Abstract
Genotyping of Mycobacterium tuberculosis isolates contributes to tuberculosis (TB) control through detection of possible outbreaks. However, 20% of U.S. cases do not have an isolate for testing, and 10% of cases with isolates do not have a genotype reported. TB outbreaks in populations with incomplete genotyping data might be missed by genotyping-based outbreak detection. Therefore, we assessed the representativeness of TB genotyping data by comparing characteristics of cases reported during January 1, 2009–December 31, 2010, that had a genotype result with those cases that did not. Of 22,476 cases, 14,922 (66%) had a genotype result. Cases without genotype results were more likely to be patients <19 years of age, with unknown HIV status, of female sex, U.S.-born, and with no recent history of homelessness or substance abuse. Although cases with a genotype result are largely representative of all reported U.S. TB cases, outbreak detection methods that rely solely on genotyping data may underestimate TB transmission among certain groups.
Since 2004, the Centers for Disease Control and Prevention (CDC) has offered routine genetic characterization (i.e., genotyping) of all U.S. tuberculosis (TB) cases with Mycobacterium tuberculosis (M. tuberculosis) isolates.1 Genotyping is a laboratory method used to determine the relatedness of isolates; although not a perfect measure of transmission,2 this tool contributes to TB control in multiple ways. Genotyping data contribute to TB control, including the detection of genotype clusters that might represent remote or recent transmission (including outbreaks).3–6 TB genotyping data are also important for defining the scope of outbreaks,7 monitoring outbreaks over time,8 distinguishing relapse from reinfection,9 detecting or confirming false-positive culture results,10,11 confirming known epidemiologic links, and finding unknown links between cases.3,4,12 The utility of genotyping is limited in populations for which few cases are genotyped, because potential transmission relationships between cases might be missed. TB genotyping is most effective when data are representative of the entire population of TB cases.13–15
Applying TB genotyping data to TB control requires that an isolate be submitted for genotyping and that the genotyping result be linked to the patient's demographic and clinical information. While some states have independent systems for generating and linking genotyping data, most states rely on the national CDC-funded system. In this system, an isolate is submitted for genotyping to the CDC-funded national genotyping laboratory, and genotyping results are linked to the patient's demographic and clinical data, which are reported to the National Tuberculosis Surveillance System (NTSS).16 This linkage is facilitated by a CDC-developed and -funded national Web-based genotyping database, which includes both NTSS and genotyping data.
First, specimens are collected from a suspected TB patient. Specimens are generally sent to a jurisdictional public health laboratory for culturing and processing and, when a specimen yields a culture that is positive for M. tuberculosis, an isolate is sent to the national genotyping laboratory. In some cases, a viable culture might not be available to be submitted for genotyping. In other cases, a viable culture might be available but not submitted to the genotyping laboratory. These latter cases represent a missed opportunity for genotyping. Although it is not possible to determine whether or not a viable culture was available for submission from nationally reported data, we can use the presence of drug susceptibility testing (DST) results, testing that requires a viable culture, to identify cases for which a viable culture was likely available to be submitted for genotyping.
Once the isolate is genotyped, the result is entered into the national Web-based genotyping database. In parallel with this process, the patient's demographic and clinical data are submitted to jurisdictional public health authorities for reporting to NTSS; these data are then uploaded into the national Web-based genotyping database. The state TB program is responsible for the critical step of linking the surveillance report to the genotyping result, using a state-assigned identification number. Failure to link the genotyping and surveillance records will result in the case appearing to have not been genotyped. Because surveillance and genotyping data are linked by the state, it is not possible at CDC to distinguish between cases that have not been genotyped and cases that have been genotyped but not linked.
National TB genotyping coverage is defined as the proportion of TB cases with a culture yielding M. tuberculosis (referred to as “culture-positive cases”) that are linked to a genotype result in the national Web-based genotyping database. In 2010, national genotyping coverage was 88%. However, approximately 20% of TB cases in the United States are not culture positive and, therefore, do not have an isolate available for genotyping.17 A case could be missing a genotype result for three general reasons: it did not have an M. tuberculosis isolate, it had an isolate that was not genotyped, or the genotyping result was not linked to NTSS data in the national Web-based genotyping database. Our aim was to characterize cases that did not have a genotype result for any of these reasons to identify populations in which outbreaks might be missed by genotype-based outbreak detection methods, and to identify opportunities to increase genotyping.
METHODS
Surveillance records for TB cases in the 50 U.S. states and the District of Columbia reported to NTSS during January 1, 2009–December 31, 2010, were linked to genotyping results in the national Web-based genotyping database using the state-assigned case identifier. Three states had technical problems with linking during 2010 and were excluded from the analysis. A case was considered to be genotyped if it was linked to a genotype record, even if the genotype result reported for that isolate was invalid (i.e., incomplete genotype result). Isolates with invalid genotype results were included as genotyped because they represented cases with an isolate that was submitted for genotyping and linked to surveillance data.
We compared demographic and clinical characteristics of genotyped cases with non-genotyped cases. We repeated the same analysis restricted to culture-positive cases, because the isolation of M. tuberculosis, which is required for genotyping, is associated with certain case characteristics (e.g., young age). For this analysis, we also compared the presence or absence of DST results. Using SAS® version 9.3,18 we conducted bivariate analyses using chi-squared statistics and risk ratios with 99% confidence intervals (CIs).
RESULTS
During 2009–2010, a total of 22,719 cases of TB were reported in the United States, of which 17,303 (76.2%) were culture positive. After excluding 243 cases from three states with technical problems linking genotyping results, 22,476 cases were used for the final analysis, including 17,125 culture-positive cases (Table). Among all TB cases, 14,922 (66%) had a linked genotype; 14,839 (87%) culture-positive cases had a linked genotype. Eighty-three cases were not reported as culture positive but had a linked genotype result, and 17 cases were linked to a genotype record with an invalid genotype result. These cases were included in the analysis.
Table.
Characteristics of genotyped cases among all reported TB cases and culture-positive TB cases, United States, 2009–2010a
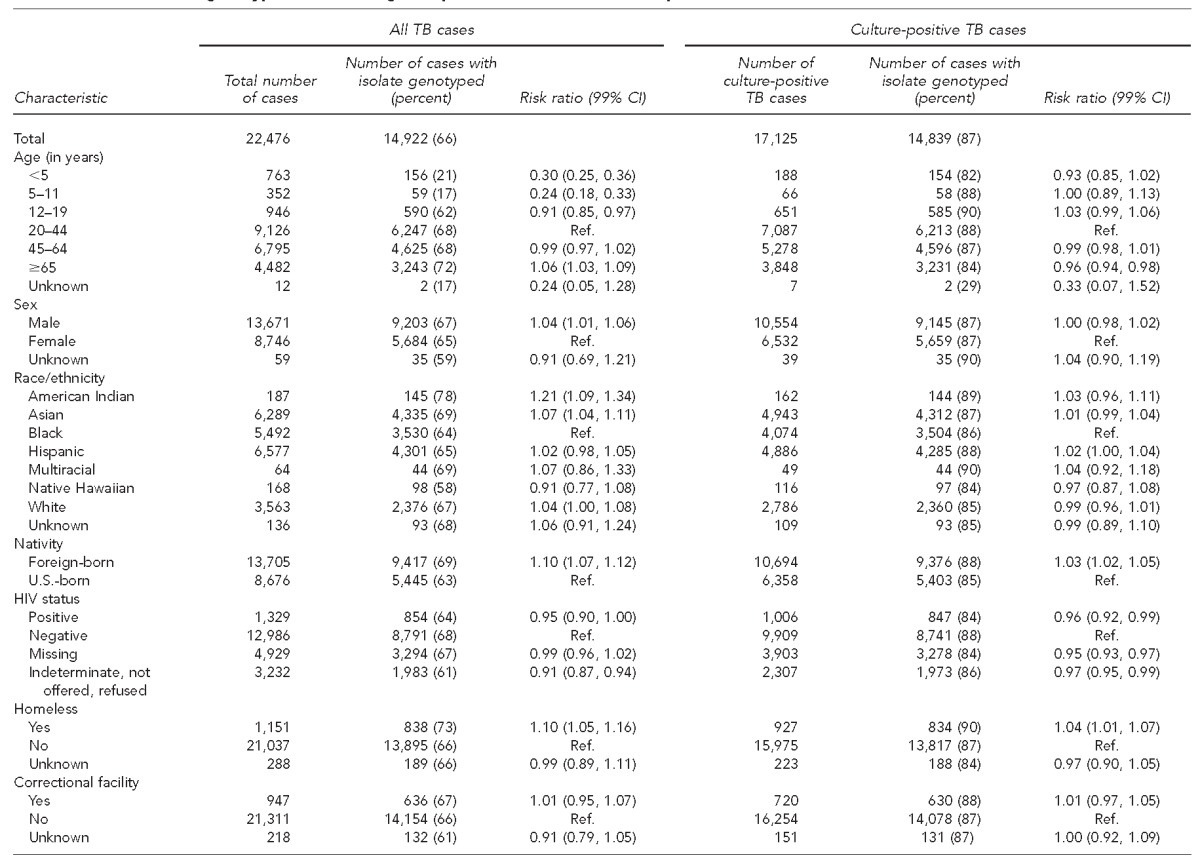
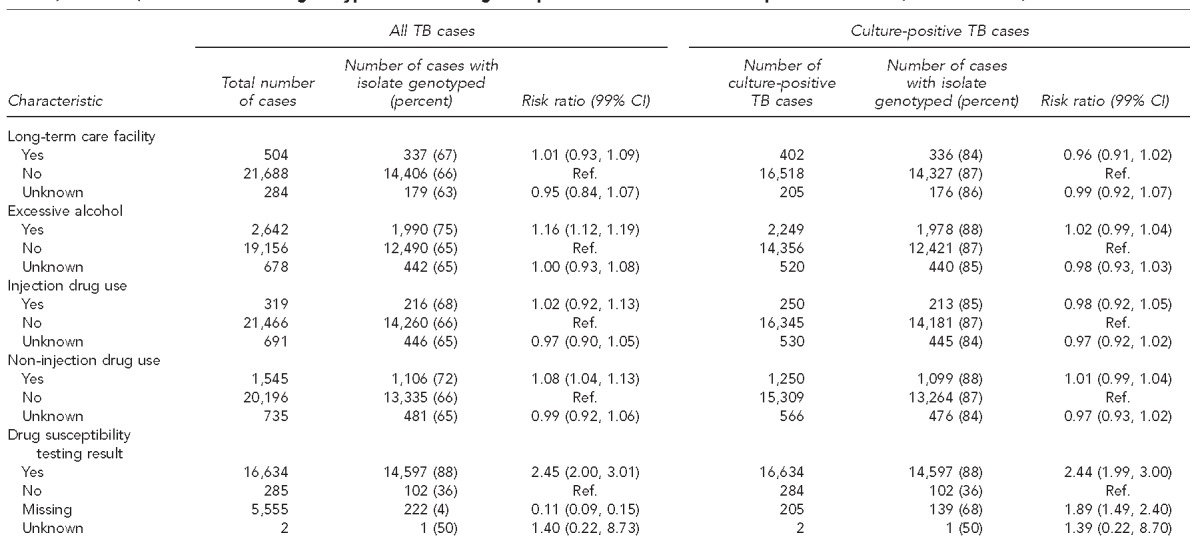
Data source: Centers for Disease Control and Prevention (US). TB genotyping information management system, January 1, 2009–December 31, 2010 [cited 2015 Jul 2]. Available from: URL: http://www.cdc.gov/tb/publications/factsheets/statistics/gims.htm
TB = tuberculosis
CI = confidence interval
Ref. = reference group
HIV = human immunodeficiency virus
Genotyping among all reported cases
Among all TB cases, regardless of culture results, the patient characteristics associated with a lower likelihood of having a linked genotype result were age <5 years, 5–11 years, or 12–19 years; female sex; being U.S.-born; having unknown human immunodeficiency virus status; and having no reported history of homelessness, excess alcohol use, or non-injecting drug use in the year before diagnosis (Table).
Genotyping among culture-positive cases
Among culture-positive cases, characteristics associated with a lower likelihood of having a linked genotype result were age ≥65 years; being U.S.-born; having positive, missing, or unknown human immunodeficiency virus status; and having no reported history of homelessness in the year before diagnosis (Table). Cases with no DST result reported were less likely to have a linked genotype result than cases with a DST result. DST results were available for 16,634 cases; among these cases, 14,597 (88%) had a linked genotype record (Table) and 2,037 (12%) did not.
DISCUSSION
U.S. TB cases with a linked genotype result are largely representative of all reported TB cases, with the likelihood of genotyping rarely differing by more than 10% between groups. However, outbreak detection methods that rely solely on genotyping surveillance data19 might underestimate transmission-related cases among groups underrepresented by genotyping data, such as children and those who are U.S.-born.
We observed several characteristics associated with having a linked genotype result among all TB cases. However, some of these differences can be explained by the likelihood that a case is culture positive. For example, TB disease in children is less likely to be culture positive than in adults and is, therefore, less likely to have an isolate available for genotyping.20 In our analysis among all TB cases, patients <5 years of age and patients 5−12 years and 12–19 years of age were less likely to have a linked genotype result than patients aged 20−44 years. However, in our analysis limited to culture-positive cases, no association between young age and risk of genotyping was observed, indicating that the overall association was driven by the lower ratio of culture-positive cases for younger age groups. Although some differences persisted after looking only at culture-positive cases, these differences were small.
While these data indicate that the genotyping data in the United States are largely representative, it is important to consider that some groups are underrepresented in genotyping data. The underrepresentation of some groups may affect routine TB-control activities, such as detecting clusters that might represent outbreaks, defining the scope of outbreaks, and monitoring outbreaks over time. However, it is reassuring that cases in patients experiencing homelessness and alcohol abuse are more likely to have a linked genotype result; in two-thirds of TB outbreaks recently investigated by CDC, more than 50% of cases exhibited these characteristics.21 Accurate identification of transmission and outbreaks in these populations is critical to prevent future cases.
A culture-positive case might not have a linked genotype result for various reasons. An isolate might not be sent for genotyping, either because of logistical and financial issues or because laboratories might be unaware of the need to refer isolates for genotyping. Although some states have statutes requiring all TB isolates to be submitted to the state public health laboratory, other states have no such regulations (Personal communication, Melisa Thombley, JD, MPH, CDC Division of Tuberculosis Elimination, April 2012). Additionally, without linking the genotype and surveillance records in the national database, a case may appear to be missing a genotype result even when one exists.
We used the result of the DST, which is often conducted at a public health laboratory, as a proxy for cases that had a viable isolate and presumably could have been genotyped. A case with a DST result but no genotype result might represent a missed opportunity to increase overall genotyping coverage. We identified a number of cases that met this criterion.
Limitations
Our study had several limitations. We might have misclassified genotyped cases as non-genotyped if they were not linked to the corresponding result. Additionally, NTSS data are limited by the quality of the data provided by reporting jurisdictions.22 For example, 88 cases that were not reported as culture positive were linked to a genotype result, suggesting either inaccuracies in reported data or incorrect linking.
CONCLUSION
Increasing genotyping coverage will further increase the representativeness of genotyping data. One potential approach to improve coverage is to target the 13% of cases with isolates of M. tuberculosis that have DST results reported but do not have linked genotype results. Continued work toward achieving comprehensive genotyping coverage in the United States is important for ensuring that outbreaks and recent transmission are not missed by routine TB-control methods. State-based versions of this analysis might also have a role in informing local TB-control efforts and improving coverage.
Footnotes
Emma B. Shak was supported by the CDC Experience, a one-year fellowship in applied epidemiology at the Centers for Disease Control and Prevention (CDC) made possible by a public/private partnership supported by a grant to the CDC Foundation from External Medical Affairs, Pfizer Inc.
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position or views of CDC.
REFERENCES
- 1.Notice to readers: new CDC program for rapid genotyping of Mycobacterium tuberculosis isolates. MMWR Morb Mortal Wkly Rep. 2005;54(02):47. [Google Scholar]
- 2.Murray M, Alland D. Methodological problems in the molecular epidemiology of tuberculosis. Am J Epidemiol. 2002;155:565–71. doi: 10.1093/aje/155.6.565. [DOI] [PubMed] [Google Scholar]
- 3.Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349:1149–56. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 4.Golub JE, Cronin WA, Obasanjo OO, Coggin W, Moore K, Pope DS, et al. Transmission of Mycobacterium tuberculosis through casual contact with an infectious case. Arch Intern Med. 2001;161:2254–8. doi: 10.1001/archinte.161.18.2254. [DOI] [PubMed] [Google Scholar]
- 5.Pevzner ES, Robison S, Donovan J, Allis D, Spitters C, Friedman R, et al. Tuberculosis transmission and use of methamphetamines in Snohomish County, WA, 1991–2006. Am J Public Health. 2010;100:2481–6. doi: 10.2105/AJPH.2009.162388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malakmadze N, Gonzalez IM, Oemig T, Isiadinso I, Rembert D, McCauley MM, et al. Unsuspected recent transmission of tuberculosis among high-risk groups: implications of universal tuberculosis genotyping in its detection. Clin Infect Dis. 2005;40:366–73. doi: 10.1086/427112. [DOI] [PubMed] [Google Scholar]
- 7.Samuel V, Benjamin C, Renwick O, Hilliard A, Arnwine S, Spike D, et al. Notes from the field: tuberculosis cluster associated with homelessness—Duval County, Florida, 2004–2012. MMWR Morb Mortal Wkly Rep. 2012;61(28):539–40. [PubMed] [Google Scholar]
- 8.Miramontes R, Winston CA, Haddad MB, Moonan PK. Use of tuberculosis genotyping for postoutbreak monitoring. J Public Health Manag Pract. 2012;18:375–8. doi: 10.1097/PHH.0b013e31823680f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Rie A, Warren R, Richardson M, Victor TC, Gie RP, Enarson DA, et al. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N Engl J Med. 1999;341:1174–9. doi: 10.1056/NEJM199910143411602. [DOI] [PubMed] [Google Scholar]
- 10.Burman WJ, Reves RR. Review of false-positive cultures for Mycobacterium tuberculosis and recommendations for avoiding unnecessary treatment. Clin Infect Dis. 2000;31:1390–5. doi: 10.1086/317504. [DOI] [PubMed] [Google Scholar]
- 11.Fitzpatrick LK, Hardacker JA, Heirendt W, Agerton T, Streicher A, Melnyk H, et al. A preventable outbreak of tuberculosis investigated through an intricate social network. Clin Infect Dis. 2001;33:1801–6. doi: 10.1086/323671. [DOI] [PubMed] [Google Scholar]
- 12.Jasmer RM, Hahn JA, Small PM, Daley CL, Behr MA, Moss AR, et al. A molecular epidemiologic analysis of tuberculosis trends in San Francisco, 1991–1997. Ann Intern Med. 1999;130:971–8. doi: 10.7326/0003-4819-130-12-199906150-00004. [DOI] [PubMed] [Google Scholar]
- 13.Borgdorff MW, van den Hof S, Kalisvaart N, Kremer K, van Soolingen D. Influence of sampling on clustering and associations with risk factors in the molecular epidemiology of tuberculosis. Am J Epidemiol. 2011;174:243–51. doi: 10.1093/aje/kwr061. [DOI] [PubMed] [Google Scholar]
- 14.Murray M. Sampling bias in the molecular epidemiology of tuberculosis. Emerg Infect Dis. 2002;8:363–9. doi: 10.3201/eid0804.000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glynn JR, Vynnycky E, Fine PE. Influence of sampling on estimates of clustering and recent transmission of Mycobacterium tuberculosis derived from DNA fingerprinting techniques. Am J Epidemiol. 1999;149:366–71. doi: 10.1093/oxfordjournals.aje.a009822. [DOI] [PubMed] [Google Scholar]
- 16.pCenters for Disease Control and Prevention (US) CDC tuberculosis surveillance data training: reports of verified case of tuberculosis (RVCT) instruction manual. 2009 [cited 2015 Jun 19] Available from: URL: http://www.cdc.gov/tb/programs/rvct/instructionmanual.pdf.
- 17.Grant J, Kammerer S, Baker B. Tuberculosis genotyping—United States, 2004–2010. MMWR Morb Mortal Wkly Rep. 2012;61(36):723–5. [PubMed] [Google Scholar]
- 18.SAS Institute, Inc. SAS®: Version 9.3 for Windows. Cary (NC): SAS Institute, Inc.; 2012. [Google Scholar]
- 19.Kammerer JS, Shang N, Althomsons SP, Haddad MB, Grant J, Navin TR. Using statistical methods and genotyping to detect tuberculosis outbreaks. Int J Health Geogr. 2013;12:15. doi: 10.1186/1476-072X-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuevas LE, Browning R, Bossuyt P, Casenghi M, Cotton MF, Cruz AT, et al. Evaluation of tuberculosis diagnostics in children: 2. Methodological issues for conducting and reporting research evaluations of tuberculosis diagnostics for intrathoracic tuberculosis in children. Consensus from an expert panel. J Infect Dis. 2012;205(Suppl 2):S209–15. doi: 10.1093/infdis/jir879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitruka K, Oeltmann JE, Ijaz K, Haddad MB. Tuberculosis outbreak investigations in the United States, 2002–2008. Emerg Infect Dis. 2011;17:425–31. doi: 10.3201/eid1703.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes S, Sodt D, Young K, Jereb J, Pratt R, Navin T, et al. Monitoring tuberculosis programs—National Tuberculosis Indicator Project, United States, 2002–2008. MMWR Morb Mortal Wkly Rep. 2010;59(10):295–8. [PubMed] [Google Scholar]


