Abstract
Background.
Mitochondrial DNA (mtDNA) heteroplasmy is a mixture of normal and mutated mtDNA molecules in a cell. High levels of heteroplasmy at specific mtDNA sites lead to inherited mitochondrial diseases with neurological, sensory, and movement impairments. Here we test the hypothesis that heteroplasmy levels in elderly adults are associated with impaired function resembling mild forms of mitochondrial disease.
Methods.
We examined platelet mtDNA heteroplasmy at 20 disease-causing sites for associations with neurosensory and mobility function among 137 participants from the community-based Health, Aging, and Body Composition Study.
Results.
Elevated mtDNA heteroplasmy at four mtDNA sites in complex I and tRNA genes was nominally associated with reduced cognition, vision, hearing, and mobility: m.10158T>C with Modified Mini-Mental State Examination score (p = .009); m.11778G>A with contrast sensitivity (p = .02); m.7445A>G with high-frequency hearing (p = .047); and m.5703G>A with 400 m walking speed (p = .007).
Conclusions.
These results indicate that increased mtDNA heteroplasmy at disease-causing sites is associated with neurosensory and mobility function in older persons. We propose the novel use of mtDNA heteroplasmy as a simple, noninvasive predictor of age-related neurologic, sensory, and movement impairments.
Key Words: Mitochondrial DNA, Heteroplasmy, Cognition, Vision, Hearing, Mobility.
Mitochondrial oxidative phosphorylation (OXPHOS), which supplies 90% of human energetic requirements, is dependent upon the coordinated expression and interaction of genes encoded in the nuclear and mitochondrial genomes. Human mitochondrial DNA (mtDNA), a maternally inherited 16,569bp loop containing genes critical to OXPHOS, commonly exhibits a mixture (heteroplasmy) of normal and mutated mtDNA molecules within a cell (1). Because mitochondria perform diverse functions in different tissues, specific mutations in mtDNA lead to mitochondrial diseases resulting in a wide spectrum of abnormalities (2). Many of these diseases, including Leber’s hereditary optic neuropathy (LHON), Leigh syndrome, deafness, and mitochondrial myopathy, result from high heteroplasmy loads (>80% burden of pathologic mtDNA mutation) and cause neuropathological impairments (3). To date, no study has investigated mtDNA heteroplasmy levels as predictors of age-related function.
Heteroplasmic mutations and rearrangements of mtDNA have been reported in various tissues of elderly individuals (4); specifically, large mtDNA deletions increase with age in skeletal muscle, heart, brain, and central nervous system (5). Elderly adults develop mtDNA mutations and rearrangements and consequently exhibit reduced activity of OXPHOS enzymes in postmitotic tissues (6). In general, organs with the highest ATP requirements and the lowest regenerative capacities, such as the brain, heart, retina, auditory neuroepithelia, and skeletal muscle, are the most sensitive to the effects of mtDNA mutations and bioenergetic defects resulting from mtDNA mutations may be critical to age-related functional decline (7–11). Thus we propose the novel use of mtDNA heteroplasmy as a simple, noninvasive predictor of neurologic, sensory, and movement impairments.
We quantified heteroplasmy at 20 disease-causing mtDNA sites in a community-based cohort of men and women older than 70 years and measured associations with cognitive function, hearing, vision, and mobility (3,12). The mtDNA sites include: seven mtDNA mutations leading to complex I deficiency and brain magnetic resonance imaging abnormalities (12) tested for associations with cognitive function, three primary LHON mutations tested for associations with vision, two nonsyndromic deafness mutations tested for associations with hearing, and eight mitochondrial myopathy mutations tested for associations with walking speed (3). We test the hypothesis that elevated heteroplasmy levels in elderly adults are associated with impaired function resembling mild forms of mitochondrial disease.
Methods
Participants
The Health, Aging, and Body Composition (Health ABC) Study is a prospective cohort of 3,075 community-dwelling men and women living in Memphis, Tennessee, or Pittsburgh, Pennsylvania, and aged 70–79 years at recruitment during 1996–1997. Participants were recruited from a random sample of white and black Medicare-eligible people within designated zip code areas. Participants had to report no difficulty with activities of daily living, walking a quarter of a mile, or climbing 10 steps without resting. They were free of life-threatening cancer diagnoses. The sample was comprised of 51% women and 41% of participants were black. Participants self-designated race or ethnicity classified as Asian or Pacific Islander, black or African American, white or Caucasian, Latino or Hispanic, and other. All eligible participants signed a written informed consent, approved by the institutional review boards at the clinical sites. This study was approved by the institutional review boards of the clinical sites and the coordinating center (University of California, San Francisco).
Mitochondrial DNA Sequencing
We studied 137 Health ABC participants of European ancestry who were part of an energetics substudy performed at the second Health ABC visit (13). Complete mtDNA was extracted from platelets collected at the year 2 (1998–1999) clinical visit using the Gentra PureGene Kit (Qiagen, Hilden, Germany) and sequenced using the Affymetrix Mitochondrial Resequencing Array 2.0 (MitoChip, Affymetrix, Santa Clara, CA) as previously described (13). We calculated quantitative estimates of heteroplasmy with established methods (14). Heteroplasmy was derived for each mtDNA nucleotide by first calculating the minor allele signal (MAS) defined as the raw signal intensity of the highest ‘‘nonexpected’’ or minor allele minus the raw signal intensity value of the smallest contributing or background allele, and the background subtracted expected allele signal (EAS) defined as the raw signal intensity of the expected allele (NC_012920.1) minus the raw signal intensity of the smallest contributing or background allele. Percent heteroplasmy was then defined as MAS/(MAS + EAS). The MitoChip has a 2% detection limit for heteroplasmy, therefore samples with heteroplasmy values <2% were excluded from analyses involving those loci. Twenty samples were repeated for concordance testing yielding >98% sequence concordance of nucleotide calls and a within-chip error rate of 0.0028%. The coefficient of variation values for the 20 repeated samples ranged from 0.09 to 0.27 for the 20 examined mtDNA mutations (Table 1).
Table 1.
Summary Statistics for 20 Candidate Mitochondrial DNA Heteroplasmic Mutations
| Heteroplasmy | ||||
|---|---|---|---|---|
| mtDNA | Mean, % (SD) | Range, % | CV* | n † |
| Cognition | ||||
| m.10158T>C | 12 (4) | 2–23 | 0.14 | 137 |
| m.10191T>C | 9 (3) | 2–18 | 0.27 | 137 |
| m.10197G>A | 5 (2) | 2–12 | 0.17 | 123 |
| m.13091T>C | 15 (3) | 8–23 | 0.14 | 137 |
| m.13513G>A | 16 (1) | 13–20 | 0.19 | 137 |
| m.13514A>G | 9 (3) | 3–18 | 0.20 | 137 |
| m.14487T>C | 26 (3) | 20–33 | 0.12 | 131 |
| Vision | ||||
| m.3460G>A | 19 (3) | 8–24 | 0.12 | 137 |
| m.11778G>A | 9 (1) | 6–13 | 0.17 | 137 |
| m.14484T>C | 32 (3) | 23–39 | 0.09 | 137 |
| Hearing | ||||
| m.7445A>G | 27 (2) | 15–34 | 0.11 | 137 |
| m.7511T>C | 12 (2) | 6–19 | 0.17 | 136 |
| Mobility | ||||
| m.3243A>G | 14 (3) | 4–24 | 0.11 | 136 |
| m.3302A>G | 18 (7) | 6–28 | 0.22 | 135 |
| m.4308G>A | 9 (3) | 3–15 | 0.27 | 137 |
| m.5650G>A | 8 (1) | 3–11 | 0.22 | 137 |
| m.5703G>A | 12 (1) | 8–15 | 0.14 | 137 |
| m.7497G>A | 10 (3) | 3–20 | 0.23 | 137 |
| m.12315G>A | 8 (3) | 2–16 | 0.26 | 137 |
| m.14709T>C | 10 (3) | 2–17 | 0.22 | 137 |
Notes: *Coefficient of variation (CV) values for 20 repeated DNA samples.
†Number of samples with heteroplasmy levels ≥2%.
Cognitive Function Testing
The Modified Mini-Mental State Examination (3MS) was administered to participants at the year 3 (1999–2000) clinical visit. The 3MS is a brief, general cognitive battery with components for orientation, concentration, language, praxis, and immediate and delayed memory. Possible scores range from 0 to 100, with higher scores indicating better cognitive function. We analyzed heteroplasmy at seven specific mtDNA mutations that lead to complex I deficiency and brain magnetic resonance imaging abnormalities (m.10158T>C, m.10191T>C, m.10197G>A, m.13091T>C, m.13513G>A, m.13514A>G, and m.14487T>C) (3,12) for associations with performance on the 3MS.
Vision
Visual testing was performed at the year 3 (1999–2000) clinical visit and included three tests: Bailey–Lovie distance visual acuity, Pelli–Robson contrast sensitivity, and Frisby stereo test. We analyzed heteroplasmy at the three primary mutations that account for over 90% of LHON cases (m.3460G>A, m.11778G>A, and m.14484T>C) (3) for associations with visual testing. The primary LHON mutations exhibit a greater penetrance in men than women (15) and we examined LHON associations in sex-stratified analyses.
Hearing
Air-conduction pure-tone hearing testing was performed at the year 5 (2001–2002) clinical visit. Hearing thresholds, measured in hearing level in decibels (dB HL), were obtained using current standard methods for manual audiometry. From the thresholds, low frequency (average of hearing thresholds at 250, 500, and 1000 Hz) and high frequency (2000, 4000, and 8000 Hz) pure-tone averages were calculated for each ear. We analyzed heteroplasmy at two confirmed heteroplasmic deafness and sensorineural hearing loss mutations (m.7445A>G and m.7511T>C) (3) for associations with high and low frequency hearing.
Mobility
A timed 400 m walk was performed at the year 2 (1998–1999) clinical visit. Participants were asked to walk 400 m after a 2-minute warm-up and time to complete the test was recorded. We analyzed heteroplasmy at the eight confirmed myopathy mutations (m.3243A>G, m.3302A>G, m.4308G>A, m.5650G>A, m.5703G>A, m.7497G>A, m.12315G>A, and m.14709T>C) (3) for associations with 400 m walking speed.
Statistical Analyses
Generalized linear models were used to analyze cognitive function, vision, hearing, and mobility as continuous outcomes and mtDNA heteroplasmy at candidate mtDNA sites from each hypothesis-based subset (complex I deficiency/brain magnetic resonance imaging abnormalities, LHON, deafness, and mitochondrial myopathy) as the independent variables. Outcome measures exhibiting significant linear associations (p < .05) with heteroplasmic mutations were compared among tertiles of heteroplasmy using analysis of variance. Adjustment for multiple comparisons was performed for each phenotype (cognition, critical α = .007; vision, critical α = .017; hearing, critical α = .025; walking speed, critical α = .006). In post hoc analyses we examined associations between each subset of mtDNA markers and the other (nonrelated) clinical measures. All analyses were adjusted for age, sex, and clinic site using SAS version 9.4 (SAS Institute Inc, Cary, NC).
Results
A total of 137 participants from the community-based Health ABC Study with mtDNA heteroplasmy were available for analysis including 63 men and 74 women aged 73.5±2.9 years. The sequenced participants were representative of the nonsequenced participants of European ancestry with regard to age, sex distribution, and phenotypes examined in the current study (Table 2). Summary metrics for the 20 candidate mutations are reported including the number of participants with heteroplasmy levels below the 2% detection limit at each locus. Heteroplasmy levels detected in this study are comparable to those from previous studies using the MitoChip (14), Illumina (16), and LS454 (16) platforms. Within each of the four subsets of mutations examined we hypothesized that heteroplasmy would be associated with related clinical measures (eg, complex I deficiency/brain magnetic resonance imaging abnormalities and cognitive function, LHON and vision) and identified statistically significant associations for each subset. The associations presented herein achieved nominal significance (p < .05); however, these results were not statistically significant after adjustment for multiple comparisons. Adjustment for smoking status and markers of oxidative stress (plasma oxidized low-density lipoprotein and urinary 8-iso-prostaglandin F2α) (17) did not alter the results. Heteroplasmy within each of the four subsets of mutations was not associated with the other (nonrelated) clinical measures assessed in this study (Supplementary Table S1).
Table 2.
Comparison of Characteristics Between Sequenced and Nonsequenced Health ABC Participants of European Ancestry
| Sequenced | Nonsequenced | |
|---|---|---|
| Total participants, n | 137 | 1,657 |
| Female participants, n (%) | 74 (54.01) | 781 (47.13) |
| Phenotypes, mean (SD) | ||
| Baseline age, years | 73.5 (2.9) | 73.8 (2.9) |
| Modified Mini-Mental State Examination | 93.1 (5.7) | 92.7 (6.3) |
| High frequency hearing (dB) | 59.0 (18.2) | 58.3 (16.8) |
| Low frequency hearing (dB) | 34.6 (18.6) | 33.4 (15.7) |
| Contrast sensitivity (log contrast) | 1.57 (0.18) | 1.57 (0.16) |
| Disparity for Frisby Stereo Test (arcmin) | 205.7 (275.6) | 206.7 (279.5) |
| Visual acuity (log minutes of arc) | 0.09 (0.15) | 0.11 (0.14) |
| 400 m walking speed (m/s) | 1.32 (0.19) | 1.32 (0.2) |
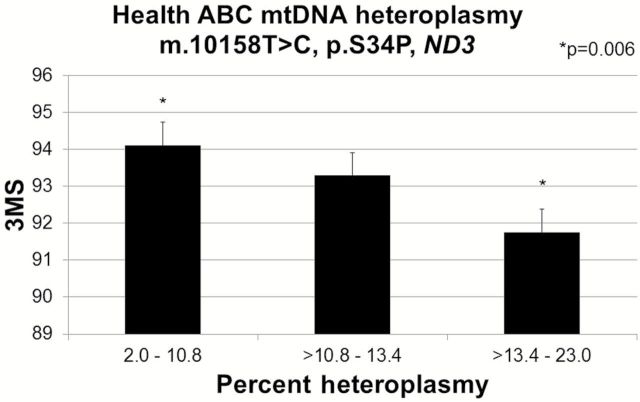
Of the seven candidate complex I mtDNA mutations examined for effects on cognitive function, increased heteroplasmy at m.10158T>C (n = 133) was significantly associated with decreased cognitive function measured by a lower 3MS score (p = .009) (Figure 1). Mean (SE) 3MS was significantly lower (p = .006) for the lowest tertile, 91.5 (0.83), when compared with the highest tertile, 94.7 (0.82) of heteroplasmy (Figure 1). The remaining candidate complex I mutations were not associated with 3MS score.
Figure 1.
Mitochondrial m.10158T>C association with Modified Mini-Mental State Examination (3MS, linear regression p = .009) among 132 Health ABC participants. 3MS was compared across tertiles of m.10158T>C heteroplasmy. Values adjusted for age, sex, and clinic site
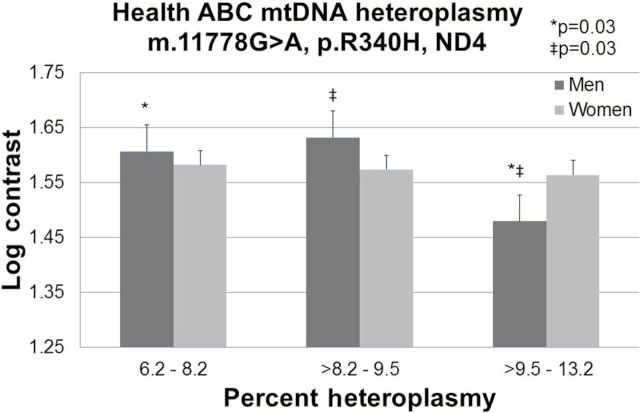
Of the three LHON mutations examined for effects on vision, increased heteroplasmy at m.11778G>A (n = 133) was significantly associated with decreased contrast sensitivity (p = .02), a measure of retinal function. Mean (SE) log contrast was significantly poorer for participants in the highest tertile of heteroplasmy, 1.53 (0.03) when compared with those in the middle, 1.60 (0.03), p = .04, and lowest tertiles, 1.60 (0.03), p = .03. The m.11778G>A mutation exhibits a higher penetrance in men than women (15) and we found that the impact was stronger in male (p = .045) than female (p = .32) participants (Figure 2). Men in the highest tertile of heteroplasmy exhibited significantly lower mean (SE) log contrast, 1.49 (0.05), when compared with those in the middle, 1.59 (0.05), p = .04, and lowest tertiles, 1.65 (0.06), p = .03. The remaining candidate LHON mutations were not associated with visual contrast sensitivity and no associations were identified for distance visual acuity or stereo test.
Figure 2.
Mitochondrial m.11778G>A, p.S34P, ND3 association with visual contrast sensitivity (linear regression p = .02) among 131 Health ABC participants including 60 male (linear regression p = .045) and 71 female participants (linear regression p = .32). Log contrast was compared across tertiles of heteroplasmy. Values adjusted for age, sex, and clinic site.
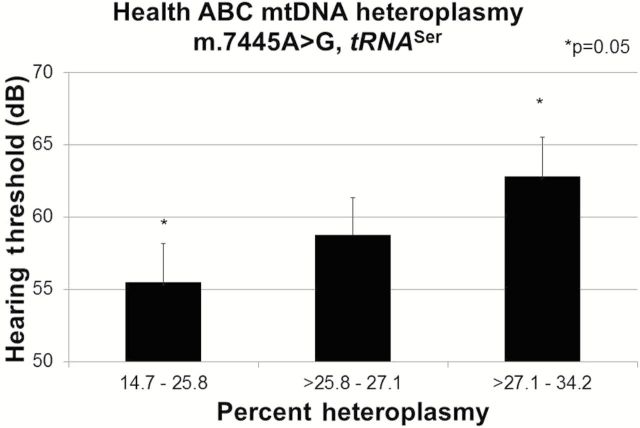
Of the two deafness mutations, increased heteroplasmy at m.7445A>G (n = 118) was significantly associated with impaired high frequency hearing (p = .047). Mean (SE) high-frequency hearing (dB HL) was significantly lower (p = .05) for the highest tertile, 62.5 (2.6) when compared with the lowest tertile, 55.5 (2.64) (Figure 3). The candidate m.7511T>C mutation was not associated with hearing and no associations were identified for low frequency hearing.
Figure 3.
Mitochondrial m.7445A>G association with high frequency hearing (linear regression p = .047) among 118 Health ABC participants. High frequency hearing (dB HL) was compared across tertiles of m.7445A>G heteroplasmy. Values adjusted for age, sex, and clinic site.
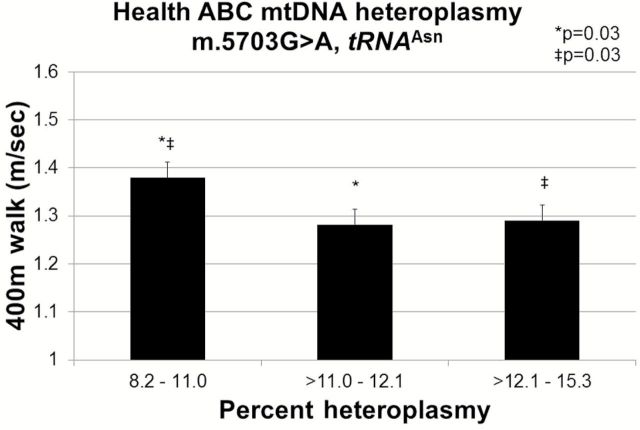
Of the eight mitochondrial myopathy mutations m.5703G>A (n = 111) heteroplasmy was significantly associated with 400 m walking speed (p = .007). Mean (SE) walking speed (m/s) was significantly faster for the lowest tertile of heteroplasmy, 1.38 (0.03) when compared with the middle, 1.28 (0.03), p = .04 and highest tertiles, 1.29 (0.03), p = .05 (Figure 4). The remaining candidate myopathy mutations were not associated with walking speed.
Figure 4.
Mitochondrial m.5703G>A association with 400 m walking speed (linear regression p = .007) among 101 Health ABC participants. 400 m walking speed (m/s) was compared across tertiles of m.5703G>A heteroplasmy. Values adjusted for age, sex, and clinic site.
Discussion
Elevated levels of heteroplasmy in mtDNA found in older adults were associated with significantly impaired cognitive function, hearing, vision, and walking speed consistent with the characteristic impairments of the associated mitochondrial diseases resulting from specific complex I and tRNA mutations. Complex I deficiency is the most frequent cause of bioenergetic dysfunction (18) and accounts for many rare clinical presentations (19). Additionally, over half of pathogenic mtDNA mutations are located in genes controlling tRNA expression (20) leading to reductions in tRNA steady-state levels, decreases in mitochondrial protein synthesis, destabilized tRNA secondary or tertiary structure (21,22), and impaired OXPHOS or oxygen consumption (22). Here we present the first study to demonstrate that heteroplasmy measured at specific mtDNA sites can be a significant predictor of neurosensory and mobility function in elderly adults.
Complex I Associations
Elevated levels of the m.10158T>C, p.S34P, ND3 (NADH dehydrogenase 3) substitution in complex I, one of three pathogenic Leigh syndrome mutations located in a 15 amino acid mutational hotspot, were associated with worse cognitive function. This mutation interacts with other mitochondrial and nuclear-encoded complex I subunits affecting overall assembly or structural stability (23). The m.10158T>C, p.S34P, ND3 substitution previously observed in mutation carriers results in reduced complex I activity as well as lower levels of fully assembled complex I (23). In support of this conclusion, a study of cybrids (experimental hybrid cells containing externally sourced mtDNA inserted into a cell with uniform nuclear DNA background) derived from m.10158T>C mutation carriers demonstrated that residual complex I enzyme activity decreased with increased load of the mutated allele (23). This previous in vitro work suggests that a complex I defect may be responsible for the lower cognitive function we observed in relation to increasing m.10158T>C mutation load.
Our results also demonstrate that elevated levels of the m.11778G>A, p.R340H, ND4 (NADH dehydrogenase 4) substitution in complex I were associated with significantly decreased contrast sensitivity. We observed that m.11778G>A heteroplasmy impacted contrast sensitivity above the 9.5% mutation burden threshold. This result may help to explain why initially asymptomatic m.11778G>A LHON mutation carriers experience progressive subclinical retinal ganglion cell dysfunction that eventually leads to permanent ganglion cell loss and subsequent blindness (24). The three primary complex I subunit LHON mutations (including m.11778G>A which accounts for up to 70% of LHON cases (3,25)) have been extensively investigated using a wide range of cell types and biochemical assays. Cybrid studies support our findings by demonstrating that carriers of either one of the three most common primary LHON mutations exhibit elevated reactive oxygen species levels (26,27), decreased mitochondrial membrane potential (26), impaired complex I-dependent ATP synthesis (15,28), hyper-fragmented mitochondrial networks (26), and increased rates of apoptotic cell death (29,30).
Two features of LHON remain largely unexplained: incomplete penetrance of mutations and the significant male bias in disease predisposition (15). Overall, 50% of male LHON mutation carriers and 10% of female mutation carriers eventually lose vision (15). We observed that the effect of m.11778G>A heteroplasmy on age-related contrast sensitivity was stronger in men than women, largely resembling the m.11778G>A LHON association pattern. This disparity may in part be explained by hormonal influence (26) or additional genetic factors involving interactions with the X-chromosome (31) or mitochondrial haplogroup (32). Supplementation of cybrids carrying of LHON mutations with 17b-estradiol mitigates many pathological features (eg, elevated reactive oxygen species levels and increased apoptosis) (26). This hormone treatment leads to more efficient mitochondrial biogenesis and an increase in cellular levels of the anti-oxidant enzyme superoxide dismutase (26), which protects LHON cells against enhanced superoxide production by mutated complex I (33).
tRNA Associations
Elevated levels of the m.7445A>G deafness mutation (3) were associated with reduced high-frequency hearing. This mutation is located on both the heavy strand COI (complex IV) and the light strand tRNA Ser precursor. On the heavy strand, m.7445A>G substitutes one stop codon for another stop codon in the COI gene and is considered essentially silent. On the light strand, m.7445A>G flanks the 3ʹ end of tRNA Ser and interferes with endonuclease cleavage resulting in a reduction in steady-state tRNASer levels (22). Interestingly, m.7445A>G was also associated with reduced ND6 (complex I) mRNA levels (22). ND6 is located approximately 7kb upstream from m.7445A>G and is cotranscribed with tRNA Ser.
In the current study, elevated levels of the m.5703G>A, tRNA Asn mutations were associated with slower walking speed. This mutation has been linked to numerous mitochondrial pathologies including myopathy (3). Cybrids carrying the m.5703G>A mutation exhibit a severe mitochondrial protein synthesis defect and impaired OXPHOS function, possibly due to a destabilized tRNAAsn structure and a reduction in the pool of functional tRNAAsn (21). Modulating the expression or dosage of nuclear-encoded factors restores normal mitochondrial function in cybrids carrying m.5703G>A (34), suggesting that nuclear gene-based strategies could compensate for this and other pathogenic tRNA mutations.
Heteroplasmy as an Indicator for Age-related Function
Our results indicate that the heteroplasmy load identified in the eighth decade of life is associated with significant preclinical loss of function. Strategies to select against heteroplasmic mtDNA may have therapeutic potential for the treatment of mitochondrial disorders and age-related diseases (35). Several advances in the development of antigenomic therapies have shown that it is possible to directly target heteroplasmic mtDNA mutations including: mitochondrially targeted peptide nucleic acid oligomers (36), zinc finger peptides (37), recombinant RNA (38), and transcription activator-like effectors (39). Modulation of nuclear-encoded (34) and mtDNA-encoded (40) factors can also compensate for pathogenic mtDNA mutations. In addition, Rapamycin-induced upregulation of mitophagy decreases levels of the m.11778G>A (LHON) mutation and partially restores cellular ATP levels (41). In contrast, heteroplasmic load may be increased by nucleoside analog antiretroviral drugs leading to the progressive accumulation of preexisting somatic mtDNA mutations (42). Treatments that enhance mtDNA replication should, therefore, be approached with caution as they could amplify the expansion of deleterious mutations with potentially detrimental long-term consequences (43).
Low-level mtDNA heteroplasmy in humans is common and may originate in early development (44) or even in the germline (1) despite protective mechanisms to minimize the maternal transmission of mutated mtDNA (45,46). Whether this early-life mutation load is the result of maternally transmitted mtDNA mutations (47) or is acquired during a critical period in childhood or early adult life (48) remains uncertain. Age-related somatic mtDNA mutations can accumulate in postmitotic tissues until a tissue-specific threshold in the ratio of mutated to normal mtDNA molecules is surpassed and cells become energetically compromised (49). The frequency of heteroplasmic variants varies considerably among different tissues of the same individual (23,50,51); however, the mechanisms leading to the unequal partitioning of mitochondrial genotypes within and among individual cells (44,47,48) are unknown and may involve selection against a particular mutation (41,52) or genetic drift (53). Understanding the origins of heteroplasmy and mechanisms influencing the expansion of deleterious mtDNA mutations will advance the development of interventions centered on the improvement of mitochondrial health.
This study had a number of strengths including the use of a chip-based mtDNA sequencing method validated for heteroplasmy assessment, a well-characterized community-based longitudinal cohort with multiple clinical assessments, and platelet mtDNA (platelets have been utilized as neuronal models (54)). In addition, we tested the hypothesis that elevated levels of mtDNA heteroplasmy at select mutations would be associated with impaired function in elderly adults consistent with mitochondrial disease impairments resulting from these mutations. Although it is not yet clear how rapidly heteroplasmy changes with aging, we identified associations with measures that took place at the same visit or after platelets were collected for sequencing, thus ensuring that the associations reported in this study are either prospective or cross-sectional. The implication of these results is that mtDNA heteroplasmy at known pathogenic sites is associated with current or later impaired functioning. While effect sizes observed for each of the clinical measures associated with heteroplasmy are moderately clinically significant, identifying additional predictors of functional decline would be important in refining the associations between these clinical measures and later disease (eg, of contrast sensitivity with age-related macular degeneration or walking speed with disability). This study is limited in that it does not include independent replication, and while the statistical associations achieved nominal significance (p < .05), these results were not statistically significant after adjustment for multiple comparisons. The lack of associations for both related and unrelated mutations and phenotypes may be due to small sample size as well as the limited tissue examined in this study (eg, relevant tissues may not have been examined for each phenotype). Indeed, further research in populations with the appropriate design, phenotypes, and biospecimens is needed to confirm these findings. The Health ABC Study cohort is a well-characterized and appropriate cohort to use for aging-related studies. The participants were healthy when entering the study and results from a single population likely cannot be generalized to all possible populations. If validated in additional studies, circulating mtDNA heteroplasmy may represent a useful peripheral biomarker for identifying those at risk of developing age-related impairments and for monitoring persons who are receiving pharmacologic agents, natural compounds, or behavioral interventions that target the mitochondria.
Supplementary Material
Supplementary material can be found at: http://biomedgerontology.oxfordjournals.org/
Funding
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging, Contracts N01-AG-6-2101, N01-AG-6-2103, and N01-AG-6-2106; National Institutes of Health grants R01-AG028050, R03-AG032498, R01-NR012459, Z01A6000932, R01-HL121023, and a grant from the Research and Education Leadership Committee of the CPMC Foundation.
Conflict of Interest
The authors declare no conflicts of interest or competing financial interests relevant to this manuscript’s subject.
Supplementary Material
References
- 1. Payne BA, Wilson IJ, Yu-Wai-Man P, et al. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013;22:384–390. 10.1093/hmg/dds435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Naviaux RK. Mitochondrial DNA disorders. Eur J Pediatr. 2000;159(suppl 3):S219–S226. 10.1007/PL00014407 [DOI] [PubMed] [Google Scholar]
- 3. MITOMAP: A Human Mitochondrial Genome Database http://www.mitomap.org. Accessed February 15, 2015.
- 4. Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. http//dx..org/10.1016/S0140-6736(89)92145-4 [DOI] [PubMed] [Google Scholar]
- 5. Arnheim N, Cortopassi G. Deleterious mitochondrial DNA mutations accumulate in aging human tissues. Mutat Res. 1992;275:157–167. 10.1016/0921-8734(92)90020-P [DOI] [PubMed] [Google Scholar]
- 6. Trounce I, Byrne E, Marzuki S. Decline in skeletal muscle mitochondrial respiratory chain function: possible factor in ageing. Lancet. 1989;1:637–639. http//dx..org/10.1016/S0140-6736(89)92143-0 [DOI] [PubMed] [Google Scholar]
- 7. Tyrrell DJ, Bharadwaj MS, Van Horn CG, Kritchevsky SB, Nicklas BJ, Molina AJ. Respirometric profiling of muscle mitochondria and blood cells are associated with differences in gait speed among community-dwelling older adults. J Gerontol A Biol Sci Med Sci. July 16, 2014. 10.1093/gerona/glu096. piiglu096. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van Remmen H, Jones DP. Current thoughts on the role of mitochondria and free radicals in the biology of aging. J Gerontol A Biol Sci Med Sci. 2009;64:171–174. 10.1093/gerona/gln058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Figueiredo PA, Powers SK, Ferreira RM, Appell HJ, Duarte JA. Aging impairs skeletal muscle mitochondrial bioenergetic function. J Gerontol A Biol Sci Med Sci. 2009;64:21–33. 10.1093/gerona/gln048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Menshikova EV, Ritov VB, Fairfull L, Ferrell RE, Kelley DE, Goodpaster BH. Effects of exercise on mitochondrial content and function in aging human skeletal muscle. J Gerontol A Biol Sci Med Sci. 2006;61:534–540. 10.1093/gerona/61.6.534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horan MP, Pichaud N, Ballard JW. Review: quantifying mitochondrial dysfunction in complex diseases of aging. J Gerontol A Biol Sci Med Sci. 2012;67:1022–1035. 10.1093/gerona/glr263 [DOI] [PubMed] [Google Scholar]
- 12. Lebre AS, Rio M, Faivre d’Arcier L, et al. A common pattern of brain MRI imaging in mitochondrial diseases with complex I deficiency. J Med Genet. 2011;48:16–23. 10.1136/jmg.2010.079624 [DOI] [PubMed] [Google Scholar]
- 13. Tranah GJ, Lam ET, Katzman SM, et al. ; Health, Aging and Body Composition Study. Mitochondrial DNA sequence variation is associated with free-living activity energy expenditure in the elderly. Biochim Biophys Acta. 2012;1817:1691–1700. 10.1016/j.bbabio.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coon KD, Valla J, Szelinger S, et al. Quantitation of heteroplasmy of mtDNA sequence variants identified in a population of AD patients and controls by array-based resequencing. Mitochondrion. 2006;6:194–210. 10.1016/j.mito.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 15. Yu-Wai-Man P, Griffiths PG, Chinnery PF. Mitochondrial optic neuropathies - disease mechanisms and therapeutic strategies. Prog Retin Eye Res. 2011;30:81–114. 10.1016/j.preteyeres.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci U S A. 2014;111:10654–10659. 10.1073/pnas.1403521111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cesari M, Kritchevsky SB, Nicklas B, et al. Oxidative damage, platelet activation, and inflammation to predict mobility disability and mortality in older persons: results from the Health Aging and Body Composition Study. J Gerontol A Biol Sci Med Sci. 2012;67A:671–676. 10.1093/gerona/glr246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smeitink J, van den Heuvel L, DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat Rev Genet. 2001;2:342–352. 10.1038/35072063 [DOI] [PubMed] [Google Scholar]
- 19. Loeffen JL, Smeitink JA, Trijbels JM, et al. Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum Mutat. 2000;15:123–134. 10.1002/(SICI)1098-1004(200002)152<123AID-HUMU1>3.0.CO;2-P [DOI] [PubMed] [Google Scholar]
- 20. Zifa E, Giannouli S, Theotokis P, Stamatis C, Mamuris Z, Stathopoulos C. Mitochondrial tRNA mutations: clinical and functional perturbations. RNA Biol. 2007;4:38–66. 10.4161/rna.4.1.4548 [DOI] [PubMed] [Google Scholar]
- 21. Hao H, Moraes CT. A disease-associated G5703A mutation in human mitochondrial DNA causes a conformational change and a marked decrease in steady-state levels of mitochondrial tRNA(Asn). Mol Cell Biol. 1997;17:6831–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guan MX, Enriquez JA, Fischel-Ghodsian N, et al. The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol Cell Biol. 1998;18:5868–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McFarland R, Kirby DM, Fowler KJ, et al. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann Neurol. 2004;55:58–64. 10.1002/ana.10787 [DOI] [PubMed] [Google Scholar]
- 24. Guy J, Feuer WJ, Porciatti V, et al. Retinal ganglion cell dysfunction in asymptomatic G11778A: Leber hereditary optic neuropathy. Invest Ophthalmol Vis Sci. 2014;55:841–848. 10.1167/iovs.13-13365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wallace DC, Singh G, Lott MT, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. 10.1126/science.3201231 [DOI] [PubMed] [Google Scholar]
- 26. Giordano C, Montopoli M, Perli E, et al. Oestrogens ameliorate mitochondrial dysfunction in Leber’s hereditary optic neuropathy. Brain. 2011;134(Pt 1):220–234. 10.1093/brain/awq276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wong A, Cavelier L, Collins-Schramm HE, et al. Differentiation-specific effects of LHON mutations introduced into neuronal NT2 cells. Hum Mol Genet. 2002;11:431–438. [DOI] [PubMed] [Google Scholar]
- 28. Baracca A, Solaini G, Sgarbi G, et al. Severe impairment of complex I-driven adenosine triphosphate synthesis in leber hereditary optic neuropathy cybrids. Arch Neurol. 2005;62:730–736. 10.1001/archneur.62.5.730 [DOI] [PubMed] [Google Scholar]
- 29. Ghelli A, Zanna C, Porcelli AM, et al. Leber’s hereditary optic neuropathy (LHON) pathogenic mutations induce mitochondrial-dependent apoptotic death in transmitochondrial cells incubated with galactose medium. J Biol Chem. 2003;278:4145–4150. [DOI] [PubMed] [Google Scholar]
- 30. Zanna C, Ghelli A, Porcelli AM, Carelli V, Martinuzzi A, Rugolo M. Apoptotic cell death of cybrid cells bearing Leber’s hereditary optic neuropathy mutations is caspase independent. Ann N Y Acad Sci. 2003;1010:213–217. 10.1196/annals.1299.037 [DOI] [PubMed] [Google Scholar]
- 31. Hudson G, Keers S, Yu Wai Man P, et al. Identification of an X-chromosomal locus and haplotype modulating the phenotype of a mitochondrial DNA disorder. Am J Hum Genet. 2005;77:1086–1091. http//dx..org/10.1086/498176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hudson G, Carelli V, Spruijt L, et al. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet. 2007;81:228–233. http://dx.doi.org/10.1086/519394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qi X, Sun L, Hauswirth WW, Lewin AS, Guy J. Use of mitochondrial antioxidant defenses for rescue of cells with a Leber hereditary optic neuropathy-causing mutation. Arch Ophthalmol. 2007;125:268–272. 10.1001/archopht.125.2.268 [DOI] [PubMed] [Google Scholar]
- 34. Hao H, Morrison LE, Moraes CT. Suppression of a mitochondrial tRNA gene mutation phenotype associated with changes in the nuclear background. Hum Mol Genet. 1999;8:1117–1124. 10.1093/hmg/8.6.1117 [DOI] [PubMed] [Google Scholar]
- 35. Russell O, Turnbull D. Mitochondrial DNA disease-molecular insights and potential routes to a cure. Exp Cell Res. 2014;325:38–43. 10.1016/j.yexcr.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor RW, Chinnery PF, Turnbull DM, Lightowlers RN. Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nat Genet. 1997;15:212–215. 10.1038/ng0297-212 [DOI] [PubMed] [Google Scholar]
- 37. Minczuk M, Papworth MA, Miller JC, Murphy MP, Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. 10.1093/nar/gkn313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Comte C, Tonin Y, Heckel-Mager AM, et al. Mitochondrial targeting of recombinant RNAs modulates the level of a heteroplasmic mutation in human mitochondrial DNA associated with Kearns Sayre syndrome. Nucleic Acids Res. 2013;41:418–433. 10.1093/nar/gks965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. 10.1126/science [DOI] [PubMed] [Google Scholar]
- 40. Rorbach J, Yusoff AA, Tuppen H, et al. Overexpression of human mitochondrial valyl tRNA synthetase can partially restore levels of cognate mt-tRNAVal carrying the pathogenic C25U mutation. Nucleic Acids Res. 2008;36:3065–3074. 10.1093/nar/gkn147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dai Y, Zheng K, Clark J, et al. Rapamycin drives selection against a pathogenic heteroplasmic mitochondrial DNA mutation. Hum Mol Genet. 2014;23:637–647. 10.1093/hmg/ddt450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Payne BA, Wilson IJ, Hateley CA, et al. Mitochondrial aging is accelerated by anti-retroviral therapy through the clonal expansion of mtDNA mutations. Nat Genet. 2011;43:806–810. 10.1038/ng.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Durham SE, Samuels DC, Chinnery PF. Is selection required for the accumulation of somatic mitochondrial DNA mutations in post-mitotic cells? Neuromuscul Disord. 2006;16:381–386. http//dx..org/10.1016/j.nmd.2006.03.012 [DOI] [PubMed] [Google Scholar]
- 44. Khrapko K, Nekhaeva E, Kraytsberg Y, Kunz W. Clonal expansions of mitochondrial genomes: implications for in vivo mutational spectra. Mutat Res. 2003;522:13–19. 10.1016/S0027-5107(02)00306-8 [DOI] [PubMed] [Google Scholar]
- 45. Stewart JB, Freyer C, Elson JL, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. 10.1371/journal.pbio.0060010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fan W, Waymire KG, Narula N, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. 10.1126/science.1147786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ross JM, Stewart JB, Hagström E, et al. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013. 10.1038/nature12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68:802–806. http://dx.doi.org/10.1086/318801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ozawa T. Mechanism of somatic mitochondrial DNA mutations associated with age and diseases. Biochim Biophys Acta. 1995;1271:177–189. 10.1016/0925-4439(95)00026-Z [DOI] [PubMed] [Google Scholar]
- 50. Crimi M, Papadimitriou A, Galbiati S, et al. A new mitochondrial DNA mutation in ND3 gene causing severe Leigh syndrome with early lethality. Pediatr Res. 2004;55:842–846. 10.1203/01.PDR.0000117844.73436.68 [DOI] [PubMed] [Google Scholar]
- 51. Lebon S, Chol M, Benit P, et al. Recurrent de novo mitochondrial DNA mutations in respiratory chain deficiency. J Med Genet. 2003;40:896–899. 10.1136/jmg.40.12.896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pyle A, Taylor RW, Durham SE, et al. Depletion of mitochondrial DNA in leucocytes harbouring the 3243A->G mtDNA mutation. J Med Genet. 2007;44:69–74. 10.1136/jmg.2006.043109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chinnery PF, Samuels DC. Relaxed replication of mtDNA: a model with implications for the expression of disease. Am J Hum Genet. 1999;64:1158–1165. 10.1086/302311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Catricala S, Torti M, Ricevuti G. Alzheimer disease and platelets: how’s that relevant. Immun Ageing. 2012;9:20. 10.1186/1742-4933-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.