Abstract
The ≥1030 bacteriophages on Earth relentlessly drive adaptive coevolution, forcing the generation of protective mechanisms in their bacterial hosts. One such bacterial phage-resistance system, ToxIN, consists of a protein toxin (ToxN) that is inhibited in vivo by a specific RNA antitoxin (ToxI); however, the mechanisms for this toxicity and inhibition have not been defined. Here we present the crystal structure of the ToxN–ToxI complex from Pectobacterium atrosepticum, determined to 2.75-Å resolution. ToxI is a 36-nucleotide noncoding RNA pseudoknot, and three ToxI monomers bind to three ToxN monomers to generate a trimeric ToxN–ToxI complex. Assembly of this complex is mediated entirely through extensive RNA-protein interactions. Furthermore, a 2′-3′ cyclic phosphate at the 3′ end of ToxI, and catalytic residues, identify ToxN as an endoRNase that processes ToxI from a repetitive precursor but is regulated by its own catalytic product.
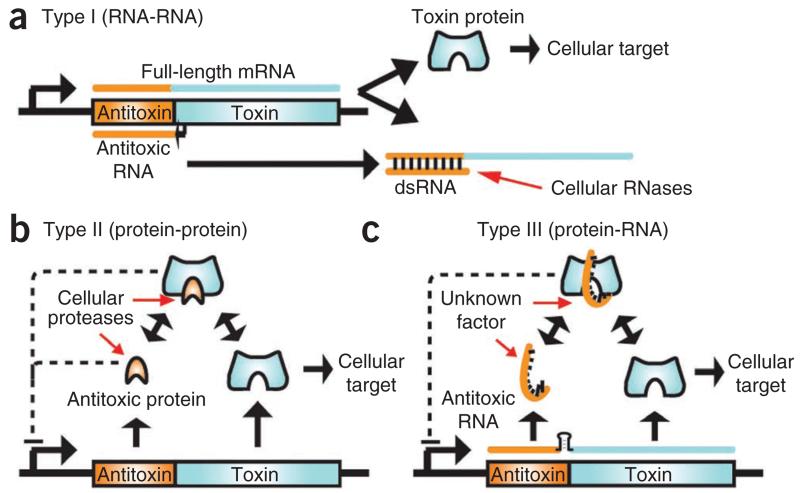
In the face of immense selection pressure from bacteriophage predation1,2, bacteria have evolved multiple phage-resistance mechanisms3. One such mechanism, abortive infection (Abi), during which a bacteriophage-infected cell altruistically commits suicide to protect the clonal bacterial population4,5, can be mediated by a toxin-antitoxin (TA) pair6,7. TA pairs are widespread throughout prokaryotes8 and have been implicated in diverse biological processes, including plasmid maintenance, stress responses and persistence9–11 (Fig. 1). They most often comprise two genes—a toxin gene preceded by an antitoxin gene—usually transcribed from a single promoter. In the cell, the antitoxin and toxin interact, thereby suppressing toxicity. The antitoxin is more labile than the toxin and is preferentially degraded in response to certain stimuli. This leads to a relative increase in toxin concentration that can induce bacteriostasis and cell death.
Figure 1.
Overview of TA systems. In general, the antitoxic gene (orange) precedes the toxin gene (cyan), as part of a bicistronic operon. (a) In type I TA systems, the short antisense antitoxic RNA forms a duplex with a short region of the full-length mRNA. This duplex prevents translation of the toxin gene and promotes degradation by a cellular RNase such as RNase III. (b) In type II TA systems, the protein antitoxin forms a complex with the protein toxin. Either as a complex or by itself the antitoxic protein often negatively regulates transcription of the operon. Cellular proteases such as Lon and Clp degrade the antitoxin, releasing the toxin. (c) In our proposed type III TA system, a proteinaceous toxin interacts with an antitoxic RNA. A transcriptional terminator (black stem-loop) between the antitoxin and toxin genes regulates relative levels of ToxI and ToxN transcript. As a complex, the toxin and antitoxin negatively regulate transcription. Cellular factor(s) that have yet to be identified trigger toxin activity by degrading the antitoxic RNA, decreasing the level of transcription from the locus or releasing the toxin from the RNA antitoxin. dsRNA, double-stranded RNA.
Type I TA systems rely on sequence complementarity between the toxin mRNA and the cis-encoded antisense antitoxin mRNA, allowing them to form a double-stranded RNA duplex that is targeted for degradation (Fig. 1a). These systems are activated when the level of antitoxin RNA decreases, allowing the toxin mRNA to be translated before it is degraded, as occurs for the hok-sok locus of plasmid R1 (refs. 12,13). In contrast, type II TA systems have components that interact as proteins (Fig. 1b). These have been more widely studied and show diverse modes of antitoxin binding to similar core toxin folds. Type II toxins generally act as endoRNases, either as free enzymes14–16 or in concert with the ribosome17,18, or as DNA gyrase inhibitors19,20. Other examples include potential proteases and phosphotransferases21,22.
ToxIN, which is encoded by a plasmid from the plant pathogen P. atrosepticum, is a powerful bicistronic Abi system that aborts infection by different phages in multiple host genera6. ToxIN is the first example of a new (type III) TA system in which a protein toxin, ToxN, is inhibited by an RNA antitoxin, ToxI6,7 (Fig. 1c). These type III systems are identified by the presence of a repetitive DNA sequence upstream of a transcriptional terminator, which is then followed by a toxin gene. Transcription of this locus is driven by a single constitutive promoter upstream of the repetitive sequence, with the transcriptional terminator acting to regulate the relative levels of repetitive antitoxic RNA to toxin mRNA6,7. Predicted homologs of ToxN are found in plasmids and the chromosomes of diverse bacteria, including human and animal pathogens, human commensals, extremophiles, and soil and marine species6. Genes for these protein homologs are usually accompanied by their own specific toxI elements, which are unique in their primary sequences and in their length and number of repeats6.
As there was no available structural information on toxins of this type III family, and to understand better how the antitoxic RNA can inhibit its cognate protein, we carried out an X-ray crystallographic study of the ToxN–ToxI complex. We present here the structure of the ToxN protein with ToxI RNA, which has allowed identification of the modes of toxin and antitoxin activity and interaction. We further confirmed our results using site-directed mutagenesis and both in vivo and in vitro functional assays.
RESULTS
The ToxIN complex is a trimeric assembly
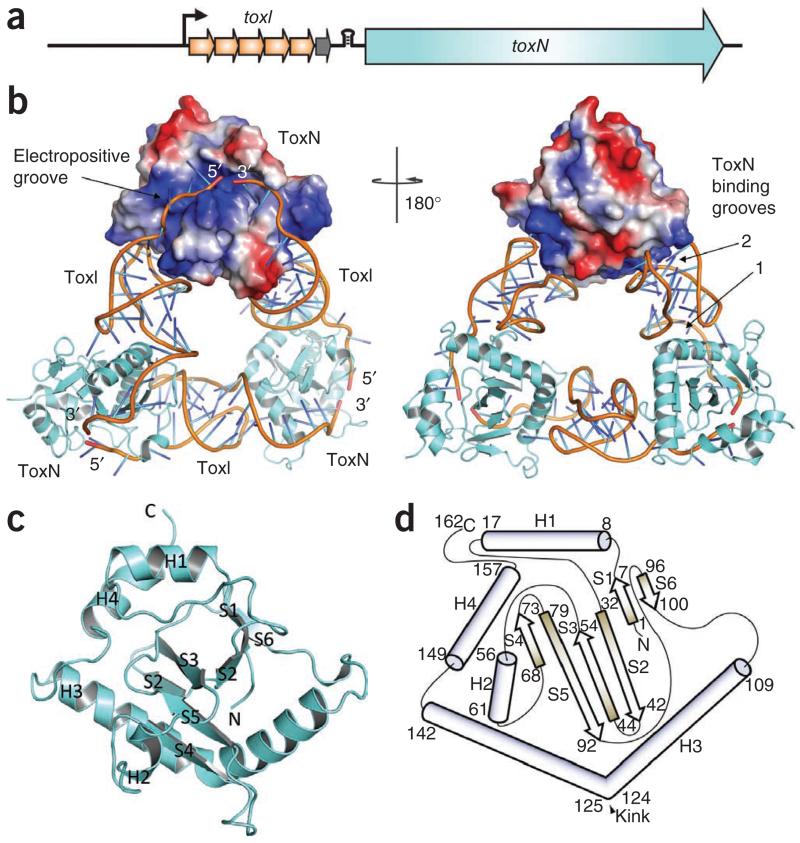
Before phage infection, the 19.7-kDa toxic Abi protein ToxN (denoting ‘toxin’) is inhibited by an upstream repetitive nucleotide sequence called toxI (for ‘ToxN inhibitor’), which acts as a noncoding RNA antitoxin6,23. ToxI RNA consists of a tandem array of 5.5 36-nt repeats that lack spacers and are therefore entirely contiguous (Fig. 2a). A single repeat is the fundamental unit that inhibits ToxN toxicity in vivo6. We carried out overexpression of the recombinant ToxN–ToxI complex in Escherichia coli, purified the complex as a stable assembly and crystallized it in both native and selenomethionine (SeMet) forms. The X-ray crystal structure was solved using SAD from the SeMet and by molecular replacement of nonisomorphous native protein crystals that diffracted to higher resolution than the derivative (Table 1).
Figure 2.
ToxIN complex structure. (a) Schematic of the toxIN locus, which is transcribed from a single promoter (black arrow). The toxN gene is downstream of a transcriptional terminator (black stem-loop) and toxI, which encodes 5.5 36-nt repeats. A whole repeat is shown as an orange arrow and the half repeat as a gray arrow. (b) An overview of the ToxIN complex structure. Two ToxN protomers are shown as cartoons (colored cyan) and the third as a surface representation, with the positively charged surface in blue and the negatively charged surface in red. The phosphate sugar backbones of the ToxI molecules are shown as orange cartoons and the bases as single sticks that are colored in a gradient from cyan to blue. Right, 180° rotation about the vertical axis. (c) Cartoon representation of monomeric ToxN, with α-helices labeled H1 to H4 and β-strands labeled S1 to S6. (d) Schematic representation of ToxN topology, with α-helices shown as cylinders and β-strands as arrows. Numbers indicate the amino acid residues at the limits of the secondary structures. ToxN monomers in c and d are orientated as in the surface representation ToxN in a (left).
Table 1. Data collection and refinement statistics.
| Native 1 | Native 2 | SeMet | |
|---|---|---|---|
| Data collection | |||
| Space group | I4132 | P212121 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 183.65, 183.65, 183.65 | 42.12, 119.13, 377.06 | 182.85, 118.13, 41.90 |
| α, β, γ(°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 92.78, 90.00 |
| Wavelength | 0.9702 | 0.87 | 0.9801 |
| Resolution (Å) | 75.00–2.75 | 35.00–3.00 | 35.00–3.20 |
| R merge a | 0.132 (0.863) | 0.107 (0.322) | 0.112 (0.376) |
| I / σIa | 16.9 (3.5) | 18.0 (3.4) | 12.1 (5.0) |
| Completeness (%)a | 100 (100) | 89.7 (66.3) | 99.9 (100.0) |
| Redundancya | 19.9 (20.6) | 6.2 (2.3) | 7.4 (7.5) |
| Refinement | |||
| Resolution (Å) | 75.00–2.75 | 35.00–3.00 | 35.00–3.20 |
| No. reflections | 32,628 | 35,161 | 28,777 |
| Rwork / Rfree | 0.176/0.212 | 0.213/0.260 | 0.212/0.273 |
| No. atoms | |||
| Protein | 1,316 | 7,896 | 3,948 |
| Ligand (RNA) | 764 | 4,584 | 2,292 |
| Ions | 10 | 0 | 8 |
| Water | 100 | 171 | 60 |
| B-factors | |||
| Protein | 60.5 | 36.4 | 51.5 |
| Ligand (RNA) | 66.6 | 62.1 | 67.6 |
| Ions | 109.2 | – | – |
| Water | 66.4 | 53.7 | 37.7 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.01 | 0.01 |
| Bond angles (°) | 1.37 | 1.67 | 2.60 |
Values in parentheses are for the highest-resolution shell.
The repeating structural unit we observed consists of a ToxN monomer in complex with a 36-nt RNA oligomer, which is derived from a full-length toxI transcript. This monomeric complex formed trimers with a triangular architecture (Fig. 2b). The 3′ end of each ToxI unit is adjacent to the 5′ end of the next unit, in a pseudo-continuous head-to-tail manner, and each ToxI oligomer interacts extensively with two ToxN molecules—one at each terminus (ToxN binding grooves 1 and 2, Fig. 2b). Every ToxN molecule thereby interacts with two ToxI molecules over an extended surface (electropositive groove, Fig. 1b). The buried surface area of ToxN at each protein-RNA interface is roughly 2,000 Å2 (Supplementary Table 1), which corresponds to an avid macromolecular interaction24 and is unlikely to occur through crystal contacts alone. Furthermore, we observed the ToxIN trimer in each of three crystal forms, by both crystallographic and noncrystallographic symmetries. We also confirmed by analytical gel filtration that ToxIN forms a high-molecular-weight complex (data not shown), indicating that the trimeric ToxIN is a biologically relevant macromolecular complex. In this complex, ToxN has a compact globular fold with a highly twisted, six-stranded, antiparallel β-sheet core surrounded by four α-helices (Fig. 2c,d), whereas ToxI forms a convoluted RNA fold that is examined below.
Noncoding, antitoxic ToxI RNA forms a pseudoknot
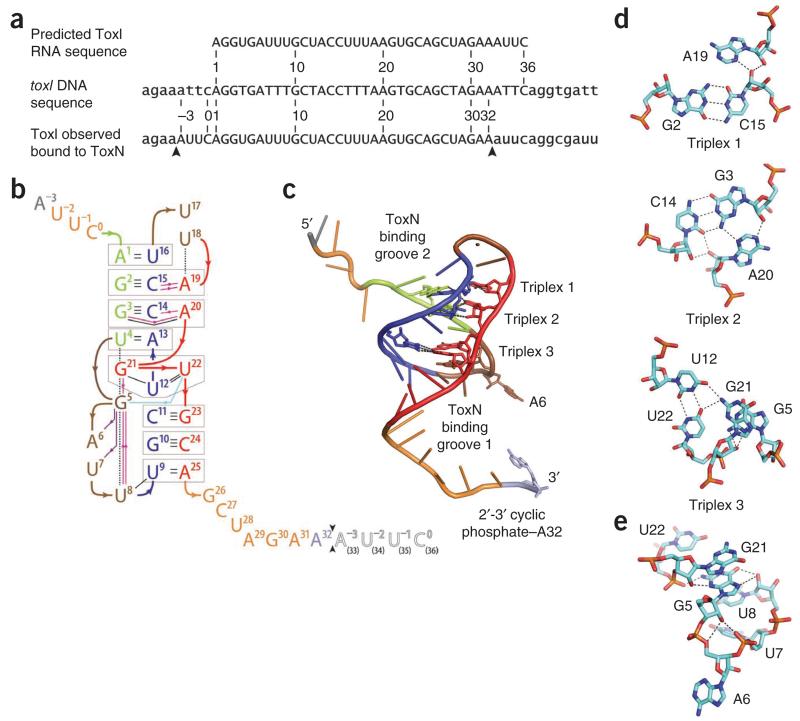
The repetitive unit in toxI DNA comprises a block of 36 nt (Fig. 3a). From our previous work6, it was predicted that the functional antitoxic ToxI RNA would comprise the transcript of these same 36 nt (Fig. 3a). In our crystal structure, we did observe a repeat of exactly 36 nt, though each individual 36-nt RNA begins 4 nt 5′ of the annotated toxI repeat start (Fig. 3a). By a succession of single cleavage events that precede each occurrence of these AUUC sequences (Fig. 3a), a single ToxI transcript of 5.5 repeats could be cut into four of these observed 36-nt ToxI RNAs. We therefore propose that the ToxIN trimer folds and assembles following, or in concert with, multiple endoRNase cutting steps that generate the observed ToxI repeat units from the full-length RNA (Fig. 3a).
Figure 3.
ToxI pseudoknot structure. (a) Sections of the toxI DNA and the predicted corresponding RNA repeat are shown with the ToxI RNA repeat that is seen in the crystal structure. Capitals indicate the 36-nt repeats. Arrows indicate the cleavage sites of a single active ToxI RNA from the longer ToxI transcript. (b) Overview of hydrogen bonding in the ToxI pseudoknot. Nucleotides −3 to 32 correspond to one 36-nt RNA oligomer in the crystal structure. Nucleotides 1 to 36 correspond to a single consensus toxI 36-nt repeat. Black arrows between nucleotides 32 and 33 indicate the putative ToxN cutting site in ToxI. The black-outlined, open letters for nucleotides 33–36 represent the 5′-most 4 nt of a second 36-nt ToxI oligomer from the crystal structure. Three interacting sections are shown in green (nucleotides 1–4), blue (nucleotides 9–16) and red (nucleotides 19–25), separated by brown loops (nucleotides 5–8 and 17–18). Duplex and triplex base pairs are highlighted by gray boxes. The single-stranded RNA tail nucleotides are shown in orange, except the termini, with A3 in gray and A32 in violet. Base-base hydrogen bonds are shown as black lines. Ribose 2′OH-base hydrogen bonds are shown as magenta lines, ribose 2′OH-phosphate hydrogen bonds as violet lines and a phosphate-base hydrogen bond as a light blue line; arrows point from the first partner. Black dashed lines depict selected stacking interactions. (c) ToxI pseudoknot structure, showing the locations of base triplexes. The main chain backbone and bases are colored as in b. (d) Detail of each of the three ToxI base triplexes, drawn as cyan sticks with oxygen in red, nitrogen in blue and phosphorus in orange. Hydrogen bonding interactions within 2.6 Å to 3.3 Å are indicated by dashed lines. (e) The network of hydrogen bonds extending from ToxI G5 supports the formation of a loop structure and facilitates the presentation of A6 for interaction with ToxN. Hydrogen bonding interactions within 2.5 Å to 3.5 Å are indicated by black dashed lines. The color scheme is the same as in d.
Each ToxI monomer folds as an interdigitated hairpin-type pseudoknot25, with two single-stranded tails (Figs. 2b and 3b). The pseudoknot is formed from three sections of ToxI that together make a central helical core of duplex and triplex base-pair interactions, interspersed by two loops (Fig. 3b,c). Three triplexes zip up the internal fold (Fig. 3d); minor-groove single-tiered triplexes 1 (G2–C15:A19) and 2 (A20:C14–G3:A20) form classical type II and type I A-minor motifs, respectively26. These A-minor motifs widen the minor groove and aid stability of the pseudoknot26,27. Major-groove triplex 3 is also single tiered: G21:U12–U22:G21 (Fig. 3d). The interdigitation of G21 is supported by base stacking between U4, G21, G5 and U8 (Fig. 3b,e) as well as an intricate hydrogen bonding network around G5 (Fig. 3d,e). The fold produces two bulged loops, exposing A6 and U17 (Fig. 3b,c,e); A6 interacts extensively with ToxN (see below). Interaction with ToxI occludes the active site of ToxN, accounting for the in vivo inhibition of ToxN activity6, to which we now turn.
ToxN acts as an endoRNase
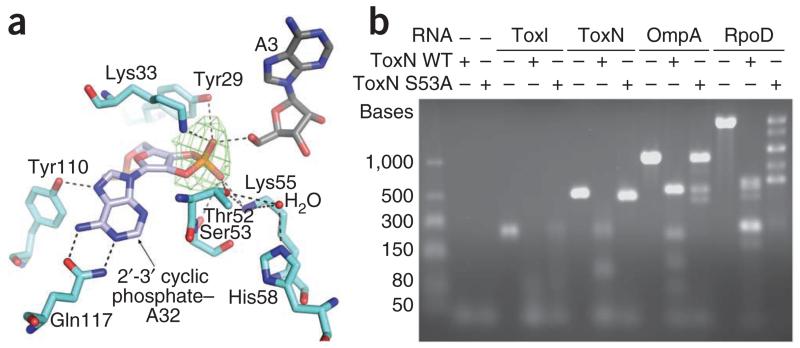
The electron density around the 3′ ribose of ToxI suggested the presence of a 2′-3′ cyclic phosphate (Fig. 4a). This cyclic group is formed between the backbone phosphate group from a longer RNA molecule and the 2′ O of the A32 ribose. The 3′ end of one ToxI oligomer is sufficiently close to the 5′ end of the next to suggest that these two RNAs were previously linked. The ToxI 2′-3′ cyclic phosphate is held in the electropositive groove crossing ToxN by an extensive hydrogen bond network that is formed with the side chains of Tyr29, Lys33, Thr52, Ser53 and Lys55, suggesting that this groove is the ToxN active site. The base of A32 is held in a bidentate interaction with the side chain of Gln117 and a single hydrogen bond interaction with the side chain of Tyr110. This precise coordination of A32 suggests base specificity in ToxN activity.
Figure 4.
ToxN has an endoRNase active site. (a) ToxN active site interactions with ToxI. A simulated annealing omit map, at 3.75σ level, was calculated using our model, omitting the proposed 2′-3′ cyclic phosphate group, and is shown in green. Hydrogen bonding interactions within 2.6 Å to 3.5 Å are indicated by black dashed lines, and a water molecule is shown as a red sphere. Coloring is as in Figure 3. (b) In vitro ToxN RNase assay. The ability of the purified wild-type (WT) ToxN protein and the ToxN S53A mutant to digest in vitro–transcribed RNAs was assessed. RNA substrates were selected to allow investigation of autoregulation (full-length ToxI RNA (5.5 36-nt repeats) and ToxN RNA) and to examine the effect on transcripts of highly active genes (OmpA and RpoD RNA). Protein was mixed with RNA at a 4:1 molar ratio and incubated. The products were analyzed by agarose gel electrophoresis.
As we showed previously that ToxN S53A was defective for both Abi and toxicity7, we expressed and purified this mutant protein for comparison with wild-type ToxN in endpoint RNase assays (Fig. 4b). We chose substrates to investigate ToxN autoregulation and the regulation of abundant cellular mRNAs: to examine autoregulation, we produced single-stranded RNA substrates by in vitro transcription from full-length toxI and the toxN gene, and we chose the ompA and rpoD genes to generate ‘highly expressed’ substrates. We incubated these substrate RNAs with the purified wild-type ToxN–ToxI and ToxN–ToxI S53A complexes and analyzed the products by agarose gel electrophoresis (Fig. 4b).
Both the wild-type ToxN and S53A protein samples digested the RNA substrates into discrete banding patterns, suggesting that this degradation was not wholly a result of background RNase contamination. These background levels were considered to be low, as preliminary control experiments using fractions from the ToxN protein purification that were devoid of ToxN showed little to no RNase activity (data not shown). Unexpectedly, S53A was not inactive, but it did have an altered RNase activity compared with wild-type ToxN and produced different banding patterns for the degradation of the rpoD and ompA RNAs (Fig. 4b). It is interesting to note that ToxI RNA was degraded by both wild-type ToxN WT and S53A, which indicates that the presence of the antitoxin does not inhibit the RNase activity in vitro (unlike for type II systems with RNase toxins). We suggest that more complex binding kinetics occur in vivo that allow ToxI to inhibit ToxN, and that this is linked to the regulation of stoichiometry that is provided by the transcriptional terminator between toxI and toxN6,7 (Fig. 2a). These results suggest that ToxN has endoRNase activity and is active independently of the ribosome, unlike the RelE family of toxins17. The specific RNA fragment patterns generated by ToxN-mediated degradation also suggest sequence-specific endoRNase activity (Fig. 4b).
ToxN forms extensive interactions with ToxI RNA
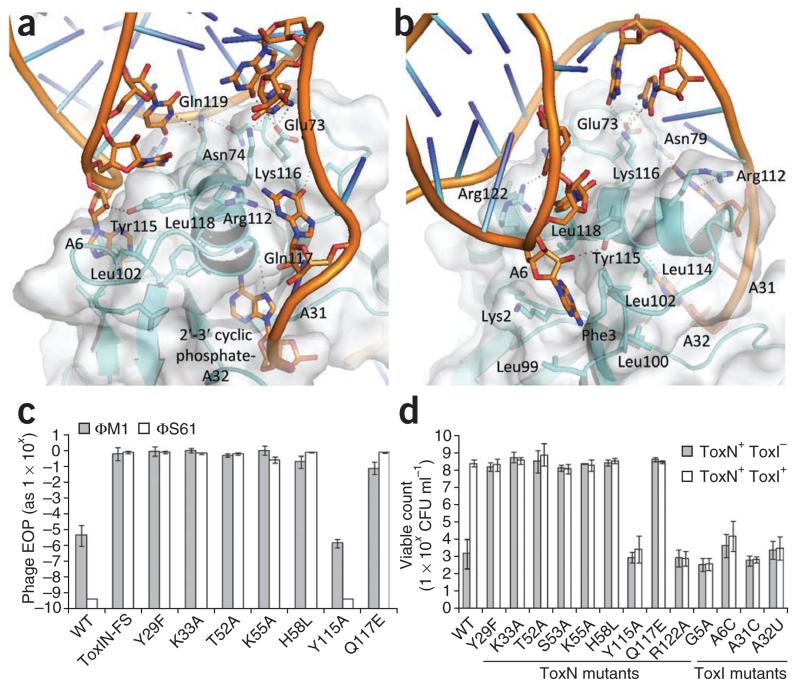
Helix H3 of ToxN kinks at an approximately 75° angle at residue Asn124, allowing it to wrap across the surface of the protein (Fig. 2d). The N-terminal section of H3 interacts with a ToxI monomer and, after the kink, the C-terminal section interacts with a second ToxI (Fig. 2b). H3 residues interacting with ToxI include Leu102, Leu114, Tyr115, Lys116, Gln117, Leu118 and Arg122 (Fig. 5a,b). ToxN loop residues Glu73 and Asn79 also contribute additional hydrogen bonds to stabilize the ToxIN complex. A further tight interaction occurs where the base of A6 inserts into a pocket formed by the backbone of Phe3 and the side chains of Lys2, Leu99, Leu100 and Leu102 (Fig. 5b).
Figure 5.
Identification of ToxN–ToxI residues that are vital for toxicity and interaction. (a) The N-terminal section of ToxN H3 forms an extensive hydrogen bonding network with ToxI groove 1 (Fig. 3c). (b) View of groove 1 after a rotation of roughly 90° about the vertical axis relative to a. ToxI A6 is held in a hydrophobic pocket on the ToxN surface. Coloring in a and b is the same as in Figure 3. (c) In vivo Abi activity of ToxN mutants. The efficiencies of plating (EOP) of phages ΦM1 and ΦS61 were assessed upon strains of P. atrosepticum 1043 carrying either wild-type (WT) toxIN or mutant toxIN plasmids. (d) In vivo toxicity of ToxN mutants and antitoxic activity of ToxI mutants to interact. Strains of E. coli DH5α carrying two independently inducible ToxN and ToxI plasmids were tested for their viability in the presence of WT ToxN or a ToxN mutant protein either with or without WT ToxI or a ToxI mutant RNA. In c and d, error bars indicate s.d.
Identification of essential toxic and antitoxic residues
Specific mutations confirmed the importance of active-site residues Tyr29, Lys33, Thr52, Lys55, His58 and Gln117 in the Abi phenotype and toxicity (Fig. 5c,d). We also found that ToxN residues Tyr115 and Arg122, together with ToxI bases A6, A31 and A32, are vital for the ability of the two components to interact in vivo and prevent toxicity (Fig. 5d). G5 is a key determinant of the pseudoknot fold, and its importance is corroborated by the loss of antitoxicity with a ToxI G5A mutation (Figs. 3e and 5d).
ToxN and ToxI structural homologs
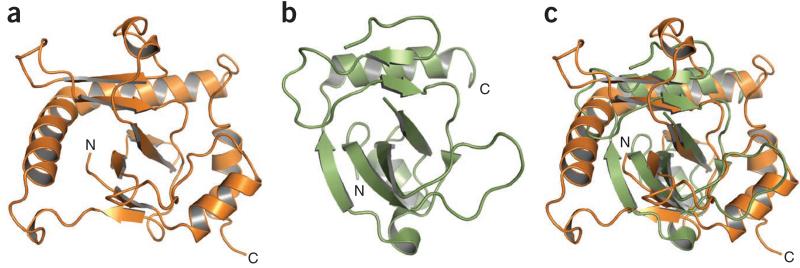
Using DALI28, we unexpectedly identified ToxN as a unique member of the Kid-like toxin family29. Three high-scoring structural homologs were toxins from type II (protein-protein) TA systems (Fig. 6 and Supplementary Figs. 1 and 2)—Kid, MazF and CcdB14,19,30—with Z-scores of 6.1, 4.8 and 6.4, respectively. This structural similarity was unexpected because the sequence similarity is low (only 11.1% identity with Kid) (Supplementary Fig. 1). Kid and MazF are endoRNases that cleave free mRNA, whereas CcdB is a DNA gyrase poison; all have proteinaceous antitoxins. The ToxN β-core regions overlap well with those of each homolog; for example, ToxN and Kid strands S1, S2 (ToxN N-terminal region only), S3 and S5 can be superimposed (Fig. 6a–c). Although the core fold is conserved, there are variations in the α-helices and loops that decorate each homologous structure (Fig. 6 and Supplementary Fig. 2). However, some regions of ToxN differ distinctly from all the structural homologs; in particular, although the H3 helices of ToxN and Kid overlay (Fig. 6c), the N-terminal section of ToxN helix H3 is greatly extended (Fig. 2c,d and Supplementary Fig. 1). It is this region of ToxN H3 that has been shown to be a principal site of ToxI RNA recognition (Fig. 5). Although Kid and ToxN share a similar fold and β-core, the proposed active site of ToxN does not overlay closely with that identified for Kid31.
Figure 6.
Structural comparisons in the ToxN family. (a) Cartoon representation of ToxN in orange. (b) Cartoon representation of Kid (PDB 1M1F14) in green. (c) Structural overlay of ToxN in orange and Kid in green. Structures in a–c are shown in the same orientation.
The ToxI pseudoknot shares similar structural features with an artificially generated vitamin B12 aptamer32 (Supplementary Fig. 3b), including the general fold and detailed interactions that stabilize non-canonical base pairing in ToxI, such as A-minor motifs26. ToxI also has a similar overall fold to a naturally occurring riboswitch that binds a precursor of queuosine33 (Supplementary Fig. 3c).
DISCUSSION
Increasing numbers of type I and type II TA systems are being identified and characterized15,34,35, and bioinformatic approaches have highlighted the widespread nature and abundance of these loci8,15,35,36. Notwithstanding this prevalence, there remains some controversy over their exact biological roles. It is necessary, therefore, to expand investigation of the variations and individual phenotypes that are provided by each TA system. Of the known TA systems, type III is predicted to be under-represented, and the structure reported in this study is the first to be solved for this new type of TA system.
Our structure greatly extends the known structural information for TA systems. We have observed the elegant inhibition of an endoRNase toxin by the RNA substrate itself. The fact that this RNA substrate, the antitoxin, folds into a complex pseudoknot, seems to be vital for inhibition of the toxin and is entirely unique throughout known examples of TA systems. After the endoRNase cleavage step, the extensive interaction between the folded RNA surface and the protein surface is thought to maintain the complex in an inert state before activation.
Because of the low sequence similarity between them, we were surprised to find that the ToxN protein is an embellished version of the type II Kid, MazF and CcdB toxins31 (Fig. 6 and Supplementary Figs. 1 and 2). Evolutionarily, this suggests that a simple endoRNase scaffold can be used for multiple purposes, at least for the reduction of plasmid loss, for responding to stress or for abortive infection. Whether the scaffold arose through convergent or extensively divergent evolution is unknown. Clearly, however, by adapting the Kid-like fold, ToxN is able to bind and be inhibited by a ToxI RNA pseudoknot. The fact that the ToxIN trimeric complex was formed entirely through RNA and protein interactions was unexpected.
The presence of a 2′-3′ cyclic phosphate indicates that ToxN is an endoRNase (Fig. 4a). This was also predicted by its structural homology with the endoRNases Kid and MazF; however, as the DNA gyrase inhibitor CcdB is also a homolog, we performed in vitro RNase assays to further investigate ToxN activity (Fig. 4b). As we were not able to specifically inhibit ToxN with ToxI in vitro, we cannot rule out RNase contamination as contributing to the RNA digestion patterns. Still, we suggest that the distinct banding patterns that resulted are indicative of an endoRNase with sequence preference, such as is found for Kid and MazF. This proposed RNase activity of ToxN relies upon specific residues to position the substrate and provide catalysis, as shown through mutagenesis studies (Figs. 4b and 5c,d). We observed some RNase activity of the S53A ToxN mutant, which carries the mutation in the active site. A similar situation is found for other Kid family enzymes, in which substitutions in the active site impair but do not eliminate catalysis, as the residues make distributive interactions to support the substrate. The RNase assay data do suggest a mode of action for ToxN: active ToxN cleaves cellular mRNAs and induces bacteriostasis, which leads to cell death.
As neither additional ToxI nor the co-purified ToxI present in the protein samples (Fig. 4b) completely inhibited ToxN in vitro, the question arises as to how stable the ToxIN complex is in vivo: the ToxN protein digests ToxI RNA in vitro and yet is inhibited by ToxI RNA in vivo, so is the same level of turnover of ToxI occurring in vivo? If so, the cellular levels of ToxI must be sufficient to overpower the activity of ToxN and thereby hold ToxN in an inactive complex. There is certainly potential for this to be the case, as indicated by our previous experiments, which showed tenfold higher transcription of toxI over toxN, owing to the transcriptional terminator between the two6,7 (Fig. 2a). However, it seems paradoxical that we have been able to crystallize this ToxIN complex, which is potentially dynamic in the cell; the high concentration of protein may have promoted complex formation in the crystallization droplets. When considering the steps to complex formation, the exact stage at which ToxI is cut by ToxN is unknown, and it is important to assess whether this event occurs during formation of the complex, whether this is co-translational and, if not, how this is influenced by folding of the two components. Further investigation is clearly warranted to fully understand the regulation of these type III systems.
It was also clear that the interaction of ToxI and ToxN is vital for Abi activity and maintenance of the complex in the cell (Fig. 5). We suggest that abortive infection may be instigated when this complex is disturbed, either through disruption of toxIN transcription or by direct molecular interaction with a phage product (as shown in Fig. 1c). Furthermore, the noncoding RNA ToxI has a similar topology to both an RNA aptamer that binds vitamin B12 (ref. 32) and a queuosine riboswitch33 (Supplementary Fig. 3). Although there is currently no biochemical evidence to support this hypothesis, we suggest that ToxI may also sense and respond to small-molecule ligands that could be allosteric modulators. In this way, the ToxIN system or homologs could acquire the capacity to respond to diverse metabolic and environmental cues. This seems a potentially exciting area for further research.
RNA pseudoknots are involved in gene regulation events and translational frameshifting in tumor viruses and retroviruses37,38, and they also form a component part of human telomerase39. Whereas pseudoknots have been artificially developed to bind specific small molecules32 and even to inhibit proteins such as the HIV-1 reverse transcriptase40, ToxI is a naturally occurring noncoding RNA pseudoknot that can bind and thereby counter the toxicity of its proteinaceous ligand. Small noncoding RNAs are encoded by a diverse and growing family of genes that are required for the regulation of bacterial physiology23,41. By acting to suppress ToxN as a pseudoknot, ToxI further highlights and extends the impact of small noncoding RNAs in diverse biological processes. Elucidating the manner in which ToxN homologs bind their cognate ToxI RNAs will provide a greater understanding of protein–noncoding RNA pseudoknot interactions, and could enable the potential exploitation of specific molecular recognition for therapeutic applications. This structure will underpin further investigations into structure-function relationships in type III TA systems. In particular, we are interested in defining the mechanisms by which ToxIN is activated by some, but not all, phages to drive a lethal, altruistic Abi response in bacteria.
ONLINE METHODS
Purification, crystallization and structure determination
The toxN gene was cloned into vector pTYB1 (New England Biolabs), and toxI, together with the natural toxIN promoter, was cloned into pACYC184 (ref. 42). These expression vectors were used to co-transform a single strain of E. coli ER2566 (New England Biolabs). The expressed native and SeMet-derivatized43 ToxIN complexes were purified with the aid of a chitin-binding domain and intein tag, followed by HiTrap Q (GE Healthcare) ion-exchange chromatography using Fast Protein Liquid Chromatography (Äkta). The protein was dialyzed against 50 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM DTT, then concentrated to ~10 mg ml−1 before use in crystallization trials.
Native and SeMet crystals were formed at 18 °C in sitting drops, and SeMet crystals were further optimized using the matrix seeding method44. Native crystals grew in conditions of 0.01 M CoCl2, 0.1 M MES, pH 6.5, and 1.8 M (NH4)2SO4. These I4132 crystals diffracted up to 2.75 Å with synchrotron X-rays produced at station I04 in the Diamond Light Source, Oxford, UK. Further native crystals grew in conditions of 0.6 M NaCl, 0.1 M MES, pH 6.5, and 20% (w/v) PEG 4000. These P212121 crystals diffracted to 3.0 Å with synchrotron X-rays focused at station ID23.1 in the European Synchrotron Research Facility, Grenoble, France. SeMet-ToxN crystals grew in condition; 0.01 M ZnSO4, 0.1 M MES pH 6.5, 25% w/v PEG 550 MME. These C2 crystals diffracted to 3.2 Å with synchrotron X-rays produced at station I03 in the Diamond Light Source. All crystals were cryoprotected in a solution containing 20–25% (v/v) glycerol in addition to the condition reservoir, before freezing at 100K.
The native data from the Native 2 P212121 crystals were processed in HKL2000 and scaled in SCALEPACK2000 (ref. 45). The native data from the Native 1 crystals and the SAD data from the SeMet crystals were processed using MOSFLM, SCALA and TRUNCATE in the CCP4 suite46 of programs (http://www.ccp4.ac.uk/). Heavy-atom sites were identified and iteratively improved by using HKL2MAP47, autoSHARP48 and PHENIX49, yielding 15 selenium and 3 zinc sites. The figure of merit to 3.2 Å from PHENIX was 0.38. After threefold non-crystallographic symmetry averaging, the auto-build mode of PHENIX produced a model with 72 main chain and 39 side chain fragments out of a total of 486 residues for a trimeric model. This model clearly showed the helical region 109–121 in both main chain and side chain, and a β-sheet including partial β-strands S1, S2, S3, S5 and S6 (Fig. 1c,d). The map produced was therefore of sufficiently high quality to permit confident interpretation and tracing of the backbone and side chains of ToxN; this was completed using COOT50.
The final refined structure of the SeMet crystal contained ToxN residues 1–162. A Fo – Fc difference map, above 3σ levels, was used to guide the modeling of ToxI RNA in COOT. This structure was used as a search model in molecular replacement calculations to determine both the Native 1 and Native 2 structures, using Phaser51. In the 2.75-Å structure of the Native 1 ToxIN complex, the final model contains ToxN residues 1–162 and 36 nt of ToxI. The stereochemistry of each model was validated using SFCHECK and PROCHECK in the CCP4 suite of programs. Ramachandran plot values (preferred, allowed, outlier), were as follows for each model: Native 1 (96.9%, 3.1%, 0.0%); Native 2 (95.3%, 4.7%, 0.0%); SeMet (91.8%, 8.2%, 0.0%). Metal ions Co2+ and Zn2+ were observed within the Native 1 and SeMet crystals, respectively, in positions suggesting that divalent cations aid ToxI binding. Figures of the ToxIN structure were generated using PyMol (http://pymol.sourceforge.net) and adapted using Adobe Illustrator (CS4; Adobe).
Analysis of ToxN structural homologs
Structural homologs of ToxN were identified using DALI28, and the amino acid sequences were aligned with ClustalW2 (ref. 52). Initial secondary-structure images were produced with ESPript53. Structural overlays were made with COOT.
In vitro endpoint RNase assays
The gene for ToxN S53A was cloned into pTYB1 and co-expressed with ToxI as for wild-type ToxN. RNAs were prepared by first cloning the necessary gene into pBluescript (Stratagene), downstream of the T7 RNA polymerase promoter. Plasmid pECA1039 was used as template DNA for toxI and toxN, whereas ompA and rpoD were amplified from E. coli DH5α genomic DNA. These recombinant plasmids were linearized and used as templates for in vitro transcription with T7 RNA polymerase (Fermentas). The resulting RNAs were mixed, when appropriate, with a ToxN protein at a 4:1 protein:RNA molar ratio. The samples were incubated at 37 °C for 1 h and then separated by electrophoresis in a 1.6% agarose, 0.5× Tris-Borate-EDTA (TBE) gel stained with ethidium bromide.
In vivo phenotypic assays
For Abi assays, the full toxIN locus was mutated by overlap extension PCR and subsequently cloned into pBR322. For toxicity and antitoxicity assays, mutant amplicons for ToxN and ToxI were first generated by overlap extension PCR, then cloned into either pBAD30 (ref. 54) or pTA100 (ref. 6), respectively. In all cases, ToxN proteins had an additional C-terminal Flag tag, which has previously been shown not to adversely affect ToxN activity6,7. Abi assays were performed as described, using phages ΦM1 and ΦS61 (ref. 7). Abi data is shown as efficiency of plating (EOP)55, which is the ratio of plaque-forming units enumerated on a test strain compared to those on a control strain. Toxicity and antitoxicity assays were also performed as described6, replacing either the ToxN or ToxI expression plasmid with the test mutant plasmid as required.
Supplementary Material
ACKNOWLEDGMENTS
We thank the beamline scientists at station ID23.1 in the European Synchrotron Research Facility, Grenoble, France, and at stations I03 and I04 in the Diamond Light Source, Oxford, UK. This work was supported by grants from the Biotechnology and Biological Sciences Research Council (UK), the Wellcome Trust (UK) and the Marsden Fund, Royal Society of New Zealand. Work with P. atrosepticum was performed under a plant health license from the Department for Environment, Food and Rural Affairs (UK). T.R.B. was supported by a Collaborative Award in Science and Engineering Studentship from UCB Ltd. F.L.S. was supported by a Commonwealth Scholarship from the Commonwealth Scholarships Commission (UK).
Footnotes
Accession codes. Protein Data Bank: Coordinates for the three ToxIN complex forms, Native 1, Native 2 and SeMet, are deposited with accession numbers 2XDB, 2XD0 and 2XDD, respectively.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lima-Mendez G, Toussaint A, Leplae R. Analysis of the phage sequence space: the benefit of structured information. Virology. 2007;365:241–249. doi: 10.1016/j.virol.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 3.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 4.Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Shub DA. Bacterial viruses. Bacterial altruism? Curr. Biol. 1994;4:555–556. doi: 10.1016/s0960-9822(00)00124-x. [DOI] [PubMed] [Google Scholar]
- 6.Fineran PC, et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blower TR, et al. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J. Bacteriol. 2009;191:6029–6039. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 11.Keren I, et al. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes K, Wagner EG. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Hargreaves D, et al. Structural and functional analysis of the Kid toxin protein from E. coli plasmid R1. Structure. 2002;10:1425–1433. doi: 10.1016/s0969-2126(02)00856-0. [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen MG, Pandey DP, Jaskolska M, Gerdes K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 2009;191:1191–1199. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miallau L, et al. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J. Biol. Chem. 2009;284:276–283. doi: 10.1074/jbc.M805061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neubauer C, et al. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu M, Zhang Y, Inouye M, Woychik NA. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc. Natl. Acad. Sci. USA. 2008;105:5885–5890. doi: 10.1073/pnas.0711949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loris R, et al. Crystal structure of CcdB, a topoisomerase poison from E. coli. J. Mol. Biol. 1999;285:1667–1677. doi: 10.1006/jmbi.1998.2395. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y, Pogliano J, Helinski DR, Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto S, et al. Novel toxin-antitoxin system composed of serine protease and AAA-ATPase homologues determines the high level of stability and incompatibility of the tumor-inducing plasmid pTiC58. J. Bacteriol. 2009;191:4656–4666. doi: 10.1128/JB.00124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinhart A, Alonso JC, Strater N, Saenger W. Crystal structure of the plasmid maintenance system ε/ζ: functional mechanism of toxin ζ and inactivation by ε2ζ2 complex formation. Proc. Natl. Acad. Sci. USA. 2003;100:1661–1666. doi: 10.1073/pnas.0434325100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 24.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 25.Puglisi JD, Wyatt JR, Tinoco I., Jr. A pseudoknotted RNA oligonucleotide. Nature. 1988;331:283–286. doi: 10.1038/331283a0. [DOI] [PubMed] [Google Scholar]
- 26.Nissen P, et al. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 29.Buts L, et al. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Kamada K, Hanaoka F, Burley SK. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell. 2003;11:875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 31.Diago-Navarro E, et al. parD toxin-antitoxin system of plasmid R1—basic contributions, biotechnological applications and relationships with closely-related toxin-antitoxin systems. FEBS J. 2010;277:3097–3117. doi: 10.1111/j.1742-4658.2010.07722.x. [DOI] [PubMed] [Google Scholar]
- 32.Sussman D, Nix JC, Wilson C. The structural basis for molecular recognition by the vitamin B12 RNA aptamer. Nat. Struct. Biol. 2000;7:53–57. doi: 10.1038/71253. [DOI] [PubMed] [Google Scholar]
- 33.Klein DJ, Edwards TE, Ferre-D’Amare AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat. Struct. Mol. Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen-Dalsgaard M, Jorgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010;75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fozo EM, et al. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of Type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su L, et al. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 40.Tuerk C, MacDougal S, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 2007;10:262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paterson NG, Riboldi-Tunnicliffe A, Mitchell TJ, Isaacs NW. Purification, crystallization and preliminary X-ray diffraction analysis of RafE, a sugar-binding lipoprotein from Streptococcus pneumoniae. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2006;62:676–679. doi: 10.1107/S1744309106021695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Arcy A, Villard F, Marsh M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 2007;63:550–554. doi: 10.1107/S0907444907007652. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor R. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Sheldrick GM. Phase annealing in SHELX-90: direct methods for larger structures. Acta Crystallogr. A. 1990;46:467–473. [Google Scholar]
- 48.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 49.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 54.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellis EL, Delbrück M. The growth of bacteriophage. J. Gen. Physiol. 1939;22:365–384. doi: 10.1085/jgp.22.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.