit) (hereafter referred to as PP2A/C), a structural subunit (PR65/A subunit) and a regulatory B subunit.2 The A subunit and C subunit each have 2 isoforms while the B subunit has 16 distinct isoforms.3 PR65/A and PP2A/C dimerize to form the PP2A core enzyme, which further interacts with a regulatory B subunit to assemble a heterotrimeric PP2A holoenzyme.4 PP2A enzymatic activity depends on PP2A/C, while the regulatory subunits determine substrate specificity and subcellular localization.2-4 The combination of each C subunit isoform and A subunit isoform with one of the 16 different B subunits forms a distinct PP2A holoenzyme, which suggests that their composition plays a key role in the regulation of PP2A activity.3 In addition to association with A and B subunits, a portion of the C subunit can bind to a protein termed α4, which prevents it from ubiquitination-mediated degradation.5-7 However, the mechanism by which PP2A/C protein is ubiquitinated remains to be fully determined.
Ubiquitin-mediated protein degradation involves a series of enzymes that transfer ubiquitin molecules to their substrates to target the latter for destruction or other cellular processes. The 3 classes of enzymes involved in this process are the E1 ubiquitin activating enzymes, E2 ubiquitin conjugating enzymes and E3 ubiquitin ligases.8 The E3 ligases determine substrate specificity and are comprised of 2 major classes, the HECT family and the RING family. In the RING family, the Cullin-RING ubiquitin ligases (CRLs) are the largest class of E3 ligases.9-11 Each E3 ligase contains a cullin subunit, a RING finger subunit and a substrate recognition subunit.12,13 The cullin-RING family ligases have been implicated in the regulation of the ubiquitination of a host of proteins.11 An issue is whether the Cullin family E3 ligase regulates PP2A/C ubiquitination in response to stimuli, including death receptor signaling.
Death receptor signaling is triggered by death ligands that belong to the tumor necrosis factor (TNF) family.14,15 TNF-related apoptosis-inducing ligand (TRAIL) is a member of the TNF family that selectively induces apoptosis in transformed or tumor cells, but not in normal cells.14,15 TRAIL induces apoptosis by binding to death receptors DR4 or DR5, which results in formation of the death-inducing signaling complex (DISC) by recruiting FADD and caspase-8, leading to activation of caspase-8 and subsequent cleavage of caspases (i.e., caspase-3, -6 and -7) and cell death.16,17 The TRAIL pathway provides a number of potential opportunities to understand death signaling and to develop therapeutic targets because the TRAIL ligand itself and TRAIL receptor-specific agonistic antibodies effectively kill transformed and cancer cells, but not most normal cells.18-20 However, not all cancer cells are susceptible to TRAIL and there appears to be a growing list of possible mechanisms by which cancer cells can evade TRAIL-induced apoptosis.21-23 A previous study indicated that the Cullin3 (Cul3) ligase targeted caspase-8 for ubiquitination and subsequent cleavage, thus facilitating TRAIL-induced cell death.24
To study the involvement of kinases and phosphatases in TRAIL-mediated apoptosis,25,26 we identify PP2A as a critical player that regulates TRAIL-induced apoptosis. We show that TRAIL treatment causes PP2A/C ubiquitination and subsequent degradation via the ubiquitin-proteasome pathway. We also show that this ubiquitination occurs at the DISC where caspase-8 is obligatory for PP2A/C degradation. We further show that PP2A phosphatase activity is increased in cells that have developed acquired TRAIL resistance and that PP2A/C protein is resistant to TRAIL-induced degradation in resistant cells. Thus, these data suggest that degradation of PP2A/C contributes to death receptor signaling-mediated apoptosis.
Results
PP2A/C is degraded in response to TRAIL treatment
To test the effect of TRAIL treatment on the stability of PP2A/C protein, SUM159 cells were treated with TRAIL in the presence and absence of cycloheximide, and the levels of PP2A/C protein were determined by western blot analysis. Figure 1A shows that TRAIL treatment decreased the half-life and levels of PP2A/C protein. To determine whether the TRAIL-induced PP2A/C decrease can be blocked by proteasome inhibition, SUM159 cells were treated with TRAIL in the presence and absence of the proteasome inhibitor MG132. Figure 1B shows that a decrease in PP2A/C protein levels induced by TRAIL treatment was blocked by MG132. To determine if PP2A/C degradation is mediated by ubiquitin, SUM159 cells treated with TRAIL were harvested to perform immunoprecipitation (IP) with an anti-PP2A/C antibody and immunoblotting with an anti-ubiquitin antibody. Figure 1C shows that TRAIL treatment caused endogenous PP2A/C ubiquitination compared to untreated control cells. These data suggest that PP2A/C is ubiquitinated and degraded by a ubiquitin-proteasome pathway in response to TRAIL signaling.
Figure 1.
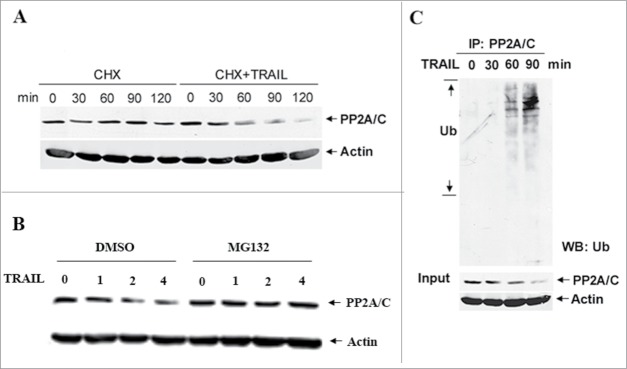
TRAIL treatment promotes PP2A/C ubiquitination and degradation. (A) TRAIL treatment decreases PP2A/C stability. SUM159 cells were pre-treated with CHX (10 μg/ml) for 30 min and then left untreated or treated with TRAIL (100 ng/ml). The levels of PP2A/C protein were detected by protein gel blots. (B) The proteasome inhibitor MG132 blocks TRAIL-induced PP2A/C degradation. SUM159 cells pretreated with MG132 (20 uM) or DMSO for 2 hr were treated with TRAIL (100 ng/ml) for indicated time points. (C) TRAIL treatment induces endogenous PP2A/C ubiquitination. SUM159 cells were treated with TRAIL (100 ng/ml) for indicated time points. Cells were lysed, immunoprecipitated with anti-PP2A/C antibody and blotted with anti-Ub antibody. Lower panel shows input.
PP2A/C ubiquitination is mediated by a Cul3 E3 ligase
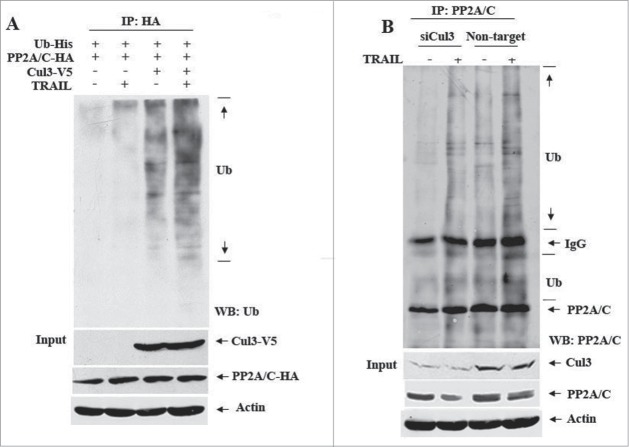
A recent study suggested that a Cul3-based E3 ligase is involved in caspase-8 ubiquitination.24 To determine whether a Cul3 ligase is involved in PP2A/C ubiquitination, HEK293T cells co-transfected with the plasmids expressing Cul3-V5, PP2A/C-HA and ubiquitin-His were left untreated or treated with TRAIL. Figure 2A shows that in the absence of Cul3, TRAIL treatment was not sufficient to induce exogenous PP2A/C ubiquitination. However, Cul3 alone was sufficient to do so in the absence of TRAIL treatment, and such a change was increased further upon TRAIL treatment. To determine whether Cul3 is required for TRAIL-induced PP2A/C ubiquitination, SUM159 cells were transfected with On-TARGET plus SMARTpool Cul3 siRNAs or non-target siRNAs and then treated with TRAIL. Figure 2B shows that Cul3 knockdown resulted in a decrease in PP2A/C ubiquitination induced by TRAIL. These data suggest that a Cul3 E3 ligase mediates PP2A/C ubiquitination.
Figure 2.
Cul3 is required for TRAIL-induced PP2A/C ubiquitination. (A) Overexpression of Cul3 enhances TRAIL-induced PP2A/C ubiquitination. HEK293T cells were co-transfected with PP2A/C-HA, His-tagged ubiquitin (Ub-His) and Cul3-V5 or vector. 48 hr later, cells were left untreated or treated with TRAIL (100 ng/ml) for 30 min. Cell lysates were prepared, immunoprecipitated with HA antibody and blotted with anti-Ub antibody. Lower panel shows input (blotted with V5 and HA antibodies). (B) Cul3 knockdown impairs TRAIL-induced PP2A/C ubiquitination. SUM159 cells were transfected with non-targeted control siRNAs (Non-target) or siRNAs against Cul3 (siCul3) for 48 hr and then treated with TRAIL (100 ng/ml) for 90 min. Cell lysates were prepared, immunoprecipitated with PP2A/C antibody and blotted with indicated antibodies.
PP2A/C interacts with caspase-8 and associates with the DISC
It is established that the formation of the DISC is a key step to initiate TRAIL signaling. Since Cul3 is involved in caspase-8 ubiquitination,24 the induction of PP2A/C degradation by TRAIL treatment suggests that PP2A/C may be associated with the DISC. To test this possibility, we first asked whether PP2A/C can physically interact with caspase-8. We transfected HEK293T cells with the plasmids expressing Cul3-V5, PP2A/C-HA, and Casp8-HA in the presence and absence of TRAIL treatment. Figure 3A shows that an anti-caspase-8 antibody was able to pull down both PP2A/C and Cul3. Importantly, we found that anti-caspase-8 antibody was able to pull down endogenous PP2A/C/ and Cul3, further confirming their associations among caspase-8, PP2A/C and Cul3 (Fig. 3B).
Figure 3.
PP2A/C is associated with caspase-8 and Cul3 in the DISC. (A) Caspase-8 interacts with Cul3 and PP2A/C in transfected cells. HEK293T cells were co-transfected with Cul3-V5, Casp8-HA and PP2A/C-HA. 48 hr later, cells were treated with TRAIL (100 ng/ml) for 30 min or left untreated. Cell lysates were prepared, immunoprecipitated with caspase-8 antibody or IgG and blotted with indicated antibodies. (B) Endogenous caspase-8 interacts with endogenous Cul3 and PP2A/C. SUM159 cells were treated with TRAIL (100 ng/ml) for 90 min. Cell lysates were prepared, immunoprecipitated with caspase-8 antibody and blotted with indicated antibodies. Input was shown in the lower panel. (C) PP2A/C is recruited to the DISC in response to TRAIL stimulation. SUM159 cells (2×107) were treated with 100 ng/ml Flag-TRAIL, 1 μg/ml Enhancer and 2 μg/ml anti-Flag (Flag/M2) for 30 min at 37°C. Cells were lysed, the DISC was immunoprecipitated with Flag-tagged Protein Immunoprecipitation Kit (Sigma) and blotted with indicated antibodies.
To determine whether TRAIL-induced associations among Cul3, caspase-8 and PP2A/C occur at the DISC, SUM159 cells were left untreated or treated with Flag-tagged TRAIL, ligand enhancer and Flag/M2 antibody and then lysed. Cell lysates were subjected to DISC IP using the Flag Immunoprecipitation assays and analyzed by immunoblotting. As expected, some components, including FADD, DR5 and caspase-8, were detected in the immunoprecipitated complex (Fig. 3C), which validated our experiments as capable of detecting proteins that are known to be recruited into the DISC upon TRAIL treatment. Importantly, we found that both PP2A/C and Cul3 proteins were detected in the DISC (Fig. 3C), suggesting that PP2A/C, Cul3 and caspase-8 are part of the DISC complex. Thus, we conclude that TRAIL stimulation leads to the recruitment of PP2A/C to the DISC where it associates with Cul3 and caspase-8, in addition to other DISC proteins.
Caspase-8 is involved in TRAIL-induced PP2A/C ubiquitination
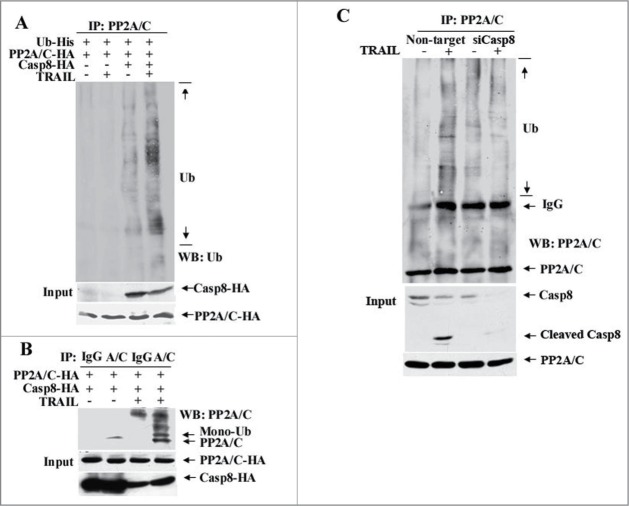
Once the associations among caspase-8, PP2A/C and Cul3 in the DISC were determined, we wanted to know whether caspase-8 is involved in PP2A/C ubiquination. To this end, HEK293T cells co-transfected with the plasmids expressing Casp8-HA, PP2A/C-HA and Ub-His were left untreated or treated with TRAIL. As shown in Fig. 4A, caspase-8 overexpression caused minimal PP2A/C ubiquitination and the latter was increased to much higher levels in combination with TRAIL treatment. In contrast, such a change was not detected in cells when caspase-8 was not overexpressed regardless of TRAIL treatment (Fig. 4A). When combining caspase-8 overexpression with TRAIL treatment, we were able to detect mono-ubiquitinated PP2A/C (Fig. 4B).
Figure 4.
Caspase-8 is required for TRAIL-induced PP2A/C ubiquitination. (A) Overexpression of caspase-8 enhances TRAIL-induced PP2A/C ubiquitination. HEK293T cells were co-transfected with PP2A/C-HA, Ub-His and Casp8-HA or vector, and then treated with TRAIL (100 ng/ml) for 30 min or left untreated. Cell lysates were prepared, immunoprecipitated with PP2A/C antibody and blotted with an anti-Ub antibody. (B) TRAIL treatment induces PP2A/C mono-ubiquitination in caspase-8 overexpressing cells. HEK293T cells co-transfected with Casp8-HA and PP2A/C-HA were treated with TRAIL (100 ng/ml) for 30 min or left alone. Cell lysates were prepared, immunoprecipitated with PP2A/C antibody (A/C) or IgG and blotted with anti-PP2A/C antibody. (C) Caspase-8 knockdown impairs TRAIL-induced PP2A/C ubiquitination. SUM159 cells were transfected with non-targeted control siRNAs or siRNAs against caspase-8 (siCasp8). After 48 hr, cells were left untreated or treated with TRAIL (100 ng/ml) for 90 min, lysed, immunoprecipitated with PP2A/C antibody and blotted with indicated antibodies.
To further determine the role of caspase-8 in PP2A/C ubiquitination, SUM159 cells were transfected with a non-targeted control siRNAs or siRNAs against caspase-8 (siCasp8), and then treated with TRAIL. Figure 4C shows that caspase-8 knockdown significantly decreased PP2A/C ubiquitination in response to TRAIL treatment. Taken together, these results suggest that caspase-8 is involved in TRAIL-induced PP2A/C ubiquitination.
Ubiquitin chains are formed through K48 and K63
To determine which residue(s) on ubiquitin is linked to the polyubiquitin chain that can attach to PP2A/C, HEK293T cells were co-transfected with PP2A/C-HA and Cul3-V5 and HA-tagged wild-type (WT) ubiquitin, lysine-null (K0), K48 mutant or K63 mutant. The resulting cells were left untreated or treated with TRAIL and then lysed to perform an IP with PP2A/C antibody. Figure 5 shows that ubiquitinated PP2A/C was detected in TRAIL-treated cells that were co-transfected with WT Ub, K48 or K63 mutant and Cul3-V5. By contrast, under the same treatment condition, PP2A/C ubiquitination was not detected in cells transfected with a K0 mutant, which served as a negative control. These data suggest that K48 and K63 of ubiquitin can be used for ubiquitin chain formation.
Figure 5.
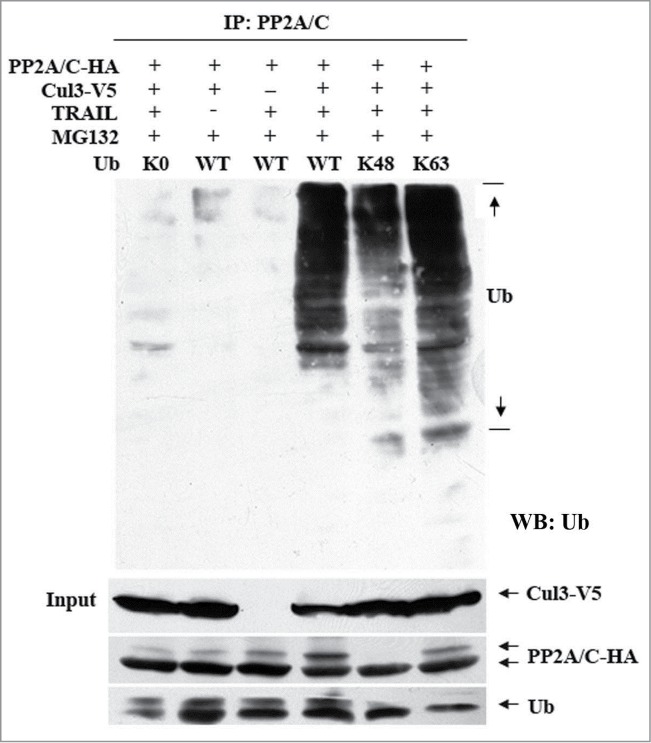
Lysine 48 (K48) and lysine 63 (K63) on ubiquitin are linked to the polyubiquitin chain. HEK293T cells transfected with PP2A/C-HA, Cul3-V5 and His-tagged wild-type (WT) ubiquitin, lysine-null (K0) or a single-lysine-containing mutant treated with TRAIL (100 ng/ml), immunoprecipitated with PP2A/C antibody, and blotted with indicated antibodies.
Resistance to PP2A/C degradation is associated with acquired TRAIL resistance
Since PP2A/C is recruited to the DISC and then degraded via the ubiquitin-proteasome pathway by a Cul3 E3 ubiquitin ligase, we suspected that PP2A/C degradation is necessary for TRAIL-induced apoptosis. To this end, we treated 2 human breast cancer cell lines, SUM159 and BT54927 with TRAIL, okadaic acid (OA) or their combination. Of note, OA is a PP2 inhibitor and the concentrations of OA (10 and 20 nM) used in these studies are selective for PP2A over other PP family members.28 The combination of TRAIL with OA significantly increased TRAIL-induced death in SUM159 and BT549 cells and the combination caused a significant increase in caspase-8 and PARP cleavage as compared to the single treatment alone (Fig. 6A and data not show). Similar results were obtained with other cancer cell lines including T47D and OV90 (data not shown).
Figure 6.
PP2A is a negative regulator in TRAIL-induced apoptosis. (A) Pharmacological effects of the PP2A inhibitor okadaic acid (OA) on TRAIL-induced cell death. SUM159 cells were treated with OA (10 nM), TRAIL (10 ng/ml) or in combination for 24 hr. Cleavage of caspase-8 and PARP was determined by western blotting. Cell survival was determined by the trypan blue exclusion assay. **, P < 0.001. (B) Knockdown of PP2A/C enhances TRAIL-induced apoptosis. SUM159 cells transfected with non-targeted control siRNAs (Non-target) or siRNAs against PP2A/C (siPP2A/C) were treated with TRAIL (10 ng/ml). The levels of PP2A/C and cleavage of caspase-8 and PARP were determined by western blots, and cell survival was determined. (C) Establishment of an acquired TRAIL resistant cell line. The TRAIL-resistant SUM159 cells (SUM159-R) were generated by exposing the parental SUM159 cells (SUM159-P) to gradually increased concentrations of TRAIL starting from 0.1 ng/ml to 120 ng/ml for over 6 months. SUM159-P and SUM159-R cells were left untreated or treated with TRAIL at 50 ng/ml (TR1) or 100 ng/ml (TR2) for 24 hr. PARP cleavage was determined by western blot and cell survival was determined by the trypan blue exclusion assay. (D) PP2A/C is resistant to TRAIL-induced degradation in resistant cells. Parental SUM159 (SUM159-P) and resistant SUM159 cells (SUM159-R) were left untreated or treated with TRAIL (100 ng/ml) for different time periods. Cell lysates were prepared and subjected to western blot with a PP2A/C antibody. (E) PP2A phosphatase activity is increased in acquired TRAIL resistant cells. SUM159-P and SUM159-R cells were tested for phosphatase activity. Relative phosphatase activity in parental cells was arbitrarily set to 100.
To investigate the direct role of PP2A in the regulation of TRAIL sensitivity, we used siRNA to knock down PP2A/C expression. Figure 6B shows that knockdown of PP2A/C increased TRAIL-induced caspase-8 and PARP cleavage and growth inhibition as compared to the same cells transfected with control siRNAs. Thus, our results indicate that PP2A acts as a negative regulator of TRAIL-induced apoptosis.
To further define the role of PP2A in TRAIL resistance, we established a TRAIL-resistant breast cancer cell line named SUM159-R. Figure 6C shows that SUM159-R cells were much more resistant to TRAIL than parental SUM159 (SUM159-P) cells. TRAIL treatment did not cause a decrease in the levels of PP2A/C protein in SUM159-R, as compared to SUM159-P, whose PP2A/C protein was degraded (Fig. 6D). Importantly, SUM159-R cells expressed higher levels of PP2A phosphatase activity than SUM159-P cells (Fig. 6E). Thus, these data suggest that impairment of PP2A/C degradation may lead to elevated PP2A phosphatase activity that contributes to TRAIL resistance.
Discussion
PP2A complexes can be degraded by the ubiquitin-proteasome pathway under both non-stress and stress conditions. In plants, stress-induced A subunit ubiquitination via the E3 ligase CHIP enhanced PP2A activity.30 In mammalian cells, on the other hand, binding of PP2A/C to α4, an evolutionarily conserved non-catalytic subunit for PP2A-like phosphatases plays a critical role for regulating PP2A/C degradation.5,6 Specifically, under normal conditions, α4 binds to and protects a portion of PP2A/C protein from ubiquitin-mediated degradation.7 Under stress conditions, the E3 ligase MID1 interacts with α4 and PP2A/C and then induces PP2A/C ubiquitination and degradation by the proteasome.31 Consistent with the regulation of PP2A/C degradation by the ubiquitin-proteasome pathway, we showed that a Cul3 E3 ligase mediates PP2A/C ubiquitination and degradation in response to TRAIL signaling. This conclusion is supported by the following lines of evidence: (1) PP2A/C is recruited to the DISC where it associates with Cul3 and caspase-8; (2) TRAIL treatment leads to PP2A/C ubiquitination and knockdown of Cul3 by siRNA decreases PP2A/C ubiquitination; and (3) TRAIL treatment leads to PP2A/C degradation and the proteasome inhibitor MG132 blocks it. Taken together, these data indicate that PP2A/C degradation is mediated by a Cul3 E3 ligase and suggest that such degradation may be an important mechanism by which PP2A phosphatase regulates TRAIL death signaling.
Cul3 is a subunit of the Cullin-RING E3 ubiquitin ligase (CRL) family that includes 7 CRLs, all of which contain a cullin subunit, a RING finger subunit and a substrate recognition subunit.10-12,32-34 A previous study indicated that in response to TRAIL stimulation, Cul3 is recruited to the DISC where it interacts with p62 to ubiquitinate and activate caspase-8.24 Our data showed that caspase-8 antibody is able to pull down both Cul3 and PP2A/C in response to TRAIL stimulation and that caspase-8, PP2A/C, and Cul3 are associated with the DISC. Thus, we speculate that caspase-8 may bring PP2A/C and Cul3 in the DISC where Cul3 mediates PP2A/C degradation.
Ubiquitin is a 76-amino-acid protein that is covalently attached to lysine residues of substrate proteins through catalytic reactions mediated by enzymes E1, E2 and E3.35 Ubiquitin itself has 7 lysine residues including K48 and K63. It is generally accepted that a K48-linked polyubiquitin chain targets the protein to the 26S proteasome for degradation, whereas a K63-linked polyubiquitin chain involves protein-protein interaction.36 We identified both K48- and K63-linked polyubiquitin in response to TRAIL signaling, which suggests that K63-linked polyubiquitin chains may facilitate an interaction among PP2A/C, caspase-8 and Cul3 while a K48-linked polyubiquitin chain targets PP2A/C for degradation.
PP2A has been shown to play important roles in the regulation of a number of signal transduction pathways, but the mechanism by which PP2A regulates apoptosis is not fully understood. A previous study indicated that the phosphatase inhibitor calyculin A, an inhibitor of PP1 and PP2A, inhibits death receptor-mediated apoptosis.37 Since calyculin can inhibit both PP1 and PP2A, the observed effects could be attributable to the inhibition of PP1. On the other hand, inhibition of UVB-induced PP2A leads to NF-κB pro-apoptotic activity,38 suggesting that PP2A acts as an apoptosis inhibitor. Consistent with the role of PP2A as a negative regulator of apoptosis, we showed that pharmacological inhibition of PP2A activity or knockdown of PP2A/C expression by siRNA increased TRAIL-induced apoptosis. Furthermore, we showed that PP2A/C is recruited into the DISC and following its recruitment in the DISC, it is targeted for degradation via a Cul3 E3 ligase. We speculate that its degradation is an important mechanism by which TRAIL signaling regulates PP2A/C levels and PP2A phosphatase activity. Therefore, the inhibition of PP2A/C degradation may contribute to TRAIL resistance in cancer cells.
TRAIL is a promising agent for cancer therapy.14-16 However, most cancer cells are resistant to TRAIL and its resistance mechanism is not fully understood. Recruitment and degradation of PP2A/C in the DISC indicate that PP2A may play a role in the regulation of TRAIL-induced apoptosis and that blocking its degradation may confer TRAIL resistance. Indeed, in TRAIL resistant cells PP2A/C is resistant to TRAIL-induced degradation and these resistant cells expressed higher levels of PP2A phosphatase activity. Consistently, pharmacological inhibition of PP2A activity or knockdown of PP2A/C expression sensitized breast cancer cells to TRAIL. Collectively, these data support a role for PP2A in negatively regulating TRAIL sensitivity.
In summary, our study reveals an important pathway by which PP2A/C protein is regulated via Cul3-mediated proteasome degradation. In response to TRAIL death receptor signaling, PP2A/C is recruited to the DISC where it interacts with caspase-8 and Cul3. The latter targets PP2A/C for ubiquitination and subsequent degradation. When PP2A/C degradation is impaired, cancer cells become resistant to TRAIL death ligand. Given that TRAIL selectively induces cancer cell death, our data suggest that targeting PP2A activity may have clinical implication for cancer therapy.
Materials and Methods
Tissue culture and experimental reagents
Neonatal kidney HEK293T cells were cultured in DMEM with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (PS). Human breast cancer cell line SUM159 was maintained in F12 supplemented with 5% FBS, 1% PS, 10 μg/ml insulin and 0.5 μg/ml hydrocortisone. Human breast cancer cell line BT549 was maintained in RPMI with 10% FBS. Recombinant human TRAIL/Apo2L was obtained from Peprotech Inc. (Rocky Hill, NJ). Okadaic acid (OA), MG132 and Cycloheximide (CHX) were obtained from Calbiochem (La Jolla, CA). Flag-tagged recombinant TRAIL and enhancer were purchased from Enzo Life Sciences (Plymouth Meeting, PA). Anti-Flag (M2) and actin antibodies were purchased from Sigma (St Louis, MO). Anti-HA and ubiquitin antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mouse anti-V5 antibody was obtained from Invitrogen (Carlsbad, CA). Mouse anti-caspase-8 and rabbit anti-PP2A/C, PARP, FADD antibodies were obtained from Cell Signaling Technology (Danvers, MA). Mouse anti-Cul3 antibody was purchased from BD Bioscience (San Diego, CA). Rabbit anti-DR5 antibody was obtained from Imgenex (San Diego, CA).
Plasmids and transfection
The full-length Cul3 cDNA was cloned into the pcDNA3.1/V5-His vector at the KpnI and XhoI sites. pcDNA3-PP2A/C-HA (PP2A/C-HA) was kindly provided by Dr. Xingming Deng (Emory University). pcDNA3-caspase-8-HA (Casp8-HA) was kindly provided by Dr. Emad Alnemri (Thomas Jefferson University). The plasmids encoding HA-tagged ubiquitin and ubiquitin with the lysine-null (K0) or a single-lysine-containing mutant (K48 and K63) were kindly provided by Dr. Yihong Ye (NIDDK/NIH). Lipofectamine 2000 (Invitrogen) was used to transfect both plasmids and siRNAs. For gene knockdown experiments, cells were transfected with Cul3 ON-TARGETplus SMARTpool siRNA, caspase-8 ON-TARGETplus SMARTpool siRNA, and ON-TARGETplus Non-targeting siRNA pool (Thermo Scientific Inc.). PP2A/C siRNA and control siRNA-F were obtained from Santa Cruz Biotechnology. Transfected cells were harvested 48 hr post-transfection.
Co-immunoprecipitation (CO-IP) and DISC-IP
SUM159 cells (2 × 107) were stimulated with 100 ng/ml Flag-TRAIL, 1 μg/ml Enhancer and 2 μg/ml anti-Flag (M2) for 30 min at 37°C. Cells were then lysed for 30 min on ice with DISC IP lysis buffer (30 mM Tris, pH7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100). In un-stimulated controls, 100 ng/ml Flag-TRAIL, 1 μg/ml Enhancer and 2 μg/ml anti-Flag (M2) were added after lysis on ice. The lysates were centrifuged twice at 4°C, 14,000 rpm for 10 min. The soluble fraction was immunoprecipitated with Flag-tagged Protein Immunoprecipitation Kit (Sigma) and analyzed by protein gel blotting. In Co-IP experiments, cells were lysed with lysis buffer (20 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3O4, 1 μg/ml leupeptin, 1 mM PMSF) on ice for 30 min. Soluble cell lysates (800 μl) were incubated with an IP antibody at 4°C overnight. Complexes were pulled down by Protein A/G agarose (Santa Cruz Biotechnology). The pellets were washed 4–5 times with lysis buffer and then subjected to western blot analysis.
Western blot analysis
Cell lysates were prepared as previously described,39 and protein concentration was determined using the Protein Assay Kit (Bio-Rad, Hercule, CA). Cell lysates were electrophoresed through denaturing polyacrylamide gels and transferred to a PVDF membrane (Millipore, Bedford, MA). The blots were probed or re-probed with the antibodies, and detected using Enhanced Chemiluminescence (ECL) or Odyssey Infrared Imaging System according to the manufacturer's protocol.
Cell viability assay
Cells were seeded in 6-well plates and treated with TRAIL, Okadaic acid or their combination. Cell viability was determined using trypan blue exclusion after 24 hr drug treatment.
PP2A activity assay
PP2A activity was determined using a Malachite Green Phosphatase assay specific for serine/threonine phosphatase activity (Millipore), as described previously.40
Statistical analysis
Statistical analysis was performed using Student's t test. The data were presented as the mean ± SD, and P ≤ 0.05 was considered significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs. Xingming Deng, Emad Alnemri and Yihong Ye for providing PP2A/C, caspase-8 and ubiquitin expressing constructs.
Funding
This work was supported in part by NIH/NCI R01CA174949–01A1.
References
- 1. Eichhorn PJ, Creyghton MP, Wilhelmsen K, van Dam H, Bernards R. A RNA interference screen identifies the protein phosphatase 2A subunit PR55gamma as a stress-sensitive inhibitor of c-SRC. PLoS Genet 2007; 3:e218; PMID:18069897; http://dx.doi.org/ 10.1371/journal.pgen.0030218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta 2009; 1795:1-15; PMID:18588945 [DOI] [PubMed] [Google Scholar]
- 3. Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serinethreonine phosphatases implicated in cell growth and signalling. Biochem J 2001; 353:417-39; PMID:11171037; http://dx.doi.org/ 10.1042/0264-6021:3530417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi Y. Serinethreonine phosphatases: mechanism through structure. Cell 2009; 139:468-84; PMID:19879837; http://dx.doi.org/ 10.1016/j.cell.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 5. Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Development 1996; 10:1904-16; PMID:8756348; http://dx.doi.org/ 10.1101/gad.10.15.1904 [DOI] [PubMed] [Google Scholar]
- 6. Murata K, Wu J, Brautigan DL. B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc Natl Acad Sci U S A 1997; 94:10624-9; PMID:9380685; http://dx.doi.org/ 10.1073/pnas.94.20.10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kong M, Ditsworth D, Lindsten T, Thompson CB. Alpha4 is an essential regulator of PP2A phosphatase activity. Mol Cell 2009; 36:51-60; PMID:19818709; http://dx.doi.org/ 10.1016/j.molcel.2009.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425-79; PMID:9759494; http://dx.doi.org/ 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 9. Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005; 6:9-20; PMID:15688063; http://dx.doi.org/ 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- 10. Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTBPOZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell 2003; 12:783-90; PMID:14527422; http://dx.doi.org/ 10.1016/S1097-2765(03)00341-1 [DOI] [PubMed] [Google Scholar]
- 11. Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 2008; 3:7; PMID:18282298; http://dx.doi.org/ 10.1186/1747-1028-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 2003; 5:1001-7; PMID:14528312; http://dx.doi.org/ 10.1038/ncb1056 [DOI] [PubMed] [Google Scholar]
- 13. Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, et al. . The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 2003; 425:311-6; PMID:13679921; http://dx.doi.org/ 10.1038/nature01959 [DOI] [PubMed] [Google Scholar]
- 14. Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, et al. . Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995; 3:673-82; PMID:8777713; http://dx.doi.org/ 10.1016/1074-7613(95)90057-8 [DOI] [PubMed] [Google Scholar]
- 15. Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem 1996; 271:12687-90; PMID:8663110; http://dx.doi.org/ 10.1074/jbc.271.22.12687 [DOI] [PubMed] [Google Scholar]
- 16. Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science 1998; 281:1305-8; PMID:9721089; http://dx.doi.org/ 10.1126/science.281.5381.1305 [DOI] [PubMed] [Google Scholar]
- 17. Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol Ther 2005; 4:139-63; PMID:15725726; http://dx.doi.org/ 10.4161/cbt.4.2.1508 [DOI] [PubMed] [Google Scholar]
- 18. Rowinsky EK. Targeted induction of apoptosis in cancer management: the emerging role of tumor necrosis factor-related apoptosis-inducing ligand receptor activating agents. J Clin Oncol 2005; 23:9394-407; PMID:16361639; http://dx.doi.org/ 10.1200/JCO.2005.02.2889 [DOI] [PubMed] [Google Scholar]
- 19. Leong S, Cohen RB, Gustafson DL, Langer CJ, Camidge DR, Padavic K, Gore L, Smith M, Chow LQ, von Mehren M, et al. . Mapatumumab, an antibody targeting TRAIL-R1, in combination with paclitaxel and carboplatin in patients with advanced solid malignancies: results of a phase I and pharmacokinetic study. J Clin Oncol 2009; 27:4413-21; PMID:19652058; http://dx.doi.org/ 10.1200/JCO.2008.21.7422 [DOI] [PubMed] [Google Scholar]
- 20. Mom CH, Verweij J, Oldenhuis CN, Gietema JA, Fox NL, Miceli R, Eskens FA, Loos WJ, de Vries EG, Sleijfer S, et al. . Mapatumumab, a fully human agonistic monoclonal antibody that targets TRAIL-R1, in combination with gemcitabine and cisplatin: a phase I study. Clin Cancer Res 2009; 15:5584-90; PMID:19690193; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-0996 [DOI] [PubMed] [Google Scholar]
- 21. Wu GS. TRAIL as a target in anti-cancer therapy. Cancer Lett 2009; 285:1-5; PMID:19299078; http://dx.doi.org/ 10.1016/j.canlet.2009.02.029 [DOI] [PubMed] [Google Scholar]
- 22. Cheng H, Hong B, Zhou L, Allen JE, Tai G, Humphreys R, Dicker DT, Liu YY, El-Deiry WS. Mitomycin C potentiates TRAIL-induced apoptosis through p53-independent upregulation of death receptors: evidence for the role of c-Jun N-terminal kinase activation. Cell cycle 2012; 11:3312-23; PMID:22895172; http://dx.doi.org/ 10.4161/cc.21670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saturno G, Valenti M, De Haven Brandon A, Thomas GV, Eccles S, Clarke PA, Workman P. Combining trail with PI3 kinase or HSP90 inhibitors enhances apoptosis in colorectal cancer cells via suppression of survival signaling. Oncotarget 2013; 4:1185-98; PMID:23852390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009; 137:721-35; PMID:19427028; http://dx.doi.org/ 10.1016/j.cell.2009.03.015 [DOI] [PubMed] [Google Scholar]
- 25. Xu J, Zhou JY, Wei WZ, Wu GS. Activation of the Akt survival pathway contributes to TRAIL resistance in cancer cells. PloS One 2010; 5:e10226; PMID:20419107; http://dx.doi.org/ 10.1371/journal.pone.0010226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu J, Xu Z, Zhou JY, Zhuang Z, Wang E, Boerner J, Wu GS. Regulation of the Src-PP2A interaction in tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. J Biol Chem 2013; 288:33263-71; PMID:24100030; http://dx.doi.org/ 10.1074/jbc.M113.508093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rahman M, Davis SR, Pumphrey JG, Bao J, Nau MM, Meltzer PS, Lipkowitz S. TRAIL induces apoptosis in triple-negative breast cancer cells with a mesenchymal phenotype. Breast Cancer Res Treat 2009; 113:217-30; PMID:18266105; http://dx.doi.org/ 10.1007/s10549-008-9924-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Favre B, Turowski P, Hemmings BA. Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. J Biol Chem 1997; 272:13856-63; PMID:9153244; http://dx.doi.org/ 10.1074/jbc.272.21.13856 [DOI] [PubMed] [Google Scholar]
- 29. Keane MM, Ettenberg SA, Nau MM, Russell EK, Lipkowitz S. Chemotherapy augments TRAIL-induced apoptosis in breast cell lines. Cancer research 1999; 59:734-41; PMID:9973225 [PubMed] [Google Scholar]
- 30. Luo J, Shen G, Yan J, He C, Zhang H. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J 2006; 46:649-57; PMID:16640601; http://dx.doi.org/ 10.1111/j.1365-313X.2006.02730.x [DOI] [PubMed] [Google Scholar]
- 31. Trockenbacher A, Suckow V, Foerster J, Winter J, Krauss S, Ropers HH, Schneider R, Schweiger S. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nate Genet 2001; 29:287-94; PMID:11685209; http://dx.doi.org/ 10.1038/ng762 [DOI] [PubMed] [Google Scholar]
- 32. Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev 1999; 13:2375-87; PMID:10500095; http://dx.doi.org/ 10.1101/gad.13.18.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003; 425:316-21; PMID:13679922; http://dx.doi.org/ 10.1038/nature01985 [DOI] [PubMed] [Google Scholar]
- 34. Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 2005; 123:1107-20; PMID:16360039; http://dx.doi.org/ 10.1016/j.cell.2005.09.033 [DOI] [PubMed] [Google Scholar]
- 35. Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nature reviews Cancer 2006; 6:369-81; PMID:16633365; http://dx.doi.org/ 10.1038/nrc1881 [DOI] [PubMed] [Google Scholar]
- 36. Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol 2004; 16:119-26; PMID:15196553; http://dx.doi.org/ 10.1016/j.ceb.2004.02.005 [DOI] [PubMed] [Google Scholar]
- 37. Harmala-Brasken AS, Mikhailov A, Soderstrom TS, Meinander A, Holmstrom TH, Damuni Z, Eriksson JE. Type-2A protein phosphatase activity is required to maintain death receptor responsiveness. Oncogene 2003; 22:7677-86; PMID:14576831; http://dx.doi.org/ 10.1038/sj.onc.1207077 [DOI] [PubMed] [Google Scholar]
- 38. Barisic S, Strozyk E, Peters N, Walczak H, Kulms D. Identification of PP2A as a crucial regulator of the NF-kappaB feedback loop: its inhibition by UVB turns NF-kappaB into a pro-apoptotic factor. Cell Death Diff 2008; 15:1681-90; PMID:18583989; http://dx.doi.org/ 10.1038/cdd.2008.98 [DOI] [PubMed] [Google Scholar]
- 39. Wang J, Zhou JY, Zhang L, Wu GS. Involvement of MKP-1 and Bcl-2 in acquired cisplatin resistance in ovarian cancer cells. Cell Cycle 2009; 8:3191-8; PMID:19755862; http://dx.doi.org/ 10.4161/cc.8.19.9751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu J, Kovach JS, Johnson F, Chiang J, Hodes R, Lonser R, Zhuang Z. Inhibition of serinethreonine phosphatase PP2A enhances cancer chemotherapy by blocking DNA damage induced defense mechanisms. Proc Natl Acad Sci U S A 2009; 106:11697-702; PMID:19564615; http://dx.doi.org/ 10.1073/pnas.0905930106 [DOI] [PMC free article] [PubMed] [Google Scholar]