Abstract
CK1 (casein kinase 1) is a family of serine/threonine protein kinase that is ubiquitously expressed in eukaryotic organism. CK1 members are involved in the regulation of many cellular processes. Particularly, CK1 was reported to phosphorylate Rec8 subunits of cohesin complex and regulate chromosome segregation in meiosis in budding yeast and fission yeast.1-3 Here we investigated the expression, subcellular localization and potential functions of CK1α, CK1δ and CK1ϵ during mouse oocyte meiotic maturation. We found that CK1α, CK1δ and CK1ϵ all concentrated at the spindle poles and co-localized with γ-tubulin in oocytes at both metaphase I (MI) and metaphase II (MII) stages. However, depletion of CK1 by RNAi or overexpression of wild type or kinase-dead CK1 showed no effects on either spindle organization or chromosome segregation during oocyte meiotic maturation. Thus, CK1 is not the kinase that phosphorylates Rec8 cohesin in mammalian oocytes, and CK1 may not be essential for spindle organization and meiotic progression although they localize at spindle poles.
Keywords: CK1, casein kinase 1, cohesion, chromosome segregation, meiosis, mouse, oocyte, rec8, spindle assembly
Introduction
It is vital for cells to keep high fidelity of chromosome segregation during cell division in both mitosis and meiosis, and any mistake in this process may result in aneuploidy. Aneuploidy in mitosis leads to the development of cancers, and aneuploidy in meiosis is one of the main causes of infertility, abortion and many genetic diseases in humans.4-10 Chromosome segregation is regulated and achieved by spindle microtubules. For accurate segregation of chromosomes, spindle organization and chromosome separation are 2 coordinated events which are orchestrated by the spindle assembly checkpoint (SAC). During the past several years, the mechanisms of spindle assembly and chromosome segregation in mitotic somatic cells have been studied intensely.
Meiotic spindles in mammalian oocytes lack typical centrosomes, and the assembly of spindle is executed by microtubule organizing centers (MTOCs) that are functional equivalents of centrosomes in mitosis. Previous study has shown that in the prophase of mouse oocyte meiosis, over 80 MTOCs form de novo from a cytoplasmic microtubule network.11 These MTOCs contain main pericentriolar material components including γ-tubulin and pericentrin which have similar microtubule nucleation properties as centrosomes.11 After germinal vesicle breakdown (GVBD), MTOCs cluster between chromosomes and promote microtubule nucleation to form a bipolar spindle.11 Many regulators of spindle assembly in mitosis are expected to play similar roles in meiosis. However, only a few of these regulators have been confirmed in meiosis. So the molecular mechanisms of spindle assembly and regulation in mammalian oocytes meiosis remain elusive.
On the other hand, unlike mitosis in somatic cells, meiosis consists of 2 consecutive rounds of chromosome separation with only one round of DNA replication. In meiosis I, the duplicated sister chromatids are held together by meiosis specific Rec8-containing cohesin complex at both chromosome arms and centromeres,12 homologous chromosomes are then paired and recombined to form chiasmatas.12 After spindle assembly, both sister kinetochores within one homolog are captured as a unit by microtubules emanating from the same spindle pole. During anaphase I, only cohesin complex along chromosome arms are cleaved and removed by the activated separase, while centromere cohesion is protected and retained to hold sister chromatids together until meiosis II.13-15 As a result, homologous chromosomes segregate each other in meiosis I. Similar to mitosis, it is believed that phosphorylation of the Rec8 subunit is necessary for the cleavage of cohesin complex by separase, and it has been found that the shugoshin-PP2A complex played important roles in the protection of Rec8-cohesin complex at centromeres by antagonisting Rec8 phosphorylation.16-20 However, little is known about the kinase that phosphorylates Rec8. Previously, it was thought that Plk1 and Aurora B may be involved in the phosphorylation of Rec8 in meiosis as in mitosis, but this has never been proven. Recently, 3 research groups reported that Casein Kinase I (CK1), and perhaps also Cdc7-Dbf4 kinase, was responsible for Rec8 phosphorylation during meiosis in budding yeast and fission yeast.1-3 However, whether these kinases are also involved in the phosphorylation of Rec8 in mammalian oocytes has not yet been explored.
CK1 is a family of monomeric serine/threonine protein kinases and is evolutionary conserved from yeast to human. So far, 7 CK1 isoforms (α, β, γ1, γ2, γ3, δ, ϵ) and their various splice variants have been characterized in mammals. All CK1 members are highly conserved within their kinase domains, but differ significantly in their N-terminal and C-terminal non-catalytic domains.21,22 CK1 family have numerous substrates and regulates diverse cellular processes including circadian rhythms, Wnt signaling, cytoskeleton maintenance, DNA replication, and DNA damage response,23-32 in which we are mostly interested in the cell division. Earlier studies have shown that CK1α was localized to the centrosomes in interphase cells and associated with spindles in mitosis.33 In Drosophila S2 cells, CK1α is a potential kinase for Drosophila kinesin 13, KLP10A.34 CK1α depletion or inhibition caused monopolar and collapsed spindles, which implied that CK1α may be involved in the regulation of spindle organization. In many types of cells, it was reported that CK1δ was localized to the centrosomes, co-localized with γ-tubulin and associated with the mitotic spindle in some cases.35-39 CK1ϵ is most like CK1δ, with greater than 98% identity within the protein kinase domain and 40% identity within the non-catalytic C-terminal regions.40-42 CK1ϵ was also reported to be localized to the centrosomes in several types of cells.43 Moreover, the CK1 specific inhibitor IC261 has been reported to lead to mitotic spindle defects, cell cycle arrest and apoptosis.38,39 Previously, it was reported that in yeast cells, CK1 was involved in Rec8 phosphorylation, as well as regulation of mitotic checkpoint and cytokinesis in mitosis and monopolar attachment of sister kinetochores in meiosis.44,45
Although numerous studies have demonstrated the potential roles of CK1 in cell division, the roles of CK1 in mammalian oocyte meiosis remain elusive. A previous study has reported that CK1α associated with the spindles at metaphase II (MII) stage in mouse oocytes.46 However, microinjection of CK1α antibodies at MII stage had no effect on the completion of second meiosis. However, another recent study found that CK1α co-localized with condensed chromosomes during mouse oocyte meiosis and early embryo development.47 CK1α knock down by morpholinos or inhibition by D4476 leaded to failure of polar body 1 (PB1) extrusion and chromosome misalignment,47 which implied that CK1α was required for chromosome alignment and segregation during oocyte meiotic maturation. Considering the current situation that lacks consistent viewpoints and sufficient evidence about the roles of CK1 in mammalian oocytes, here we investigated the expression, localization and potential roles of CK1α, CK1δ and CK1ϵ in mouse oocyte meiosis. We found that all of these 3 CK1 members, either endogenous or exogenous, were localized to spindle poles at both MI and MII stages and co-localized with γ-tubulin. The specific localization pattern of these 3 CK1 members in mouse oocytes is different from that in yeast meiosis, which implies that they may be functionally different from CK1 in yeast. We then investigated the roles of CK1 by RNAi and overexpression of exogenous wild type or kinase-dead CK1. We found that CK1 knock down by siRNAs had little to no effect on either spindle assembly or chromosome segregation. Moreover, overexpression of wild type or kinase dead CK1 displayed little to no effect, as well. Our results indicated that CK1 was not the kinase that phosphorylates Rec8 in mammalian oocyte meiosis; and CK1 may not be essential for spindle organization and meiotic maturation although they are localized at MTOCs during mouse oocytes meiosis.
Results
Expression of CK1δ and CK1ϵ during mouse oocyte meiotic maturation
To examine the expression level of CK1 during mouse oocyte meiosis, mouse oocytes were cultured for 0h, 8h and 12h, corresponding to germinal vesicle (GV), MI and MII stages of meiotic maturation respectively, and then collected for immunoblot analysis. The immunoblotting results showed that the expression level of CK1δ and CK1ϵ were relatively low at GV stages and then gradually increased at MI and MII stages (Fig. 1A, B).
Figure 1.
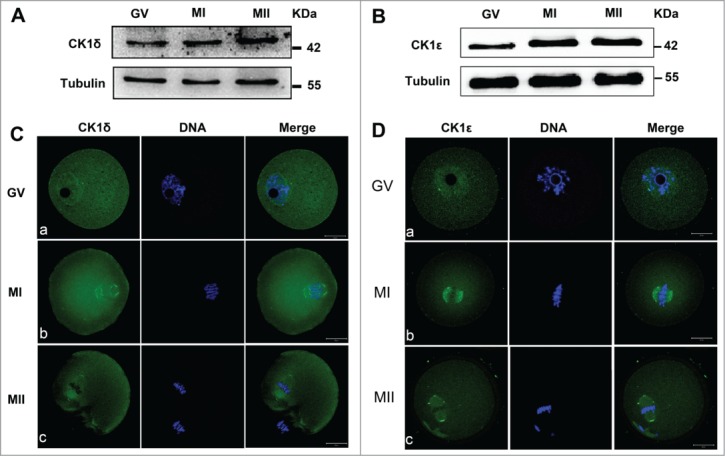
Expression and subcellular localization of CK1δ and CK1ϵ during mouse oocyte meiotic maturation. (A, B) Expression of CK1δ (A) and CK1ϵ (B) were measured by western blotting. Samples (100 oocytes) were collected after oocytes had been cultured for 0, 8h and 12h, corresponding to GV, MI and MII stages respectively. Samples were immunostained for CK1δ or CK1ϵ, α-tubulin was stained as loading control. (C, D) Confocal microscopy showing the subcellular localization of CK1δ (A, green) and CK1ϵ (B, green) in mouse oocytes at GV, MI and MII stages. Oocytes at various stages were fixed and stained with anti-CK1δ or anti-CK1ϵ antibody, DNA (blue) was counterstained with Hoechst 33342. Bar=20 μm.
Localization of endogenous CK1 during mouse oocyte meiotic maturation
To investigate the subcellular localization of endogenous CK1 during oocyte meiotic maturation, oocytes at different stages were collected for immunofluorescent analysis with CK1δ and CK1ϵ specific antibodies. As shown in Figure 1C, at GV stage, CK1δ was distributed in both cytoplasm and GV, with slightly more concentrating in the GV. Interestingly, there were usually one or 2 point signals adjacent to or around the GV. At MI and MII stages, when chromosomes were aligned at the equatorial plates, CK1δ was localized to the 2 sides of aligned chromosomes (Fig. 1C). The localization pattern of CK1ϵ was similar to that of CK1δ, which distributed in both cytoplasm and GV at GV stage, with one or 2 point signals adjacent to or around the GV. At MI and MII stages, CK1ϵ was localized at the 2 sides of aligned chromosomes (Fig. 1D).
Since the CK1 localization in our experiment is different from that reported previously, we further confirmed the localization of CK1 by micro-injection of myc-tagged CK1 mRNA into mouse oocytes and detecting the distribution of myc-CK1 at different stages. First, we constructed expression plasmids of CK1α, CK1δ and CK1ϵ which were linked by myc tags and then transcribed corresponding mRNAs in vitro. These mRNA (in a low concentration, about 0.1μg/μl) were micro-injected into mouse oocytes at GV stage, the injected oocytes were cultured for different times and then fixed for examination of exogenous CK1 by anti-myc antibodies. As shown in Figure 2, exogenous myc-CK1α, myc-CK1δ and myc-CK1ϵ were all localized to the 2 sides of aligned chromosomes at MI and MII stages, which were the samilar to the localization patterns of endogenous CK1.
Figure 2.
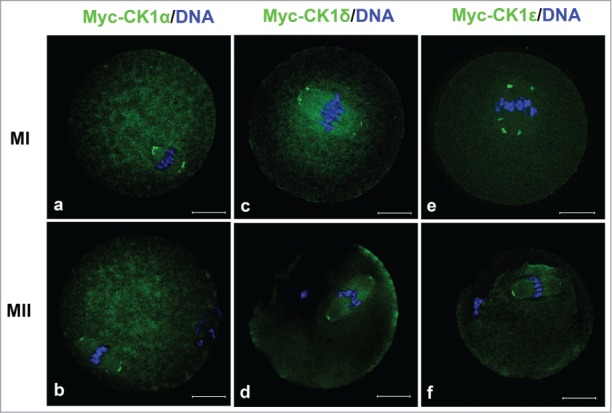
The localization of myc-CK1α (a, b), myc-CK1δ (c, d) and myc-CK1ϵ (e, f) at MI and MII stages. GV oocytes were microinjected with myc-CK1α, myc-CK1δ or myc-CK1ϵ mRNA respectively and cultured to MI and MII stage, then fixed and stained with anti-myc antibody (green) and Hoechst 33342 (blue). Bar=20 μm.
To further define the localization of CK1, myc-CK1α, myc-CK1δ or myc-CK1ϵ was counterstained with α-tubulin and γ-tubulin. As shown in Figure 3, myc-CK1α, myc-CK1δ and myc-CK1ϵ were all co-localized with γ-tubulin at the 2 poles of spindles.
Figure 3.
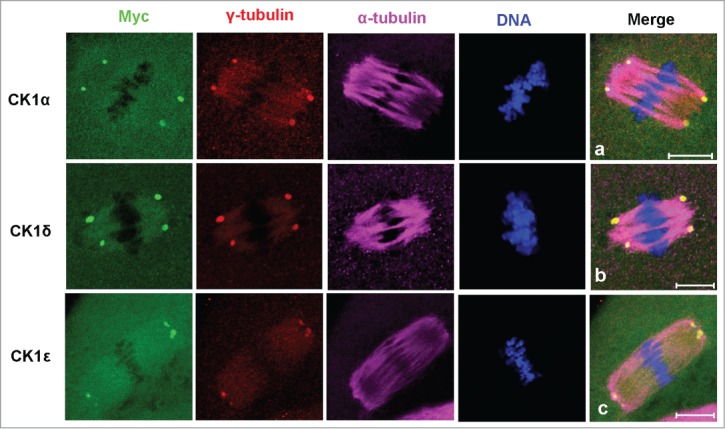
Co-localization of myc-CK1α (a), myc-CK1δ (b) and myc-CK1ϵ (c) with γ-tubulin at spindle poles. Oocytes were microinjected with myc-CK1α, myc-CK1δ or myc-CK1ϵ mRNA and cultured to MI or MII stages, then fixed and stained with anti-myc (green), anti-γ-tubulin (red) and anti-α-tubulin (purple) antibodies. DNA (blue) was counterstained with Hoechst 33342. Bar=10 μm.
CK1 knock down had little effect on the meiosis process
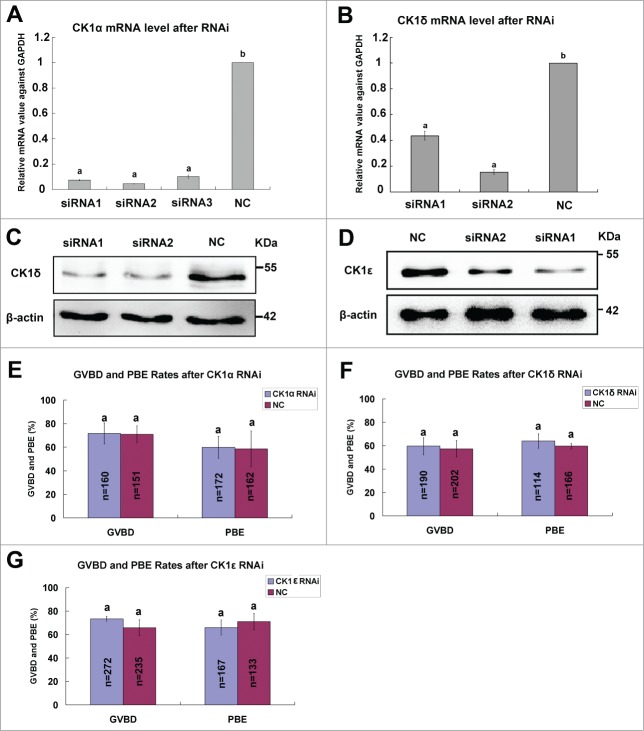
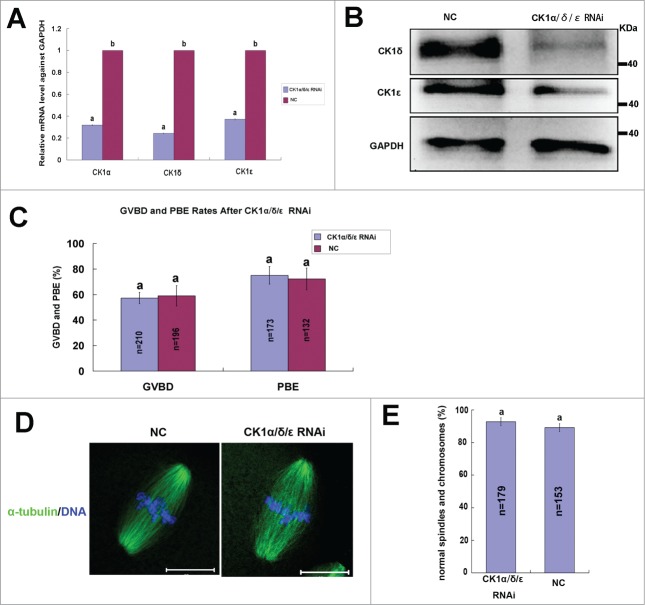
To explore the roles of CK1 in mouse oocyte meiosis, we depleted the expression of CK1 by 2-3 specific siRNAs to CK1α, CK1δ and CK1ϵ, respectively. The knock down efficiency of each siRNA was detected by real time PCR or western blot (Fig. 4A-D), and siRNAs (CK1α-siRNA2, CK1δ-siRNA2 and CK1ϵ-siRNA1) with the highest knock down efficiency were selected for further experiments. The selected siRNAs against CK1α, CK1δ or CK1ϵ were microinjected individually into mouse oocytes at GV stage, the same amount of negative control (NC) siRNAs was injected as controls. These oocytes were arrested at GV stage in M16 medium containing 2.5μM milrinone for 24 hours and then released to fresh M16 medium to resume meiosis. The GVBD rates and polar body extrusion (PBE) rates were calculated after culturing for 2 hours and 14 hours. No significant differences were observed in the GVBD rates and PBE rates for individual depletion of CK1α, CK1δ or CK1ϵ, respectively (Fig. 4E-G). We then mixed these 3 siRNAs and microinjected simultaneously into GV oocytes. After 24 hours of arresting in 2.5μM milrinone, real time PCR and protein gel blot analysis confirmed marked reduction of endogenous CK1α, CK1δ and CK1ϵ at both mRNA level and protein level (Fig. 5A, B). After simultaneous depletion of CK1α, CK1δ and CK1ϵ, we still did not observe any significant differences in the GVBD rates and PBE rates between CK1 RNAi group and NC group (Fig. 5C). We then investigated the spindle organization and chromosome congression after CK1 depletion and found that both spindle organization and chromosome alignment were normal (Fig. 5D). No significant abnormalities had been found as compared to NC group (Fig. 5E).
Figre 4.
Individual effect of CK1α, CK1δ or CK1ϵ knock down on oocyte meiotic progression. Oocytes at GV stage were microinjected with siRNAs (30 μM) against CK1α, CK1δ and CK1ϵ, respectively. The same amount of negative control siRNA (NC) was injected as control. After 24 hours of arrest at GV stage, oocytes were collected and used for RNAi efficiency measurement at mRNA level or protein level. (A, B) The knockdown efficiency of CK1α and CK1δ siRNAs at mRNA level. The relative mRNA level of CK1α (A) or CK1δ (B) compared to NC group was measured by real-time quantitative PCR. Different superscript letters indicate statistical difference (p < 0.05). (C, D) Western blot analysis showed the knockdown efficiency of CK1δ (C) and CK1ϵ (D) siRNAs at protein level. A total of 100 oocytes were collected per sample. (E-G) 30μM CK1α-siRNA2 (E), CK1δ-siRNA2 (F) and CK1ϵ-siRNA1 (G) was microinjected into GV oocytes, respectively. The same amount of NC siRNA was microinjected as control. After 24 hours arrest at GV stage, oocytes were released to fresh M16 medium and cultured for 14 hours. The GVBD rates and PBE rates were calculated, respectively. Data are presented as mean±s.e.m. The same superscript letters indicate no statistical difference (p > 0.05).
Figure 5.
Effect of CK1α, CK1δ and CK1ϵ RNAi on the meiotic maturation process. (A) Oocytes at GV stages were injected with a mixture containing 30 μM CK1α-siRNA2, 30μM CK1δ-siRNA2 and 30μM CK1ϵ-RNAi1, control oocytes were injected with 30 μM NC siRNA. After 24 hours arresting at GV stage, the relative mRNA level of CK1α, CK1δ and CK1ϵ in RNAi group compared to NC group were measured by realtime PCR. Different superscript letters indicate statistical difference (p < 0.05). (B) CK1 RNAi significantly decreased the expression of CK1δ and CK1ϵ. Samples (100 oocytes) of CK1 RNAi group and NC group were collected and immunostained for CK1δ, CK1ϵ and GAPDH. (C) Oocytes of CK1 RNAi group and NC group were arrested at GV stage for 24 hours, then released and cultured for 14h. The rates of GVBD and PBE were calculated respectively. Data are presented as mean±s.e.m. Same superscript letters indicate no statistical difference (p>0.05). (D) Representative images of spindles and chromosomes in RNAi group and NC group. Oocytes of CK1α/δ/ϵ RNAi group and NC group were cultured to MII stage, then fixed and stained for α-tubulin (green) and DNA (blue). Bar=20μm. (E) Percentage of oocytes with normal spindles and chromosomes in CK1 RNAi group and NC group. The same superscript letters indicate no statistical difference (p > 0.05).
Overepxression of wild type CK1 had little effects on the meiotic maturation
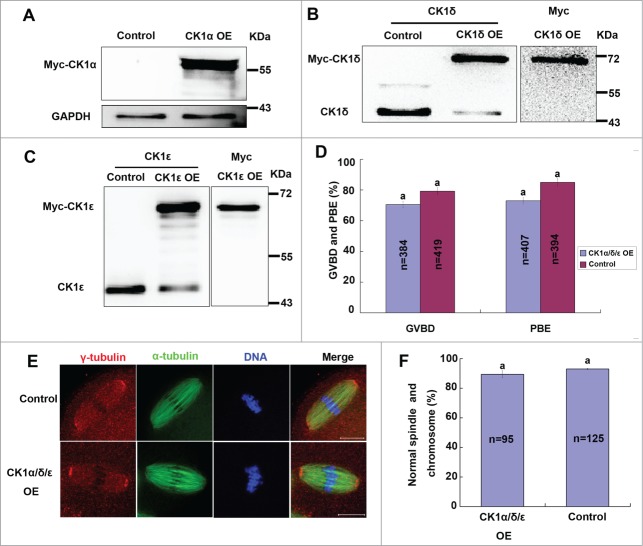
We next investigated the roles of CK1 in mouse oocyte meiosis by protein overexpression. First, we overexpressed wild type myc-CK1α, myc-CK1δ or myc-CK1ϵ alone in mouse oocytes. Immunoblot analysis confirmed successful overexpression of exogenous myc-CK1α, myc-CK1δ and myc-CK1ϵ, respectively (Fig. 6A-C). However, CK1 overexpression individually showed little effect on meiosis process (Data not shown), so we then overexpressed these 3 CK1 isoforms simultaneously. Unexpectedly, simultaneously overexpression of CK1α, CK1δ and CK1ϵ still had little effect on meiotic maturation (Fig. 6D). Moreover, we also examined the spindle organization and chromosome congression of CK1 overexpression group and control group, yet we still did not find any significant abnormalities in both groups (Fig. 6E, F).
Figure 6.
Effect of wild type CK1 overexpression on mouse oocyte meiosis. (A-C) Oocytes were injected with 1 μg/μl myc-CK1α, myc-CK1δ or myc-CK1ϵ mRNA, respectively. Control oocytes were injected with Rnase-free water. After 12 hours arresting at GV stage, these oocytes were collected for protein gel blot analysis of the expression of endogenous and/or exogenous protein. (D) Oocytes were injected with a mixture of 1 μg/ul myc-CK1α, 1 μg/μl myc-CK1δ and 1 μg/μl myc-CK1ϵ, control oocytes were injected with Rnase-free water. After 24 hours of arresting at GV stage, oocytes were released and cultured for 14h. The rates of GVBD and PBE were calculated respectively. Data are presented as mean±s.e.m. The same superscript letters indicate no statistical difference (p > 0.05). (E) Representative images of spindles and chromosomes in CK1 overexpression group and control group. Oocytes of CK1 overexpression group and control group were cultured to MII stage, then fixed and stained for γ-tubulin (red), α-tubulin (green) and DNA (blue). Bar=20 μm. (F) Percentage of oocytes with normal spindles and chromosomes in CK1 overexpression group and control group. The same superscript letters indicate no statistical difference (p > 0.05).
Overexpression of kinase-dead CK1 did not affect meiotic progression
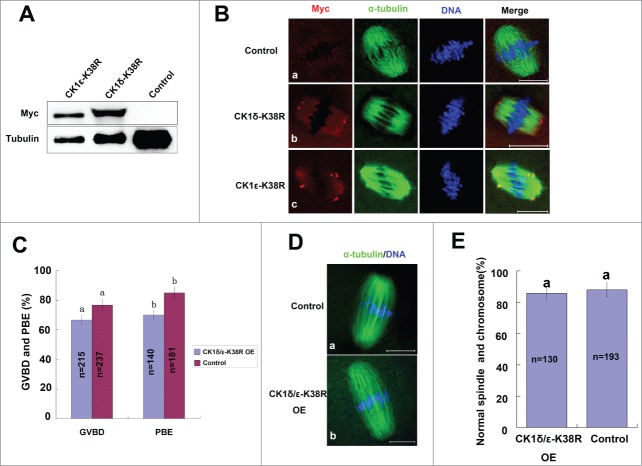
Previous studies have reported that the kinase activity of CK1δ and CK1ϵ was severely compromised by mutating the lysine at residue 38 into arginine (K38R);25,29 we then used this dominant negative approach to disrupt the activity of CK1. We constructed myc-CK1δ-K38R and myc-CK1ϵ-K38R expression plasmids, and transcribed their mRNAs in vitro. After mRNAs of myc-CK1δ-K38R or myc-CK1ϵ-K38R were microinjected into mouse oocytes, exogenous myc-CK1δ-K38R and myc-CK1ϵ-K38R proteins were confirmed by western blot analysis (Fig. 7A). Furthermore, immunofluorescent analysis showed that exogenous myc-CK1δ-K38R and myc-CK1ϵ-K38R were concentrated at the spindle poles (Fig. 7B), which were similar to the localization of endogenous and exogenous wild type CK1, indicating that the localization of CK1 was not dependent on their kinase activity. We next overexpressed both myc-CK1δ-K38R and myc-CK1ϵ-K38R simultaneously and found that the GVBD rate and PBE rate of overexpression group were only slightly decreased as compared to the control group, but without statistical significance (P>0.05) (Fig. 7C). We also examined the spindle assembly and chromosome alignment but still found no significant differences (Fig. 7D, E).
Figure 7.
Effect of overexpression of kinase-dead CK1-K38R on mouse oocyte meiosis. (A, B) Oocytes were injected with 1 μg/μl myc-CK1δ-K38R or myc-CK1ϵ-K38R mRNA, control oocytes were injected with RNAse-free water. After 12 hours of arresting at GV stage, these oocytes were either collected for western blot analysis of the expression of exogenous protein (A), or cultured to MI and immunostained with anti-myc (green) and anti-α-tubulin (red) antibodies (B), DNA (blue) was counterstained with Hoechst 33342. Bar =20 μm. (C) Oocytes were injected with a mixture of 1 μg/ul myc-CK1δ-K38R and myc-CK1ϵ-K38R, control oocytes were injected with Rnase-free water. After 12 hours of arresting at GV stage, oocytes were released and cultured for 14 hours. The rates of GVBD and PBE were calculated respectively. Data are presented as mean±s.e.m. The same superscript letters indicate no statistical difference (p > 0.05). (E) Representative images of spindles and chromosomes in CK1-K38R overexpression group and control group. Oocytes of CK1 overexpression group and control group were cultured to MII stage, then fixed and stained for α-tubulin (green) and DNA (blue). Bar = 20 μm. (F) Percentage of oocytes with normal spindles and chromosomes in CK1-K38R overexpression group and control group. The same superscript letters indicate no statistical difference (p > 0.05).
Discussion
In this study, for the first time, we investigated the expression, localization and potential roles of CK1α, CK1δ and CK1ϵ in mouse oocyte meiosis, but differing results were obtained from other researchers. Unexpectedly, our results showed that CK1α, CK1δ and CK1ϵ were not essential for mouse oocyte meiotic progression, although they all have specific localizations at the spindle poles.
According to studies of meiosis in both fission yeast and budding yeast, CK1 was localized along chromosomes and co-localized with Rec8 subunits, which makes it spatially possible for the phosphorylation of Rec8.1,3 However, our results showed that CK1α, CK1δ and CK1ϵ were not distributed on chromosomes but all concentrated at spindle poles and co-localized with γ-tubulin. The different subcellular localization patterns of CK1 indicated that they might not be involved in Rec8 phosphorylation and chromosome segregation in mammalian oocyte meiosis. Indeed, our subsequent functional analysis confirmed that CK1 was not essential for chromosome separation, as loss-of-function of CK1 by RNAi or dominant-negative approach had no effect on homologous chromosome segregation and polar body extrusion.
In mammalian cells, previous studies have reported that CK1α was localized to centrosomes in interphase cells and was associated with spindles during mitosis;33 CK1δ was localized to centrosomes, co-localized with γ-tubulin and was associated with the mitotic spindles under DNA damage; and CK1ϵ was concentrated at centrosomes.35-39,43 The localization patterns of CK1 on centrosomes or spindles make them possible candidates for spindle assembly regulators. Moreover, mitotic spindle defects and cell cycle arrest caused by IC261 treatment indicate that CK1 activity may be related to spindle assembly.38,39 However, as we know, the spindle assembly process in meiosis is not always identical to that in mitosis. In meiosis, the spindles have their own specific organization and regulation mechanisms. Focusing on CK1 in mammalian oocyte meiosis, a previous study has reported that CK1α was associated with the spindle at MII stage in mouse oocyte but was not essential for the completion of second meiosis.46 But a recent study reported that CK1α distributed on the condensed chromosomes and was required for chromosome alignment and segregation during oocyte meiotic maturation,47 which was inconsistent with the previous study.46 We therefore investigated the localization of CK1δ and CK1ϵ, both endogenous and exogenous, and found that they both concentrated at spindle poles, co-localized with γ-tubulin, which was consistent with the previously reported localization pattern in mitotic somatic cells.35-42 We also detected the localization of exogenous CK1α and found that exogenous CK1α was mainly localized to the spindle poles in mouse oocytes at both MI and MII stages.. However, 2 previous studies in mouse oocytes showed different localization patterns. Gross et al. found that CK1α was associated with the spindles at MII stage in mouse oocytes, but Wang et al. found that CK1α was co-localized with condensed chromosomes during mouse oocyte meiosis.46,47 The different localizations observed by Gross et al. and Wang et al. may be due to the different specificities of the anti-CK1α antibodies that were used, as polyclonal antibodies could give non-specific signals. In contrast, our results of CK1α localization obtained by both a monoclonal anti-myc antibody staining and myc-taged mRNA injection were more reliable, as the specificity of the anti-myc antibody has been proven by numerous previous studies.48,49
The specific localizations of CK1 prompted us to further explore its functions in mouse oocyte meiosis. However, knock down of CK1α, CK1δ and CK1ϵ by RNAi, either individually or simultaneously, caused little effect on either the meiosis process, spindle organization or chromosome alignment. There were 2 possible explanations for this situation. Firstly, different members of the CK1 family were highly conserved within their kinase domains and there may be various splice variants, and these CK1 isoforms and variants may be functionally redundant. Knock down of one isoform may be compensated by other isoforms. Secondly, it was also probable that CK1 was not essential for the spindle organization and meiotic maturation of mouse oocytes. In addition, our results were in accordance with those obtained by Gross et al. who found that microinjection of CK1α antibodies at MII stage had no effect on the completion of second meiosis.46
We next applied protein overexpression to investigate their effects on meiosis, but again obtained negative results. Overexpression of exogenous CK1α, CK1δ and CK1ϵ, both individually and simultaneously, caused little effect on either the meiosis process, spindle organization or chromosome alignment. We also overexpressed kinase-dead CK1 mutants to disturb their kinase activities, yet obtained negative results. The overexpressed CK1δ-K38R and CK1ϵ-K38R also concentrated at spindle poles like wild type CK1, which indicated that their localizations and functions did not depend on their kinase activity. Moreover, the overexpression results of both wild type and mutated CK1 also supported our conclusion that CK1 was not essential for mouse oocyte meiotic progression.
In summary, several lines of evidence in our report demonstrate that although CK1α, CK1δ and CK1ϵ are all localized to MTOCs or spindle poles, they are not the kinase responsible for Rec8 phosphorylation during mouse oocyte meiosis, indicating that they are probably not essential for mouse oocyte meiotic maturation.
Materials and Methods
Oocyte collection and culture
Mouse oocyte collection and culture were performed as previously described.49,50 Animal care and handling were conducted in accordance with Animal Research Committee policies of the Institute of Zoology, Chinese Academy of Sciences.
Plasmids construction and mRNA synthesis
For the construction of wild type myc-CK1α-pCS2+, myc-CK1δ-pCS2+ and myc-CK1ϵ-pCS2+, mouse oocyte total cDNA was used as the template for PCR amplification of the full length of mouse CK1α, CK1δ and CK1ϵ CDS, respectively. The primers used for amplification are listed in Table 1. After two rounds of nest PCR, the PCR products were purified and digested using Fse I and Asc I (NEB), then subcloned into the myc-pCS2+ vectors. Myc-CK1δ-K38R-pCS2+ and myc-CK1ϵ-K38R-pCS2+ mutants were generated by PCR using primers CK1δ-FM/CK1δ-RM and CK1ϵ-FM/CK1ϵ-RM to induce the point mutation at residue 38 (AAG->AGG), respectively. Wild type myc-CK1δ-pCS2+ and myc-CK1ϵ-pCS2+ plasmids were used as templates for PCR amplification. All plasmids were sequenced to ensure the accuracy of coding sequences.
Table 1.
Primers for PCR amplification of CK1α/δ/ϵ and CK1δ/ϵ-K38R
| Genes | Primers (5′- 3′) |
|---|---|
| CK1α-F1 | CGACAGAGCCTTGGGTTTAG |
| CK1α-R1 | CTCAGGATCCGCAGTGCTAC |
| CK1α-F2 | GTTGGCCGGCCGATGGCGAGCAGCAG |
| CK1α-R2 | GTTGGCGCGCCTGCTTAGAAACCTGTG |
| CK1δ-F1 | GGAAAGCGATGGTGAAA |
| CK1δ-R1 | CGGTGGGATCATCAGG |
| CK1δ-F2 | GTTGGCCGGCCCATGGAGCTGAGGGTC |
| CK1δ-R2 | GTTGGCGCGCCTCATCGGTGCACGACAG |
| CK1ϵ-F1 | GAATCCTCTGGCATCCG |
| CK1ϵ-R1 | CATCTCACCGCACCCT |
| CK1ϵ-F2 | GTTGGCCGGCCCATGGAGTTGCGTGTG |
| CK1ϵ-R2 | GTTGGCGCGCCTCATTTCCCAAGATGG |
| CK1δ-FM | GACACATTCAAGCCTGATGGCAACT |
| CK1δ-RM | AGTTGCCATCAGGCTTGAATGTGTC |
| CK1ϵ-FM | TCACACATTCGAGCCTGATGGCTAC |
| CK1ϵ-RM | GTAGCCATCAGGCTCGAATGTGTGA |
For mRNA synthesis, plasmids were linearized by Sal I (Takara) and used as templates for in vitro transcription with Sp6 mMESSAGE mMACHINE kit (Ambion). The capped mRNAs were then purified by RNeasy clean up kit (Qiagen) and stored at −80°C.
Microinjection of mRNA and siRNA
Microinjection of mRNAs and siRNAs were performed as previously described.51,52 The corresponding mRNA concentration was 0.1 μg/μl for protein localization and 1 μg/μl for protein overexpression. The same amount of RNase-free water was injected as control. After microinjection, oocytes were arrested at GV stage for 12 hours for protein translation. For each individual RNAi injection, siRNA was used at 30μM. For simultaneous RNAi, injection, siRNA concentration was adjusted to make the same final concentration of 30μM. The same amount of NC siRNA was microinjected as control. After siRNA microinjection, oocytes were arrested at the GV stage for 24 hours for the depletion of endogenous mRNA and protein. The siRNAs used for knock down of CK1α, CK1δ and CK1ϵ are listed inTable 2.
Table 2.
siRNA sequences for CK1α, CK1δ and CK1ϵ RNAi
| siRNA | sequence | |
|---|---|---|
| sense(5′-3′) | antisense(5′-3′) | |
| CK1α siRNA1 | 5′ GCCAUGUACUUAAAUUACUTT 3′ | 5′ AGUAAUUUAAGUACAUGGCTT 3′ |
| CK1α siRNA2 | 5′ GGGCGUCACUGUAAUAAGUTT 3′ | 5′ ACUUAUUACAGUGACGCCCTT 3′ |
| CK1α siRNA3 | 5′ GCCUUGAAGACCUCUUCAATT 3′ | 5′ UUGAAGAGGUCUUCAAGGCTT 3′ |
| CK1δ siRNA1 | 5′ GGGACUACAAUGUCAUGGUTT 3′ | 5′ACCAUGACAUUGUAGUCCCTT3′ |
| CK1δ siRNA2 | 5′ GGAUCAGUGAGAAGAAGAUTT 3′ | 5′AUCUUCUUCUCACUGAUCCTT3′ |
| CK1ϵ siRNA1 | 5′ GUAGCCAUCAAGCUCGAAUTT 3' | 5' AUUCGAGCUUGAUGGCUACTT 3' |
| CK1ϵ siRNA2 | 5' CCAACAAACUGGCAAUACUTT 3' | 5' AGUAUUGCCAGUUUGUUGGTT 3' |
Western blotting
Mouse oocytes were collected and lysed with equal volume of 2×SDS loading buffer (CW biotech) and boiled for 5min. Samples were separated by SDS-PAGE and then transferred to PVDF (Millipore) membranes. After transfer, the membranes were washed briefly with TBST buffer and blocked with TBST containing 5% skimmed milk for 1 hour at room temperature, then incubated with primary antibody overnight at 4˚C. After washing 3 times in TBST, 10 min each, the membranes were incubated with HRP-conjugated secondary antibody for 2 hours at room temperature. Finally, the membranes were washed and processed using the enhanced chemiLuminescence detection system (Bio-Rad, CA). If a second protein was needed to be detected, the membrane was then incubated in stripping buffer (CW biotech, Beijing) for 30 min at room temperature and washed thoroughly, then blocked again and incubated with another antibody. The following antibodies were used for protein gel blotting: Rabbit anti-CK1δ (1:1000, Proteintech, 14388-1-AP); Mouse anti-CK1ϵ (1:1000, BD Biosciences, 610445); Rabbit anti-α-tubulin(1:1000, Cell Signaling Technology, #2125); Rabbit anti-GAPDH(1:1000, Cell Signaling Technology, #2118); Mouse anti-myc (1:1000, Invitrogen, R950-25); Mouse anti-β-actin (1:1000, ZSGB-BIO, TA-09); HRP-conjugated goat anti-rabbit IgG (1:2000, ZSGB-BIO, ZB-2301), HRP-conjugated goat anti-mouse IgG (1:2000, ZSGB-BIO, ZB-2305).
Immunofluorescent analysis
Oocytes were fixed with 4% paraformaldehyde in PBS containing 0.5% Triton X-100 for 30 minutes, blocked with blocking buffer (1% BSA, 0.1% Tween 20 in PBS) for 1 hour at room temperature, then incubated with primary antibody overnight at 4≡°C. The oocytes were washed 3 times with washing buffer (0.1% Tween 20, 0.01% Triton X-100 in PBS), then incubated with secondary antibody for 2 hours at room temperature, then washed another 3 times. For double staining or triple staining, oocytes were first stained with one antibody and its corresponding secondary antibody, after 3 washes, oocytes were blocked again and incubated with another antibody as described before. Finally, DNA was stained with Hoechst 33342 (10μg/μl, Sigma) for 30 minutes, and mounted on glass slides for detection by Confocal microscopy (Zeiss LSM 780, Germany). Primary antibodies used were goat anti-CK1δ (1:50, Santa Cruz, sc-6474); rabbit anti-CK1ϵ (1:50, BD Biosciences, 610445); FITC-conjugated mouse anti-α-tubulin (1:200, Sigma, F2168); mouse anti-γ-tubulin (1:200, Sigma, T6557); rabbit anti-α-tubulin (1:100, Cell Signaling Technology, #2125); FITC-conjugated mouse anti-myc (1:100, Invitrogen, R953-25); mouse anti-myc (1:100, Invitrogen, R950-25); The following secondary antibodies were used at a 1:200 dilution: FITC-conjugated goat anti-rabbit IgG (ZSGB-BIO, ZF-0311), FITC-conjugated goat anti-mouse IgG (ZSGB-BIO, ZF-0312), FITC-conjugated rabbit anti-goat IgG (ZSGB-BIO, ZF-0314), TRITC-conjugated goat anti-mouse IgG (ZSGB-BIO, ZF-0313), TRITC-conjugated goat anti-rabbit IgG (ZSGB-BIO, ZF-0316) and Cy5-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratory, 111-177-003).
Real time PCR analysis
Total RNA was extracted from 50 oocytes using RNeasy micro purification kit (Qiagen); the first strand cDNA was generated with M-MLV first strand cDNA synthesis kit (Invitrogen) using oligo(dT) primers. Realtime PCR was performed using UltraSYBR Mixture (CW biotech) in triplicate on a LightCycler→ 480 II Real-Time PCR System (Roche). GAPDH was selected as a reference gene. The primers used for real time PCR are listed in Table 3.
Table 3.
Primers for realtime PCR
| Genes | Primers (5'- 3') |
|---|---|
| CK1α-F3 | ATGGGTATTGGGCGTCACTG |
| CK1α-R3 | TCGGCGACTCTGCTCAATAC |
| CK1δ-F3 | GACCTGTTCAACTTCTGT |
| CK1δ-R3 | GTTATCTGGCTTCACATCT |
| CK1ϵ-F3 | GGTTGAAGCATGGAGTTGCG |
| CK1ϵ-R3 | TGTTGGCACCCAGGTAGATG |
| GAPDH-F | TGGCAAAGTGGAGATTGTTGCC |
| GAPDH-R | AAGATGGTGATGGGCTTCCCG |
Statistical analysis
All experiments were repeated at least 3 times. All data are expressed as means ±s.e.m. and the number of oocytes observed (n) given in parentheses. Data were analyzed by independent-sample t-tests with SPSS software (SPSS Inc..,Chicago, IL). P<0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Shi-wen Li and Hua Qin for their technical assistance.
Funding
This study was supported by National Basic Research Program of China (No. 2012CB944404, 2011CB944501) and the National Natural Science Foundation of China (No. 30930065, 31371451).
Author Contributions
S-TQ, Q-YS, and W-HW conceived and designed the experiments. S-TQ, Z-BW, LH, and Y-CO performed the experiments. S-TQ, L-FL, and Y-XX analyzed the data. YH contributed reagents, materials and analysis tools. S-TQ, Q-YS, and W-HW wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, Ammerer G, Mechtler K, Gregan J. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle 2010; 9:2657–62; PMID:20581463; http://dx.doi.org/ 10.4161/cc.9.13.12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev cell 2010; 18:397–409; PMID:20230747; http://dx.doi.org/ 10.1016/j.devcel.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol 2010; 12:500–6; PMID:20383139; http://dx.doi.org/ 10.1038/ncb2052 [DOI] [PubMed] [Google Scholar]
- 4.Cowchock FS, Gibas Z, Jackson LG. Chromosome errors as a cause of spontaneous abortion: the relative importance of maternal age and obstetric history. Fertility Sterility 1993; 59:1011–4; PMID:8486167 [DOI] [PubMed] [Google Scholar]
- 5.Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, Millie E, Saker D, Shen J, Zaragoza M. Human aneuploidy: incidence, origin, and etiology. Environmental Mol Mutagenesis 1996; 28:167–75; PMID:8908177; http://dx.doi.org/ 10.1002/(SICI)1098-2280(1996)28:3%3c167::AID-EM2%3e3.0.CO;2-B [DOI] [PubMed] [Google Scholar]
- 6.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001; 2:280–91; PMID:11283700; http://dx.doi.org/ 10.1038/35066065 [DOI] [PubMed] [Google Scholar]
- 7.Mailhes JB, Young D, London SN. Postovulatory ageing of mouse oocytes in vivo and premature centromere separation and aneuploidy. Biol Reprod 1998; 58:1206–10; PMID:9603254; http://dx.doi.org/ 10.1095/biolreprod58.5.1206 [DOI] [PubMed] [Google Scholar]
- 8.Zenzes MT, Casper RF. Cytogenetics of human oocytes, zygotes, and embryos after in vitro fertilization. Hum Genet 1992; 88:367–75; PMID:1740312; http://dx.doi.org/ 10.1007/BF00215667 [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Wang L, Ma M, Song Z, Zhang J, Xu G, Fan J, Li N, Cram DS, Yao Y. A PGD pregnancy achieved by embryo copy number variation sequencing with confirmation by non-invasive prenatal diagnosis. J Genet Genomics 2014; 41:453–6; PMID:25160978; http://dx.doi.org/ 10.1016/j.jgg.2014.06.007 [DOI] [PubMed] [Google Scholar]
- 10.Jiang H, Wang L, Cui Y, Xu Z, Guo T, Cheng D, Xu P, Yu W, Shi Q. Meiotic chromosome behavior in a human male t(8;15) carrier. J Genet Genomics 2014; 41:177–85; PMID:24656237; http://dx.doi.org/ 10.1016/j.jgg.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 11.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 2007; 130:484–98; PMID:17693257; http://dx.doi.org/ 10.1016/j.cell.2007.06.025 [DOI] [PubMed] [Google Scholar]
- 12.Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K. A central role for cohesions in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 1999; 98:91–103; PMID:10412984; http://dx.doi.org/ 10.1016/S0092-8674(00)80609-1 [DOI] [PubMed] [Google Scholar]
- 13.Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 2000; 103:387–98; PMID:11081626; http://dx.doi.org/ 10.1016/S0092-8674(00)00131-8 [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Iwai T, Yokota T, Yamashita M. Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J Cell Sci 2003; 116:2781–90; PMID:12759374; http://dx.doi.org/ 10.1242/jcs.00495 [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Okada K, Ogushi S, Miyano T, Miyake M, Yamashita M. Loss of Rec8 from chromosome arm and centromere region is required for homologous chromosome separation and sister chromatid separation, respectively, in mammalian meiosis. Cell Cycle 2006; 5:1448–55; PMID:16855401; http://dx.doi.org/ 10.4161/cc.5.13.2903 [DOI] [PubMed] [Google Scholar]
- 16.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 2004; 427:510–7; PMID:14730319; http://dx.doi.org/ 10.1038/nature02312 [DOI] [PubMed] [Google Scholar]
- 17.Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 2006; 441:46–52; PMID:16541025; http://dx.doi.org/ 10.1038/nature04663 [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol 2008; 10:42–52; PMID:18084284; http://dx.doi.org/ 10.1038/ncb1667 [DOI] [PubMed] [Google Scholar]
- 19.Yin S, Ai JS, Shi LH, Wei L, Yuan J, Ouyang YC, Hou Y, Chen DY, Schatten H, Sun QY. Shugoshin1 may play important roles in separation of homologous chromosomes and sister chromatids during mouse oocyte meiosis. PloS One 2008; 3:e3516; PMID:18949044; http://dx.doi.org/ 10.1371/journal.pone.0003516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wassmann K. Sister chromatid segregation in meiosis II: deprotection through phosphorylation. Cell Cycle 2013; 12:1352–9; PMID:23574717; http://dx.doi.org/ 10.4161/cc.24600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal 2005; 17:675–89; PMID:15722192; http://dx.doi.org/ 10.1016/j.cellsig.2004.12.011 [DOI] [PubMed] [Google Scholar]
- 22.Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. Intl J Biochem Cell Biol 2011; 43:465–9; PMID:21145983; http://dx.doi.org/ 10.1016/j.biocel.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 23.Li L, Zhao D, Wei H, Yao L, Dang Y, Amjad A, Xu J, Liu J, Guo L, Li D, et al.. REGgamma deficiency promotes premature aging via the casein kinase 1 pathway. Proc Natl Acad Sci U S A 2013; 110:11005–10; PMID:23766372; http://dx.doi.org/ 10.1073/pnas.1308497110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, et al.. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF β-TRCP ubiquitin ligase. Cancer Cell 2010; 18:147–59; PMID:20708156; http://dx.doi.org/ 10.1016/j.ccr.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc Natl Acad Sci U S A 2009; 106:21359–64; PMID:19948962; http://dx.doi.org/ 10.1073/pnas.0906651106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakaguchi K, Saito S, Higashimoto Y, Roy S, Anderson CW, Appella E. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase – Effect on Mdm2 binding. J Biol Chem 2000; 275:9278–83; PMID:10734067; http://dx.doi.org/ 10.1074/jbc.275.13.9278 [DOI] [PubMed] [Google Scholar]
- 27.Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, et al.. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A 2009; 106:15744–9; PMID:19805222; http://dx.doi.org/ 10.1073/pnas.0908733106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huart AS, MacLaine NJ, Meek DW, Hupp TR. CK1alpha plays a central role in mediating MDM2 control of p53 and E2F-1 protein stability. J Biol Chem 2009; 284:32384–94; PMID:19759023; http://dx.doi.org/ 10.1074/jbc.M109.052647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature 1999; 401:345–50; PMID:10517632; http://dx.doi.org/ 10.1038/43830 [DOI] [PubMed] [Google Scholar]
- 30.Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, et al.. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol 2009; 29:3853–66; PMID:19414593; http://dx.doi.org/ 10.1128/MCB.00338-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, Yin H, Kuret J. Casein kinase 1 delta phosphorylates tau and disrupts its binding to microtubules. J Biol Chem 2004; 279:15938–45; PMID:14761950; http://dx.doi.org/ 10.1074/jbc.M314116200 [DOI] [PubMed] [Google Scholar]
- 32.Yasojima K, Kuret J, DeMaggio AJ, McGeer E, McGeer PL. Casein kinase 1 delta mRNA is upregulated in alzheimer disease brain. Brain Res 2000; 865:116–20; PMID:10814741; http://dx.doi.org/ 10.1016/S0006-8993(00)02200-9 [DOI] [PubMed] [Google Scholar]
- 33.Brockman JL, Gross SD, Sussman MR, Anderson RA. Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proc Natl Acad Sci U S A 1992; 89:9454–8; PMID:1409656; http://dx.doi.org/ 10.1073/pnas.89.20.9454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mennella V, Tan DY, Buster DW, Asenjo AB, Rath U, Ma A, Sosa HJ, Sharp DJ. Motor domain phosphorylation and regulation of the drosophila kinesin 13, KLP10A. J Cell Biol 2009; 186:481–90; PMID:19687256; http://dx.doi.org/ 10.1083/jcb.200902113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milne DM, Looby P, Meek DW. Catalytic activity of protein kinase CK1 delta (casein kinase 1delta) is essential for its normal subcellular localization. Exp Cell Res 2001; 263:43–54; PMID:11161704; http://dx.doi.org/ 10.1006/excr.2000.5100 [DOI] [PubMed] [Google Scholar]
- 36.Greer YE, Rubin JS. Casein kinase 1 delta functions at the centrosome to mediate Wnt-3a-dependent neurite outgrowth. J Cell Biol 2011; 192:993–1004; PMID:21422228; http://dx.doi.org/ 10.1083/jcb.201011111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zyss D, Ebrahimi H, Gergely F. Casein kinase I delta controls centrosome positioning during T cell activation. J Cell Biol 2011; 195:781–97; PMID:22123863; http://dx.doi.org/ 10.1083/jcb.201106025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoter M, Bamberger AM, Aslan B, Kurth M, Speidel D, Loning T, Frank HG, Kaufmann P, Lohler J, Henne-Bruns D, et al.. Inhibition of casein kinase I delta alters mitotic spindle formation and induces apoptosis in trophoblast cells. Oncogene 2005; 24:7964–75; PMID:16027726; http://dx.doi.org/ 10.1038/sj.onc.1208941 [DOI] [PubMed] [Google Scholar]
- 39.Behrend L, Milne DM, Stoter M, Deppert W, Campbell LE, Meek DW, Knippschild U. IC261, a specific inhibitor of the protein kinases casein kinase 1-delta and -epsilon, triggers the mitotic checkpoint and induces p53-dependent postmitotic effects. Oncogene 2000; 19:5303–13; PMID:11103931; http://dx.doi.org/ 10.1038/sj.onc.1203939 [DOI] [PubMed] [Google Scholar]
- 40.Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal 1998; 10:699–711; PMID:9884021; http://dx.doi.org/ 10.1016/S0898-6568(98)00042-4 [DOI] [PubMed] [Google Scholar]
- 41.Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM. Isolation and characterization of human casein kinase I epsilon (CKI), a novel member of the CKI gene family. J Biol Chem 1995; 270:14875–83; PMID:7797465; http://dx.doi.org/ 10.1074/jbc.270.25.14875 [DOI] [PubMed] [Google Scholar]
- 42.Graves PR, Haas DW, Hagedorn CH, DePaoli-Roach AA, Roach PJ. Molecular cloning, expression, and characterization of a 49-kilodalton casein kinase I isoform from rat testis. J Biol Chem 1993; 268:6394–401; PMID:8454611 [PubMed] [Google Scholar]
- 43.Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW. Centrosomal anchoring of the protein kinase CK1 delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol 2002; 322:785–97; PMID:12270714; http://dx.doi.org/ 10.1016/S0022-2836(02)00857-4 [DOI] [PubMed] [Google Scholar]
- 44.Johnson AE, Chen JS, Gould KL. CK1 is required for a mitotic checkpoint that delays cytokinesis. Curr Biol 2013; 23:1920–6; PMID:24055157; http://dx.doi.org/ 10.1016/j.cub.2013.07.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell 2006; 126:1049–64; PMID:16990132; http://dx.doi.org/ 10.1016/j.cell.2006.07.029 [DOI] [PubMed] [Google Scholar]
- 46.Gross SD, Simerly C, Schatten G, Anderson RA. A casein kinase I isoform is required for proper cell cycle progression in the fertilized mouse oocyte. J Cell Sci 1997; 110:3083–90; PMID:9365278 [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Lu A, Zhou HX, Sun R, Zhao J, Zhou CJ, Shen JP, Wu SN, Liang CG. Casein kinase 1 α regulates chromosome congression and separation during mouse oocyte meiotic maturation and early embryo development. PloS One 2013; 8:e63173; PMID:23690993; http://dx.doi.org/ 10.1371/journal.pone.0063173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M, Li S, Yuan J, Wang ZB, Sun SC, Schatten H, Sun QY. Bub3 is a spindle assembly checkpoint protein regulating chromosome segregation during mouse oocyte meiosis. PloS One 2009; 4:e7701; PMID:19888327; http://dx.doi.org/ 10.1371/journal.pone.0007701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qi ST, Wang ZB, Ouyang YC, Zhang QH, Hu MW, Huang X, Ge Z, Guo L, Wang YP, Hou Y, et al.. Overexpression of SETbeta, a protein localizing to centromeres, causes precocious separation of chromatids during the first meiosis of mouse oocytes. J Cell Sci 2013; 126:1595–603; PMID:23444375; http://dx.doi.org/ 10.1242/jcs.116541 [DOI] [PubMed] [Google Scholar]
- 50.Lin F, Ma XS, Wang ZB, Wang ZW, Luo YB, Huang L, Jiang ZZ, Hu MW, Schatten H, Sun QY. Different fates of oocytes with DNA double-strand breaks in vitro and in vivo. Cell Cycle 2014; 13:2674–80; PMID:25486355; http://dx.doi.org/ 10.4161/15384101.2015.945375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L, Ge ZJ, Wang ZB, Sun T, Ouyang YC, Sun QY, Sun YP. TGN38 is required for the metaphase I/anaphase I transition and asymmetric cell division during mouse oocyte meiotic maturation. Cell Cycle 2014; 13:2723–32; PMID:25486359; http://dx.doi.org/ 10.4161/15384101.2015.945828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo YB, Ma JY, Zhang QH, Lin F, Wang ZW, Huang L, Schatten H, Sun QY. MBTD1 is associated with Pr-Set7 to stabilize H4K20me1 in mouse oocyte meiotic maturation. Cell Cycle 2013; 12:1142–50; PMID:23475131; http://dx.doi.org/ 10.4161/cc.24216 [DOI] [PMC free article] [PubMed] [Google Scholar]