Abstract
Background.
Mitochondrial dysfunction is a prominent hallmark of many sensory neuropathies. The purpose of this study was to assess the influence of mitochondrial DNA sequence variation on peripheral nerve function in the population-based Health, Aging, and Body Composition Study.
Methods.
We investigated the role of common mitochondrial DNA variation (n = 1,580) and complete mitochondrial DNA sequences (n = 138) on peroneal motor nerve conduction velocity and amplitude, average vibration detection threshold, and monofilament sensitivity.
Results.
Nominal associations among common mitochondrial DNA variants and haplogroups were identified but were not statistically significant after adjustment for multiple comparisons. Sequence-based approaches were used to identify aggregate variant associations across the 16S rRNA (weighted-sum, p = 2E-05 and variable threshold, p = 9E-06) for nerve conduction velocity. Several of these rare 16S variants occurred at or near sites with earlier disease associations and are also in close proximity to the peptidyl transferase center, which is the catalytic center of the 16S rRNA
Conclusions.
These results suggest that sequence variation related to mitochondrial protein synthesis/assembly is associated with peripheral nerve function and may provide insight into targets for intervention or new clinical strategies to preserve nerve function in late life.
Key Words: Sensory, Genetics, Epidemiology, Functional Performance.
Age-related peripheral neuropathy is one of the most common sensorimotor deficits in the elderly and has severe negative influences on the quality of life, physical function, and psychosocial well-being of older adults (1–6). The incidence and prevalence of poor peripheral nerve function is higher in older than younger adults, even those without diabetes (2,4,5). In the United States for 1999–2000, 28% of adults aged 70–79 years and 35% of adults aged >80 years had neuropathy symptoms and loss of touch sensation, with a higher prevalence in diabetic adults (4). The factors that determine the development of peripheral neuropathy vary between persons (7,8). Sex and race appear to play a role in the development of peripheral neuropathy in older people with men exhibiting poorer sensory and motor nerve function than women and European-Ancestry adults having worse vibration threshold sensitivity than those with African-Ancestry (1,3).
Mitochondrial dysfunction is a common characteristic of many neuropathies (9,10). Several mitochondrial diseases feature a spectrum of abnormalities which often include significant neuropathological impairments and sensory neuropathies (11) (eg, Leber’s hereditary optic neuropathy, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes [MELAS], and neuropathy, ataxia, and retinitis pigmentosa). Mitochondrial oxidative phosphorylation supplies the vast majority (90%) of the energy needs of the human body and is dependent upon the coordinated expression and interaction of genes encoded in both the nuclear and mitochondrial genomes. The prevalence of neuronal diseases associated with mutations in mitochondrial DNA (mtDNA)-encoded genes indicates the important functional relationship between mitochondria and neurons (12,13). Human mtDNA is a maternally inherited 16,569 base pair loop containing genes critical to oxidative phosphorylation (14), and bioenergetic defects resulting from acquired and inherited mtDNA mutations (15–17) may be critical for both age-related neuropathy and associated neuropathological changes (9,10).
The influence of inherited and acquired mtDNA mutations on neuropathological impairments has been established for many mitochondrial and age-related diseases. While mitochondrial dysfunction resulting from inherited and acquired mtDNA mutations is a hallmark of several neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (18), the role of mtDNA sequence variation in age-related peripheral neuropathy has not been assessed. For this study, we investigated the role of common and rare mtDNA sequence variations on peripheral nerve function in the Health ABC Study. Because human mtDNA has a mutation rate, that is 10–20 times higher than that of nuclear DNA and up to one-third of sequence variants found in the general population may be functionally important (19), it is likely that most of the mtDNA variation that impacts function is rare in frequency and only detectable by direct sequencing. Indeed, we have shown earlier that aggregated common and rare variants and the accumulation of singleton variants may be more important contributors to dementia and cognitive decline (20,21), cancer risk (22), and human energy expenditure (23,24) than common polymorphisms. Uncovering mtDNA variants impacting peripheral nerve function may lead to the development of new clinical strategies for improving mitochondrial function as well as genetic tests for identifying individuals who may benefit from mitochondria-directed therapies.
Methods
Population
Health ABC is a prospective cohort study of 3,075 community-dwelling black and white men and women living in Memphis, Tennessee, or Pittsburgh, Pennsylvania, and aged 70–79 years at recruitment in 1996–1997. Participants were recruited from a random sample of white and black Medicare-eligible people within designated zip code areas. Participants had to report no difficulty with activities of daily living, walking a quarter of a mile, or climbing 10 steps without resting, and be free of life-threatening cancer diagnoses. The sample was 51% women and 41% of participants were black. Participants self-designated race/ethnicity classified as Asian/Pacific Islander, black/African American, white/Caucasian, Latino/Hispanic, and other. All eligible participants signed a written informed consent, approved by the institutional review boards at the clinical sites. This study was approved by the institutional review boards of the clinical sites and the coordinating center (University of California, San Francisco).
Peripheral Nerve Function
At the 2000–2001 clinical follow-up visit, 2,208 Health ABC participants underwent peripheral nerve function evaluations (1). Peripheral nerve function measurements included: (i) peroneal motor nerve conduction velocity (m/s) and amplitude (mV) from the popliteal fossa to ankle; (ii) average vibration detection threshold (μ) on the bottom of the great toe; and (iii) monofilament sensitivity using 1.4-g and 10-g monofilaments. Clinic examiners with training and certification in the nerve function measures performed the tests on the right leg unless it was contraindicated because of knee replacement, amputation, ulcer, trauma, or surgery. Nerve conduction (NC) measures have moderate to high intraobserver reproducibility in older adults and are not affected by diabetes, race, or gender (25).
Genotyping
Genomic DNA was extracted from buffy coat collected using PUREGENE DNA Purification Kit during the baseline exam. Genotyping was performed by the Center for Inherited Disease Research using the Illumina Human1M-Duo BeadChip system. This platform includes 138 mtDNA SNPs including the majority of haplogroup-defining variants. Samples were excluded from the data set for the reasons of sample failure, genotypic sex mismatch, and first-degree relative of an included individual based on genotype data as described earlier (24). Genotyping of 138 mtDNA SNPs (including 137 variant sites) was successful for 1,631 unrelated individuals of European genetic ancestry. The major European haplogroups (Table 1) were defined using PhyloTree (26).
Table 1.
Characteristics of Genotyped and Sequenced Health ABC Participants From the 2000–2001 Clinical Follow-Up Visit
| Genotyped | Sequenced | |
|---|---|---|
| N | 1,580 | 135 |
| Age, mean (SD) | 76.7 (2.8) | 76.5 (2.9) |
| Sex, n (%) | ||
| Male | 819 (52) | 63 (47) |
| Female | 761 (48) | 72 (53) |
| Haplogroup, n* (%) | ||
| H | 589 (45) | 60 (47) |
| U | 183 (14) | 11 (9) |
| K | 145 (11) | 16 (12) |
| T | 140 (11) | 18 (14) |
| J | 103 (8) | 13 (10) |
| V | 51 (4) | 2 (2) |
| I | 35 (3) | 5 (4) |
| W | 27 (2) | 1 (1) |
| X | 27 (2) | 3 (2) |
| Nerve conduction velocity (m/s), mean (SD) | 43.5 (5.2) | 42.5 (5.8) |
| Nerve conduction amplitude (mV), mean (SD) | 3.3 (1.85) | 3.0 (2.0) |
| Vibration sensitivity (µ), mean (SD) | 52.9 (35.2) | 49.9 (35.5) |
| Monofilament test, n (%) | ||
| Good function | 760 (55) | 75 (60%) |
| Impaired function | 494 (36) | 38 (30%) |
| Poor function | 124 (9) | 12 (12%) |
Notes: SD = standard deviation.
*Numbers do not add up to total due to missing information for haplogroups.
MtDNA Sequencing
MtDNA was extracted from platelets and sequenced with the Affymetrix Mitochondrial Resequencing Array 2.0 (MitoChip, Affymetrix, Santa Clara, CA) in 138 Health ABC participants of European ancestry as described earlier (23). Ten samples were repeated for concordance testing. For samples passing initial filtering, ResqMi 1.2 (27) was used for reanalysis of bases originally called as “N” by GSEQ. Data was extracted from gene regions as defined by NCBI annotations for the revised Cambridge Reference Sequence (NC_012920.1).
Statistical Analyses: Common mtDNA Haplogroups and Individual Variants
We analyzed differences in nerve conduction velocity, nerve conduction amplitude, and vibration sensitivity for the European haplogroups and 137 mtDNA variants.
Each haplogroup was compared to individual and collapsed haplogroups using the pairwise differences (PDIFF) and PDIFF = analysis of means (ANOM) options of the generalized linear model in SAS version 9.2 (SAS Institute Inc., Cary, North Carolina [NC]). All analyses were adjusted for age, sex, and clinic site. For the 137 mtDNA variants, comparisons between alternate mtDNA alleles were made using the PDIFF option of the generalized linear model and all analyses were adjusted for age, sex, clinic site, and six eigenvectors of mitochondrial genetic ancestry derived from principal component analysis (28).
Statistical Analyses: MtDNA Sequences
The joint effects of all mitochondrial variants on nerve conduction velocity, nerve conduction amplitude, and vibration sensitivity were evaluated using several rare variant burden tests using variable threshold test (29) in R. All analyses were adjusted for age, sex, and study site using residuals from linear regression and then normalized to Z scores prior to conducting analyses. We applied these approaches to continuous values for each nerve conduction test and computed statistical significance for each test using 10,000 independent simulations. Variant aggregations were tested across the following regions: (i) the four individual oxidative phosphorylation complexes; (ii) all rRNAs combined; (iii) all tRNAs combined; and (iv) the HV 2–3 region. The 16S rRNA secondary structure was visualized using VARNA (30).
Results
Among 1,631 genotyped Health ABC participants of European ancestry, 1,580 (97%) attended the 2000–2001 clinical follow-up visit. Of these, 1,080 (68%) had nerve conduction amplitude, 1,056 of these (67%) had nerve conduction velocity obtained as well, 1,355 (86%) completed vibration sensitivity (μ) testing, and 1,378 (87%) completed monofilament sensitivity testing (Table 1). A total of 135 Health ABC participants yielded sequence data of sufficient quality for analysis including: 97 (72%) who completed nerve conduction amplitude testing, 95 (70%) who completed nerve conduction velocity, and 125 (93%) who completed both vibration sensitivity (μ) testing and monofilament sensitivity testing (Table 1). Of these, 63 were men and 72 were women, with mean (SD) age of 73.4 (2.9) years and are representative of the larger cohort with regard to haplogroup frequencies and peripheral nerve function. Sequencing of 16,544 mtDNA bases (positions 12–16,555) from 135 participants yielded a cumulative total of 449 variants including: 56 common (minor allele frequency [MAF] ≥ 5%), 160 low-frequency variants (MAF 1%–5%), and 233 singletons. The 10 duplicate samples had >98% sequence concordance (the majority of discordant calls resulted from positions successfully called in one but called as “N” in another). The within-chip error rate was 0.0028%, which is comparable to the earlier published rates of 0.0025% and 0.0021% (31).
Common mtDNA Haplogroups and Individual Variants
Haplogroup differences in nerve conduction velocity, nerve conduction amplitude, and vibration sensitivity are reported in Table 2. Haplogroup I participants exhibited poor conduction amplitude (p < .05), whereas haplogroup W participants exhibited higher conduction amplitude (p < .05) and vibration sensitivity (p < .05) when compared with other European haplogroups. However, these results were not statistically significant after adjustment for multiple comparisons (36 tests among nine haplogroups, critical α = .0014). Additional analyses comparing each haplogroup to the mean of the remaining haplogroups did not identify statistically significant differences. None of the 137 individual mtDNA variants available on the Illumina Human1M-Duo BeadChip system were associated with nerve conduction velocity, nerve conduction amplitude, or vibration sensitivity after adjustment for multiple comparisons (137 variants, critical α = .0004).
Table 2.
Nerve Conduction Velocity, Amplitude, and Vibration Average Among Haplogroup N Subgroups (n = 1,549)
| Haplogroup | Nerve Conduction Velocity (m/s) | Nerve Conduction Amplitude (mV) | Vibration Sensitivity (µ) |
|---|---|---|---|
| Mean (SE)* | Mean (SE)* | Mean (SE)* | |
| H | 43.1 (0.23) | 3.1 (0.09) | 53.7 (1.42) |
| U | 43.5 (0.42) | 3.4 (0.15) | 50.4 (2.56) |
| K | 43.8 (0.46) | 3.2 (0.17) | 52.2 (2.85) |
| T | 43.2 (0.48) | 3 (0.18) | 54.2 (2.92) |
| J | 43.3 (0.57) | 3.2 (0.21) | 56 (3.46) |
| V | 43.4 (0.77) | 3.1 (0.29) | 51.3 (4.89) |
| I | 44.3 (0.98) | 2.4 (0.36)† | 57.1 (5.79) |
| W | 43.6 (1.05) | 4 (0.40)‡ | 68.3 (6.72)§ |
| X | 43.6 (1.13) | 3.4 (0.43) | 49.4 (6.59) |
Notes: *Least square means and standard errors (SE) adjusted for age, sex, and clinic site.
†Haplogroup I vs. W, H, J, T, U, and K (p < .05).
‡Haplogroup W vs. I, H, and T (p < .05).
§Haplogroup W vs. X, H, V, U, and K (p < .05).
MtDNA Sequences
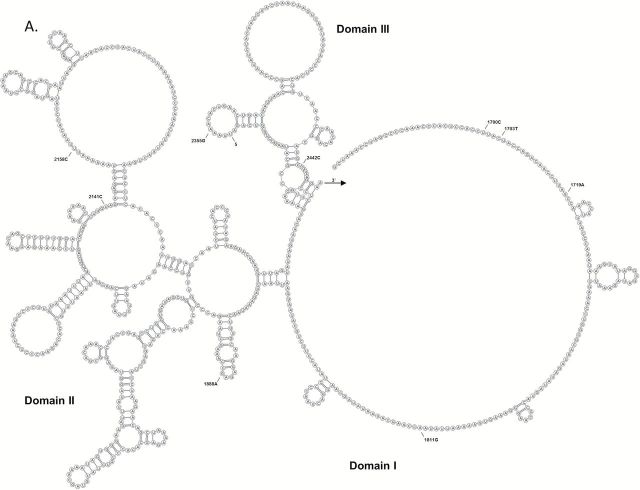
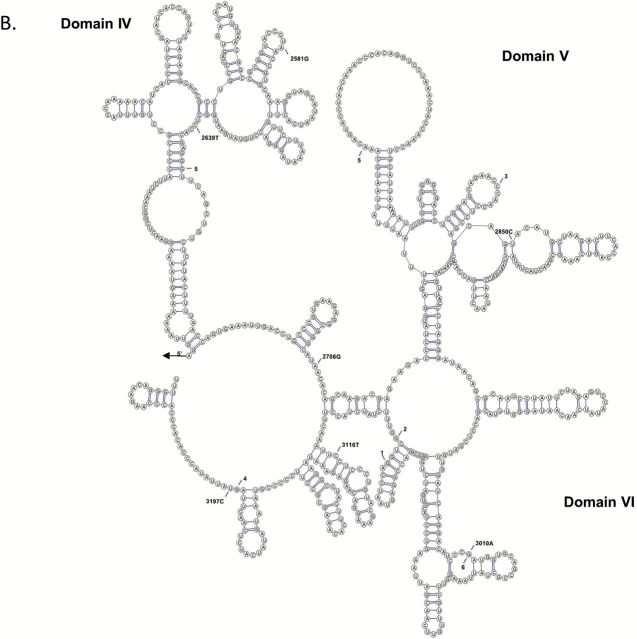
The joint effects of all mitochondrial variants within each mtDNA region on nerve function were evaluated using several rare variant burden tests (29). Statistically significant pooled variant associations across the 16S rRNA were observed for nerve conduction velocity using the weighted-sum and variable threshold weighted-sum (WE, p = 2E-05) and variable threshold (p = 9E-06) methods. Statistical significance accounted for multiple testing (seven mtDNA regions, critical α = .007) and was based on 10,000 independent simulations. Of the 95 participants with both full mtDNA sequences nerve conduction velocity measurements, 85 carried at least one 16S rRNA variant. Among these, 66 participants carried a single 16S rRNA variant and 19 participants carried two-to-three variants. Of the fourteen 16S rRNA variants detected by sequencing (Table 3), three individual rare variants were associated with significantly higher nerve conduction velocity levels after adjustment for multiple comparisons (14 tests, critical α = .004).Two participants carrying the m.2850C allele had a significantly higher (p < .001) nerve conduction velocity (48.9 m/s) when compared with the remaining participants carrying the m.2850T allele (42.4 m/s). The m.1703C>T and m.2639C>T variants are in complete LD and participants carrying the m.1703T/m.2639T alleles had a significantly higher (p = .004) nerve conduction velocity (53.9m/s) when compared with the remaining participants carrying the m.1703C/m.2639C alleles (42.3 m/s). We also compared nerve conduction velocities among the participants carrying two or three 16S rRNA variants. Participants carrying both m.2706A and m.3010A alleles had significantly lower nerve conduction velocity (42.5 m/s) when compared with participants carrying both m.1888A and m.2850C alleles (48.9 m/s, p = .03) or participants carrying the m.1703T, m.1811A, m.2639T alleles (53.9 m/s, p = .01). All 16S rRNA variants detected by sequencing were identified on the predicted secondary structure (Figure 1). Several of these rare variants occur at or near sites with earlier disease associations (32) and are also in close proximity to the peptidyl transferase center, which is the catalytic center of the 16S rRNA (33).
Table 3.
16S rRNA Sequence Variant Effects on Nerve Conduction Velocity (m/s) (n = 135)
| Common Allele | n | Mean (SE) | Variant Allele | n | Mean (SE) | t Test p Value |
|---|---|---|---|---|---|---|
| m.1703C | 92 | 42.3 (0.88) | m.1703T | 2 | 53.9 (6.9) | .004 |
| m.1719G | 86 | 42.1 (0.90) | m.1719A | 8 | 46.9 (3.6) | .023 |
| m.1811A | 78 | 42.4 (0.97) | m.1811G | 15 | 43.0 (2.2) | .726 |
| m.1888G | 83 | 42.3 (0.95) | m.1888A | 11 | 44.0 (2.6) | .370 |
| m.2141T | 93 | 42.5 (0.90) | m.2141C | 1 | 43.1 | — |
| m.2158T | 92 | 42.5 (0.90) | m.2158C | 2 | 45.5 (5.5) | .469 |
| m.2355A | 91 | 42.3 (0.90) | m.2355G | 3 | 48.1 (5) | .087 |
| m.2442T | 91 | 42.3 (0.90) | m.2442C | 3 | 48.1 (5) | .087 |
| m.2639C | 92 | 42.3 (0.88) | m.2639T | 2 | 53.9 (6.9) | .004 |
| m.2706G | 57 | 43.6 (1.2) | m.2706A | 37 | 40.9 (1.3) | .02 |
| m.2850T | 92 | 42.4 (0.90) | m.2850C | 2 | 48.9 (4.9) | <.001 |
| m.3010G | 76 | 42.4 (1.0) | m.3010A | 18 | 42.9 (1.8) | .77 |
| m.3116C | 93 | 42.4 (0.89) | m.3116T | 1 | 51.2 | — |
| m.3197T | 93 | 42.5 (0.90) | m.3197C | 1 | 44.6 | — |
Note: SE = standard error.
Figure 1.
Predicted secondary structure of the mitochondrial 16S rRNA containing domains I–III (A) and IV–VI (B). 16S rRNA variants detected by sequencing are indicated by nucleotide position. Previously identified disease associations are indicated: (1) Myopathy, encephalopathy, lactic acidosis, and stroke-like episodes syndrome (MELAS); (2) Myopathy; (3) Rett syndrome; (4) Alzheimer’s and Parkinson’s diseases; (5) Left ventricular noncompaction; 6) Migraine headache and cyclic vomiting syndrome.
Discussion
In older adults, approximately half of the incident and prevalent clinical neuropathy (34) is attributable to diabetes with the remaining ~50% being of uncertain etiology. In this study, we genotyped and sequenced the entire mtDNA in a large, population-based, longitudinal study of elderly participants to examine the role of common mtDNA haplogroups and genetic variants, rare variants, and aggregate mtDNA sequence variation in peripheral nerve function. Research to identify genetic factors that contribute to complex phenotypes must account for the many ways that genetic variations operate. Common genetic variants play a much smaller role in mediating phenotypic expression and disease risk than initially hypothesized (35) and identification of causative variants requires comprehensive resequencing of genomic loci in multiple subjects (36). Because collections of variants within genes or genomic regions do not work in isolation and are likely to influence phenotypic expression in important ways (37), we considered how multiple sequence variants impact peripheral nerve function. Analytic approaches testing the combined effect of multiple variants have been used to resolve genetic associations for several complex traits (38) including mtDNA variation (23,39). We evaluated several variant pooling approaches and identified highly significant associations between aggregate 16S rRNA sequence variation and nerve conduction velocity. In addition to three highly significant individual variant associations with elevated nerve conduction velocity, we identified novel associations among participants carrying two or three 16S rRNA variants. The effects of the two or three variant haplotypes were greater than those observed for the individual variants contributing to these haplotypes, suggesting an epistatic effect of mtDNA on nerve conduction velocity. Several of the 16S rRNA variants including m.2706 A>G, m.3116 C>T, m.3010 G>A, and m.3197 T>C occur at or near sites associated with earlier disease associations (40–50) including MELAS, Alzheimer’s and Parkinson’s diseases, and migraine headache and cyclic vomiting syndrome which is often accompanied by cognitive delay and myopathy (51). These variants are in close proximity to the peptidyl transferase center, which is the catalytic center of the 16S rRNA (33). The peptidyl transferase center is the main target site for many antibiotics which bind to functional centers and inhibit protein synthesis possibly by hindering peptide chain development and promoting dissociation of peptidyl RNA from the ribosome (10027979). Although it is unknown how disease-associated variants located within this catalytic center are impacting function of the 16S rRNA, it is possible that they mimic drug-induced mechanisms of protein inhibition.
In this study we sequenced the mtDNA from platelets, which are classified as terminally differentiated, anucleate blood cells containing fully functional mitochondria (52). In addition to being easily accessible, platelets have numerous similarities with neurons and have been frequently utilized as neuronal models (53–58). Platelets and neurons both contain mitochondria with the enzyme monoamine oxidase (59); receptors and transport mechanisms for the uptake and storage of serotonin (60); as well as amyloid precursor protein (APP) and the secretases necessary to produce all APP metabolites including amyloid β (Aβ) (61). In fact, platelet APP is synthesized by the megakaryocyte platelet precursor in the bone marrow (62) and may account for 90% of the circulating APP (63,64). Platelets show concentrations of APP isoforms equivalent to those found in brain (65) and it has been suggested that platelet APP contributes to the accumulation of Aβ in the brain and its vasculature through the blood–brain barrier (66,67).
This study had a number of strengths: a well-characterized population-based longitudinal cohort with several state of the art measures of nerve function, a large sample size for assessing peripheral nerve function among mtDNA haplogroups and genotyped variants, complete mtDNA sequencing allowing for an unbiased assessment of mitochondrial genomic variation, an analytic approach including both individual and pooled sequence variants, and in silico prediction and structural modeling that enabled detailed interpretation of sequence-based findings. Small sample size for sequence-based analyses and limited power to detect individual variant effects of rare variants are acknowledged. These results are based on a single cohort and further studies are needed to confirm these findings.
Mitochondrial dysfunction is a common characteristic of many neuropathies (9,10). Neurons require adenosine triphosphate (ATP) for axonal transport and maintenance of ionic gradients to generate action potentials and synaptic activity (15). Axons are the largest consumer of mitochondria-derived ATP in peripheral nerve fibers, and the vast majority of mitochondria (~84% in humans) are located within unmyelinated and myelinated axons (9). In most eukaryotic cells, including neurons, mitochondria are essential for providing energy and managing oxidative stress. Mitochondria are primarily assembled in the neuronal cell body and subsequently transported along the long axons (9). The transport and localization of mitochondria to particular axonal segments is critical for proper neuronal function (68). MtDNA deletion mutations occur more frequently in distal nerve segments and have been associated with impaired mitochondrial function and loss of respiratory chain function (9) implicated in the demyelination and axonal degeneration (10) and found in most peripheral neuropathies (9). MtDNA deletions resulting in defective oxidative phosphorylation have also been observed in axonal and hypomyelinating sensory-motor neuropathy (15), polyneuropathy (17), and sensorineural hearing loss (17). Our results help to uncover mtDNA sequence variation that impacts age-related peripheral nerve function, suggesting specific mitochondrial functions that may affect nerve conduction velocity. In particular, the 16S rRNA region may affect mitochondrial assembly by impacting the rate or efficiency of mitochondrial biogenesis. Mitochondrial biogenesis (the increase in mitochondrial numbers and/or mass) involves a complex interaction between mitochondrial and nuclear encoded genes. Mitochondrial DNA encodes a small number of proteins consisting mostly of hydrophobic subunits of the respiratory chain localized in the inner mitochondrial membrane. Nuclear encoded proteins include structural proteins, enzymes, enzyme subunits, chaperones, and components responsible for import, replication, transcription, and translation that are imported into mitochondria. The rate of turnover may decline with age (69) which may allow for defective mitochondria to accumulate, especially in older, postmitotic cells. Although the underlying mechanism of the accumulation of defective mitochondria is still unclear, the individual and collective 16S rRNA variation identified in this study may affect mitochondrial function by impacting the rate or efficiency of mitochondrial biogenesis. Impaired ability to turnover may allow the accumulation of defective mitochondria resulting from oxidative injury (70,71) and/or mtDNA damage (72) in neurons leading to impaired respiratory capacity (73). Identifying neuropathy-associated variants related to mitochondrial biogenesis may lead to targeted interventions that impact mitochondrial function (74,75). For example, several lines of evidence show that mitochondrial biogenesis is affected by pharmacologic agents (76–81), natural compounds (82), and behavioral interventions such as caloric restriction and exercise (83–86). Further studies confirming our findings may confirm degenerative mechanisms related to specific mitochondrial variants and ultimately contribute to the development of genotype-specific therapeutic interventions for delaying the onset of peripheral nerve dysfunction and disability in old age.
Funding
This research was supported by National Institute on Aging (NIA) contracts N01-AG-6-2101, N01-AG-6-2103, N01-AG-6-2106; NIA grants R01-AG028050, R01-AG032098, and R03-AG032498; NINR grant R01-NR012459 and Z01A6000932. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. Y.Z. was supported by NLM grant LM009722. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
Data analyses for this study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Maryland (http://biowulf.nih.gov).
References
- 1. Strotmeyer ES, Cauley JA, Schwartz AV, et al. Health ABC Study. Reduced peripheral nerve function is related to lower hip BMD and calcaneal QUS in older white and black adults: the Health, Aging, and Body Composition Study. J Bone Miner Res. 2006;21:1803–1810. :10.1359/jbmr.060725 [DOI] [PubMed] [Google Scholar]
- 2. Strotmeyer ES, de Rekeneire N, Schwartz AV, et al. The relationship of reduced peripheral nerve function and diabetes with physical performance in older white and black adults: the Health, Aging, and Body Composition (Health ABC) study. Diabetes Care. 2008;31:1767–1772. :10.2337/dc08-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Strotmeyer ES, de Rekeneire N, Schwartz AV, et al. Health ABC Study. Sensory and motor peripheral nerve function and lower-extremity quadriceps strength: the health, aging and body composition study. J Am Geriatr Soc. 2009;57:2004–2010. :10.1111/j.1532-5415.2009.02487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gregg EW, Sorlie P, Paulose-Ram R, et al. Prevalence of lower-extremity disease in the US adult population >=40 years of age with and without diabetes: 1999-2000 National Health and Nutrition Examination Survey. Diabetes Care. 2004;27:1591–1597. :10.2337/diacare.27.7.1591 [DOI] [PubMed] [Google Scholar]
- 5. Franklin GM, Kahn LB, Baxter J, Marshall JA, Hamman RF. Sensory neuropathy in non-insulin-dependent diabetes mellitus. The San Luis Valley Diabetes Study. Am J Epidemiol. 1990;131:633–643. [DOI] [PubMed] [Google Scholar]
- 6. Rivner MH, Swift TR, Malik K. Influence of age and height on nerve conduction. Muscle Nerve. 2001;24:1134–1141. :10.1002/mus.1124 [DOI] [PubMed] [Google Scholar]
- 7. Robinson LR, Rubner DE, Wahl PW, Fujimoto WY, Stolov WC. Influences of height and gender on normal nerve conduction studies. Arch Phys Med Rehabil. 1993;74:1134–1138. [PubMed] [Google Scholar]
- 8. Soudmand R, Ward LC, Swift TR. Effect of height on nerve conduction velocity. Neurology. 1982;32:407–410. :10.1212/WNL.32.4.407 [DOI] [PubMed] [Google Scholar]
- 9. Lehmann HC, Chen W, Borzan J, Mankowski JL, Höke A. Mitochondrial dysfunction in distal axons contributes to human immunodeficiency virus sensory neuropathy. Ann Neurol. 2011;69:100–110. :10.1002/ana.22150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viader A, Golden JP, Baloh RH, Schmidt RE, Hunter DA, Milbrandt J. Schwann cell mitochondrial metabolism supports long-term axonal survival and peripheral nerve function. J Neurosci. 2011;31:10128–10140. :10.1523/JNEUROSCI.0884-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naviaux RK. Mitochondrial DNA disorders. Eur J Pediatr. 2000;159(Suppl 3):S219–S226. :10.1007/PL00014407 [DOI] [PubMed] [Google Scholar]
- 12. Wallace DC. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found Symp. 2001;235:247–263; discussion 263–264. :10.1146/annurev.genet.39.110304.095751 [DOI] [PubMed] [Google Scholar]
- 13. Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. :10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wallace DC. Colloquium paper: bioenergetics, the origins of complexity, and the ascent of man. Proc Natl Acad Sci USA. 2010;107(Suppl 2):8947–8953. :10.1098/rstb.2012.0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Santoro L, Manganelli F, Lanzillo R, et al. A new POLG1 mutation with peo and severe axonal and demyelinating sensory-motor neuropathy. J Neurol. 2006;253:869–874. :10.1007/s00415-006-0082-6 [DOI] [PubMed] [Google Scholar]
- 16. Di Fonzo A, Bordoni A, Crimi M, et al. POLG mutations in sporadic mitochondrial disorders with multiple mtDNA deletions. Hum Mutat. 2003;22:498–499. :10.1002/humu.9203 [DOI] [PubMed] [Google Scholar]
- 17. Mancuso M, Filosto M, Bellan M, et al. POLG mutations causing ophthalmoplegia, sensorimotor polyneuropathy, ataxia, and deafness. Neurology. 2004;62:316–318. :10.1016/j.nmd.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 18. Reddy PH. Role of mitochondria in neurodegenerative diseases: mitochondria as a therapeutic target in Alzheimer’s disease. CNS Spectr. 2009;14(8 Suppl 7):8–13; discussion 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. :10.1146/annurev.pathol.4.110807.092314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tranah GJ, Nalls MA, Katzman SM, et al. Mitochondrial DNA sequence variation associated with dementia and cognitive function in the elderly. J Alzheimers Dis. 2012;32:357–372. :10.3233/JAD-2012-120466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tranah GJ, Yokoyama JS, Katzman SM, et al. ; Health, Aging and Body Composition Study. Mitochondrial DNA sequence associations with dementia and amyloid-β in elderly African Americans. Neurobiol Aging. 2014;35:442.e1–442.e8. :10.1016/j.neurobiolaging.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lam ET, Bracci PM, Holly EA, et al. Mitochondrial DNA sequence variation and risk of pancreatic cancer. Cancer Res. 2012;72:686–695. :10.1158/0008-5472.CAN-11-1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tranah GJ, Lam ET, Katzman SM, et al. Health, Aging and Body Composition Study. Mitochondrial DNA sequence variation is associated with free-living activity energy expenditure in the elderly. Biochim Biophys Acta. 2012;1817:1691–1700. :10.1016/j.bbabio.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tranah GJ, Manini TM, Lohman KK, et al. Mitochondrial DNA variation in human metabolic rate and energy expenditure. Mitochondrion. 2011;11:855–861. :10.1016/j.mito.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ward RE, Boudreau RM, Vinik AI, et al. Reproducibility of peroneal motor nerve conduction measurement in older adults. Clin Neurophysiol. 2013;124:603–609. :10.1016/j.clinph.2012.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. :10.1002/humu.20921 [DOI] [PubMed] [Google Scholar]
- 27. Symons S, Weber K, Bonin M, Nieselt K. ResqMi—a versatile algorithm and software for resequencing microarrays. In: Beyer A, Schroeder M, eds. Proceedings of the German Conference on Bioinformatics. Dresden, Germany: GI; 2008:10–20. [Google Scholar]
- 28. Biffi A, Anderson CD, Nalls MA, et al. Principal-component analysis for assessment of population stratification in mitochondrial medical genetics. Am J Hum Genet. 2010;86:904–917. :10.1016/j.ajhg.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Price AL, Kryukov GV, de Bakker PI, et al. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:832–838. :10.1016/j.ajhg.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Darty K, Denise A, Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. :10.1093/bioinformatics/btp250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maitra A, Cohen Y, Gillespie SE, et al. The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 2004;14:812–819. :10.1101/gr.2228504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.http://www.mitomap.org. http://www.mitomap.org MITOMAP: A Human Mitochondrial Genome Database. Accessed October 10, 2014.
- 33. Schlünzen F, Zarivach R, Harms J, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. :10.1038/35101544 [DOI] [PubMed] [Google Scholar]
- 34. Baldereschi M, Inzitari M, Di Carlo A, Farchi G, Scafato E, Inzitari D; ILSA Working Group. Epidemiology of distal symmetrical neuropathies in the Italian elderly. Neurology. 2007;68:1460–1467. :10.1212/01.wnl.0000260606.36443.29 [DOI] [PubMed] [Google Scholar]
- 35. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. :10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. :10.1126/science.1156409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–219. :10.1016/j.gde.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. :10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tranah GJ, Nalls MA, Katzman SM, et al. Mitochondrial DNA sequence variation associated with dementia and cognitive function in the elderly. J Alzheimers Dis. 2012;32:357–372. :10.3233/JAD-2012-120466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsieh RH, Li JY, Pang CY, Wei YH. A novel mutation in the mitochondrial 16S rRNA gene in a patient with MELAS syndrome, diabetes mellitus, hyperthyroidism and cardiomyopathy. J Biomed Sci. 2001;8:328–335. :10.1007/BF02258374 [DOI] [PubMed] [Google Scholar]
- 41. Li JY, Hsieh RH, Peng NJ, et al. A follow-up study in a Taiwanese family with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes syndrome. J Formos Med Assoc. 2007;106:528–536. :http://dx.doi.org/10.1016/S0929-6646(07)60003-5 [DOI] [PubMed] [Google Scholar]
- 42. Coulbault L, Deslandes B, Herlicoviez D, et al. A novel mutation 3090 G>A of the mitochondrial 16S ribosomal RNA associated with myopathy. Biochem Biophys Res Commun. 2007;362:601–605. :10.1016/j.bbrc.2007.08.040 [DOI] [PubMed] [Google Scholar]
- 43. Tang J, Qi Y, Bao XH, Wu XR. Mutational analysis of mitochondrial DNA of children with Rett syndrome. Pediatr Neurol. 1997;17:327–330. :10.1016/S0887-8994(97)00151-3 [DOI] [PubMed] [Google Scholar]
- 44. Cardaioli E, Dotti MT, Hayek G, Zappella M, Federico A. Studies on mitochondrial pathogenesis of Rett syndrome: ultrastructural data from skin and muscle biopsies and mutational analysis at mtDNA nucleotides 10463 and 2835. J Submicrosc Cytol Pathol. 1999;31:301–304. [PubMed] [Google Scholar]
- 45. Brown MD, Shoffner JM, Kim YL, et al. Mitochondrial DNA sequence analysis of four Alzheimer’s and Parkinson’s disease patients. Am J Med Genet. 1996;61:283–289. :10.1002/(SICI)1096-8628(19960122)61:3<283::AID-AJMG15>3.0.CO;2-P [DOI] [PubMed] [Google Scholar]
- 46. Shoffner JM, Brown MD, Torroni A, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. :10.1006/geno.1993.1299 [DOI] [PubMed] [Google Scholar]
- 47. Tanaka N, Goto Y, Akanuma J, et al. Mitochondrial DNA variants in a Japanese population of patients with Alzheimer’s disease. Mitochondrion. 2010;10:32–37. :10.1016/j.mito.2009.08.008 [DOI] [PubMed] [Google Scholar]
- 48. Tang S, Batra A, Zhang Y, Ebenroth ES, Huang T. Left ventricular noncompaction is associated with mutations in the mitochondrial genome. Mitochondrion. 2010;10:350–357. :10.1016/j.mito.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 49. Boles RG, Zaki EA, Lavenbarg T, et al. Are pediatric and adult-onset cyclic vomiting syndrome (CVS) biologically different conditions? Relationship of adult-onset CVS with the migraine and pediatric CVS-associated common mtDNA polymorphisms 16519T and 3010A. Neurogastroenterol Motil. 2009;21:936–e972. :10.1111/j.1365-2982.2009.01305.x [DOI] [PubMed] [Google Scholar]
- 50. Zaki EA, Freilinger T, Klopstock T, et al. Two common mitochondrial DNA polymorphisms are highly associated with migraine headache and cyclic vomiting syndrome. Cephalalgia. 2009;29:719–728. :10.1111/j.1468-2982.2008.01793.x [DOI] [PubMed] [Google Scholar]
- 51. Boles RG, Adams K, Ito M, Li BU. Maternal inheritance in cyclic vomiting syndrome with neuromuscular disease. Am J Med Genet A. 2003;120A:474–482. :10.1002/ajmg.a.20126 [DOI] [PubMed] [Google Scholar]
- 52. Schwertz H, Köster S, Kahr WH, et al. Anucleate platelets generate progeny. Blood. 2010;115:3801–3809. :10.1182/blood-2009-08-239558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pletscher A, Laubscher A. Blood platelets as models for neurons: uses and limitations. J Neural Transm Suppl. 1980;16:7–16. :10.1007/978-3-7091-8582-7_2 [DOI] [PubMed] [Google Scholar]
- 54. Pletscher A, Affolter H. The 5-hydroxytryptamine receptor of blood platelets. J Neural Transm. 1983;57:233–242. :10.1007/BF01248995 [DOI] [PubMed] [Google Scholar]
- 55. Da Prada M, Cesura AM, Launay JM, Richards JG. Platelets as a model for neurones? Experientia. 1988;44:115–126. :10.1007/BF01952193 [DOI] [PubMed] [Google Scholar]
- 56. Pletscher A. Platelets as models: use and limitations. Experientia. 1988;44:152–155. :10.1007/BF01952200 [DOI] [PubMed] [Google Scholar]
- 57. Barradas MA, Mikhailidis DP. The use of platelets as models for neurons: possible applications to the investigation of eating disorders. Biomed Pharmacother. 1993;47:11–18. :10.1016/0753-3322(93)90031-F [DOI] [PubMed] [Google Scholar]
- 58. Bakken AM, Staeffler A, Jørgensen HA, Holmsen H. Glycerophospholipid molecular species in platelets and brain tissues—are platelets a good model for neurons? Platelets. 2006;17:484–492. :10.1080/09537100600759196 [DOI] [PubMed] [Google Scholar]
- 59. Murphy DL, Donnelly CH. Monoamine oxidase in man: enzyme characteristics in platelets, plasma, and other human tissues. Adv Biochem Psychopharmacol. 1974;12:71–85. [PubMed] [Google Scholar]
- 60. Buus Lassen J, Lund J, Bechgaard E, Søndergaard I. Inhibition of 5-HT uptake into neurons and platelets in mice treated chronically with chlorimipramine and femoxetine. Psychopharmacology (Berl). 1979;64:149–153. :10.1007/BF00496055 [DOI] [PubMed] [Google Scholar]
- 61. Catricala S, Torti M, Ricevuti G. Alzheimer disease and platelets: how’s that relevant. Immun Ageing. 2012;9:20. :10.1186/1742-4933-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bush AI, Martins RN, Rumble B, et al. The amyloid precursor protein of Alzheimer’s disease is released by human platelets. J Biol Chem. 1990;265:15977–15983. :10.1016/j.exger.2009.08.004 [PubMed] [Google Scholar]
- 63. Li QX, Berndt MC, Bush AI, et al. Membrane-associated forms of the beta A4 amyloid protein precursor of Alzheimer’s disease in human platelet and brain: surface expression on the activated human platelet. Blood. 1994;84:133–142. :10.1182/blood-2005-11-007799 [PubMed] [Google Scholar]
- 64. Chen M, Inestrosa NC, Ross GS, Fernandez HL. Platelets are the primary source of amyloid beta-peptide in human blood. Biochem Biophys Res Commun. 1995;213:96–103. :10.1006/bbrc.1995.2103 [DOI] [PubMed] [Google Scholar]
- 65. Selkoe DJ, Podlisny MB, Joachim CL, et al. Beta-amyloid precursor protein of Alzheimer disease occurs as 110- to 135-kilodalton membrane-associated proteins in neural and nonneural tissues. Proc Natl Acad Sci USA. 1988;85:7341–7345. :10.1073/pnas.85.19.7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Deane R, Zlokovic BV. Role of the blood–brain barrier in the pathogenesis of Alzheimer’s disease. Curr Alzheimer Res. 2007;4:191–197. :10.2174/156720507780362245 [DOI] [PubMed] [Google Scholar]
- 67. Roher AE, Esh CL, Kokjohn TA, et al. Amyloid beta peptides in human plasma and tissues and their significance for Alzheimer’s disease. Alzheimers Dement. 2009;5:18–29. :http://dx.doi.org/10.1016/j.jalz.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baloh RH. Mitochondrial dynamics and peripheral neuropathy. Neuroscientist. 2008;14:12–18. :10.1177/1073858407307354 [DOI] [PubMed] [Google Scholar]
- 69. Donati A, Cavallini G, Paradiso C, et al. Age-related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J Gerontol A Biol Sci Med Sci. 2001;56:B288–B293. :10.1093/gerona/56.7.B288 [DOI] [PubMed] [Google Scholar]
- 70. de la Monte SM, Luong T, Neely TR, Robinson D, Wands JR. Mitochondrial DNA damage as a mechanism of cell loss in Alzheimer’s disease. Lab Invest. 2000;80:1323–1335. [DOI] [PubMed] [Google Scholar]
- 71. Anglade P, Vyas S, Hirsch EC, Agid Y. Apoptosis in dopaminergic neurons of the human substantia nigra during normal aging. Histol Histopathol. 1997;12:603–610. [PubMed] [Google Scholar]
- 72. Migliore L, Fontana I, Trippi F, et al. Oxidative DNA damage in peripheral leukocytes of mild cognitive impairment and AD patients. Neurobiol Aging. 2005;26:567–573. :10.1016/j.neurobiolaging.2004.07.016 [DOI] [PubMed] [Google Scholar]
- 73. de Grey AD. A proposed refinement of the mitochondrial free radical theory of aging. Bioessays. 1997;19:161–166. :10.1002/bies.950190211 [DOI] [PubMed] [Google Scholar]
- 74. Brand MD. The sites and topology of mitochondrial superoxide production. Exp Gerontol. 2010;45:466–472. :10.1016/j.exger.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li N, Ragheb K, Lawler G, et al. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003;278:8516–8525. :10.1074/jbc.M210432200 [DOI] [PubMed] [Google Scholar]
- 76. Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. :10.1038/nature05354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Davis JM, Murphy EA, Carmichael MD, Davis B. Quercetin increases brain and muscle mitochondrial biogenesis and exercise tolerance. Am J Physiol Regul Integr Comp Physiol. 2009;296:R1071–R1077. :1152/ajpregu.90925.2008 [DOI] [PubMed] [Google Scholar]
- 78. Liu Z, Sun L, Zhu L, et al. Hydroxytyrosol protects retinal pigment epithelial cells from acrolein-induced oxidative stress and mitochondrial dysfunction. J Neurochem. 2007;103:2690–2700. :10.1111/j.1471-4159.2007.04954.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rasbach KA, Schnellmann RG. Isoflavones promote mitochondrial biogenesis. J Pharmacol Exp Ther. 2008;325:536–543. :10.1124/jpet.107.134882 [DOI] [PubMed] [Google Scholar]
- 80. Stites T, Storms D, Bauerly K, et al. Pyrroloquinoline quinone modulates mitochondrial quantity and function in mice. J Nutr. 2006;136:390–396. [DOI] [PubMed] [Google Scholar]
- 81. Chowanadisai W, Bauerly KA, Tchaparian E, Wong A, Cortopassi GA, Rucker RB. Pyrroloquinoline quinone stimulates mitochondrial biogenesis through cAMP response element-binding protein phosphorylation and increased PGC-1alpha expression. J Biol Chem. 2010;285:142–152. :10.1074/jbc.M109.030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Timmers S, Konings E, Bilet L, et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011;14:612–622. :10.1016/j.cmet.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Guarente L. Mitochondria–a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Civitarese AE, Carling S, Heilbronn LK, et al. CALERIE Pennington Team. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. :10.1371/journal.pmed.0040076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Menshikova EV, Ritov VB, Fairfull L, Ferrell RE, Kelley DE, Goodpaster BH. Effects of exercise on mitochondrial content and function in aging human skeletal muscle. J Gerontol A Biol Sci Med Sci. 2006;61:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Johnston AP, De Lisio M, Parise G. Resistance training, sarcopenia, and the mitochondrial theory of aging. Appl Physiol Nutr Metab. 2008;33:191–199. :10.1139/H07-141 [DOI] [PubMed] [Google Scholar]