Abstract
Motivated by differential co-expression analysis in genomics, we consider in this paper estimation and testing of high-dimensional differential correlation matrices. An adaptive thresholding procedure is introduced and theoretical guarantees are given. Minimax rate of convergence is established and the proposed estimator is shown to be adaptively rate-optimal over collections of paired correlation matrices with approximately sparse differences. Simulation results show that the procedure significantly outperforms two other natural methods that are based on separate estimation of the individual correlation matrices. The procedure is also illustrated through an analysis of a breast cancer dataset, which provides evidence at the gene co-expression level that several genes, of which a subset has been previously verified, are associated with the breast cancer. Hypothesis testing on the differential correlation matrices is also considered. A test, which is particularly well suited for testing against sparse alternatives, is introduced. In addition, other related problems, including estimation of a single sparse correlation matrix, estimation of the differential covariance matrices, and estimation of the differential cross-correlation matrices, are also discussed.
Keywords: Adaptive thresholding, covariance matrix, differential co-expression analysis, differential correlation matrix, optimal rate of convergence, sparse correlation matrix, thresholding
1 Introduction
Statistical inference on the correlation structure has a wide array of applications, ranging from gene co-expression network analysis (Carter et al., 2004; Lee et al., 2004; Zhang et al., 2008; Dubois et al., 2010; Fuller et al., 2007) to brain intelligence analysis (Shaw et al., 2006). For example, understanding the correlations between the genes is critical for the construction of the gene co-expression network. See Kostka and Spang (2004), Lai et al. (2004), and Fuller et al. (2007). Driven by these and other applications in genomics, signal processing, empirical finance, and many other fields, making sound inference on the high-dimensional correlation structure is becoming a crucial problem.
In addition to the correlation structure of a single population, the difference between the correlation matrices of two populations is of significant interest. Differential gene expression analysis is widely used in genomics to identify disease-associated genes for complex diseases. Conventional methods mainly focus on the comparisons of the mean expression levels between the disease and control groups. In some cases, clinical disease characteristics such as survival or tumor stage do not have significant associations with gene expression, but there may be significant effects on gene co-expression related to the clinical outcome (Shedden and Taylor (2005); Hudson et al. (2009); Bandyopadhyay et al. (2010)). Recent studies have shown that changes in the correlation networks from different stages of disease or from case and control groups are also of importance in identifying dysfunctional gene expressions in disease. See, for example, de la Fuente (2010). This differential co-expression network analysis has become an important complement to the original differential expression analysis as differential correlations among the genes may reflect the rewiring of genetic networks between two different conditions (See Shedden and Taylor (2005); Bandyopadhyay et al. (2010); de la Fuente (2010); Ideker and Krogan (2012); Fukushima (2013)).
Motivated by these applications, we consider in this paper optimal estimation of the differential correlation matrix. Specifically, suppose we observe two independent sets of p-dimensional i.i.d. random samples with mean μt, covariance matrix Σt, and correlation matrix Rt, where t = 1 and 2. The goal is to estimate the differential correlation matrix D = R1 − R2. A particular focus of the paper is on estimating an approximately sparse differential correlation matrix in the high dimensional setting where the dimension is much larger than the sample sizes, i.e., p ≫ max(n1, n2). The estimation accuracy is evaluated under both the spectral norm loss and the Frobenius norm loss.
A naive approach to estimating the differential correlation matrix D = R1 − R2 is to first estimate the covariance matrices Σ1 and Σ2 separately and then normalize to obtain estimators R̂1 and R̂2 of the individual correlation matrices R1 and R2, and finally take the difference D̂ = R̂1 − R̂2 as the estimator of the differential correlation matrix D. A simple estimate of a correlation matrix is the sample correlation matrix. However, in the high-dimensional setting, the sample correlation matrix is a poor estimate. Significant advances have been made in the last few years on optimal estimation of a high-dimensional covariance matrix. Regularization methods such as banding, tapering, and thresholding have been proposed. In particular, Cai et al. (2010) established the optimal rate of convergence and Cai and Yuan (2012) developed an adaptive estimator of bandable covariance matrices. For sparse covariance matrices where each row and each column has relatively few nonzero entries, Bickel and Levina (2008) introduced a thresholding estimator and obtained rates of convergence; Cai and Liu (2011) proposed an adaptive thresholding procedure and Cai and Zhou (2012) established the minimax rates of convergence for estimating sparse covariance matrices.
Structural assumptions on the individual correlation matrices R1 and R2 are crucial for the good performance of the difference estimator. These assumptions, however, may not hold in practice. For example, gene transcriptional networks often contain the so-called hub nodes where the corresponding gene expressions are correlated with many other gene expressions. See, for example, (Barabási and Oltvai, 2004; Barabási et al., 2011). In such settings, some of the rows and columns of R1 and R2 have many nonzero entries which mean that R1 and R2 are not sparse. In genomic applications, the correlation matrices are rarely bandable as the genes are not ordered in any particular way.
In this paper, we propose a direct estimation method for the differential correlation matrix D = R1 − R2 without first estimating R1 and R2 individually. This direct estimation method assumes that D is approximately sparse, but otherwise does not impose any structural assumptions on the individual correlation matrices R1 and R2. An adaptive thresholding procedure is introduced and analyzed. The estimator can still perform well even when the individual correlation matrices cannot be estimated consistently. For example, direct estimation can recover the differential correlation network accurately even in the presence of hub nodes in R1 and R2 as long as the differential correlation network is approximately sparse. The key is that sparsity is assumed for D and not for R1 or R2.
Theoretical performance guarantees are provided for direct estimator of the differential correlation matrix. Minimax rates of convergence are established for the collections of paired correlation matrices with approximately sparse differences. The proposed estimator is shown to be adaptively rate-optimal. In comparison to adaptive estimation of a single sparse covariance matrix considered in Cai and Liu (2011), both the procedure and the technical analysis of our method are different and more involved. Technically speaking, correlation matrix estimators are harder to analyze than those of covariance matrices and the two-sample setting in our problem further increases the difficulty.
Numerical performance of the proposed estimator is investigated through simulations. The results indicate significant advantage of estimating the differential correlation matrix directly. The estimator outperforms two other natural alternatives that are based on separate estimation of R1 and R2. To further illustrate the merit of the method, we apply the procedure to the analysis of a breast cancer dataset from the study by van de Vijver et al. (2002) and investigate the differential co-expressions among genes in different tumor stages of breast cancer. The adaptive thresholding procedure is applied to analyze the difference in the correlation alternation in different grades of tumor. The study provides evidence at the gene co-expression level that several genes, of which a subset has been previously verified, are associated with the breast cancer.
In addition to optimal estimation of the differential correlation matrix, we also consider hypothesis testing of the differential correlation matrices, H0 : R1−R2 = 0 versus H1 : R1−R2 ≠ 0. We propose a test which is particularly well suited for testing again sparse alternatives. The same ideas and techniques can also be used to treat other related problems. We also consider estimation of a single sparse correlation matrix from one random sample, estimation of the differential covariance matrices as well as estimation of the differential cross-correlation matrices.
The rest of the paper is organized as follows. Section 2 presents in detail the adaptive thresholding procedure for estimating the differential correlation matrix. The theoretical properties of the proposed estimator are analyzed in Section 3. In Section 4, simulation studies are carried out to investigate the numerical performance of the thresholding estimator and Section 5 illustrates the procedure through an analysis of a breast cancer dataset. Hypothesis testing on the differential correlation matrices is discussed in Section 6.1, and other related problems are considered in the rest of Section 6. All the proofs are given in the Appendix.
2 Estimation of Differential Correlation Matrix
We consider in this section estimation of the differential correlation matrix and introduce a data-driven adaptive thresholding estimator. The theoretical and numerical properties of the estimator are investigated in Sections 3 and 4 respectively.
Let be a p-variate random vector with mean μt, covariance matrix Σt = (σijt)1≤i,j≤p, and correlation matrix Rt = (rijt)1≤i,j≤p, for t = 1 and 2. Suppose we observe two i.i.d. random samples, { } from X(1) and { } from X(2), and the two samples are independent. The goal is to estimate the differential correlation matrix D = R1 − R2 under the assumption that D is approximately sparse.
Given the two random samples, the sample covariance matrices and sample correlation matrices are defined as, for t = 1 and 2,
| (1) |
| (2) |
where and diag(Σ̂t) is the diagonal matrix with the same diagonal as Σ̂t. We propose a thresholding estimator of the differential correlation matrix D by individually thresholding the entries of the difference of the two sample correlation matrices R̂1 − R̂2 with the threshold adaptive to the noise level of each entry. A key to the construction of the procedure is the estimation of the noise levels of the individual entries of R̂1 − R̂2, as these entries are random variables themselves.
We first provide some intuition before formally introducing the estimate of the noise levels of the individual entries of R̂1 − R̂2. Note that and . Define
| (3) |
Then one can intuitively write
| (4) |
where zijt is approximately normal with mean 0 and variance 1. Hence, θijt/nt measures the uncertainty of the sample covariance σ̂ijt. Based on the first order Taylor expansion of the 3-variate function for x ∈ ℝ, and y, z > 0,
| (5) |
the entries r̂ijt of the sample correlation matrix R̂t = (r̂ijt) can be approximated by
| (6) |
where we denote
It then follows from (6) that
| (7) |
where the random variables zij1 and zij2 are approximately normal with mean 0 and variance 1, but not necessarily independent for 1 ≤ i, j ≤ p.
Equation (7) suggests that estimation of rij1 − rij2 is similar to the sparse covariance matrix estimation considered in Cai and Liu (2011), where it is proposed to adaptively threshold entries according to their individual noise levels. However, the setting here is more complicated as r̂ij1 − r̂ij2 is not an unbiased estimate of rij1 − rij2 and the noise levels are harder to estimate. These make the technical analysis more involved. The noise levels are unknown here but can be estimated based on the observed data. Specifically, we estimate θijt and ξijt by the following data-driven quantities,
| (8) |
| (9) |
We are now ready to introduce the adaptive thresholding estimator of R1 − R2 using data-driven threshold levels. Let sλ(z) be a thresholding function satisfying the following conditions:
-
(C1)
|sλ(z)| ≤ c|y| for all z, y satisfying |z − y| ≤ λ for some c > 0;
-
(C2)
sλ(z) = 0 for |z| ≤ λ;
-
(C3)
|sλ(z) − z| ≤ λ, for all z ∈ ℝ.
Note that the commonly used soft thresholding function sλ(z) = sgn(z)(z−λ)+ and the adaptive lasso rule sλ = z(1 − |λ/z|η)+ with η ≥ 1 satisfy these three conditions. See Rothman et al. (2009) and Cai and Liu (2011). Although the hard thresholding function sλ(z) = z · 1{|z|≥λ} does not satisfy Condition (C1), the technical arguments given in this paper still work with very minor changes.
We propose to estimate the sparse differential correlation matrix D by the entrywise thresholding estimator defined as
where sλ(z) is a thresholding function satisfying (C1)–(C3) and the threshold level λij is given by λij = λij1 + λij2 with
| (10) |
Here ξ̂ijt are given by (9) and the thresholding constant τ can be chosen empirically through cross-validation. See Section 4.1 for more discussions on the empirical choice of τ.
3 Theoretical Properties
We now analyze the theoretical properties of the data-driven thresholding estimator D̂* proposed in the last section. We will establish the minimax rate of convergence for estimating the differential correlation matrix D over certain classes of paired correlation matrices (R1, R2) with approximately sparse difference D = R1 − R2 under the spectral norm loss. The results show that D̂* is rate-optimal under mild conditions.
3.1 Rate Optimality of the Thresholding Estimator
We consider the following class of paired correlation matrices in ℝp×p with approximately sparse difference
| (11) |
for some 0 ≤ q < 1. Here R1, R2 ⪰ 0 and diag(R1) = diag(R2) = 1 mean that R1 and R2 are symmetric, semi-positive definite, and with all diagonal entries 1. For (R1, R2) ∈
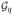 (s0(p)), their difference R1 − R2 is approximately sparse in the sense that each row vector of R1 − R2 lies in the ℓq ball with radius s0(p) and 0 ≤ q < 1. When q = 0, this constraint becomes the commonly used exact sparsity condition.
(s0(p)), their difference R1 − R2 is approximately sparse in the sense that each row vector of R1 − R2 lies in the ℓq ball with radius s0(p) and 0 ≤ q < 1. When q = 0, this constraint becomes the commonly used exact sparsity condition.
Let
We assume that for each i, Yi is sub-Gaussian distributed, i.e. there exist constants K, η > 0 such that for all 1 ≤ i ≤ p and t = 1, 2,
| (12) |
In addition, we assume for some constant ν0 > 0
| (13) |
The following theorem provides an upper bound for the risk of the thresholding estimator D̂* under the spectral norm loss.
Theorem 3.1 (Upper bound)
Suppose log p = o(min(n1, n2)1/3) and (12) and (13) hold. Suppose the thresholding function sλ(z) satisfy Conditions (C1)–(C3). Then the thresholding estimator D̂* defined in (2) and (10) with τ > 4 satisfies
| (14) |
| (15) |
| (16) |
for some constant C > 0 that does not depend on n1, n2 or p.
Remark 3.1
Condition (13) holds naturally when X(t) are jointly Gaussian. To see this point, we suppose ρijt is the correlation between and . Then one can write , where , W are independently standard Gaussian. It is easy to calculate that , which implies (13) holds for ν0 = 1. Condition (13) is used in Lemma 6.1 to show that θ̂ijt is a good estimate of θijt and |σ̂ijt − σijt| can be controlled by C(θ̂ijt log p/nt)1/2 with high probability.
Theorem 3.1 gives the rate of convergence for the thresholding estimator D̂*. The following result provides the lower bound for the minimax risk of estimating the differential correlation matrix D = R1 − R2 with (R1, R2) ∈
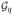 (s0(p)).
(s0(p)).
Theorem 3.2 (Lower Bound)
Suppose log p = o(min(n1, n2)) and s0(p) ≤ M min(n1, n2)(1−q)/2 × (log p)−(3−q)/2 for some constant M > 0. Then minimax risk for estimating D = R1 − R2 satisfies
| (17) |
| (18) |
| (19) |
for some constant c > 0.
Theorems 3.1 and 3.2 together yield the minimax rate of convergence
for estimating D = R1 − R2 with (R1, R2) ∈
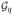 (s0(p)) under the spectral norm loss, and show that the thresholding estimator D̂* defined in (2) and (10) is adaptively rate-optimal.
(s0(p)) under the spectral norm loss, and show that the thresholding estimator D̂* defined in (2) and (10) is adaptively rate-optimal.
Remark 3.2
The technical analysis here for the different of two correlation matrices is more complicated in comparison to the problem of estimating a sparse covariance matrix considered in Cai and Liu (2011). It can be seen in (7), i.e. the “signal + noise” expression of r̂ij1 − r̂ij2, the difference of the sample correlation matrices has six “noise terms”. It is necessary to deal with all these six terms in the theoretical analysis of Theorem 3.1.
4 Numerical Studies
We investigate in this section the numerical performance of the adaptive thresholding estimator of the differential correlation matrix through simulations. The method is applied to the analysis of a breast cancer dataset in the next section.
In the previous sections, we proposed the entrywise thresholding method for estimating R1 − R2 and then studied the theoretical properties of D̂* with a fixed τ > 4. However, the theoretical choice of τ may not be optimal in finite sample performance, as we can see in the following example. Let R1 and R2 be 200 × 200-dimensional matrices such that R1,ij = (−1)|i−j|×max(1−|i−j|/10, 0)×(1{i=j}+fifj1{i≠j}) and R2,ij = max(1−|i−j|/10, 0)×(1{i=j}+fifj1{i≠j}). Here 1{·} is the indicator function, f1, ···, f200 are i.i.d. random variables that are uniformly distributed on [0, 1]. In this setting, both R1 and R2 are sparse, but their difference is even more sparse. We set Σt = Rt and generate 200 independent samples from X(1) ~ N(0, Σ1) and 200 independent samples from X(2) ~ N(0, Σ2). For various values of τ ∈ [0, 5], we implement the proposed method with hard thresholding and repeat the experiments for 100 times. The average loss in spectral, ℓ1 and Frobenious norms are shown in Figure 1. Obviously in this example, τ > 4 is not the best choice.
Figure 1.
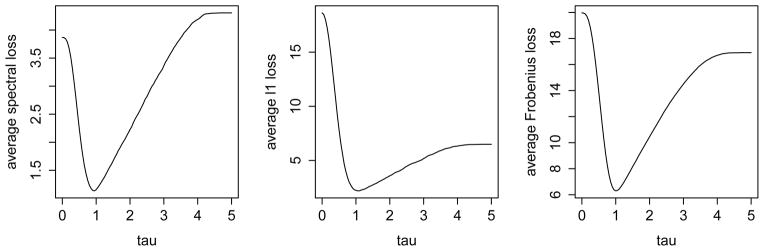
Average (Spectral, ℓ1, Frobenious) norm losses for τ ∈ [0, 5]. p = 100, n1 = n2 = 50.
Empirically, we find that the numerical performance of the estimator can often be improved by using a data-driven choice of τ based on cross-validation. We thus begin by introducing the following K-fold cross-validation method for the empirical selection of τ.
4.1 Empirical Choice of τ
For an integer K ≥ 2, we first divide both samples and randomly into two groups for H times as and . Here h = 1, …, H represents the h-th division. For t = 1 and 2, the size of the first group is approximately (K − 1)/K · nt and the size of the second group is approximately nt/K. We then calculate the corresponding sample correlation matrices as and for all four sub-samples. Partition the interval [0, 5] into an equi-spaced grid {0, }. For each value of τ ∈ {0, }, we obtain the thresholding estimator defined in (2) and (10) with the thresholding constant τ based on the subsamples and . Denote the average loss for each τ for the second sub-samples and as
We select
as our empirical choice of the thresholding constant τ, and calculate the final estimator D̂*(τ̂) with the thresholding constant τ̂ based on the whole samples X(1) and X(2).
4.2 Estimation of Differential Correlation Matrix
The adaptive thresholding estimator is easy to implement. We consider the following two models under which the differential correlation matrix is sparse.
-
Model 1 (Random Sparse Difference) R1 and R2 are p-dimensional symmetric positive definite matrices such that is a fixed matrix, where with B1,ij = 1 if i = j and B1,ij = 0·2 if i ≠ j, is the identity matrix, and R2 is randomly generated as , where with
and λ is a constant that ensures the positive definiteness of R2.
Model 2 (Banded Difference) In this setting, p-dimensional matrices R1 and R2 satisfy R1,ij =0·2×1{i=j}+0·8×(−1)|i−j| × max(1 − |i − j|/10, 0) and R2,ij = R1,ij+0·2×1{i≠j} × max(1 − |i − j|/3, 0). Here 1{·} is the indicator function.
In each of the two settings, we set Σt = diag(|ωt|1/2)Rtdiag(|ωt|1/2) for both t = 1, 2, where ω1, ω2 ∈ ℝp are two i.i.d. samples from N(0, Ip). These operations make the covariance matrices Σ1 and Σ2 have different values along the diagonals.
We generate i.i.d. samples from X(1) ~ N(0, Σ1) and X(2) ~ N(0, Σ2) for various values of p, n1, and n2 and then apply the proposed algorithm with 5-fold cross-validation for the selection of the thresholding constant τ. For each setting, both the hard thresholding and adaptive-Lasso thresholding (Rothman et al. (2009)),
| (20) |
are used. For comparison, we also implement three natural estimators of D.
-
The covariance matrices Σ1 and Σ2 are estimated individually by the adaptive thresholding method proposed in Cai and Liu (2011) with 5-fold cross-validation and then and are normalized to yield estimators of R1 and R2,
and finally D = R1 − R2 is estimated by the difference .
The correlation matrices and are estimated separately using the method proposed in Section 6.2 and then take the difference.
D is estimated directly the difference of the sample correlation matrices R̂1 − R̂2.
The numerical results are summarized in Tables 1 and 4.2 for the two models respectively. In each case, we compare the performance of the three estimators D*, and R̂1 − R̂2 under the spectral norm, matrix ℓ1 norm, and Frobenius norm losses. For both models, it is easy to see that the direct thresholding estimator D* significantly outperforms and R̂1 − R̂2. Under Model 1, the individual correlation matrices R1 and R2 are “dense” in the sense that half of the rows and columns contain many non zeros entries, but their difference D is sparse. In this case, R1 and R2 cannot be estimated consistently and the two difference estimators and R̂1 − R̂2 based on the individual estimators of R1 and R2 perform very poorly, while the direct estimator D* performs very well. Moreover, the numerical performance of the thresholding estimators does not depend on the specific thresholding rules in a significant way. Different thresholding rules including hard thresholding and adaptive Lasso behave similarly.
Table 1.
Comparison of D̂* with and R̂1 − R̂2 under Model 1.
| p | n1 | n2 | Hard
|
Adaptive Lasso
|
Sample
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| D̂* |
|
|
D̂* |
|
|
R̂1 − R̂2 | ||||
| Spectral Norm | ||||||||||
| 100 | 50 | 50 | 0·50(0·41) | 1·75(1·37) | 6·94(1·07) | 0·33(0·31) | 1·51(1·75) | 6·17(0·98) | 7·28(0·93) | |
| 100 | 100 | 100 | 0·34(0·21) | 3·79(3·17) | 4·74(0·55) | 0·28(0·23) | 3·53(2·71) | 4·49(0·71) | 5·02(0·65) | |
| 100 | 200 | 200 | 0·29(0·19) | 4·22(2·14) | 3·23(0·47) | 0·24(0·13) | 4·52(1·86) | 3·14(0·54) | 3·55(0·47) | |
| 100 | 500 | 500 | 0·24(0·10) | 1·72(0·35) | 2·07(0·32) | 0·22(0·08) | 1·82(0·35) | 1·87(0·26) | 2·23(0·25) | |
| 500 | 50 | 50 | 0·56(0·77) | 3·02(2·76) | 31·88(4·04) | 0·40(0·65) | 3·47(4·63) | 29·15(4·26) | 34·66(3·84) | |
| 500 | 100 | 100 | 0·41(0·48) | 8·09(11·02) | 23·38(4·52) | 0·34(0·39) | 12·99(13·26) | 21·82(3·23) | 24·19(2·77) | |
| 500 | 200 | 200 | 0·32(0·40) | 22·22(13·06) | 15·67(3·30) | 0·26(0·34) | 21·31(9·29) | 14·61(2·24) | 16·50(1·97) | |
| 500 | 500 | 500 | 0·20(0·19) | 7·80(1·37) | 7·80(1·29) | 0·18(0·14) | 8·21(1·69) | 8·70(1·39) | 10·46(1·31) | |
|
| ||||||||||
| Matrix ℓ1 Norm | ||||||||||
| 100 | 50 | 50 | 0·89(0·68) | 3·63(2·97) | 18·91(1·55) | 0·78(0·80) | 3·14(3·20) | 16·88(1·42) | 21·33(1·61) | |
| 100 | 100 | 100 | 0·64(0·25) | 7·34(4·85) | 13·42(1·08) | 0·70(0·87) | 7·03(4·14) | 12·72(1·18) | 14·97(1·14) | |
| 100 | 200 | 200 | 0·64(0·34) | 9·63(1·85) | 9·22(0·75) | 0·61(0·37) | 9·37(1·77) | 8·67(0·87) | 10·60(0·81) | |
| 100 | 500 | 500 | 0·58(0·22) | 4·54(0·56) | 5·92(0·54) | 0·56(0·21) | 4·85(0·61) | 5·33(0·45) | 6·69(0·44) | |
| 500 | 50 | 50 | 1·69(3·09) | 7·85(8·14) | 97·28(6·64) | 1·37(2·87) | 9·49(11·93) | 87·02(7·31) | 112·40(8·97) | |
| 500 | 100 | 100 | 1·06(1·28) | 20·12(19·50) | 64·98(5·93) | 1·17(1·47) | 27·95(22·75) | 65·60(5·07) | 79·66(5·37) | |
| 500 | 200 | 200 | 0·97(1·48) | 51·64(10·13) | 46·74(5·01) | 0·95(1·24) | 47·87(8·50) | 45·54(3·70) | 55·77(3·70) | |
| 500 | 500 | 500 | 0·68(0·54) | 23·19(2·86) | 23·47(2·56) | 0·69(0·52) | 24·67(2·92) | 27·21(2·09) | 35·32(2·04) | |
|
| ||||||||||
| Frobenious Norm | ||||||||||
| 100 | 50 | 50 | 1·40(1·32) | 4·34(2·51) | 19·01(0·37) | 1·06(1·04) | 3·26(2·61) | 16·60(0·42) | 19·87(0·38) | |
| 100 | 100 | 100 | 0·96(0·59) | 7·14(3·67) | 13·38(0·23) | 0·94(0·81) | 6·23(3·31) | 12·10(0·27) | 14·05(0·25) | |
| 100 | 200 | 200 | 0·89(0·57) | 9·09(1·03) | 9·30(0·20) | 0·84(0·50) | 8·15(1·07) | 8·42(0·25) | 9·94(0·18) | |
| 100 | 500 | 500 | 0·85(0·32) | 4·37(0·27) | 5·92(0·11) | 0·82(0·30) | 4·43(0·25) | 5·26(0·11) | 6·39(0·10) | |
| 500 | 50 | 50 | 3·33(5·63) | 11·18(7·71) | 95·05(0·91) | 2·27(3·92) | 9·57(9·31) | 83·40(1·24) | 99·97(0·93) | |
| 500 | 100 | 100 | 2·18(2·98) | 20·17(14·55) | 61·37(2·09) | 2·10(2·47) | 22·54(16·69) | 60·58(0·85) | 70·40(0·67) | |
| 500 | 200 | 200 | 1·77(2·39) | 45·06(5·89) | 42·11(1·67) | 1·63(1·96) | 39·52(5·14) | 41·88(0·73) | 49·53(0·49) | |
| 500 | 500 | 500 | 1·27(1·09) | 20·17(0·99) | 21·74(0·65) | 1·22(0·84) | 20·48(0·91) | 25·47(0·41) | 31·34(0·33) | |
5 Analysis of A Breast Cancer Dataset
Identifying gene expression networks can be helpful for conducting more effective treatment based to the condition of patients. de la Fuente (2010) demonstrated that the gene expression networks can vary in different disease states and the differential correlations in gene expression (i.e. co-expression) are useful in disease studies.
In this section, we consider the dataset “70pathwaygenes-by-grade” from the study by van de Vijver et al. (2002) and investigate the differential co-expressions among genes in different tumor stages of breast cancer. In this dataset, there are 295 records of patients with 1624 gene expressions, which are categorized into three groups based on the histological grades of tumor (“Good”, “Intermediate” and “Poor”) with 74, 101 and 119 records, respectively. We denote these three groups of samples as X(1), X(2) and X(3). In order to analyze the difference in the correlation alternation in different grades of tumor, we apply our adaptive thresholding method with cross-validation to estimate the differential correlation matrices among those gene expressions from different stages.
The number of gene pairs with significant difference in correlation are listed in Table 3. The results show that the correlation structures between the “Good” and “Intermediate” groups are similar and there is some significant changes between the “Good” and “Poor” group.
Table 3.
The number of gene pairs that have significant differential correlation betweens two groups of different tumor grades
| Good v.s. Intermediate | Intermediate v.s. Poor | Good v.s. Poor | |
|---|---|---|---|
| # of selected pairs | 0 | 2 | 152 |
More interestingly, by combining the “Good” and “Intermediate” sub-samples and comparing with the “Poor” group, we find significant differences between their correlation structure. There are 4526 pairs of genes that have significantly different correlations between the “Good + Intermediate” and “Poor” groups. For each given gene, we count the number of the genes whose correlation with this gene is significantly different between these two groups, and rank all the genes by the counts. That is, we rank the genes by the size of the support of D̂* in each row. The top ten genes are listed in Table 4.
Table 4.
The top ten genes that appear for most times in the selected pairs in “Good + Intermediate” v.s. “Poor”
| Gene | number of pairs |
|---|---|
| growth differentiation factor 5 (GDF5) | 67 |
| transcription factor 7-like 1 (TCF7L1) | 64 |
| 3′-phosphoadenosine 5′-phosphosulfate synthase 1 (PAPSS1) | 51 |
| secreted frizzled-related protein 1(SFRP1) | 43 |
| gamma-aminobutyric acid A receptor, pi (GABRP) | 41 |
| mannosidase, alpha, class 2B, member 2 (MAN2B2) | 37 |
| desmocollin 2 (DSC2) | 36 |
| transforming growth factor, beta 3 (TGFB3) | 35 |
| CRADD | 35 |
| ELOVL fatty acid elongase 5(ELOVL5) | 32 |
Among these ten genes, six of them, GDF5, TCF7L1, PAPSS1, SFRP1, GABRP, TGFB1, have been previously studied and verified in the literature that are associated with the breast cancer (See Margheri et al. (2012), Shy et al. (2013), Xu et al. (2012), Klopocki et al. (2004), Zafrakas et al. (2006), and Ghellal et al. (2000), respectively). Take for example GDF5 and TCF7L1, the overproduction of Transforming growth factor beta-1 (TGFβ), a multifunctional cytokine, is an important characteristic of late tumor progression. Based on the study by Margheri et al. (2012), TGFβ produced by breast cancer cells brings about in endothelial cells expression of GDF5. The findings in (Shy et al. (2013)) suggested the important role played by TCF7L1 in breast cancer. Although these biological studies mainly focus on the the behavior of the single gene expression, our study provides evidence in the gene co-expression level that these gene expressions are related with the breast cancer.
We should point out that the two well-known genes related to the breast cancer, BRCA1 and BRCA2, were not detected by our method. This is mainly due to the fact that our method focus on the differential gene co-expressions, not the changes in the gene expression levels.
6 Other Related Problems
We have so far focused on optimal estimation of the differential correlation matrix. In addition to optimal estimation, hypothesis testing of the differential correlation matrix is also an important problem. In this section we consider testing the hypotheses H0 : R1 − R2 = 0 versus H1 : R1 − R2 ≠ 0 and propose a test which is particularly well suited for testing again sparse alternatives.
Similar ideas and techniques can also be used to treat several other related problems, including estimation of a single sparse correlation matrix from one random sample, estimation of the differential covariance matrices, and estimation of the differential cross-correlation matrices. We also briefly discuss these problems in this section.
6.1 Testing Differential Correlation Matrices
Suppose we are given two sets of independent and identical distributed samples with the mean μt, covariance matrix Σt and correlation matrix Rt, where t = 1 and 2, and wish to test the hypotheses
| (21) |
This testing problem is similar to, but also different from, testing the equality of two high-dimensional covariance matrices, which has been considered in several recent papers. See, for example, Schott (2007), Srivastava and Yanagihara (2010), Li et al. (2012), and Cai et al. (2013). Here we are particularly interested in testing against sparse alternatives and follow similar ideas as those in Cai et al. (2013).
To construct the test statistic, we need more precise understanding of the sample correlation coefficients r̂ijt. It follows from (5) that
Since , we introduce
Then asymptotically as n, p → ∞,
The true value of ηijt is unknown but can be estimated by
We define the test statistic by
where
Under regularity conditions (similar to (C1)–(C3) in Cai et al. (2013)), the asymptotic distribution of Tn can be shown to be the type I extreme value distribution. More precisely,
| (22) |
for any given t ∈ ℝ.
The asymptotic null distribution (22) can then be used to construct a test for testing the hypothesis H0 : R1 − R2 = 0. For a given significance level 0 < α < 1, define the test Ψα by
| (23) |
where τα = −log(8π) − 2 log log(1 − α)−1 is the 1 − α quantile of the type I extreme value distribution with the cumulative distribution function exp(−(8π)−1/2 exp(−x/2)). The hypothesis H0 : R1 − R2 = 0 is rejected whenever Ψα = 1. As the test proposed in Cai et al. (2013) for testing the equality of two covariance matrices, the test Ψα defined in (23) can also be shown to be particularly well suited for testing H0 : R1 − R2 = 0 against sparse alternatives.
6.2 Optimal Estimation of a Sparse Correlation Matrix
The ideas and technical tools can also be used for estimation of a single correlation matrix from one random sample, which is a simpler problem. Suppose we observe an independent and identical distributed sample X = (X1, …, Xn) from a p-dimensional distribution with mean μ ∈ ℝp, covariance matrix Σ, and correlation matrix R ∈ ℝp×p. When R is approximately sparse, it can be naturally estimated by a thresholding estimator R̂ as follows. Let . Define the sample covariance matrix Σ̂ = (σ̂ij)1≤i,j≤p and the sample correlation matrix R̂ = (r̂ij)1≤i,j≤p respectively by
Same as in (8) and (9), we define
| (24) |
| (25) |
| (26) |
where τ is the thresholding constant that can be chosen empirically through cross-validation. The correlation matrix R is then estimated by with
We consider the following class of approximately sparse correlation matrices
The following theoretical results for R̂* can be established using a similar analysis.
Proposition 6.1
Suppose log p = o(n1/3) and X satisfies (12), (13). For τ > 6, there exists some constant C does not depend on n or p such that
| (27) |
| (28) |
| (29) |
Moreover, when log p = o(n), s0(p) ≤ Mn(1−q)/2(log p)−(3−q)/2 for some constant M > 0, the rate in (27) is optimal as we also have the lower bound
| (30) |
| (31) |
| (32) |
Remark 6.1
Cai and Liu (2011) proposed an adaptive thresholding estimator Σ̂* of a sparse covariance matrix Σ. This estimator leads naturally to an estimator R̃ = (r̃ij) of a sparse correlation matrix R by normalizing via . The correlation matrix estimator R̃ has similar properties as the estimator introduced above. For example, R̃ and R̂* achieve the same rate of convergence.
6.3 Optimal Estimation of Sparse Differential Covariance Matrices
Our analysis can also be used for estimation of sparse differential covariance matrices, Δ = Σ1 − Σ2. Define θijt as in (3) and its estimate θ̂ijt as in (8). Similar to the estimation of the differential correlation matrix D = R1 − R2, we estimate Δ = Σ1 − Σ2 by adaptive entrywise thresholding. Specifically, we define the thresholding estimator by
| (33) |
where γij is the thresholding level given by
| (34) |
Same as in the last section, here sλ(z) belongs to the class of thresholding functions satisfying Conditions (C1)–(C3) and the thresholding constant τ can be taken chosen empirically by cross-validation.
We consider the following class of paired covariance matrices with approximately sparse differences, for 0 ≤ q < 1,
| (35) |
Under the same conditions as those in Theorems 3.1 and 3.2, a similar analysis can be used to derive the minimax upper and lower bounds. It can be shown that the estimator Δ̂* given in (33) with τ > 4 satisfies
| (36) |
for some constant C > 0. Furthermore, the following minimax lower bound holds,
| (37) |
for some constant c > 0. Equations (36) and (37) together show that the thresholding estimator Δ̂* defined in (33) and (34) is rate-optimal.
6.4 Estimate Differential Cross-Correlation Matrices
In many applications such as phenome-wide association studies (PheWAS) which aims to study the relationship between a set of genomic markers X and a range of phenotypes Y, the main focus is on the cross-correlations between the components of X and those of Y. That is, the object of interest is a submatrix of the correlation matrix of the random vector . More specifically, let X = (X1, …, Xp1)′ be a p1-dimensional random vector and Y = (Y1, …, Yp2)′ be a p2-dimensional random vector. In the PheWAS setting, X may be all phenotypic disease conditions of interest and Y is a vector of genomic markers.
Suppose we have two independent and identical distributed samples of the (X, Y) pairs, one for the case group and one for the control group,
Here for , k = 1, …, n1 are independent and identical distributed samples generated from some distribution with mean μt, covariance matrix Σt and correlation matrix Rt given by
In applications such as PheWAS, it is of special interest to estimate the differential cross-correlation matrix of X and Y, i.e. DXY = RXY1 − RXY2 ∈ ℝp1×p2. Again, we introduce the following set of paired correlation matrices with sparse cross-correlations,
The thresholding procedure proposed in Section 2 can be applied to estimate DXY,
| (38) |
where R̂XY is sample cross-correlation matrix of X and Y; λij is given by (10). Similar to Theorem 3.1, the following theoretical results hold for the estimator .
Proposition 6.2
Suppose p = p1 + p2, log(p) = o(min(n1, n2)1/3) and (12) and (13) hold. Suppose the thresholding function sλ(z) satisfies Conditions (C1)–(C3). Then D̂* defined in (38) with the thresholding constant τ > 4 satisfies
| (39) |
| (40) |
| (41) |
for some constant C > 0 that does not depend on n1, n2 or p.
The proof of Proposition 6.2 is similar to that of Theorem 3.1 by analyzing the block D̂XY − (RXY1 − RXY2) instead of the whole matrix D* − (R1 − R2). We omit the detailed proof here.
Table 2.
Comparison of D̂* with and R̂1 − R̂2 under Model 2.
| p | n1 | n2 | Hard
|
Adaptive Lasso
|
Sample
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| D̂* |
|
|
D̂* |
|
|
R̂1 − R̂2 | ||||
| Spectral Norm | ||||||||||
| 100 | 50 | 50 | 0·98(1·00) | 4·61(1·49) | 7·25(0·87) | 0·71(0·70) | 4·47(1·44) | 6·05(0·74) | 8·29(0·98) | |
| 100 | 100 | 100 | 0·70(0·51) | 2·88(0·81) | 5·01(0·57) | 0·62(0·47) | 2·93(0·87) | 4·25(0·52) | 5·83(0·59) | |
| 100 | 200 | 200 | 0·60(0·35) | 1·93(0·55) | 3·53(0·42) | 0·48(0·24) | 1·98(0·57) | 2·98(0·37) | 4·07(0·47) | |
| 100 | 500 | 500 | 0·47(0·14) | 1·23(0·27) | 2·32(0·23) | 0·46(0·17) | 1·30(0·36) | 1·98(0·21) | 2·66(0·27) | |
| 500 | 50 | 50 | 0·97(0·99) | 5·03(1·00) | 20·61(1·07) | 0·80(0·75) | 4·55(0·96) | 16·91(0·90) | 24·96(1·16) | |
| 500 | 100 | 100 | 0·79(0·62) | 3·17(0·49) | 13·64(0·59) | 0·59(0·41) | 3·14(0·63) | 11·13(0·53) | 16·39(0·71) | |
| 500 | 200 | 200 | 0·60(0·36) | 2·13(0·30) | 9·12(0·42) | 0·51(0·31) | 2·11(0·35) | 7·44(0·37) | 10·94(0·50) | |
| 500 | 500 | 500 | 0·51(0·20) | 1·34(0·16) | 5·63(0·23) | 0·49(0·20) | 1·35(0·22) | 4·65(0·21) | 6·78(0·29) | |
|
| ||||||||||
| Matrix ℓ1 Norm | ||||||||||
| 100 | 50 | 50 | 1·84(2·66) | 10·61(3·48) | 19·11(1·55) | 1·26(1·86) | 9·88(3·14) | 16·18(1·55) | 21·92(1·61) | |
| 100 | 100 | 100 | 1·18(1·44) | 6·73(2·24) | 13·62(1·08) | 1·10(1·26) | 6·73(2·12) | 11·73(1·20) | 15·87(1·11) | |
| 100 | 200 | 200 | 0·98(0·98) | 4·53(1·47) | 9·79(0·85) | 0·71(0·71) | 4·68(1·46) | 8·39(0·89) | 11·37(0·97) | |
| 100 | 500 | 500 | 0·67(0·48) | 2·95(0·89) | 6·44(0·56) | 0·65(0·59) | 3·08(1·03) | 5·58(0·47) | 7·47(0·53) | |
| 500 | 50 | 50 | 1·79(2·65) | 11·03(2·80) | 79·71(2·64) | 1·64(2·26) | 10·38(3·02) | 64·46(2·73) | 97·88(2·55) | |
| 500 | 100 | 100 | 1·45(1·75) | 7·66(1·79) | 56·52(2·16) | 1·02(1·35) | 7·73(2·40) | 45·65(1·86) | 69·42(1·76) | |
| 500 | 200 | 200 | 1·02(1·18) | 4·97(1·09) | 39·86(1·39) | 0·83(1·14) | 5·03(1·27) | 31·90(1·15) | 49·11(1·33) | |
| 500 | 500 | 500 | 0·81(0·70) | 3·15(0·72) | 25·34(0·77) | 0·82(0·77) | 3·27(1·00) | 20·39(0·77) | 31·36(0·79) | |
|
| ||||||||||
| Frobenious Norm | ||||||||||
| 100 | 50 | 50 | 3·36(2·53) | 13·82(1·83) | 18·46(0·81) | 2·66(1·47) | 12·13(2·00) | 15·87(0·79) | 19·92(0·94) | |
| 100 | 100 | 100 | 2·67(1·19) | 9·46(1·28) | 13·26(0·55) | 2·54(1·10) | 8·77(1·18) | 11·51(0·54) | 14·32(0·58) | |
| 100 | 200 | 200 | 2·43(0·69) | 6·94(0·72) | 9·75(0·39) | 2·26(0·51) | 6·68(0·76) | 8·59(0·37) | 10·49(0·43) | |
| 100 | 500 | 500 | 2·24(0·34) | 5·29(0·36) | 6·96(0·19) | 2·25(0·46) | 5·28(0·44) | 6·33(0·17) | 7·39(0·19) | |
| 500 | 50 | 50 | 6·77(4·86) | 34·24(3·33) | 91·09(0·85) | 6·18(3·86) | 27·39(3·39) | 75·97(0·83) | 100·71(0·92) | |
| 500 | 100 | 500 | 6·19(2·98) | 22·76(1·92) | 64·37(0·56) | 5·30(1·79) | 20·12(2·21) | 53·72(0·56) | 71·23(0·58) | |
| 500 | 200 | 200 | 5·32(1·49) | 16·34(1·18) | 45·79(0·44) | 5·10(1·36) | 15·01(1·15) | 38·36(0·42) | 50·61(0·43) | |
| 500 | 500 | 500 | 5·00(0·62) | 12·14(0·59) | 29·77(0·27) | 4·99(0·69) | 11·76(0·70) | 25·27(0·24) | 32·80(0·25) | |
Appendix: Proofs
We prove the main theorems in the Appendix. Throughout the Appendix, we denote by C a constant which does not depend on p, n1 and n2, and may vary from place to place.
Proof of Theorem 3.1
To prove this theorem, we consider the following three events separately,
| (42) |
| (43) |
| (44) |
Here ε is the fixed constant which satisfies 0 < ε < ν0/2 where ν0 is introduce in (13); C1 and C3 are constants which do not depends on p, n1, n2 and shall be specified later in Lemma 6.1.
1. First we would like to show that under the event A1,
| (45) |
| (46) |
| (47) |
In fact,
for all 1 ≤ i ≤ p, so
| (48) |
So by the definition of A1, we have
| (49) |
| (50) |
Hence,
| (51) |
| (52) |
Suppose x = σiit/σ̂iit, y = σjjt/σ̂jjt. By (51) and , we have when nt is large enough. Thus for large nt, we obtain
| (53) |
It then follows from the assumption that for large nt,
| (54) |
and
We shall note the difference between and above. Next, we rearrange the inequality above and write it into an inequality for |r̂ijt − rijt|,
| (55) |
(55) implies
| (56) |
Next, by (56) and (C1) and (C3) of sλ(z),
| (57) |
| (58) |
which implies
| (59) |
| (60) |
where 0 ≤ q < 1. Hence,
which yields to (46). (45) also holds due to the fact that ||A||2 ≤ ||A||L1 for any symmetric matrix A. Similarly,
which implies (47).
2. For A2, we wish to prove,
| (61) |
| (62) |
| (63) |
In order to prove these probability bounds, we introduce the following lemma, which revealed the relationship between θ̂ijt, θijt and σ̂ijt, σijt.
Lemma 6.1
For any τ > 0,
| (64) |
There exist constants C1, C2, C3 which do not depend on p, n1, n2 such that
| (65) |
For any ε > 0 and M > 0,
| (66) |
The proof of Lemma 6.1 is given later. Note that (64) immediately leads to
| (67) |
By the definition of A2 (43), we still have (49). Besides, by the definition of A2, , which leads to σ̂iit ≥ 0.5σiit. Thus,
| (68) |
For convenience, we denote the random variable
| (69) |
Under A2, we have T ≤ 0.5. Then for all 1 ≤ i, j ≤ p, t = 1, 2,
Similarly calculation also leads to r̂ijt − rijt ≥ −4T. Then, by (C3) of sλij (z),
| (70) |
In addition, due to ||·||ℓ1 ≥ ||·||, we also have . Similarly,
| (71) |
Therefore,
| (72) |
Similarly, we have
which finishes the proof of (61), (62) and (63) when we choose M > τ /4 − 1.
3. For A3, (66) and log p = o(n1/3) leads to
| (73) |
Besides, since rijt, r̂ijt are the population and sample correlations, |rijt| ≤ 1, |r̂ijt| ≤ 1. By (C1) of thresholding sλ(z), we have |sλ(x) − x| ≤ c|x| for all x ∈ ℝ. Thus,
which yields
| (74) |
Similarly, ||D̂* − (R1 − R2)||2 ≤ (2 + 2c)2p2, . Therefore,
| (75) |
| (76) |
| (77) |
Finally, we combine the situations of A1, A2 and A3. When τ > 4 and M > 2, we have
| (78) |
which has proved (14). (15) and (16) can be proved similarly by (46), (62), (76) and (47), (63), (77).
Proof of Lemma 6.1
(64) is directly from (25) in Cai and Liu (2011). For (66), the proof is essentially the same as the proof of (26) in Cai and Liu (2011) as long as we use x = ((M + 2) log p + log n)1/2 in stead of x = ((M + 2) log p)1/2 in their proof. Now we mainly focus on the proof of (65). Without loss of generality, we can translate X and assume that μ1 = μ2 = 0. Note that we have the following formulation,
| (79) |
Since
where C4 is a constant which does not depend on n1, n2, p. Thus, we set ; based on lemma 1 in Cai and Liu (2011), we have
| (80) |
for all , where Cη/2 = η/2 + 2/η. Next for , we similarly apply Lemma 1 in Cai and Liu (2011) and get
| (81) |
for all . Combining (80) and (81),
| (82) |
Proof of Theorem 3.2
Without loss of generality, we assume n1 ≤ n2. For (R1,
R2) ∈
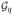 (s0(p)), set Σ2 = R2 = Ip×p and we have already known this information. The estimation of sparse difference immediately becomes the estimation of the sparse correlation matrix R1. Then the lower bound result for estimating single sparse covariance matrix can be used to prove this theorem.
(s0(p)), set Σ2 = R2 = Ip×p and we have already known this information. The estimation of sparse difference immediately becomes the estimation of the sparse correlation matrix R1. Then the lower bound result for estimating single sparse covariance matrix can be used to prove this theorem.
We follow the idea of Cai and Zhou (2012) and define the set of diagonal-1 covariance matrices as
We have {(R1,
I) : R1 ∈
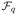 (s0(p))} ⊆
(s0(p))} ⊆
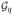 (s0(p)). Besides, the proof of Theorem 2 in Cai and Zhou (2012) shows that
(s0(p)). Besides, the proof of Theorem 2 in Cai and Zhou (2012) shows that
| (83) |
Since the correlation matrix equals to covariance matrix (i.e. R = Σ) when diag(Σ) = 1, then
| (84) |
which implies (17). By ||·||ℓ1 ≥ ||·|| for symmetric matrices, (18) also follow immediately.
Similarly, (19) follows from Theorem 4 of Cai and Zhou (2012).
Proof of Proposition 6.1
The proof of Proposition 6.1 is similar to Theorem 3.1. For the upper bound, again, we split the whole events into three,
| (85) |
| (86) |
| (87) |
Here ε is the fixed constant which satisfies 0 < ε < ν0/2 where ν0 was introduced in (13); C1, C3 are constants specified in Lemma 6.1. Similarly to the proof of Theorem 3.1, we can prove the following statements.
- Under A1,
- For A2,
- For A3,
The rest of proof, including the lower bound results, are omitted here as they are essentially the same as Theorem 3.1.
Footnotes
The research was supported in part by NSF Grant DMS-1208982 and NIH Grant R01 CA127334.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, Fiedler D, Dutkowski J, Guénolé A, van Attikum H, Shokat KM, Kolodner RD, Huh WK, Aebersold R, Keogh MC, Krogan NJ, Ideker T. Rewiring of genetic networks in response to DNA damage. Sci Signal. 2010;330(6009):1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Bickel P, Levina E. Covariance regularization by thresholding. The Annals of Statistics. 2008;26:879–921. [Google Scholar]
- Cai TT, Liu W. Adaptive thresholding for sparse covariance matrix estimation. Journal of American Statistical Association. 2011;106:672–684. [Google Scholar]
- Cai TT, Liu W, Xia Y. Two-sample covariance matrix testing and support recovery in high-dimensional and sparse settings. Journal of American Statistical Association. 2013;108:265–277. [Google Scholar]
- Cai TT, Yuan M. Adaptive covariance matrix estimation through block thresholding. The Annals of Statistics. 2012;40:2014–2042. [Google Scholar]
- Cai TT, Zhang CH, Zhou HH. Optimal rates of convergence for covariance matrix estimation. The Annals of Statistics. 2010;38:2118–2144. [Google Scholar]
- Cai TT, Zhou HH. Optimal rates of convergence for sparse covariance matrix estimation. The Annals of Statistics. 2012;40:2389–2420. [Google Scholar]
- Carter SL, Brechbühler CM, Griffin M, Bond AT. Gene co-expression network topology provides a framework for molecular characterization of cellular state. Bioinformatics. 2004;20(14):2242–2250. doi: 10.1093/bioinformatics/bth234. [DOI] [PubMed] [Google Scholar]
- de la Fuente A. From “differential expression” to “differential networking” identification of dysfunctional regulatory networks in diseases. Trends in Genetics. 2010;26:326–333. doi: 10.1016/j.tig.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, Zhernakova A, Heap GA, Ádány R, Aromaa A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nature genetics. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A. Diffcorr: an r package to analyze and visualize differential correlations in biological networks. Gene. 2013;518(1):209–214. doi: 10.1016/j.gene.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Fuller TF, Ghazalpour A, Aten JE, Drake TA, Lusis AJ, Horvath S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mammalian Genome. 2007;18(6–7):463–472. doi: 10.1007/s00335-007-9043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghellal A, Li C, Hayes M, Byrne G, Bundred N, Kumar S. Prognostic significance of tgf beta 1 and tgf beta 3 in human breast carcinoma. Anticancer Rec. 2000;20:4413–4418. [PubMed] [Google Scholar]
- Hudson NJ, Reverter A, Dalrymple BP. A differential wiring analysis of expression data correctly identifies the gene containing the causal mutation. PLoS Comput Biol. 2009;5(5):e1000382. doi: 10.1371/journal.pcbi.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Krogan NJ. Differential network biology. Molecular systems biology. 2012;8(1):565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki E, Kristiansen G, Wild PJ, Klaman I, Castanos-Velez E, Singer G, Sthr R, Simon R, Sauter G, Leibiger H, Essers L, Weber B, Hermann K, Rosenthal A, Hartmann A, Dahl E. Loss of sfrp1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol. 2004;25:641–649. [PubMed] [Google Scholar]
- Kostka D, Spang R. Finding disease specific alterations in the co-expression of genes. Bioinformatics. 2004;20(suppl 1):i194–i199. doi: 10.1093/bioinformatics/bth909. [DOI] [PubMed] [Google Scholar]
- Lai Y, Wu B, Chen L, Zhao H. A statistical method for identifying differential gene-gene co-expression patterns. Bioinformatics. 2004;20:3146–3155. doi: 10.1093/bioinformatics/bth379. [DOI] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome research. 2004;14(6):1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen SX, et al. Two sample tests for high-dimensional covariance matrices. The Annals of Statistics. 2012;40(2):908–940. [Google Scholar]
- Margheri F, Schiavone N, Papucci L, Magnelli L, Serrat S, Chill A, Laurenzana A, Bianchini F, Calorini L, Torre E, Dotor J, Feijoo E, Fibbi G, Del Rosso M. Gdf5 regulates tgf-dependent angiogenesis in breast carcinoma mcf-7 cells: In vitro and in vivo control by anti-tgf peptides. PloS ONE. 2012;7:e50342. doi: 10.1371/journal.pone.0050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A, Levina E, Zhu J. Generalized threshholding of large covariance matrices. Journal of the American Statistical Association. 2009;104:177–186. [Google Scholar]
- Schott JR. A test for the equality of covariance matrices when the dimension is large relative to the sample sizes. Computational Statistics & Data Analysis. 2007;51(12):6535–6542. [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, Evans A, Rapoport J, Giedd J. Intellectual ability and cortical development in children and adolescents. Nature. 2006;440(7084):676–679. doi: 10.1038/nature04513. [DOI] [PubMed] [Google Scholar]
- Shedden K, Taylor J. Methods of Microarray Data Analysis. Springer; 2005. Differential correlation detects complex associations between gene expression and clinical outcomes in lung adenocarcinomas; pp. 121–131. [Google Scholar]
- Shy B, Wu C, Khramtsova G, Zhang J, Olopade O, Goss K, Merrill B. Regulation of tcf7l1 dna binding and protein stability as principal mechanisms of wnt/b-catenin signaling. Cell Reports. 2013;4:1–9. doi: 10.1016/j.celrep.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava MS, Yanagihara H. Testing the equality of several covariance matrices with fewer observations than the dimension. Journal of Multivariate Analysis. 2010;101(6):1319–1329. [Google Scholar]
- van de Vijver M, He Y, van’t Veer L, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, vander Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. New England Journal of Medicine. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu X, Guo F, Ning Y, Zhi X, Wang X, Chen S, Yin L, Li X. Effect of estrogen sulfation by sult1e1 and papss on the development of estrogen-dependent cancers. Cancer science. 2012;103:1000–1009. doi: 10.1111/j.1349-7006.2012.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrakas M, Chorovicer M, Klaman I, Kristiansen G, Wild PJ, Heindrichs U, Knüchel R, Dahl E. Systematic characterisation of gabrp expression in sporadic breast cancer and normal breast tissue. International Iournal of Cancer. 2006;118(6):1453–1459. doi: 10.1002/ijc.21517. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li J, Deng H. Class-specific correlations of gene expressions: identification and their effects on clustering analyses. The American Journal of Human Genetics. 2008;83(2):269–277. doi: 10.1016/j.ajhg.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]


