Abstract
We report the effects of the interaction of two camelid antibody fragments, generally called nanobodies, namely cAb-HuL5 and a stabilized and more aggregation-resistant variant cAb-HuL5G obtained by protein engineering, on the properties of two amyloidogenic variants of human lysozyme, I56T and D67H, whose deposition in vital organs including the liver, kidney, and spleen is associated with a familial non-neuropathic systemic amyloidosis. Both NMR spectroscopy and X-ray crystallographic studies reveal that cAb-HuL5 binds to the α-domain, one of the two lobes of the native lysozyme structure. The binding of cAb-HuL5/cAb-HuL5G strongly inhibits fibril formation by the amyloidogenic variants; it does not, however, suppress the locally transient cooperative unfolding transitions, characteristic of these variants, in which the β-domain and the C-helix unfold and which represents key early intermediate species in the formation of amyloid fibrils. Therefore, unlike two other nanobodies previously described, cAb-HuL5/cAb-HuL5G does not inhibit fibril formation via the restoration of the global cooperativity of the native structure of the lysozyme variants to that characteristic of the wild-type protein. Instead, it inhibits a subsequent step in the assembly of the fibrils, involving the unfolding and structural reorganization of the α-domain. These results show that nanobodies can protect against the formation of pathogenic aggregates at different stages in the structural transition of a protein from the soluble native state into amyloid fibrils, illustrating their value as structural probes to study the molecular mechanisms of amyloid fibril formation. Combined with their amenability to protein engineering techniques to improve their stability and solubility, these findings support the suggestion that nanobodies can potentially be developed as therapeutics to combat protein misfolding diseases.
INTRODUCTION
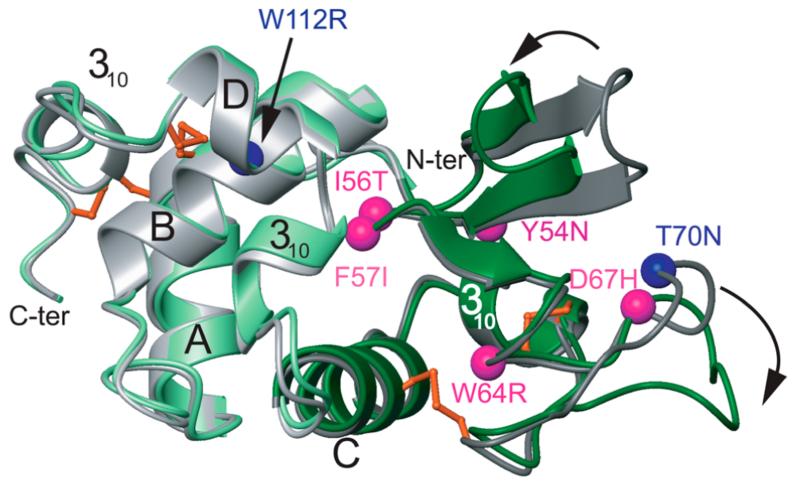
Human lysozyme is a 130-residue protein belonging to the c-type class of lysozymes and is present at high concentrations in various tissues and fluids including liver, articular cartilage, saliva, and tears. Since its discovery in 1922 by Alexander Fleming, this family of proteins has represented one of the most popular and important model systems for understanding the complexity of protein structure and function.1-3 In the early 1990s, mutational variants of human lysozyme were found to be associated with a familial systemic non-neuropathic amyloidosis in which large quantities of the variant protein accumulate in a range of tissues and organs, including the liver, spleen, and kidney.4 To date, nine naturally occurring variants of human lysozyme have been identified (Y54N, I56T, F57I, W64R, D67H, F57I/T70N, T70N, W112R, and T70N/W112R),4-9 seven of which are associated with disease (Y54N, I56T, F57I, W64R, D67H, F57I/T70N and T70N/W112R) (Figure 1). The extensive knowledge that has been accumulated on wild-type lysozyme (WT-HuL) (for reviews, see refs 1, 2 and 10) has enabled detailed studies of the effects of both amyloidogenic (I56T and D67H),11-20 non-amyloidogenic (T70N)21,22 and non-naturally occurring (I59T)23 mutations on the properties of the protein including its folding, stability and in vitro aggregation behavior. These studies have shed light on the molecular basis of amyloid formation by a globular protein, and revealed the link between normal folding behavior that leads to biological function and aberrant misfolding events that can lead to disease.24-26
Figure 1.
Overlay of ribbon diagrams representing the structures of WT-HuL (gray) and the D67H variant (green); the structure of the I56T variant is indistinguishable from that of WT-HuL. The α-helices are labeled A–D, and the three 310 helices are also indicated. The four disulfide bonds are shown in orange, and the Cα atoms of the amyloidogenic mutations 54, 56, 57, 64, and 67 are represented as magenta spheres. The T70N and W112R mutations, colored in blue, are not associated with disease unless they are in combination with a substitution at another position (F57I/T70N and T70N/W112R). The black arrows show the regions of the molecule (i.e., the two β-strands (residues 42–55) and the long loop (residues 66–77) of the β-domain) that are significantly distorted in the D67H variant compared to those of the WT-HuL and I56T proteins. The regions of the I56T and D67H variants that unfold transiently in a cooperative manner are colored in dark gray and dark green, respectively. The lysozyme structures were generated from coordinates determined by X-ray diffraction studies [PDB 1LYY (D67H), 1LOZ (I56T), and 1LZ1 (WT-HuL)] and produced using MOLMOL.
These investigations have allowed the identification of at least two factors that regulate the amyloidogenicity of lysozyme. The first is a significant decrease in the stability of the protein; for example, at pH 5.0, the two amyloidogenic variants, I56T and D67H, are ~10 ± 2 °C less thermally stable than WT-HuL,11,16,18,27,28 while the naturally occurring non-amyloidogenic T70N variant is destabilized by only ~4 °C.21,22 The second factor is a reduction in the global cooperativity under native conditions, which allows each amyloidogenic variant to significantly populate transient, partially unfolded species at physiologically relevant temperature and pH, where the wild-type protein is otherwise stable.4,12,27 Within these transient species, the region of the protein structure involving the C-helix and the β-domain (i.e., residues 42–100) is cooperatively unfolded, whereas the remainder of the α-domain remains largely native-like.12,27,28 This locally cooperative unfolding event is thought to be critical for initiating the self-association of the lysozyme variants into oligomeric species and subsequently into amyloid fibrils via intermolecular interactions between the locally unfolded segments of several molecules.12,27,28 A study using limited proteolysis has shown that the core region of human lysozyme fibrils formed in vitro by incubation at low pH and moderate temperature involves residues 38–102 of the protein,29 which corresponds to the β-sheet and the C-helix, the region that undergoes the cooperative unfolding event.15,20 These results support the conclusion that unfolding of these segments of the chain triggers the formation of the amyloid structure.
We have shown previously that interactions with the N-terminal fragment (VHH or nanobody) of two camelid heavy-chain antibodies (HCAb),30 denoted cAb-HuL6 and cAb-HuL22, which specifically recognize two different epitopes on the lysozyme structure, inhibit significantly in vitro fibril formation by both the I56T and D67H variants.27,28,31 cAb-HuL6 interacts with three residues of the loop between the A- and B-helices within the α-domain (L15, G16, and Y20), four residues of the long loop within the β-domain (A76, C77, H78, and L79), and most residues of the C-helix (A90, D91, A94, C95, K97, R98, and R101).28 Although no crystal structure of the complex between cAb-HuL22 and WT-HuL is available, competition binding experiments with a substrate analogue, as well as WT-HuL enzymatic activity measurements in the presence of cAb-HuL22, show that it binds to residues located in the active site of WT-HuL.31 Interestingly, despite the different location of their epitopes, the binding of each nanobody efficiently suppresses the locally cooperative unfolding event of the β-domain and C-helix for both the I56T and D67H variants under physiologically relevant conditions, thus restoring the global cooperativity of these variants to that characteristic of WT-HuL. The results of these studies strongly support the mechanism of lysozyme aggregation in which the reduction of both native stability and global cooperativity of the amyloidogenic variants are crucial initial events on the pathway of aggregation.2
In the present work, we have isolated a novel nanobody, referred to as cAb-HuL5, which binds to WT-HuL and the I56T and D67H amyloidogenic variants. In contrast to the previously reported nanobodies, cAb-HuL5 does not interact with any of the residues of HuL that are involved in the cooperative unfolding of the β-domain and C-helix of amyloidogenic lysozyme variants. This finding provides us with a unique opportunity to evaluate the effects of binding to non-amyloidogenic regions of HuL on the formation of partially unfolded species, and consequently of amyloid fibrils, by amyloidogenic variants of HuL. We show that the binding of the nanobody does not inhibit the formation of partially unfolded intermediates of amyloidogenic HuL variants under native conditions, highlighting the importance of the location of the epitope for restoring the loss of global cooperativity due to destabilizing mutations in the amyloidogenic variants of HuL. Nevertheless, the binding of cAb-HuL5G (a very stable chimeric nanobody generated by grafting the three complementarity determining regions (CDRs) of cAb-HuL5 onto the framework of cAb-HuL6) suppresses amyloid fibril formation very effectively, revealing that the involvement of the α-domain is a crucial step in the formation of amyloid fibrils by human lysozyme. These investigations demonstrate further the versatility of nanobodies for isolating different species present along the aggregation pathway and confirm that these biomolecules can act as very valuable structural tools to uncover the underlying molecular mechanisms of protein misfolding and aggregation.32 We further show that nanobodies are particularly attractive moieties compared to other antibody fragments due to their simplicity and amenability to modification by protein engineering techniques, allowing them to be tailored towards improved stability and potency to inhibit amyloid fibril formation; these characteristics are likely to be highly beneficial for therapeutic lead development.
MATERIALS AND METHODS
Generation and Selection of cAb-HuL5
A dromedary was immunized with wild-type human lysozyme, as described previously.33 A phage library containing the genes coding for the variable domains of the heavy-chain antibodies (nanobodies) was then generated using mRNA extracted from the lymphocytes of the immunized dromedary.34 A novel nanobody specific for human lysozyme, denoted cAb-HuL5, was isolated by biopanning from this phage library. cAb-HuL5 was expressed in Escherichia coli and purified to homogeneity as described previously.35
Generation of cAb-HuL5G
cAb-HuL5G is a chimeric nanobody obtained by grafting the three complementarity determining regions (CDRs) of cAb-HuL5 onto the framework of cAb-HuL6, a nanobody found to be highly stable and resistant to aggregation under the conditions used to induce in vitro fibril formation of the amyloidogenic variants of lysozyme.27,28,35 CDRs 1 and 2 of cAb-HuL5 were grafted onto the framework of cAb-HuL6, using a PCR method36 and the following primers: CDR1HuL5_HuL6_F (5′-CTGTTCAGCCTCCGGCCTTAGTACTACTGTCATGGCCTGGTTCCGCCAGG-3′) and CDR2HuL5_HuL6_R (5′-GGAGTCGGCATAGTATGGGAAACCATCACCAGTATAAATAGCTGCGACCCCCTCGCG-3′). CDR 3 of cAb-HuL5 was grafted onto the CDR1/2 grafted-cAb-HuL6 using PfuI polymerase and the primers CDR3HuL5_HuL6_R (5′-GACATCCACCACAAAGAACCGTACGAGAAAGCACCTGTCTTCGCTGCACAGTAGTA-3′) and CDR3HuL5_HuL6_F (5′-CCGGGCGTATAATCACTGGGGCCAGGGGAC-3′). The linear PCR fragment was inserted into the pHEN6 expression vector33 by ligation with T4 DNA ligase, transformed into E. coli WK6 (Δ(lac-proAB)galEstrA [F′ lacIq lacZΔM15 proAB+]) CaCl2 competent cells and its sequence was confirmed by DNA sequencing analysis. The cAb-HuL5 loop-grafted variant, denoted cAb-HuL5G, was expressed and purified in the same way as cAb-HuL5.
Expression and Purification of Human Lysozyme
Wild-type human lysozyme, including uniformly 15N-labeled protein, was expressed in Pichia pastoris and purified as previously described.22 The I56T and D67H lysozyme variants, including uniformly 15N-labeled proteins, were expressed in Aspergillus niger and purified following established protocols.35,37 The lysozyme concentration was determined by absorbance measurements using a calculated molar extinction coefficient of 36 940 cm−1 M−1.38
Surface Plasmon Resonance Measurements
The affinity of the nanobodies for the various lysozyme variants was determined using surface plasmon resonance following protocols described previously.35 The nanobody concentrations were determined using a calculated extinction coefficient of 34 045 cm−1 M−1 for both cAb-HuL5 and cAb-HuL5G.38
Thermal Denaturation Monitored by Circular Dichroism (CD) Spectroscopy
The thermal denaturation of cAb-HuL5 and cAb-HuL5G was followed at 237 nm, since at this wavelength there is a large difference between their far UV-CD spectra recorded at 25 and 95 °C, respectively. Thermal unfolding was monitored in 0.1 M sodium citrate at pH 5.5 containing 3M urea, the buffer used to initiate aggregation of the amyloidogenic lysozyme variants. The temperature was increased monotonically from 25 to 90 °C at a rate of 0.5 °C· min−1. The data obtained from the buffer alone was subtracted from the melting curves of the samples containing a nanobody. The resulting data were then fitted to a two-state unfolding model, using eq 1:
| (1) |
where y is the CD signal at 237 nm, yN and yD are the CD signals for the native and denatured states of the protein at 0 °C, respectively, T is the temperature in °C, R is the gas constant in J·mol−1·°C−1, Tm is the midpoint of the heat-induced unfolding in °C, ΔHm is the Van’t Hoff enthalpy at Tm, and mN and mD are the slopes for the pre- and post-unfolding baselines, respectively. Nonlinear regression analysis was performed using the program Origin 7.0 (MicroCal, Northampton, MA, USA).
Kinetics of Aggregation of D67H Lysozyme in the Presence of cAb-HuL5G
Protein samples containing the D67H variant alone (6.8 μM), the D67H/cAb-HuL5G complex (both proteins at 6.8 μM or in some cases with a 2-fold excess of cAb-HuL5G, i.e., 13.6 μM), and the cAb-HuL5G fragment alone (14 μM) were prepared in 0.1 M sodium citrate buffer pH 5.5 with 3 M urea and passed through 0.22 μm filters. Samples were placed in quartz cuvettes of 1 cm path length and stirred vigorously at 48 °C. Right angle light scattering at 430 nm was recorded every 1 min for each sample using a Cary 400 scan UV–visible spectrophotometer (Varian, CA, USA) with a slit width of 5 nm. 50 μL aliquots from each sample were taken at various time points throughout the reaction to be analyzed by transmission electron microscopy (TEM). To determine the effects of the cAb-HuL5G:D67H ratio on the kinetics of aggregation of the D67H variant, samples were prepared using 14 μM cAb-HuL5G and either 3.4 or 1.24 μM D67H lysozyme (i.e., corresponding to 4:1 and 11.2:1 molar ratios of cAb-HuL5G:D67H) in 3 M urea, 0.1 M sodium citrate buffer at pH 5.5, and incubated at 48 °C under stirring conditions in quartz cuvettes. The light scattering at 430 nm was measured at right angles in a Cary Eclipse fluorimeter (Varian, Walnut Creek, CA, USA).
Transmission Electron Microscopy
Samples were applied to formvar carbon-coated grids (Agar Scientific, Stansted, UK) and then stained with 2% uranyl acetate, and examined using a Philips CEM 100 transmission electron microscope operating at 80 kV.
Mass Spectrometry
The I56T and D67H variants were deuterated at exchangeable sites by unfolding in 6 M deuterated guanidinium chloride solutions, followed by dilution with 10 volumes of deuterated buffer (50 mM deuterated acetic acid pH* 5.0) to refold the proteins. The proteins were concentrated and subsequently diluted with D2O for eight cycles to remove guanidinium chloride, using Centri-pre-3 concentrators (Amicon, Millipore, Watford, UK). For the experiments in the presence of cAb-HuL5, the deuterated lysozyme variant and the protonated cAb-HuL5 fragment were combined immediately before the H/D exchange experiments were performed to give a stoichiometric molar ratio of lysozyme:cAb-HuL5 = 1:2 (to ensure all the lysozyme variants were in the antibody-bound state). H/D exchange was initiated by diluting 1 volume of the protein solution into 15 volumes of 100 mM ammonia/formic acid buffer in H2O at pH 8.0 and 37 °C, and exchange was allowed to proceed for various time points between 5 and 300 s. The H/D exchange was quenched by the addition of 7 volumes of 1 M acetic acid in H2O to generate a final pH of 3.5. The samples were immediately placed on ice before being analyzed by electrospray mass spectrometry; samples were analyzed at the base pressure of a LC-ToF spectrometer (Waters, Milford MA, UK) with a capillary voltage of 1600 V and a cone voltage of 80 V. Under these conditions, the antibody/lysozyme complexes dissociate in the mass spectrometer, enabling the mass distributions of the lysozyme molecules alone to be determined readily and directly.28 No adjustment was made to the total number of exchanged proton atoms for the remaining 6% deuterium present in the sample, and the mass spectra shown in Figure 4 represent the convolution of the +8, +9, and +10 charge states with minimal smoothing, converted to a mass scale.12,28
NMR Studies of the WT-HuL/cAb-HuL5 and I56T/cAb-HuL5 Complexes
Two-dimensional [15N–1H]-HSQC spectra of wild-type human lysozyme (uniformly 15N-labeled) in the presence and absence of the unlabeled cAb-HuL5 fragment were recorded at 35 °C using a Bruker Biospin Avance 700 MHz NMR spectrometer equipped with a cryo-platform (Bruker, Coventry, UK). Samples of the nanobody/lysozyme complex were made up with a 2-fold molar excess of the antibody fragment to effectively ensure that all the lysozyme molecules were in the bound state, in 20 mM phosphate buffer pH 6.5 in 95% H2O/5% D2O. The HSQC spectra were collected with 2048 and 256 complex points in t1 (1H) and t2 (15 N), and with sweep widths of 8389 and 2483 Hz in the 1H and 15N dimensions, respectively. The 1H and 15N resonances of wild-type human lysozyme in complex with the antibody fragment were assigned by 15N-edited 3D NOESY-HSQC measurements. These spectra were collected with 2048, 74, and 138 complex points in t1 (1H), t2 (15N), and t3 (1H), respectively, and the observed NOE data for the antibody/lysozyme complex were interpreted by using the assignments of the free protein. All the NMR spectra were processed with NMRPipe39 and Sparky (http://www.cgl.ucsf.edu/home/sparky/). Similar NMR measurements were also performed with the I56T variant and the I56T/cAb-HuL5 complex in 20 mM sodium acetate buffer pH 5.0 in 95% H2O/5% D2O at 37 °C.
Crystallization and Structure Solution of the cAb-HuL5/WT-HuL Complex
Single crystals of the cAb-HuL5/WT-HuL complex were obtained by hanging drop vapor diffusion at 3.9 mg/mL, in 4.8 M NaCl, 0.1 M HEPES pH 7, and 3% glycerol. X-ray diffraction data from frozen crystals were obtained using the EMBL BW7B beamline equipped with a MAR CCD 165 mm detector at the DESY synchrotron facility (Hamburg, Germany). The diffraction data were processed by DENZO and SCALEPACK40 to determine the space group and to scale and merge the data. Molecular replacement was performed by the program Phaser41 using the structures of cAb-HuL6 and WT-HuL in PDB entry 1OP9 as search models for, respectively, cAb-HuL5 and WT-HuL, to obtain the phase information associated with the structure factors. Model building and refinement were achieved using the programs Phenix Suite42 and Refmac5 as implemented in the CCP4 suite.43 The graphics program Coot44 was used to interpret the electron density maps and for rebuilding of the structure. Data collection and refinement statistics are listed in Table 1.
Table 1. Data Collection and Refinement Statistics.
| Dataset | cAb-HuL5/WT-HuL |
|---|---|
| Data Collection | |
| space group | P 43 21 2 |
| cell dimensions | |
| a, b, c (Å) | 96.32, 96.32, 156.75 |
| α, β, γ (deg) | 90.00, 90.00, 90.00 |
| resolution (Å) | 1.95 |
| R sym | 11.5 (54.0)a |
| I/σI | 23.3 (4.3)a |
| completeness (%) | 99.9 (99.9) |
| redundancy | 8.6 (7.6)a |
| Refinement | |
| resolution range (Å) | 19.90–1.95 (2.10–1.95) |
| no. reflections | 54358 |
| Rwork/Rfree | 18.9/21.9 |
| B-factors (Å2) | |
| protein | 24.6 |
| water | 28.1 |
| rms deviations | |
| bond lengths (Å) | 0.007 |
| bond angles (deg) | 1.014 |
The values in parentheses represent those of the highest resolution shell.
The structural and chemical properties of the cAb-HuL5/WT-HuL interface were analyzed using the PDBePISA server at the EMBL (http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver).45 The Lee and Richards algorithm, using a probe radius of 1.4 Å, was used to calculate the change in solvent accessible surface area (ΔASA) of both cAb-HuL5 and WT-HuL upon complex formation.46 The residues of WT-HuL and cAb-HuL5 that have atoms within 4 Å of each other in the complex were considered to be interface residues. These interatom distances were calculated using the program Contact, implemented in the CCP4 software.47 The HBPLUS program,48 was used to calculate the number of hydrogen bonds formed in the interface. Figures were prepared with the program Pymol (www.pymol.org).
Binding of cAb-HuL5 to Amyloid Fibrils Monitored by Tryptophan Fluorescence Measurements
Fibrils of the D67H variant were prepared by incubating the D67H variant protein (6.8 μM) in 0.1 M sodium citrate buffer pH 5.5 containing 3 M urea at 48 °C with stirring for approximately 12 h. The fibrils were collected by centrifugation at 353 200 g and 25 °C for 1 h using an Optima TLX ultracentrifuge (Beckman Coulter, High Wycomb, UK), and then purified by three cycles of washing with 1 mL of 0.1 M sodium acetate buffer, pH 5.5, and further centrifuged at 353 200 g and 25 °C for 1 h. The fibrils were resuspended at a concentration of 0.4 mg/mL in 0.1 M sodium acetate buffer pH 5.5 containing 0.4 mg/mL of cAb-HuL5. After incubation for 1 h at 25 °C, the sample was centrifuged at 353 200 g and 25 °C for 1 h. The fluorescence emission spectrum of the supernatant was recorded on a Cary Eclipse spectrofluorimeter at 25 °C between 300 and 440 nm. The excitation and emission slit widths were 5 and 10 nm, respectively, and the excitation wavelength was 295 nm. For comparison purposes, similar fluorescence measurements were also performed with a solution of 0.4 mg/mL cAb-HuL5 in 0.1 M sodium acetate, pH 5.5.
FTIR Measurements
Fibrils of the D67H variant were prepared and purified as described above. The fibrils were resuspended, at ~10 mg/mL, in 10 μL of 0.1 M sodium acetate buffer pH 5.5. A total of 256 interferograms for each sample were recorded on a Bruker Equinox 55 FTIR spectrometer (Bruker, Coventry, UK) (purged with N2) by means of attenuated total reflection (ATR) at 25 °C, and then subjected to Fourier transformation. These signals were subtracted from those of the buffer (0.1 M sodium acetate, pH 5.5) recorded under the same conditions. The amide I region (1580–1720 cm−1) of each IR spectrum was subjected to Fourier self-deconvolution and subsequently fitted to Gaussian functions to determine component peaks using Grams/32 (Galactic Industries Co., NH, USA). Similar IR measurements and data analysis were also performed with the monomeric D67H variant (10 mg/mL) under similar conditions. The percentage area of the amide I component was obtained by integrating the area under each deconvoluted band; the area corresponding to side chain contributions was not considered.
Isothermal Calorimetry Measurements
The interaction of cAb-HuL5G with WT-HuL was measured by isothermal calorimetry using a VP-ITC instrument (GE Healthcare, Buckinghamshire, UK) with a cell volume of 1.4 mL. The assay was performed in 100 mM sodium citrate buffer pH 5.5 containing 3M urea at 50 °C. Before analysis, lyophilized cAb-HuL5G and WT-HuL were dissolved in water and extensively dialyzed in 100 mM sodium citrate buffer pH 5.5 containing 3M urea to ensure matching buffer compositions. Protein concentrations were measured spectrophotometrically, using molar extinction coefficients at 280 nm of 34 045 M−1 cm−1 for the nanobody and 36 940 M−1 cm−1 for WT-HuL.38 After degassing both protein solutions, cAb-HuL5G at 700 μM was loaded into the syringe and WT-HuL at 10 μM was loaded in the ITC cell. After an initial 4 μL injection of cAb-HuL5G, 27 injections of 10 μL of nanobody were performed. The heat change for the last three injections, for which binding reached saturation, was used to estimate the heat of dilution and mixing, by subtraction of a straight line through the last three points. The integrated heats for each injection were then plotted against the molar ratio of nanobody:WT-HuL, discarding the data for the first two injections. Data were processed using MicroCal Origin 7.0 software.
RESULTS
cAb-HuL5 Binds to the α-Domain of the Lysozyme Structure
A new nanobody, cAb-HuL5, with high affinity for human lysozyme has been selected from a library of nanobody genes cloned from the blood of a dromedary immunized with WT-HuL according to a previously described procedure.34 This antibody fragment was expressed in E. coli35 and purified to homogeneity using previously described procedures.35 Its binding properties to WT-HuL and the I56T and D67H amyloidogenic variants were characterized by surface plasmon resonance (SPR) measurements (Table 2).35
Table 2. Kinetic and Equilibrium Parameters for the Interaction of the cAb-HuL5 and cAb-HuL5G with WT-HuL and the I56T and D67H Amyloidogenic Variants Determined by SPR.
| WT-HuL |
D67H |
I56T |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
kon( M−1 s−1) 10−5 |
koff ((s−1) | KD (M) 107 |
kon (M−1 s−1) 10−5 |
koff (s−1) | KD (M) 107 |
kon(M−1 s−1) 10−5 |
koff (s−1) | KD (M) 107 | |
| cAb-HuL5 | 1.9 ± 0.2 | 0.088 ± 0.009 | 4.6 ± 0.1 | 2.0 ± 0.2 | 0.095 ± 0.001 | 4.6 ± 0.05 | 1.8 ± 0.02 | 0.073 ± 0.007 | 4.1 ± 0.43 |
| cAb-HuL5G | 4.10 ± 0.06 | 0.128 ± 0.001 | 3.1 ± 0.05 | Nda | Nda | Nda | Nda | Nda | Nda |
Not determined.
This antibody fragment displays an affinity for the WT-HuL (KD = 460 nM) that is lower than what is typically found for other in vivo affinity matured nanobodies (low nM range).35,49 The KD values measured for the cAb-HuL5/I56T and cAb-HuL5/D67H variant complexes are, however, closely similar to those for WT-HuL (410 and 460 nM for I56T and D67H variants, respectively); this observation suggests that cAb-HuL5 recognizes a region of the protein structure that is essentially the same in both variants and the wild-type structure (i.e., it does not involve the β-strands (residues 42–55) or the long loop (residues 66–77) within the β-domain; Figure 1).11
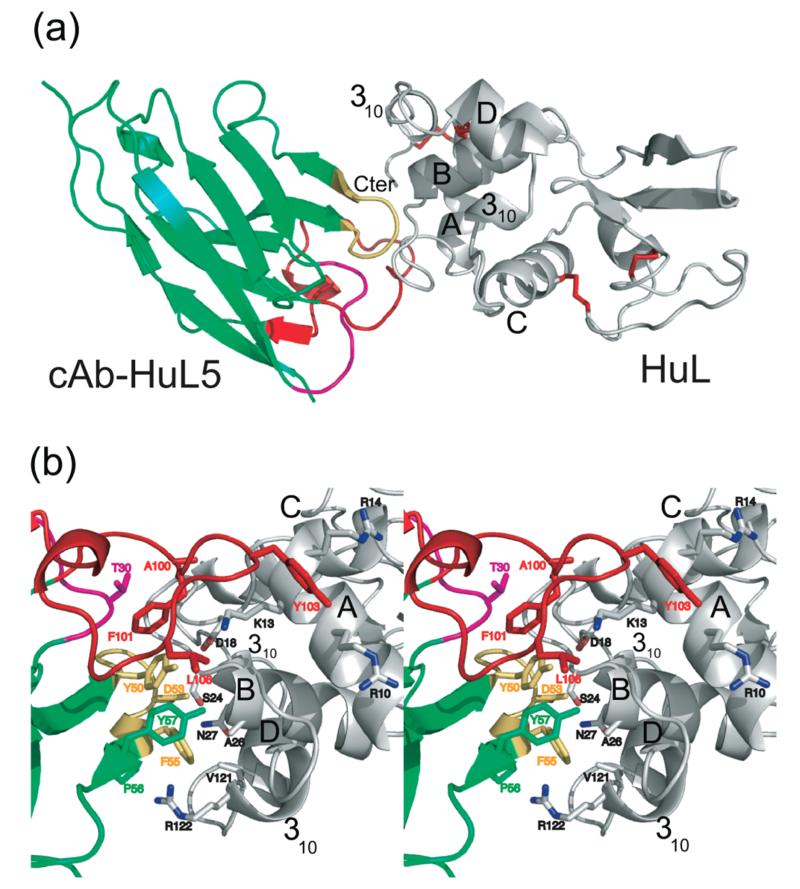
To define the epitope in detail, cAb-HuL5 was crystallized in complex with WT-HuL and the structure of the complex was solved at a resolution of 1.95 Å by using X-ray diffraction (Figure 2).
Figure 2.
X-ray structure of the complex between WT-HuL and cAb-HuL5. (a) Ribbon representation of the overall structure of the complex between cAb-HuL5 (green) and WT-HuL (gray). The CDRs are colored in magenta (CDR1), yellow (CDR2), and red (CDR3), respectively. The disulfide bonds in WT-HuL are represented as red sticks. (b) Stereo drawing of the details of the cAb-HuL5/WT-HuL interaction. The side chains of the residues of both proteins that are within 4 Å of each other are represented by sticks and are labeled; the labels for the antibody fragment are color-coded according to the antigen-binding loop to which they belong. (Figures were prepared using Pymol, www.pymol.org.)
The formation of the cAb-HuL5/WT-HuL complex buries a total of 661 Å2 of the accessible surface area (ASA) of WT-HuL and 679 Å2 of the ASA of the antibody fragment, with 10 direct hydrogen bonds being formed between the two proteins. These interface properties are typical for nanobody–antigen and antibody–antigen interfaces.50-52 There is, however, an interesting difference in the polarities of the surface-binding patches on the two proteins, as that of the nanobody is more apolar (71%) than the corresponding patch of WT-HuL (61%). The residues in the interface region of the complex (defined as residues of WT-HuL and cAb-HuL5 that have atoms which are within 4 Å of each other in the complex) are R10, K13, R14, G16, D18, G19, G22, I23, S24, A26, N27, V121, R122, and V130 for WT-HuL and T30, Y50, T51, G52, D53, F55, P56, Y57, A100, F101, S102, Y103, S105, and L106 for cAb-HuL5. The epitope of cAb-HuL5 therefore consist of 14 residues of the lysozyme molecule located primarily in the loop between the helices A and B but also including some residues located at the beginning of helix B and the C-terminal 310 helix and at the end of helix A.
Analysis of this interface reveals that cAb-HuL5, unlike the previously characterized nanobodies raised against WT-HuL, does not bind to any of the residues involved in the locally cooperative unfolding of the amyloidogenic variants, that is thought to be the trigger of amyloid fibril formation.12,27,28 Moreover, comparing the structural alignment of lysozyme in complex with cAb-HuL5 with structures of WT-HuL, previously deposited in the protein databank, reveals that the binding of the nanobody does not induce any conformational changes which would cause the structure within the complex to deviate significantly from those found in other lysozyme crystal structures (Figure S1 and Table S1, Supporting Information); the average RMSD of the Cα atoms of lysozyme in the complex compared to those of the structures listed (see Table S1, Supporting Information) is 0.42 Å, while all the structures of the free lysozyme molecules have averaged RMSD values between 0.1 and 0.2 Å; the slightly larger RMSD value found for the lysozyme molecule within the cAb-HuL5:HuL complex is due to a small difference in the conformation of the lysozyme protein backbone in the region of residue 22 and residue 120, which are both part of the binding site of cAb-HuL5 (Figure S1, Supporting Information).
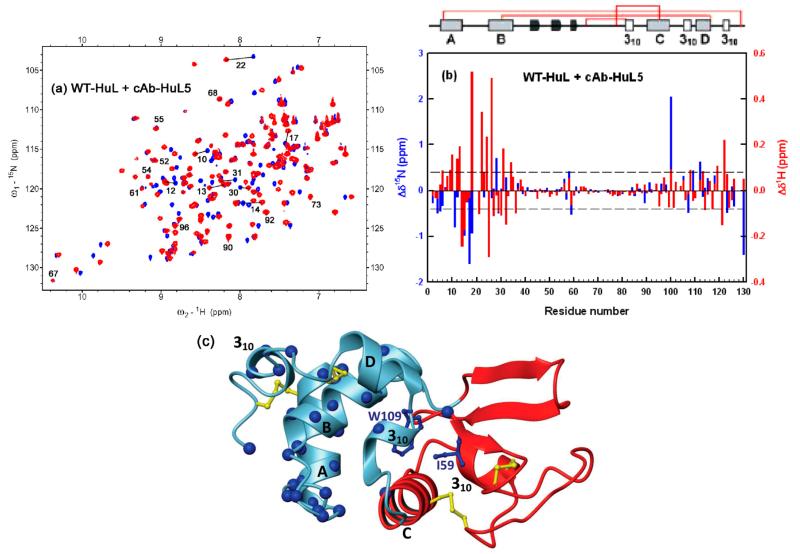
Previous work has, however, shown that the dynamical properties of regions far from the epitope can be affected by the binding of an antibody or antibody fragment via the long-range propagation of very subtle structural perturbations.53 We therefore studied the long-range effects of the binding of cAb-HuL5 by carrying out a series of NMR experiments. We first mapped the effects on WT-HuL resulting from the binding of cAb-HuL5 by comparing the HSQC spectrum of 15N-labeled free WT-HuL with that of the labeled protein within the complex with unlabeled cAb-HuL5.27,28 The chemical shift perturbations of the amide resonances of WT-HuL were then analyzed to identify those that are significantly affected by the interactions with cAb-HuL5 (Figure 3a). Analysis of these data indicates that 35 residues of WT-HuL show significant chemical shift perturbations (|Δδ1H| ≥ 0.1 ppm or (|Δδ15N| ≥ 0.4 ppm) upon binding to the antibody fragment (Figure 3b). Resonances of most of the residuesfound to be in direct contact with cAb-HuL5 in the X-ray structure are significantly shifted relative to their position in the unbound protein. Most of the additional residues with perturbed chemical shift are located primarily in the A- and B-helices and, additionally, in the 310 helix, which is adjacent to the C-helix within the α-domain. These results strongly suggest that the nanobody binds to WT-HuL in solution and in the crystal in the same way.
Figure 3.
Overlaid [15N–1H]-HSQC NMR spectra of (a) WT-HuL (pH 6.5 and 35 °C) in the absence (blue) and presence (red) of cAb-HuL5. (b) Chemical shift perturbations of WT-HuL induced by the binding of cAb-HuL5; red and blue bars represent the perturbations of the 1H and 15N resonances, respectively. Residues experiencing a chemical shift change ≥ |0.4| ppm for 15N or ≥ |0.1| ppm for 1H resonances are considered to be affected significantly by the binding of the nanobody. (c) Ribbon diagram of WT-HuL showing the Cα atoms of the residues affected significantly by the binding of cAb-HuL5 (blue spheres). The four disulfide bonds are shown in yellow. The β-domain and the adjacent C-helix that are unfolded in the aggregation-prone intermediates (see text) are colored red. The peaks corresponding to G19 and Y20 (which are located in the loop between the A- and B-helices) in the complex have not been assigned; their chemical shift differences are therefore not shown in the histograms. No unassigned peaks are visible in the HSQC spectra of the complexes within 0.1 ppm (1H) and 0.4 ppm (15N) of the peak positions of G19 and Y20 in the HSQC spectrum of the unbound lysozyme protein, suggesting that there are very significant chemical shift perturbations for these residues upon binding to the antibody fragment. Note that I59 and W109 (shown at blue sticks) whose side chains point toward the interface between the α- and β-domains are significantly affected by the binding of cAb-HuL5. The structure of WT-HuL was generated from its X-ray coordinates (PDB 1LZ1) and the diagram produced using MOLMOL.
Similar NMR experiments were carried out with the I56T variant (Figure S2, Supporting Information), which indicate that cAb-HuL5 binds to this species in a closely similar manner to that of WT-HuL. This observation is completely consistent with the findings from SPR studies, which show that the affinities of cAb-HuL5 for the WT-HuL (KD = 4.6 × 10−7 M), I56T (KD = 4.1 × 10−7 M), and D67H (KD = 4.6 × 10−7 M) are essentially identical (Table 1). In addition, none of the residues of the locally unfolded region of the amyloidogenic intermediate (the C-helix and the β-domain) show significant chemical shift perturbations upon binding to the nanobody except for Q58, I59 (located in the β-domain at its interface with the α-domain), and V100 (located in the C-terminal end of the C-helix) (Figure 3c); again, this observation is in marked contrast to the findings with the other nanobodies that interact with human lysozyme.27,28,31 Indeed, the chemical shift of 11 and 26 residues from the region that unfolds cooperatively were affected upon binding of respectively cAb-HuL6 and cAb-HuL22.
The Binding of cAb-HuL5 Does Not Restore the Global Cooperativity of the I56T and D67H Variants
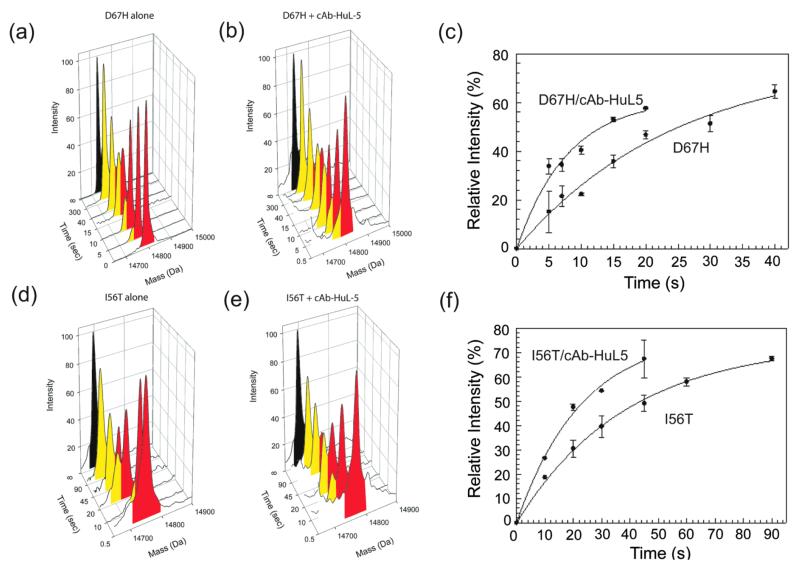
In order to investigate the effects of the binding of cAb-HuL5 on the structural cooperativity of the I56T and D67H variants, pulse-labeling H/D exchange experiments coupled with ESI-MS were performed.11,12,27,28,31 Briefly, the deuterated I56T and D67H variants were first subjected to exchange with protons of the solvent H2O at pH 8.0 and 37 °C, and the exchange was subsequently quenched at various time intervals by decreasing the pH and the temperature of the sample. Similar experiments were performed with both variants in the presence of a 2-fold molar excess of cAb-HuL5. In the absence of cAb-HuL5, the lysozyme variants show a clear bimodal distribution of mass peaks over the time course of exchange (Figure 4a and c).
Figure 4.
H/D exchange behavior of the I56T and D67H variants of human lysozyme in the absence and presence of cAb-HuL5. Electrospray mass spectra of the D67H (a, b) and I56T (d, e) variants following H/D exchange in the absence (a, d) and presence (b, e) of cAb-HuL5 at pH 8.0 and 37 °C. For the lysozyme/nanobody mixtures, a 2-fold molar excess of cAb-HuL5 was added to ensure that effectively all the lysozyme molecules were in the bound state. The mass peaks colored red result from the gradual loss of deuterium atoms through an EX2 mechanism due to local independent structural fluctuations. The mass peaks colored yellow arise from locally cooperative unfolding events that give rise to H/D exchange through an EX1 mechanism. Note that the I56T and D67H variants exhibit bimodal distributions of mass peaks even in the presence of cAb-HuL5. Time courses of the relative intensities of the lower mass species (i.e., the yellow peaks) of the (c) D67H and (f) I56T variants in the absence and presence of cAb-HuL5. The data are the average of two experiments, and the bars show the error estimated from these measurements. These data were fitted to exponential functions to determine the time constants (τ) of the locally cooperative unfolding of the lysozyme variants; τD67H = 25.2 ± 5.1 s; τD67H/cAb-HuL5 = 7.9 ± 0.05 s; τI56T = 36.8 ± 3.2 s; τI56T/cAb-HuL5 = 24.4 ± 8.6 s.
The peaks colored red arise from the lysozyme variants in their native states undergoing H/D exchange through an EX2 mechanism as a result of local independent fluctuations, while those colored yellow result from the variant protein molecules which have accessed the transient partially unfolded state at least once and have therefore undergone exchange through an EX1 mechanism.11,12 In the presence of cAb-HuL5, this pattern of behavior is qualitatively unchanged, indicating that the binding of cAb-HuL5 does not suppress the locally cooperative unfolding process, and hence does not restore the global cooperativity to the lysozyme variants (Figure 4b and d). The relative intensities of the mass peaks corresponding to the two species were determined and the fractions of the lower mass species at the different time intervals calculated. The resulting time course of the relative intensity of the peak corresponding to the lower mass species provides a direct measure of the opening rate of the locally cooperative unfolding event of the D67H and I56T variants.12,27 Interestingly, the time constants for both variants in complex with cAb-HuL5 (τD67H/cAb-HuL5 = 7.9 ± 0.05 s; τI56T/cAb-HuL5 = 24.4 ± 8.6 s) are significantly smaller than those of the corresponding free variant proteins (τD67H = 25.2 ± 5.1 s; τI56T = 36.8 ± 3.2 s), indicating that binding to the nanobody in fact increases the rate of formation of the intermediate species for both variants (Figure 4c and f).
The Binding of cAb-HuL5G Inhibits Fibril Formation by the Amyloidogenic Lysozyme Variants
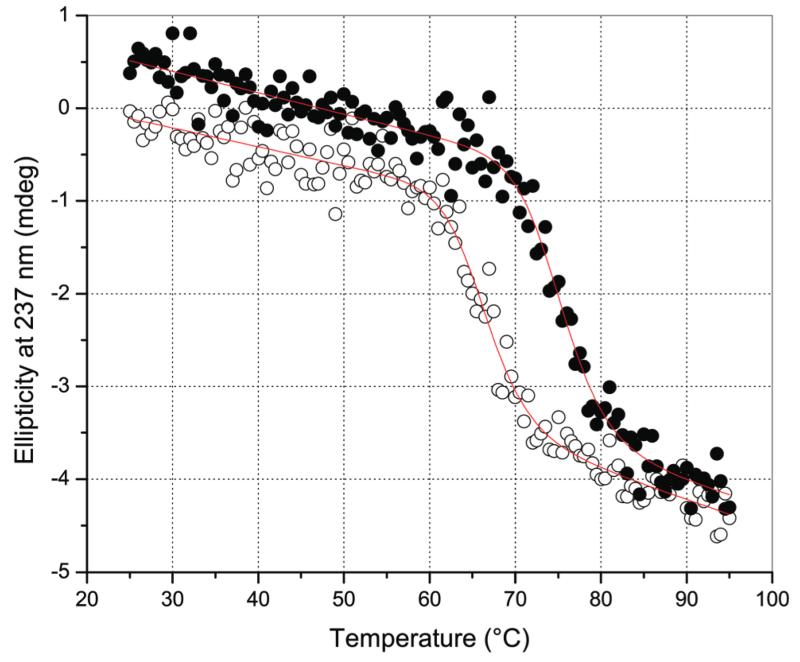
The effect of the cAb-HuL5 fragment on the propensity of the amyloidogenic variants to form fibrils was investigated by light scattering measurements of the D67H variant in 0.1 M sodium citrate buffer pH 5.5 containing 3M urea and 48 °C, conditions shown previously to give highly reproducible kinetics.28,31 Since the properties of the I56T and D67H variants are identical,12,13,27,28 only one of them (i.e. the D67H) was chosen as a model protein to test the ability of the nanobody to inhibit lysozyme formation. When incubated under these conditions, uncomplexed D67H aggregates within 60–100 min. In addition, however, despite being much more stable than the lysozyme variant under these conditions, cAb-HuL5 was itself found to aggregate significantly into amyloid fibrils, on the same time scale as D67H alone (data not shown). Thus, the effects of binding cAb-HuL5 on the lysozyme aggregation could not be investigated reliably under these conditions. We therefore engineered a less aggregation prone version of cAb-HuL5, referred to as cAb-HuL5G (see the Methods section), by grafting its CDRs54 onto the extremely stable and non-aggregation-prone scaffold of cAb-HuL6. The binding parameters of cAb-HuL5G to WT-HuL are similar to those measured for the cAb-HuL5 at pH 7.5 and 25 °C (Table 2). The midpoint of thermal unfolding (Tm) of cAb-HuL5G in 0.1 M sodium citrate buffer pH 5.5 containing 3 M urea is increased by ~9.5 °C compared to that of cAb-HuL5 (i.e., 75.5 ± 0.5 °C versus 66.0 ± 0.5 °C) (Figure 5).
Figure 5.
Thermal denaturation of cAb-HuL5 and cAb-HuL5G monitored by far UV-CD spectroscopy. Ellipticity at 237 nm measured as a function of temperature T (°C) for cAb-HuL5 (open circles) and cAb-HuL5G (filled black circles) after subtraction of the buffer signal. The red solid lines represent a nonlinear least-squares fit to the data using a two-state denaturation model described by eq 1.
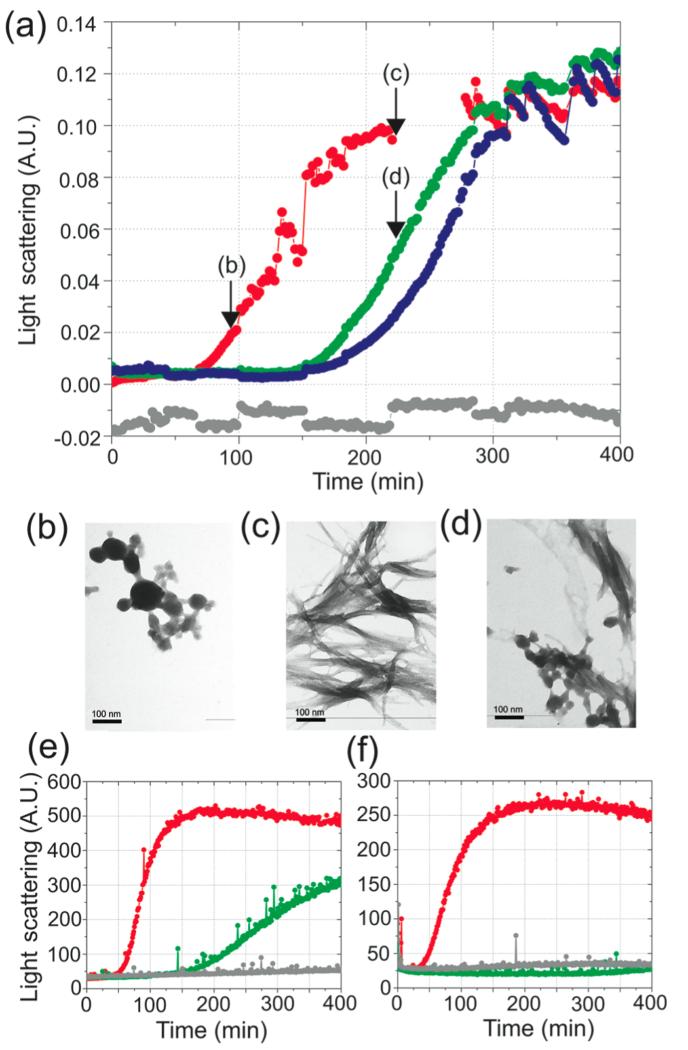
The increased thermal stability of cAb-HuL5G results in a greatly reduced tendency for the nanobody to aggregate under these conditions, enabling the aggregation of D67H to be monitored in the presence of a variety of molar equivalents of cAb-HuL5G (Figure 6). These experiments show that binding of the nanobody inhibits lysozyme aggregation, and in the presence of one equivalent of cAb-HuL5G, the lag time prior to the observation of light scattering of a 6.8 μM solution of D67H was increased by ~100 min, whereas with 2 equiv of cAb-HuL5G it was increased by ~125 min. Samples of the aggregation reaction mixtures in the presence and absence of cAb-HuL5G at identical time points (220 min) were taken for TEM analysis. The fibrils formed by D67H in the presence of 1 equiv of cAb-HuL5G are similar in morphology to those formed by D67H alone (Figure 6b–d). The samples with cAb-HuL5G, however, also contained small globular species, morphologically similar to species present at early time points (t = 98 min) in the samples of D67H alone. Given that at t = 220 min the reaction of the D67H variant in the presence of cAbHuL5G is close to its midpoint, these species probably represent prefibrillar aggregates which subsequently convert into mature fibrils, as observed in similar circumstances with many other systems.55-60
Figure 6.
Fibril formation by the D67H lysozyme variant in the absence and presence of cAb-HuL5G. (a) Time course of the aggregation in 0.1 M sodium citrate buffer pH 5.5 containing 3M urea at 48 °C with stirring of the D67H variant at 6.8 μM in the absence (red) and the presence of 1 equiv (green) and 2 equiv (blue) of cAb-HuL5G. The trace corresponding to the cAb-HuL5G alone and at a concentration that is equivalent to a 2-fold molar excess of the complex is shown in gray. Light scattering at 430 nm was recorded using a UV–visible spectrophotometer with a slit width of 5 nm. Arrows on the plot indicate the time points when samples were taken for TEM analysis. (b–d) TEM images of the aggregation reaction of (b) the D67H variant at 98 min, (c) the D67H variant at 220 min, and (d) the cAb-HuL5/D67H complex at 220 min. The scale bar representing 100 nm is shown in each panel. (e, f) Time course of the aggregation reactions of D67H at 3.4 μM (e) and 1.24 μM (f), respectively, in the presence of 14 μM cAb-HuL5G, resulting in the ratios 1:4 and 1:11.2 of D67H:cAb-HuL5G. The aggregation trace for the D67H variant alone is shown in red, and the trace corresponding to the D67H variant in the presence of cAb-HuL5G is shown in green. The traces for the cAb-HuL5G alone are shown in gray. Light scattering at 430 nm was recorded using a Cary eclipse fluorimeter with excitation and emission slit width of 5 nm.
As reported for the antibody fragment cAb-HuL22,31 the affinity constant of the nanobody:HuL interaction could be significantly perturbed as a result of the presence of the chemical denaturant and the elevated temperature that are used in the aggregation assay. The affinity constant for the interaction of the antibody fragment with WT-HuL in 0.1 M sodium citrate buffer, pH 5.0, containing 3M urea and at 50 °C, was therefore measured by ITC; the KD value obtained is 2.5 μM, which is similar to the protein concentrations used in the aggregation experiment (Figure S3, Supporting Information). A similar analysis could not be performed for the cAb-HuL5G/D67H interaction, as the D67H variant forms aggregates within the time frame of the experiment in the ITC cell. When adding 1 (6.8 μM) or 2 equiv (13.6 μM) of nanobody to the 6.8 μM D67H, we calculate using the law of mass action that 3.1 and 1.5 μM D67H, respectively, remains non-complexed in the aggregation experiments described above. Therefore, in both cases, a significant amount of D67H is still free to form fibrils, which eventually drives the dissociation of the cAb-HuL5G/D67H complex toward completion as D67H gets quasi-irreversibly sequestered into fibrillar aggregates. Subsequent experiments were therefore designed to test the effect of increasing the cAb-HuL5G/D67H stoichiometry on the kinetics of aggregation of the D67H variant. We chose to reduce the concentration of D67H, while keeping that of cAb-HuL5G constant at 14 μM, as at 28 μM cAb-HuL5G shows signs of aggregation under the experimental conditions used here. The results show that the lag phase increases and the rate of elongation of the fibrils decreases at higher ratios of cAb-HuL5G/D67H; indeed, when an excess of between 4.0 and 11.2 equiv of cAbHuL5G was present, the aggregation of the D67H variant was found to be completely inhibited, at least within the time scales monitored in these experiments (up to 400 min).
The α-Domain of the D67H Variant Substantially Unfolds upon Forming Amyloid Fibrils
As the epitope of cAb-HuL5 is essentially located in the loop between the A- and B-helices of the α-domain in its native state (Figure 2), this nanobody can serve as a structural probe to explore whether or not some elements of the native structure are still present in the α-domain of the D67H variant once it has converted into amyloid fibrils. In order to test this hypothesis, a sample containing fibrils of the D67H variant (~0.4 mg/mL) was first incubated with cAb-HuL5 (0.4 mg/mL) at pH 5.5, ultra-centrifuged to remove the fibrils and any cAb-HuL5 with which it is associated. The tryptophan fluorescence emission spectrum of the supernatant was then recorded, and for comparison, a control sample containing only the antibody fragment (0.4 mg/mL) was prepared and subjected to the same procedure. No difference in the two spectra could be observed (Figure S4, Supporting Information), indicating that the nanobody does not bind tightly to the fibrils. The results suggest, therefore, that the epitope region does not maintain its native structure within the fibrils.
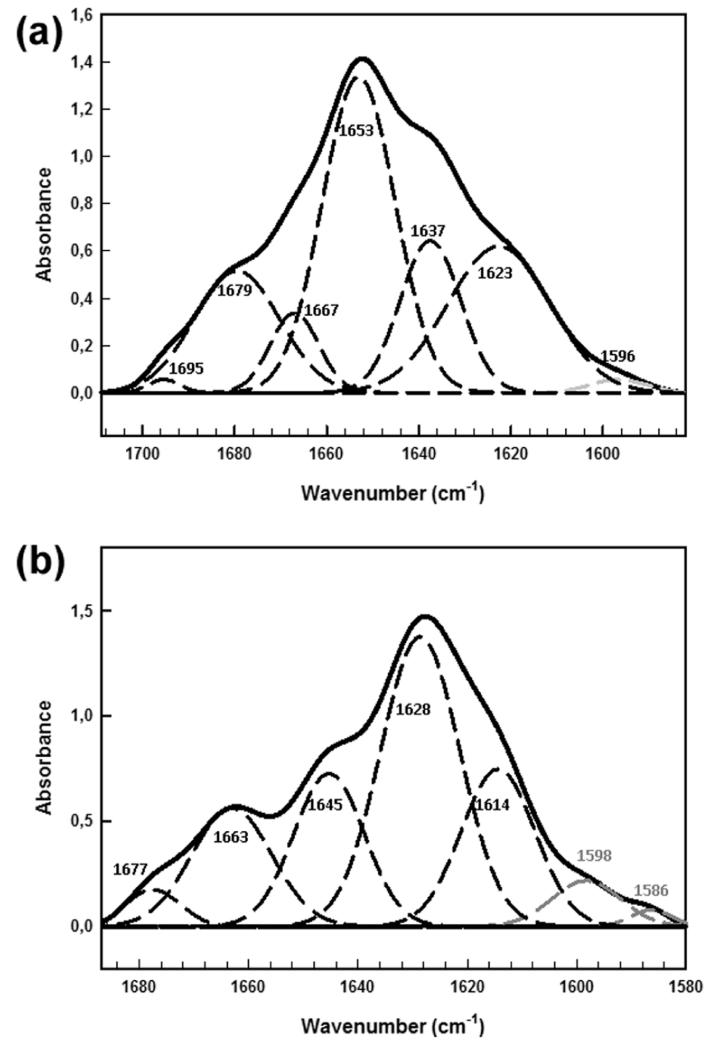
This conclusion is strongly supported by FTIR spectra of native lysozyme and of the lysozyme fibrils in the amide I region (Figure 7 and Table 3). The results highlight the fact that D67H lysozyme undergoes profound conformational changes upon fibril formation (Figure 7 and Table 3); native D67H lysozyme contains significant quantities of both α and β structure, but in the fibrillar form, it exhibits ~64% β-sheet structure. Note that the decrease in α-helical structure is not attributable to the cleavage of the α-domain from the D67H fibrils, since SDS-PAGE analysis of the protein after the fibrils were dissolved in DMSO corresponds to that of the full-length lysozyme protein (data not shown).
Figure 7.
Fourier deconvoluted and fitted IR spectra in the amide I region with component peaks of (a) the native state of the D67H lysozyme variant and (b) fibrils of the D67H variant measured both at pH 5.5 and 25 °C. The ATR-FTIR spectra are reported as thick lines, whereas the dashed lines correspond to the individual components obtained by curve fitting. Within the fitted spectra, the black lines are assigned to signals corresponding to the main chain and the gray lines are assigned to signals corresponding to the side chains.
Table 3. Secondary-Structure Content of Monomeric and Fibrillar D67H HuL as Determined by Curve Fitting of the ATR-FTIR Spectra Shown in Figure 7.
| monomeric D67H | % | fibrils of D67H lysozyme | % | ||
|---|---|---|---|---|---|
| 1623 cm−1 | β-sheet | 24 | 1614 cm−1 | β-sheet | 20 |
| 1637 cm−1 | disordered | 15 | 1628 cm−1 | β-sheet | 41 |
| 1653 cm−1 |
α-helix + disordered |
37 | 1645 cm−1 |
α-helix + disordered |
19 |
| 1667 cm−1 | turn | 6 | 1663 cm−1 | turn | 17 |
| 1679 cm−1 | turn | 17 | 1677 cm−1 | turn | 3 |
| 1695 cm−1 | β-sheet | 1 | |||
DISCUSSION
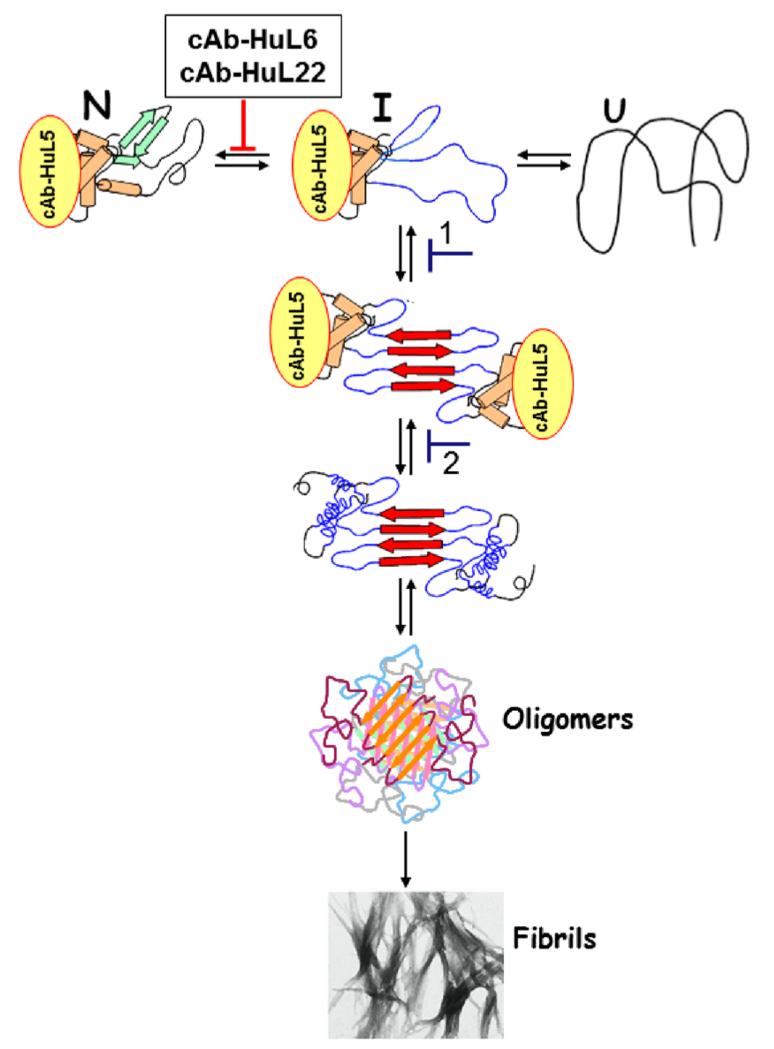
Seven mutational variants of human lysozyme are associated with familial non-neuropathic systemic amyloid disease in which large quantities of the variant proteins can be deposited in vital organs, notably the liver, spleen, and kidney.2 Detailed comparison of the properties of the amyloidogenic variants, in particular I56T and D67H, with those of wild-type lysozyme and a naturally occurring and non-amyloidogenic variant (T70N) has shown that both amyloidogenic variants have essentially identical reductions in native state stability and in global cooperativity relative to the wild-type protein. These changes are therefore likely to be determinant features underlying the intrinsic propensity of these variants to misfold and form amyloid fibrils.11,12,27 Studies of the amyloidogenic variants have shown that partially folded species in which the β-domain and the adjacent C-helix of the protein are unfolded, but the remainder of the α-domain remains in its native state, can form transiently under physiologically relevant conditions.12,15,20,27,28 It is thus very likely that intermolecular interactions between the unfolded regions of the protein in such intermediate species are responsible for the initial association process that eventually leads to fibril formation (Figure 8). In accord with this mechanism, we have previously shown that the stabilization of the native state of the variant proteins consequent upon binding to two camelid antibody fragments, referred to as cAb-HuL6 and cAb-HuL22, is an extremely effective way to restore global cooperativity and thus to inhibit fibril formation.27,28,31 Such a strategy has proved to be very successful for another system, namely, the suppression of amyloidosis and pathology associated with the tetrameric protein transthyretin (TTR).61-64 TTR amylodosis has been successfully inhibited by the design of small organic compounds, which bind to the interface between monomeric subunits within the tetramer, thereby stabilizing kinetically the TTR tetramer architecture and inhibiting misfolding and fibril formation. Indeed, such a strategy has resulted in the successful development of a drug for the treatment of TTR amyloid disease.61-64
Figure 8.
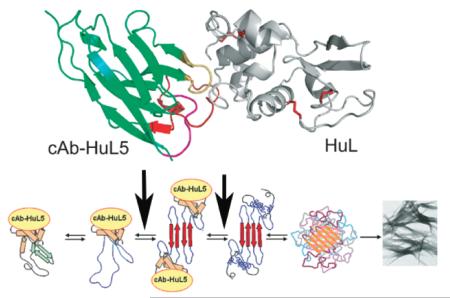
Possible mechanism for lysozyme fibril formation and its inhibition by nanobodies. Partially unfolded intermediate species of the protein (I) in which the C helix and the β-domain are cooperatively unfolded self-associate through the unfolded regions to initiate oligomer formation. This provides a template for the further aggregation of the protein and for the development of the stable, mainly β-sheet, core structure of the protofilaments within an amyloid fibril. The binding of cAb-HuL5 to the loop connecting the A and B helices is likely to inhibit the formation of the first intermolecular interaction (1) or the unfolding and reorganization of the regions of the α-domain that are native-like in the amyloidogenic intermediate (2) but are disorganized in the fibrils. The stage at which aggregation is inhibited by the two nanobodies studied previously, cAb-HuL6 and cAb-HuL22,28,31 is also shown for comparison. Note that the native disulfide bridges, though not represented in this scheme, are present in all species including the fibrils.
In the present study, we have investigated the effects of a third nanobody targeted to human lysozyme, denoted cAb-HuL5, along with a stabilized analogue denoted cAb-HuL5G, on the stability and global cooperativity of the I56T and D67H variants and on the propensity of one of these variants, D67H, to form fibrils. H/D exchange experiments monitored by mass spectrometry reveal that, under physiologically relevant conditions, cAb-HuL5 does not suppress the transient and cooperative unfolding of the β-domain and C-helix of the I56T and D67H variants (Figure 4). Moreover, MS data in fact show that the rate of this local unfolding process in both the I56T and D67H variants is increased by a factor of 1.5–3.0 upon binding to cAb-HuL5. The structure of the complex between cAb-HuL5 and WT-HuL shows that the epitope of this nanobody is primarily located in the loop between helices A and B of native lysozyme. Thus, the epitope of cAb-HuL5 does not encompass any of the residues that are transiently and cooperatively denatured in the amyloidogenic intermediates of the I56T and D67H variants.11
As the integrity of the interface between the α- and β-domains is a key element in the maintenance of the global cooperativity, these results suggest that cAb-HuL5 disrupts interface interactions via long-range conformational effects and thus facilitates the formation of the intermediate species. In support of this hypothesis, we found that the amide resonances of two residues (I59 and W109) of the cAb-HuL5/WT-HuL complex and one residue (I59) of the cAb-HuL5/I56T complex, whose side chains point toward the interface between the α- and β-domains, exhibit significant chemical shift perturbations upon nanobody binding (Figure 3c and Figure S2, Supporting Information). The structural basis for these effects is, however, likely to be extremely subtle, as no significant conformational deviations from the WT-HuL structure are detectable for any of these residues in the crystal structure of WT-HuL in complex with cAbHuL5 (Figure S1, Supporting Information). The observed increased rate of partially folded intermediate formation also suggests that the transition state, or the ensemble of partially unfolded HuL species, provides extra, albeit transient, interactions with the nanobody, leading to a reduced kinetic barrier for the formation of the intermediate species.
These results contrast sharply with those obtained previously with cAb-HuL6 and cAb-HuL22, which inhibit the cooperative unfolding of the β-domain and C-helix of amyloidogenic variants of HuL.27,28,31 This inhibition might readily be explained by the direct binding of the nanobodies to residues of the β-domain and C-helix.27,28,31 However, in the case of cAb-HuL6, only 11 of the nearly 60 residues involved in the transient unfolding of the I56T and D67H variants are in direct contact with the nanobody, suggesting that this nanobody does not suppress unfolding simply by masking the region that is destabilized by the mutation, but in fact restores the cooperativity of the lysozyme structure that is disrupted by the mutation through long-range structural perturbations. This model is again supported by the fact that the amide resonances of the residues in the interface between the α- and β-domains, which are located far from the nanobody epitope, including those at the positions of the amyloidogenic mutations, have significant chemical shift perturbations upon binding to the nanobody cAb-HuL6.27,28 Taking together the results for cAb-HuL5, cAb-HuL6, and cAb-HuL22, we conclude that the effects of nanobody binding on the properties of the interface between the α- and β-domains, and thus on the global cooperativity of the amyloidogenic lysozyme variants, are highly dependent on the location of the epitope.
By contrast, we do not observe a clear correlation between the location of the epitopes of the nanobodies and their apparent ability to interfere with the formation of fibrillar material by lysozyme; indeed we observe that all three lysozyme-specific nanobodies that we have studied, cAb-HuL5, cAb-HuL2231 and cAb-HuL6,27,28 are capable of inhibiting aggregation while binding to different epitopes. Their efficiency to suppress fibril formation, on the other hand, does correlate strongly with the measured affinity of the nanobody:lysozyme interaction. Under the conditions used to initiate lysozyme fibril formation (involving chemical denaturants, elevated temperature and low pH), the binding of cAb-HuL5 to D67H is significantly weaker than it is under more physiological conditions; thus an excess of nanobody is necessary to suppress fibril formation fully (Figure 6). Similarly, the KD values for the cAb-HuL22:WT-HuL and cAb-HuL6:WT-HuL interactions are, respectively, increased from, respectively, 35 nM and 0.7 nM under physiologically relevant conditions to ~1 μM31 and ~10 nM (M.D. personal communication) under the conditions used to trigger lysozyme fibril formation; moreover, the KD value for the cAb-HuL22:D67H interaction is even further increased, by a factor of five, relative to that of cAb-HuL22:WT-HuL.31 Consequently, under the conditions used in these studies, the affinity of cAb-HuL6 is still sufficiently high to observe full inhibition of amyloid fibril by the amyloidogenic variants of HuL at equimolar concentrations27,28 whereas an excess of cAb-HuL22 is needed to achieve this effect. We note, however, that an excess of either cAb-HuL5 or cAb-HuL22 would not be necessary to inhibit lysozyme fibril formation under physiological conditions, where the dissociation constants for their interactions with the lysozyme variants are significantly lower (Table 2 and refs 27, 28).
As the binding of cAb-HuL5G does not suppress the local unfolding of the D67H variant and strongly inhibits its fibril formation, the nanobody must influence a subsequent step in the fibril formation pathway of HuL amyloidogenic variants. This step could range from the initial dimer formation to the formation of small oligomeric structures, preceding their conversion into protofilaments and amyloid fibrils (Figure 8). We have previously shown that the extracellular chaperone clusterin strongly inhibits the fibril formation by the I56T lysozyme variant when present at sub-stoichiometric amounts,65 and demonstrated that the mechanism of inhibition by clusterin is via interactions with oligomeric species that are generated during the lag phase of the fibril formation reaction.65 Although it is possible that cAb-HuL5 binds to similar species and prevents them from further conversion into amyloid protofibrils and fibrils, the mechanism for inhibition is clearly different as, unlike clusterin, cAb-HuL5 binds to the monomeric form of lysozyme and a saturating amount of nanobody to lysozyme is needed to suppress fibril formation. This result suggests that the mechanism of inhibition most probably involves prevention of the unfolding of the α-domain upon conversion to one of the soluble species during this process.
In support of this hypothesis, we have found using IR spectroscopy that the α-helical content of D67H is significantly reduced in the D67H fibrils relative to that of the native state (Figure 7). Furthermore, proteolytic studies of human lysozyme fibrils formed at low pH and moderate temperature have shown that the fragments corresponding to residues 32–108, essentially consisting of residues in the β-domain, the C-helix and much of the α-domain, are likely to be involved in the formation of the core of the amyloid cross-β structure, whereas the residues forming the A, B, and D helices in the native state of lysozyme are essentially disordered.29 These findings are also consistent with the fact that cAb-HuL5 does not detectably bind to D67H fibrils (Figure S4, Supporting Information). This observation contrasts strongly with those with nanobodies that we have recently reported to interact with the intrinsically unfolded protein α-synuclein. In these cases, the nanobodies bind to a linear epitope in the C-terminal region of α-synuclein (the key mediator of Parkinson’s disease), and consequently also binds to α-synuclein in its fibrillar form.66,67
We can therefore conclude from all these findings that nanobodies are excellent reagents to probe conformational changes as well as epitope accessibility in species populated during amyloid fibril formation pathways.32,66,67 In this report, we have shown that cAb-HuL5 can effectively probe the existence of native-like structure of the α-domain in HuL, thereby rendering it an extremely useful reagent in conjunction with biophysical methods, such as AFM, fluorescence microscopy and single molecule techniques,55 for probing the conformation and accessibility of the α-domain in the variety of forms of species of HuL that are populated during the process of fibril formation.
CONCLUSIONS
Understanding the general principles of protein folding68-74 and misfolding24,75,76 is crucial to our understanding of the factors that enable biological systems to self-assemble and function normally, and to our knowledge of the origins of a wide range of increasingly prevalent protein deposition diseases.77 Studies of the lysozyme family of proteins by a wide range of experimental and theoretical studies1,78,79 have provided key information about both of these phenomena and the nature of the link between them.
Our earlier studies have shown that the nanobodies, cAb-HuL6 and cAb-HuL22, can strongly inhibit fibril formation by the amyloidogenic variants of human lysozyme through the suppression of the population of an early aggregation-prone intermediate. In the present study, we have shown that lysozyme fibril formation can be inhibited by intervention at a stage of the aggregation process that is subsequent to the formation of an aggregation-prone intermediate. This finding, therefore, provides further evidence of the potential of specific antibody-based therapies directed against different types of protein misfolding diseases.28,32
The present findings also highlight the benefits of nanobodies as probes to provide a deeper knowledge of the underlying mechanism of human lysozyme amyloid formation as well as a better understanding of the structural features of species populated on the fibril formation pathway of human lysozyme. We have also shown by replacing the sequence of the antigen binding loops of the extremely stable and aggregation-resistant nanobody cAb-HuL6 with the sequences of those of cAb-HuL5, that nanobodies are very amenable to modifications by the method of protein engineering to enhance their properties. Further improvement, via in vitro evolution techniques, to enhance the affinity of cAb-HuL5G for lysozyme, can be envisaged to improve the power of the nanobody to inhibit HuL fibril formation. The results that have been obtained in the work for lysozyme should in addition be beneficial for fundamental experimental investigations on protein misfolding and aggregation and they indicate the potential for the development of rational therapeutic strategies to combat the family of diseases involving the misfolding of a wide variety of proteins into amyloid fibrils.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Archer, Andrew Spencer, and Marcos Alcocer (University of Nottingham) for assistance in expression and purification of human lysozymes. E.D.G. is supported by the Medical Research Council (MRC G1002272). We wish to express our gratitude for the receipt of an EMBO long-term Fellowship and a Marie-Curie Intra-European Fellowship (for E.D.G.), and a scholarship from the Gates Cambridge Trust (for P.-H.C). M.D. is a Research Associate of the FRS-FNRS. S.-T.D.H. is a recipient of the Career Development Award (CDA - 00025/2010-C) from the International Human Frontier Science Program and is supported by funding from the National Science Council (NSC99-2911-I-007-034, 100-2113-M-001-031-MY2, and 101-2627-M-001-004) and Academia Sinica, Taiwan. D.Y.C. is funded by the Wellcome Trust and by the Crystallographic X-ray Facility, Department of Biochemistry, University of Cambridge. J.C. is supported by the Wellcome Trust, J.R.K. was supported in part by an NSERC (Canada) postdoctoral fellowship and a BBSRC project grant (BB/E019927/1), and C.V.R. by a BBSRC Advanced Research Fellowship. C.V.R. is a Royal Society University Research Professor. This work was also supported by a Programme Grant from the Wellcome Trust (to C.M.D.) and by BBSRC grants (to C.M.D. and C.V.R.), the Belgian Program of Interuniversity Attraction Poles administered by the Federal Office for Scientific Technical and Cultural Affairs (PAI number P6/19 to L.W., C.M.D., M.D., and A.M. and P7/44 to M.D. and A.M.), by the Onderzoeksraad VUB (to S.M.) and the EU’s Sixth Framework Program (Project LSHM-CT-2006-037525 to C.M.D. and M.D.), and by FRS-F.N.R.S. (FRC 2.4581.12F to M.D.). We are also very grateful to Vincent Raussens for his help with the FTIR analysis.
Footnotes
ASSOCIATED CONTENT
Three-dimensional alignment of human lysozyme bound to cAb-HuL5 with known human lysozyme structures and a table of the Cα RMSD values compared to an average structure. [1H-15N] HSQC NMR spectra of I56T free and in complex with cAb-HuL5, a graph showing the magnitude of the chemical shift perturbations of the 1H and 15N resonances of the residues of I56T as a result of cAb-HuL5 binding and an image showing the structural location of the residues of I56T that have the largest chemical shift perturbations. ITC binding isotherm for cAb-HuL5 binding to lysozyme in conditions that enhance lysozyme fibril formation. Fluorescence spectra, of the cAb-HuL5 fragment free or pre-incubated with the fibrils of the D67H amyloidogenic variant of human lysozyme. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Dumoulin M. In: Protein Misfolding Diseases: Basis of Protein Misfolding, Pathophysiology, Current, and Emerging Therapies. Ramirez-Alvarado M, Kelley JW, Dobson CM, editors. John Wileys and Sons, Inc.; Hoboken, NJ: 2012. pp. 867–884. [Google Scholar]
- (2).Dumoulin M, Kumita JR, Dobson CM. Normal and Aberrant Biological Self-Assembly: Insights from Studies of Human Lysozyme and Its Amyloidogenic Variants. Acc. Chem. Res. 2006;39:603–610. doi: 10.1021/ar050070g. [DOI] [PubMed] [Google Scholar]
- (3).Radford SE. Protein Folding: Progress Made and Promises Ahead. Trends Biochem. Sci. 2000;25:611–618. doi: 10.1016/s0968-0004(00)01707-2. [DOI] [PubMed] [Google Scholar]
- (4).Pepys MB, Hawkins PN, Booth DR, Vigushin DM, Tennent GA, Soutar AK, Totty N, Nguyen O, Blake CC, Terry CJ, et al. Human Lysozyme Gene Mutations Cause Hereditary Systemic Amyloidosis. Nature. 1993;362:553–557. doi: 10.1038/362553a0. [DOI] [PubMed] [Google Scholar]
- (5).Booth DR, Pepys MB, Hawkins PN. A Novel Variant of Human Lysozyme (T70N) Is Common in the Normal Population. Hum. Mutat. 2000;16:180. doi: 10.1002/1098-1004(200008)16:2<180::AID-HUMU20>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- (6).Röcken C, Becker K, Fändrich M, Schroeckh V, Stix B, Rath T, Kähne T, Dierkes J, Roessner A, Albert FW. ALys Amyloidosis Caused by Compound Heterozygosity in Exon 2 (Thr70Asn) and Exon 4 (Trp112Arg) of the Lysozyme Gene. Hum. Mutat. 2006;27:119–120. doi: 10.1002/humu.9393. [DOI] [PubMed] [Google Scholar]
- (7).Valleix S, Drunat S, Philit JB, Adoue D, Piette JC, Droz D, MacGregor B, Canet D, Delpech M, Grateau G. Hereditary Renal Amyloidosis Caused by a New Variant Lysozyme W64R in a French Family. Kidney Int. 2002;61:907–912. doi: 10.1046/j.1523-1755.2002.00205.x. [DOI] [PubMed] [Google Scholar]
- (8).Yazaki M, Farrell SA, Benson MD. A Novel Lysozyme Mutation Phe57Ile Associated with Hereditary Renal Amyloidosis. Kidney Int. 2003;63:1652–1657. doi: 10.1046/j.1523-1755.2003.00904.x. [DOI] [PubMed] [Google Scholar]
- (9).Girnius S, Skinner M, Spencer B, Prokaeva T, Bartholomew C, O’Hara C, Seldin DC, Connors LH. A New Lysozyme Tyr54Asn Mutation Causing Amyloidosis in a Family of Swedish Ancestry with Gastrointestinal Symptoms. Amyloid. 2012;19:182–185. doi: 10.3109/13506129.2012.723074. [DOI] [PubMed] [Google Scholar]
- (10).Dumoulin M, Johnson JR, Bellotti V, Dobson CM. In: Protein Misfolding, Aggregation, and Conformational Diseases. Uversky VN, Fink A, editors. Vol. 6. Springer; New York: 2007. pp. 285–308. [Google Scholar]
- (11).Booth DR, Sunde M, Bellotti V, Robinson CV, Hutchinson WL, Fraser PE, Hawkins PN, Dobson CM, Radford SE, Blake CC, et al. Instability, Unfolding and Aggregation of Human Lysozyme Variants Underlying Amyloid Fibrillogenesis. Nature. 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- (12).Canet D, Last AM, Tito P, Sunde M, Spencer A, Archer DB, Redfield C, Robinson CV, Dobson CM. Local Cooperativity in the Unfolding of an Amyloidogenic Variant of Human Lysozyme. Nat. Struct. Mol. Biol. 2002;9:308–315. doi: 10.1038/nsb768. [DOI] [PubMed] [Google Scholar]
- (13).Canet D, Sunde M, Last AM, Miranker A, Spencer A, Robinson CV, Dobson CM. Mechanistic Studies of the Folding of Human Lysozyme and the Origin of Amyloidogenic Behavior in Its Disease-Related Variants. Biochemistry. 1999;38:6419–6427. doi: 10.1021/bi983037t. [DOI] [PubMed] [Google Scholar]
- (14).Chamberlain AK, Receveur V, Spencer A, Redfield C, Dobson CM. Characterization of the Structure and Dynamics of Amyloidogenic Variants of Human Lysozyme by Nmr Spectroscopy. Protein Sci. 2001;10:2525–2530. doi: 10.1110/ps.28101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dhulesia A, Cremades N, Kumita JR, Hsu ST, Mossuto MF, Dumoulin M, Nietlispach D, Akke M, Salvatella X, Dobson CM. Local Cooperativity in an Amyloidogenic State of Human Lysozyme Observed at Atomic Resolution. J. Am. Chem. Soc. 2010;132:15580–15588. doi: 10.1021/ja103524m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Funahashi J, Takano K, Ogasahara K, Yamagata Y, Yutani K. The Structure, Stability, and Folding Process of Amyloidogenic Mutant Human Lysozyme. J. Biochem. 1996;120:1216–1223. doi: 10.1093/oxfordjournals.jbchem.a021544. [DOI] [PubMed] [Google Scholar]
- (17).Morozova-Roche LA, Zurdo J, Spencer A, Noppe W, Receveur V, Archer DB, Joniau M, Dobson CM. Amyloid Fibril Formation and Seeding by Wild-Type Human Lysozyme and Its Disease-Related Mutational Variants. J. Struct. Biol. 2000;130:339–351. doi: 10.1006/jsbi.2000.4264. [DOI] [PubMed] [Google Scholar]
- (18).Takano K, Funahashi J, Yutani K. The Stability and Folding Process of Amyloidogenic Mutant Human Lysozymes. Eur. J. Biochem. 2001;268:155–159. doi: 10.1046/j.1432-1327.2001.01863.x. [DOI] [PubMed] [Google Scholar]
- (19).Wain R, Smith LJ, Dobson CM. Oxidative Refolding of Amyloidogenic Variants of Human Lysozyme. J. Mol. Biol. 2005;351:662–671. doi: 10.1016/j.jmb.2005.06.035. [DOI] [PubMed] [Google Scholar]
- (20).Buell AK, Dhulesia A, Mossuto MF, Cremades N, Kumita JR, Dumoulin M, Welland ME, Knowles TP, Salvatella X, Dobson CM. Population of Nonnative States of Lysozyme Variants Drives Amyloid Fibril Formation. J. Am. Chem. Soc. 2011;133:7737–7743. doi: 10.1021/ja109620d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Esposito G, Garcia J, Mangione P, Giorgetti S, Corazza A, Viglino P, Chiti F, Andreola A, Dumy P, Booth D, et al. Structural and Folding Dynamics Properties of T70N Variant of Human Lysozyme. J. Biol. Chem. 2003;278:25910–25918. doi: 10.1074/jbc.M211000200. [DOI] [PubMed] [Google Scholar]
- (22).Johnson RJ, Christodoulou J, Dumoulin M, Caddy GL, Alcocer MJ, Murtagh GJ, Kumita JR, Larsson G, Robinson CV, Archer DB, et al. Rationalising Lysozyme Amyloidosis: Insights from the Structure and Solution Dynamics of T70N Lysozyme. J. Mol. Biol. 2005;352:823–836. doi: 10.1016/j.jmb.2005.07.040. [DOI] [PubMed] [Google Scholar]
- (23).Hagan CL, Johnson RJ, Dhulesia A, Dumoulin M, Dumont J, De Genst E, Christodoulou J, Robinson CV, Dobson CM, Kumita JR. A Non-Natural Variant of Human Lysozyme (I59T) Mimics the in Vitro Behaviour of the I56T Variant That Is Responsible for a Form of Familial Amyloidosis. Protein Eng., Des. Sel. 2010;23:499–506. doi: 10.1093/protein/gzq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Dobson CM. Protein Folding and Misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- (25).Chiti F, Dobson CM. Amyloid Formation by Globular Proteins under Native Conditions. Nat. Chem. Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- (26).Dobson CM. Principles of Protein Folding, Misfolding and Aggregation. Semin. Cell Dev. Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- (27).Dumoulin M, Canet D, Last AM, Pardon E, Archer DB, Muyldermans S, Wyns L, Matagne A, Robinson CV, Redfield C, et al. Reduced Global Cooperativity Is a Common Feature Underlying the Amyloidogenicity of Pathogenic Lysozyme Mutations. J. Mol. Biol. 2005;346:773–788. doi: 10.1016/j.jmb.2004.11.020. [DOI] [PubMed] [Google Scholar]
- (28).Dumoulin M, Last AM, Desmyter A, Decanniere K, Canet D, Larsson G, Spencer A, Archer DB, Sasse J, Muyldermans S, et al. A Camelid Antibody Fragment Inhibits the Formation of Amyloid Fibrils by Human Lysozyme. Nature. 2003;424:783–788. doi: 10.1038/nature01870. [DOI] [PubMed] [Google Scholar]
- (29).Frare E, Mossuto MF, Polverino de Laureto P, Dumoulin M, Dobson CM, Fontana A. Identification of the Core Structure of Lysozyme Amyloid Fibrils by Proteolysis. J. Mol. Biol. 2006;361:551–561. doi: 10.1016/j.jmb.2006.06.055. [DOI] [PubMed] [Google Scholar]
- (30).Muyldermans S. Single Domain Camel Antibodies: Current Status. J. Biotechnol. 2001;74:277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- (31).Chan P-H, Pardon E, Menzer L, De Genst E, Kumita JR, Christodoulou J, Saerens D, Brans A, Bouillenne F, Archer DA, et al. Engineering a Camelid Antibody Fragment That Binds to the Active Site of Human Lysozyme and Inhibits Its Conversion into Amyloid Fibrils. Biochemistry. 2008;47:11041–11054. doi: 10.1021/bi8005797. [DOI] [PubMed] [Google Scholar]
- (32).De Genst E, Dobson CM. Nanobodies as Structural Probes of Protein Misfolding and Fibril Formation. Methods Mol. Biol. 2012;911:533–558. doi: 10.1007/978-1-61779-968-6_34. [DOI] [PubMed] [Google Scholar]
- (33).Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. Beta-Lactamase Inhibitors Derived from Single-Domain Antibody Fragments Elicited in the Camelidae. Antimicrob. Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and Identification of Single Domain Antibody Fragments from Camel Heavy-Chain Antibodies. FEBS Lett. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- (35).Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LG, Muyldermans S, Wyns L, Matagne A. Single-Domain Antibody Fragments with High Conformational Stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chen Z, Ruffner DE. Amplification of Closed Circular DNA in Vitro. Nucleic Acids Res. 1998;26:1126–1127. [PMC free article] [PubMed] [Google Scholar]
- (37).Spencer A, Morozova-Roche LA, Noppe W, MacKenzie DA, Jeenes DJ, Joniau M, Dobson CM, Archer DB. Expression, Purification, and Characterization of the Recombinant Calcium-Binding Equine Lysozyme Secreted by the Filamentous Fungus Aspergillus Niger: Comparisons with the Production of Hen and Human Lysozymes. Protein Expression Purif. 1999;16:171–180. doi: 10.1006/prep.1999.1036. [DOI] [PubMed] [Google Scholar]
- (38).Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to Measure and Predict the Molar Absorption Coefficient of a Protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe: A Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- (40).Borek D, Minor W, Otwinowski Z. Measurement Errors and Their Consequences in Protein Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003;59:2031–2038. doi: 10.1107/s0907444903020924. [DOI] [PubMed] [Google Scholar]
- (41).McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. Phenix: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- (44).Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- (45).Jones S, Thornton JM. Principles of Protein-Protein Interactions. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lee B, Richards FM. The Interpretation of Protein Structures: Estimation of Static Accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- (47).Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the Ccp4 Suite and Current Developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).McDonald IK, Thornton JM. Satisfying Hydrogen Bonding Potential in Proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- (49).Transue TR, De Genst E, Ghahroudi MA, Wyns L, Muyldermans S. Camel Single-Domain Antibody Inhibits Enzyme by Mimicking Carbohydrate Substrate. Proteins. 1998;32:515–522. doi: 10.1002/(sici)1097-0134(19980901)32:4<515::aid-prot9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- (50).Davies J, Riechmann L. Single Antibody Domains as Small Recognition Units: Design and in Vitro Antigen Selection of Camelized, Human VH Domains with Improved Protein Stability. Protein Eng., Sel. Des. 1996;9:531–537. doi: 10.1093/protein/9.6.531. [DOI] [PubMed] [Google Scholar]
- (51).De Genst E, Saerens D, Muyldermans S, Conrath K. Antibody Repertoire Development in Camelids. Dev. Comp. Immunol. 2006;30:187–198. doi: 10.1016/j.dci.2005.06.010. [DOI] [PubMed] [Google Scholar]
- (52).Muyldermans S, Cambillau C, Wyns L. Recognition of Antigens by Single-Domain Antibody Fragments: The Superfluous Luxury of Paired Domains. Trends Biochem. Sci. 2001;26:230–235. doi: 10.1016/s0968-0004(01)01790-x. [DOI] [PubMed] [Google Scholar]
- (53).Ermolenko DN, Zherdev AV, Dzantiev BB. Antibodies as Specific Chaperones. Biochemistry. 2004;69:1233–1238. doi: 10.1007/s10541-005-0069-4. [DOI] [PubMed] [Google Scholar]
- (54).Saerens D, Pellis M, Loris R, Pardon E, Dumoulin M, Matagne A, Wyns L, Muyldermans S, Conrath K. Identification of a Universal Vhh Framework to Graft Non-Canonical Antigen-Binding Loops of Camel Single-Domain Antibodies. J. Mol. Biol. 2005;352:597–607. doi: 10.1016/j.jmb.2005.07.038. [DOI] [PubMed] [Google Scholar]
- (55).Cremades N, Cohen SI, Deas E, Abramov AY, Chen AY, Orte A, Sandal M, Clarke RW, Dunne P, Aprile FA, et al. Direct Observation of the Interconversion of Normal and Toxic Forms of α-Synuclein. Cell. 2012;149:1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Breydo L, Wu JW, Uversky VN. α-Synuclein Misfolding and Parkinson’s Disease. Biochim. Biophys. Acta. 2012;1822:261–285. doi: 10.1016/j.bbadis.2011.10.002. [DOI] [PubMed] [Google Scholar]
- (57).Narayan P, Orte A, Clarke RW, Bolognesi B, Hook S, Ganzinger KA, Meehan S, Wilson MR, Dobson CM, Klenerman D. The Extracellular Chaperone Clusterin Sequesters Oligomeric Forms of the Amyloid-Beta(1–40) Peptide. Nat. Struct. Mol. Biol. 2012;19:79–83. doi: 10.1038/nsmb.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Rochet JC, Lansbury PT., Jr. Amyloid Fibrillogenesis: Themes and Variations. Curr. Opin. Struct. Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- (59).Lee J, Culyba EK, Powers ET, Kelly JW. Amyloid-Beta Forms Fibrils by Nucleated Conformational Conversion of Oligomers. Nat. Chem. Biol. 2011;7:602–609. doi: 10.1038/nchembio.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Giehm L, Svergun DI, Otzen DE, Vestergaard B. Low-Resolution Structure of a Vesicle Disrupting α-Synuclein Oligomer That Accumulates during Fibrillation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3246–3251. doi: 10.1073/pnas.1013225108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Grimster NP, Connelly S, Baranczak A, Dong J, Krasnova LB, Sharpless KB, Powers ET, Wilson IA, Kelly JW. Aromatic Sulfonyl Fluorides Covalently Kinetically Stabilize Transthyretin to Prevent Amyloidogenesis while Affording a Fluorescent Conjugate. J. Am. Chem. Soc. 2013;135:5656–5668. doi: 10.1021/ja311729d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Bulawa CE, Connelly S, Devit M, Wang L, Weigel C, Fleming JA, Packman J, Powers ET, Wiseman RL, Foss TR, et al. Tafamidis, a Potent and Selective Transthyretin Kinetic Stabilizer That Inhibits the Amyloid Cascade. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Alhamadsheh MM, Connelly S, Cho A, Reixach N, Powers ET, Pan DW, Wilson IA, Kelly JW, Graef IA. Potent Kinetic Stabilizers That Prevent Transthyretin-Mediated Cardiomyocyte Proteotoxicity. Sci. Transl. Med. 2011;3:97ra81. doi: 10.1126/scitranslmed.3002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Connelly S, Choi S, Johnson SM, Kelly JW, Wilson IA. Structure-Based Design of Kinetic Stabilizers That Ameliorate the Transthyretin Amyloidoses. Curr. Opin. Struct. Biol. 2010;20:54–62. doi: 10.1016/j.sbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Kumita JR, Poon S, Caddy GL, Hagan CL, Dumoulin M, Yerbury JJ, Stewart EM, Robinson CV, Wilson MR, Dobson CM. The Extracellular Chaperone Clusterin Potently Inhibits Human Lysozyme Amyloid Formation by Interacting with Prefibrillar Species. J. Mol. Biol. 2007;369:157–167. doi: 10.1016/j.jmb.2007.02.095. [DOI] [PubMed] [Google Scholar]
- (66).Guilliams T, El-Turk F, Buell A, O’Day E, Aprile F, Esbjorner E, Vendruscolo M, Cremades N, Pardon E, Wyns L, et al. Nanobodies Raised against Monomeric α-Synuclein Distinguish between Fibrils at Different Maturation Stages. J. Mol. Biol. 2013;425:2397–2411. doi: 10.1016/j.jmb.2013.01.040. [DOI] [PubMed] [Google Scholar]
- (67).De Genst EJ, Guilliams T, Wellens J, O’Day EM, Waudby CA, Meehan S, Dumoulin M, Hsu ST, Cremades N, Verschueren KH, et al. Structure and Properties of a Complex of α-Synuclein and a Single-Domain Camelid Antibody. J. Mol. Biol. 2010;402:326–343. doi: 10.1016/j.jmb.2010.07.001. [DOI] [PubMed] [Google Scholar]
- (68).Wolynes PG, Onuchic JN. Thirumalai, D. Navigating the Folding Routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- (69).Wolynes PG. Folding Funnels and Energy Landscapes of Larger Proteins within the Capillarity Approximation. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6170–6175. doi: 10.1073/pnas.94.12.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Wolynes PG, Eaton WA, Fersht AR. Chemical Physics of Protein Folding. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17770–17771. doi: 10.1073/pnas.1215733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Dobson CM, Šali A, Karplus M. Protein Folding: A Perspective from Theory and Experiment. Angew. Chem., Int. Ed. Engl. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- (72).Panchenko AR, Luthey-Schulten Z, Wolynes PG. Foldons, Protein Structural Modules, and Exons. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2008–2013. doi: 10.1073/pnas.93.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Brooks CL, 3rd, Gruebele M, Onuchic JN, Wolynes PG. Chemical Physics of Protein Folding. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11037–11038. doi: 10.1073/pnas.95.19.11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Onuchic JN, Wolynes PG. Theory of Protein Folding. Curr. Opin. Struct. Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- (75).Ong DS, Kelly JW. Chemical and/or Biological Therapeutic Strategies to Ameliorate Protein Misfolding Diseases. Curr. Opin. Cell Biol. 2011;23:231–238. doi: 10.1016/j.ceb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Zheng W, Schafer NP, Wolynes PG. Frustration in the Energy Landscapes of Multidomain Protein Misfolding. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1680–1685. doi: 10.1073/pnas.1222130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Chiti F, Dobson CM. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- (78).Shoemaker BA, Wolynes PG. Exploring Structures in Protein Folding Funnels with Free Energy Functionals: The Denatured Ensemble. J. Mol. Biol. 1999;287:657–674. doi: 10.1006/jmbi.1999.2612. [DOI] [PubMed] [Google Scholar]
- (79).Dinner AR, Šali A, Smith LJ, Dobson CM, Karplus M. Understanding Protein Folding via Free-Energy Surfaces from Theory and Experiment. Trends Biochem. Sci. 2000;25:331–339. doi: 10.1016/s0968-0004(00)01610-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.