Summary
Hepatocytes and cholangiocytes self renew following liver injury. Following severe injury hepatocytes are increasingly senescent, whether Hepatic Progenitor Cells (HPCs) then contribute to liver regeneration is unclear. Here, we describe a mouse model where Mdm2 is inducibly deleted in over 98% of hepatocytes, causing apoptosis, necrosis and senescence with nearly all hepatocytes expressing p21. This results in florid HPC activation, which is necessary for survival, followed by complete, functional liver reconstitution. HPCs isolated from genetically normal mice, using cell surface markers, were highly expandable and phenotypically stable in vitro. These HPCs were transplanted into adult mouse livers where hepatocyte Mdm2 was repeatedly deleted, creating a non-competitive repopulation assay. Transplanted HPCs contributed significantly to restoration of liver parenchyma, regenerating hepatocytes and biliary epithelia, highlighting their in vivo lineage potency. HPCs are therefore a potential future alternative to hepatocyte or liver transplantation for liver disease.
Introduction
Liver transplantation is the only effective cure for advanced liver disease. However a global shortage in donor organs means many patients die waiting for suitable organs. Hepatocyte cell therapy is an attractive alternative to liver transplantation1, 2. Hepatocytes have a high replicative potential in vivo3, 4, however hepatocyte isolation is technically demanding, requires cadaveric liver donation, and hepatocytes cannot be expanded effectively in vitro5.
Ductular reactions are activated in chronic liver injury and are thought to contain putative hepatic progenitor cells (HPCs) with the potential to regenerate hepatocytes and cholangiocytes6, 7. However, the significance of this regenerative pathway is unclear. Recent studies have sought to identify HPCs using surface markers8, 9 or in vivo lineage tracing10-13. These studies demonstrated that HPCs arise within the liver, engraft the parenchyma14 and may have some differentiation capacity but did not regenerate significant quantities of liver parenchyma11-13. Only if HPCs have regeneration capacity, are expandable in vitro and transplantable would they be a potential future therapeutic target. Many dietary and chemical models of injury have been exploited to investigate the biology of HPCs15-17. Experiments utilising hepatocyte lineage tracing in mice have shown in various liver injury models that hepatocytes regenerate themselves without any significant contribution from HPCs18, 19. This calls into question the nature and role of HPCs in liver injury and regeneration20. Further experiments in mice have shown that hepatocytes can change into a biliary ductular phenotype21, 22 then later re-differentiate into hepatocytes23.
In advanced human liver disease there is often widespread hepatocyte senescence i.e. an irreversible block to hepatocyte replication, indicated by p21 or p16 positivity. In this setting ductular reactions develop, however the functional role of putative HPCs in human liver disease is difficult to discern in the absence of lineage tracing24. The question arises as to whether mouse models of liver injury adequately reflect human disease. In the rat complete suppression of hepatocyte proliferation can be achieved using chemical toxins which provokes an extensive ductular/HPC response which is thought to regenerate parenchyma, although lineage tracing studies are required to formally prove this25. The transdifferentiation of hepatocytes into biliary ductules is damage dependent and negligible unless significant injury is induced26.
To model the human (and rat) situation we have utilised a genetic means of inducing hepatocyte injury and senescence in adult mouse liver. We have exploited an AhCre system27 with an Mdm2loxp 28, which remains inactive until induced with β-napthoflavone (βNF). Following induction with βNF, Cre recombinase is expressed in >98% of hepatocytes where it renders Mdm2 inactive. Mdm2 is an E3 ubiquitin-protein ligase that functions to degrade TRP53 (p53). Inactivation of Mdm2 results in upregulation of p53 and induces p53 mediated hepatocyte death and senescence. This results in rapid activation of HPCs throughout the liver, which proliferate, differentiate into hepatocytes, and completely restore architecture and function.
A highly purified population of HPCs were isolated, using surface antigen profile and expanded in vitro. HPCs were transplanted in vivo in a non-competitive model of liver regeneration where they expand massively and differentiate, reconstituting the liver, significantly improving liver function and architecture.
Results
Transgenic targeted hepatocellular injury as a model of whole organ repair
To determine whether endogenous ductular cells give rise to hepatocytes we analysed a lineage tracing system using the CDE (choline deficient ethionine supplemented) diet - recovery model11 (Supplementary figure 1a). To label biliary/ductular cells we used the Krt19CreERTLSLtdTomato mouse and found labelling strictly limited to ducts in the un-injured liver with zero hepatocyte labelling (Supplementary figure 1b). Following CDE - recovery there was a minor degree of tracing into hepatocytes (Supplementary figure 1b, c), indicating that in this model the ductular–hepatocyte regeneration pathway is insignificant. We found minimal hepatocyte senescence (Supplementary figure 1b), in contrast to advanced human diseases and rat models of HPC activation where high levels of hepatocyte senescence occur29. To analyse liver regeneration in advanced injury we therefore studied a model of induced hepatocytes senescence in mouse.
The activation and proliferation of HPCs in vivo requires both hepatocellular injury and inhibition of hepatocyte replication. To achieve this we utilised the AhCre transgenic line, which contains the rat Cyp1A1 promoter cloned upstream of Cre recombinase, we combined this line with a transgenic Mdm2 locus in which exons 5 and 6 are flanked with loxP sites (Mdm2flox/flox), to generate AhCre+Mdm2flox/flox mice18, 19. Following βNF induction (Figure 1a) Cre recombinase is expressed in hepatocytes and Mdm2 is rendered inactive (ΔMdm2).
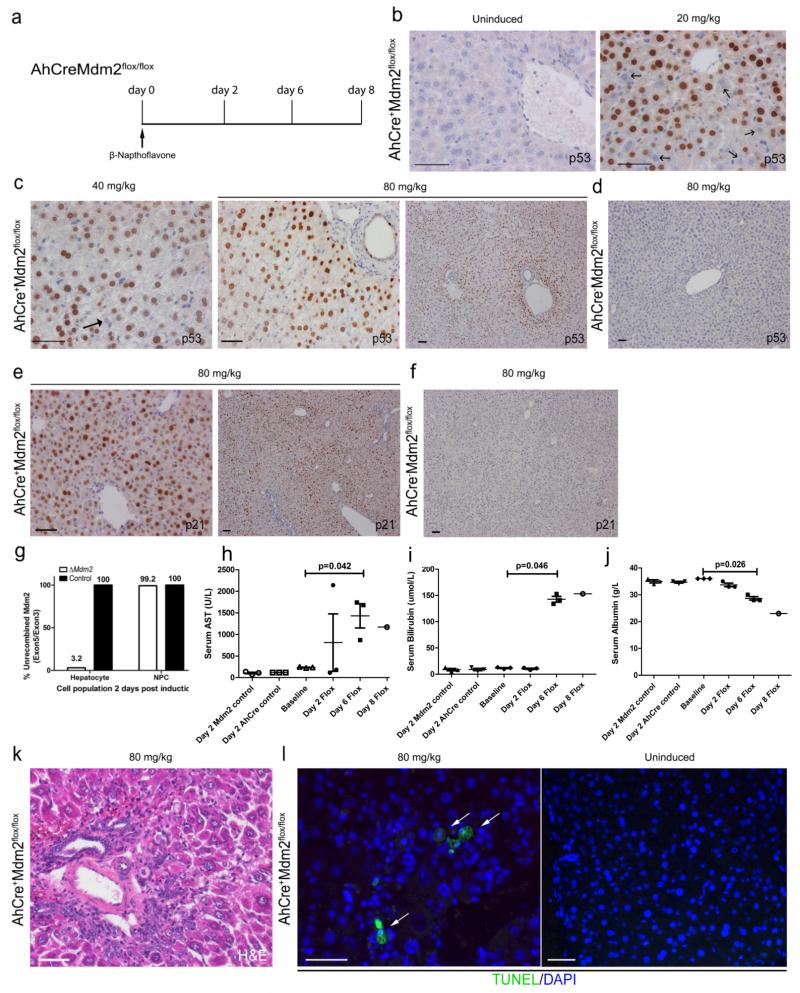
Figure 1. Induction of hepatocyte damage following AhCre mediated loss of Mdm2.
(a) Schematic representation of the AhCreMdm2flox/flox time course used in this study. (b) Immunohistochemistry for p53 mice not induced with βNF (b-c) Immunohistochemistry for p53 in induced AhCre+Mdm2flox/flox mice with a 20, 40 and 80mg/Kg dose of βNF, Black arrows denote p53 negative cells. (d) low power photomicrograph of p53 staining in the AhCre−Mdm2flox/flox control. (e) Immunohistochemistry for p21 in induced AhCre+Mdm2flox/flox mice with 80mg/Kg dose of βNF. (f) low power photomicrograph of p21 staining in the AhCre−Mdm2flox/flox control. (g) Genomic PCR of Mdm2 exon 5/ exon 3 in hepatocytes and Non-parenchymal cells (NPCs) from ΔMdm2 versus control; n = 3 biological replicates. (h-j) Serum AST, bilirubin and albumin levels over the time course in ΔMdm2 mice compared to AhCre−, Mdm2WT/WT and uninduced controls (mean ± s.e.m , (h) P = 0.042 (i) P = 0.046 (j) P = 0.026 one-way ANOVA; n = 3 mice each group, except day 8 in which n = 1 due to mortality). (k) H&E staining for ΔMdm2 following induction with 80mg/Kg βNF. (l) Apoptosis identified by TUNEL staining in ΔMdm2 mice following induction with 80mg/Kg βNF. White arrows show TUNEL positive hepatocytes. The representative images shown here are representative for 3 experiments with 3-5 mice each group per experiments. Data are represented as mean ± s.e.m. Scale bars = 50μm.
Uninduced AhCreMdm2flox/flox mice have normal hepatic architecture and do not express detectable p53 protein in hepatocytes (Figure 1b). Two days following induction, and Cre activation, with 20, 40 or 80 mg/kg of βNF, rapid expression of p53 protein was observed in hepatocyte nuclei in a dose dependent manner (Figure 1 b and c); however following βNF administration to Mdm2flox/flox animals lacking the AhCre, we found no expression of p53 protein confirming p53 expression is a consequence of Cre activity and Mdm2 loss (Figure 1d). We also observed expression of p21 protein in hepatocytes following βNF administration (Figure 1e) but not in Cre− animals (Figure 1f). To validate Mdm2 recombination in this system following Cre activation, we isolated a highly purified population of hepatocytes and non-parenchymal cells (NPC) two days following high dose (80mg/Kg) βNF administration. We evaluated the presence of both exon 5 (which would be lost following Cre mediated recombination) and exon 3 (which would remain intact following Cre mediated recombination), thereby offering an internal control. In hepatocytes there was a 96.8% reduction in the quantity of genomic Mdm2 exon 5 following recombination, however only a 0.8% reduction in NPCs from the same cohort (Figure 1g, supplementary figure 2a-d) indicating the AhCre is highly specific in recombining ΔMdm2 in hepatocytes.
Following βNF administration, ΔMdm2 mice developed jaundice after three days and required humane euthanasia by day 8 (Supplementary Table 1). Serum markers for liver injury (AST, bilirubin, alkaline phosphatase and ALT) were all elevated and serum albumin levels fell, signifying injury and impaired synthetic function of the livers of ΔMdm2 mice (Figure 1h-j, Supplementary figure 2e and f). This reduction in functionality is associated with hepatocyte necrosis by H&E staining which shows clear disruption of the hepatic architecture as well as increased positivity of lactate dehydrogenase staining (Figure 1k, Supplementary figure 2h). The levels of apoptosis in ΔMdm2 mice was determined through TUNEL which was absent in uninduced AhCre+Mdm2flox/flox animals however we found clear positive cells throughout the parenchyma in AhCre+Mdm2flox/flox mice which had received βNF (Figure 1l). Further to this we found increased mRNA expression of the p53-dependent apoptotic gene Bax1 (Supplementary figure 2g) throughout the 8 days time course in ΔMdm2 mice.
Widespread hepatocyte injury promotes a ductular reaction
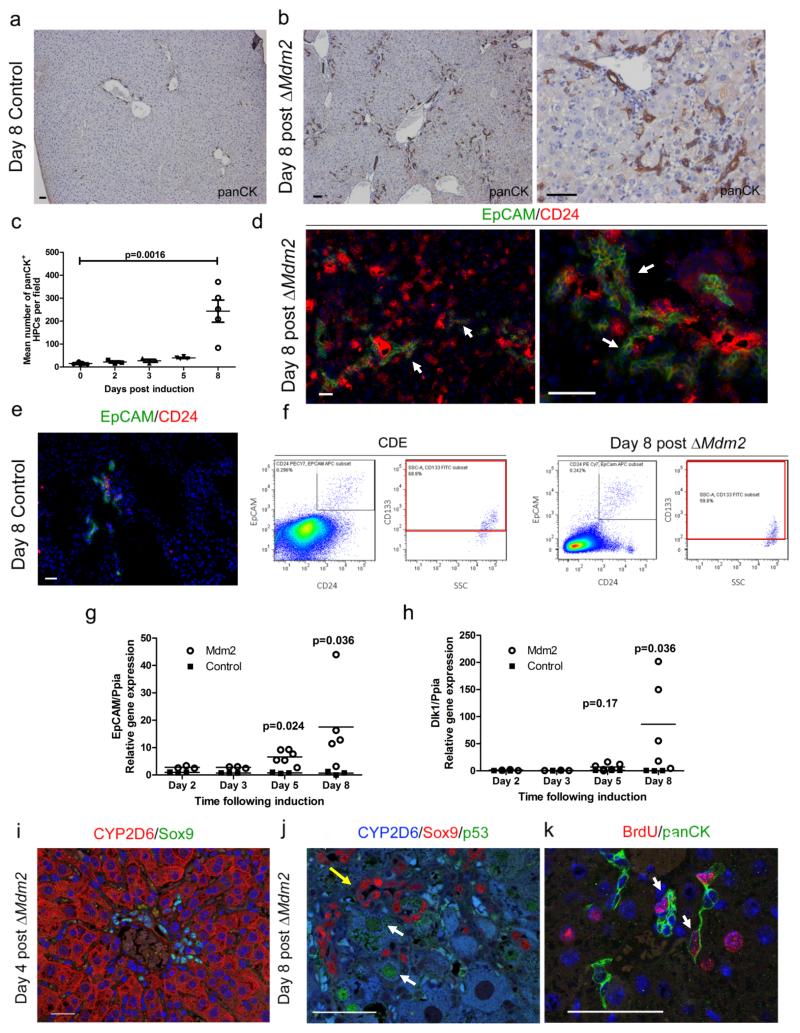
Following βNF administration we found small cells appearing in the livers of ΔMdm2 mice that did not upregulate p53. We examined the ductular reaction in ΔMdm2 livers to ascertain whether HPCs arise in this model. In the un-induced liver, panCK is expressed throughout the biliary tree and is restricted to the portal tract (Figure 2a). Eight days following induction of the AhCre transgene the number of panCK positive ductular reactions containing putsignificantly increases and are found migrating as cords throughout the liver parenchyma (Figure 2b and c, Supplementary figure 3a-c). Following AhCre induction the migrating cords of HPCs also express the HPC markers EpCAM and CD24 (Figure 2d, Supplementary figure 3d). In contrast, we found that the cell population that express EpCAM and CD24 are restricted to the biliary epithelium in the healthy liver (Figure 2e). We compared our AhCre ΔMdm2 progenitor cell profile to those that are generated in the widely studied CDE model of liver damage and regeneration using FACS. Here we found an expansion of a cell population that expresses the HPC markers EpCAM, CD24, and CD133 (Figure 2f) in CDE; critically our AhCre ΔMdm2 HPCs express the same surface antigens at comparable levels. In line with this increase in HPC number we observed a significant increase in mRNA expression of HPC markers EpCAM, Dlk1, CK19 and Ascl2 over 8 days in the AhCre ΔMdm2 line (Figure 2g and h, Supplementary figure 3e and f). The extent of the ductular reaction in the ΔMdm2 was significantly greater than in the CDE diet (Supplementary figure 3g). In the ΔMdm2 livers Sox9 positive HPCs did not express CYP2D6 (Figure 2i). Furthermore, HPCs were not affected by the Cre system in response to the βNF induction, as they lacked upregulation of p53 (Figure 2j), and are proliferative following βNF administration and recombination of the Mdm2loxP locus in hepatocytes (Figure 2k). Proliferative Bromodeoxiuridine (BrdU) positive HPCs were often found closely association with BrdU positive hepatocytes 8 days following AhCre induction, suggesting that hepatocytes arise from BrdU positive HPCs in this model. Importantly we also found that 2 days after induction there are no BrdU positive hepatocytes adjacent to BrdU positive HPCs, suggesting that proliferation of HPCs themselves does not lead to hepatocyte proliferation. (Supplementary figure 3h).
Figure 2. Hepatocyte Mdm2 loss results in rapid activation of HPCs.
(a) Immunohistochemistry for HPCs (panCK, brown) in un-induced AhCre+Mdm2flox/flox mice. (b) Immunohistochemistry for HPCs (panCK, brown) in ΔMdm2 mice 8 days following induction with βNF. (c) Quantification of mean number of panCK+ cells per field over the 8 day time course following induction with βNF. (mean ± s.e.m, Kruskal Wallis test. P = 0.0016; n = 7,3,3,4,5 mice for days 0,2,3,5,8 respectively). (d) Immunohistochemistry for EpCAM+ CD24+ HPCs (EpCAM, green; CD24, red) in ΔMdm2 mice 8 days following induction with βNF. White arrows show EpCAM+ CD24+ HPCs (e) Immunohistochemistry for EpCAM+ CD24+ HPCs (EpCAM, green; CD24, red) in un-induced AhCre+Mdm2flox/flox mice. (f) FACS analysis of EPCAM+ CD24+ CD133+ HPCs in both 12 days CDE treated mice and βNF induced AhCre+Mdm2flox/flox mice. (g - h) mRNA expression of EpCAM and Dlk1 over the experimental time course following induction with βNF. (mean ± s.e.m, (g) P = 0.024 on day 6, P = 0.036 on day 8 (h) P = 0.036 on day 8, One-way ANOVA with Bonferroni correction; n = 3,3,6,5 and 3,3,3,3 mice for days 2,3,5,8 for experimental and controls respectively). (i) Immunohistochemistry of 4 days post ΔMdm2 showing hepatocytes, CYP2D6 (red) and HPCs, Sox9 (green). (j) Immunohistochemistry of day 8 ΔMdm2 showing hepatocytes (CYP2D6, blue), HPCs (Sox9, red) and p53 (green). White arrows CYP2D6+, p53+ hepatocytes. Yellow arrow Sox9+ HPCs adjacent to hepatocytes. (k) Immunohistochemistry of HPC proliferation, HPCs (panCK, green) and proliferation (BrdU, red). Arrows mark BrdU labelled HPCs. BrdU labelled hepatocytes are associated with HPCs. The representative images shown here are representative for 2 experiments with 3-5 mice each group per experiments. Scale bars = 50μm
Large scale hepatocyte replacement can arise from endogenous HPCs
The AhCre+Mdm2flox/flox model results in massive hepatocyte injury, where more than 96% of hepatocytes upregulate p53, and activates a florid HPC response. In order to investigate how significant the HPC contribution is to regeneration we sought to induce the Mdm2 transgene and follow progenitor cells out to 6 months, to evaluate whether they can reconstitute the liver efficiently.
To avoid the early mortality observed in AhCre+Mdm2flox/flox mice, we examined the long-term effects of Mdm2 recombination in AhCre+Mdm2flox/− mice. AhCre+Mdm2flox/− mice had a somatic heterozygotic deficiency of the Mdm2 gene, however are phenotypically normal until the second Mdm2 allele is lost. Recombination was as efficient in the AhCre+Mdm2flox/− as in AhCre+Mdm2flox/flox counterparts (Supplementary Table 1). In AhCre+Mdm2flox/− mice the onset of liver injury and clinical signs of acute disease, demonstrated through necrosis and apoptosis as demonstrated through H&E and TUNEL staining (Figure 3a and b), together with tachypnoea, hunching, prostration, and bilirubinuria occur between and 14 and 21 days following AhCre activation all of which are delayed in the Mdm2flox/− compared with the Mdm2flox/flox given the same induction regime. Concurrently HPC activation was more gradual than in the AhCre+Mdm2flox/flox counterparts and arose from d14 post Mdm2 deletion (Figure 3c). The most likely explanation for this delay is that compensatory changes occur in the hepatocytes that already lack one allele, when the second allele is removed the effect is less dramatic. A similar effect of Mdm2 haploinsufficiency has been described in the context of tumour development30. Furthermore, no evidence of stable, nuclear p53 could be found in HPCs arising in the AhCre+Mdm2flox/− indicating this long-term model faithfully recapitulates what we see in Mdm2flox/flox mice (Figure 3d and e).
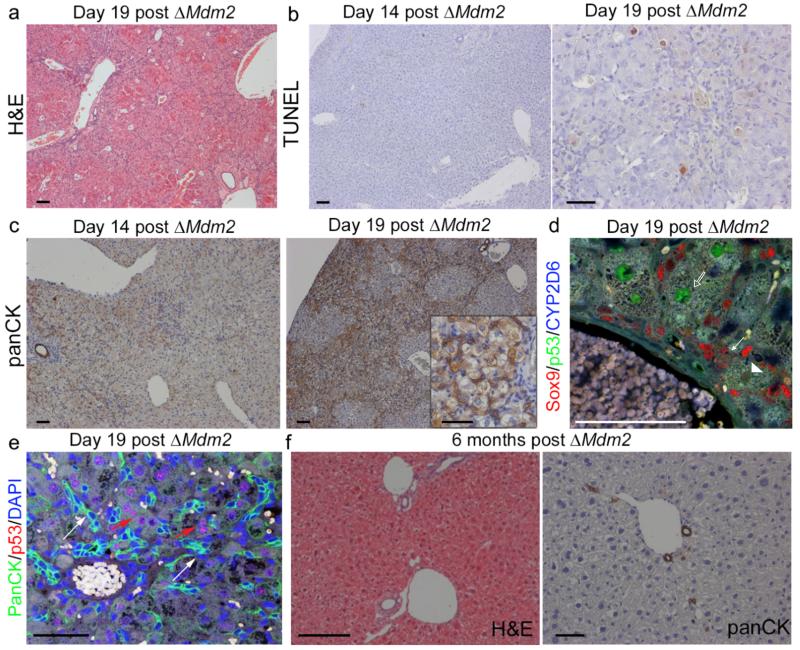
Figure 3. Mdm2 deletion in Mdm2flox/− model leads to HPC expansion and subsequent recovery.
(a) H&E staining of AhCre+Mdm2flox/− 19 days following βNF administration. (b) Apoptosis stained for by TUNEL in AhCre+Mdm2flox/− 14 and 19 days following βNF administration. (c) HPCs (panCK) in AhCre+Mdm2flox/− 14 and 19 days following βNF administration (inset, higher magnification). (d) Immunohistochemistry for HPCs (Sox9, red), hepatocytes (CYP2D6, blue) and p53 (green). White arrows denote HPCs, open arrows p53 high hepatocytes and arrowheads p53 low hepatocytes. (e) Immunohistochemistry of HPCs (PanCK, green), p53 (red) and DNA (DAPI, blue). White arrows show p53 low PanCK+ HPCs. Red arrows denote p53 high hepatocytes. (f) H&E and immunohistochemistry for HPCs (panCK) 6 months following ΔMdm2 in the AhCre+Mdm2flox/−. The representative images shown here are representative for 2 experiments with 3-5 mice each group per experiment. Scale bars = 50μm.
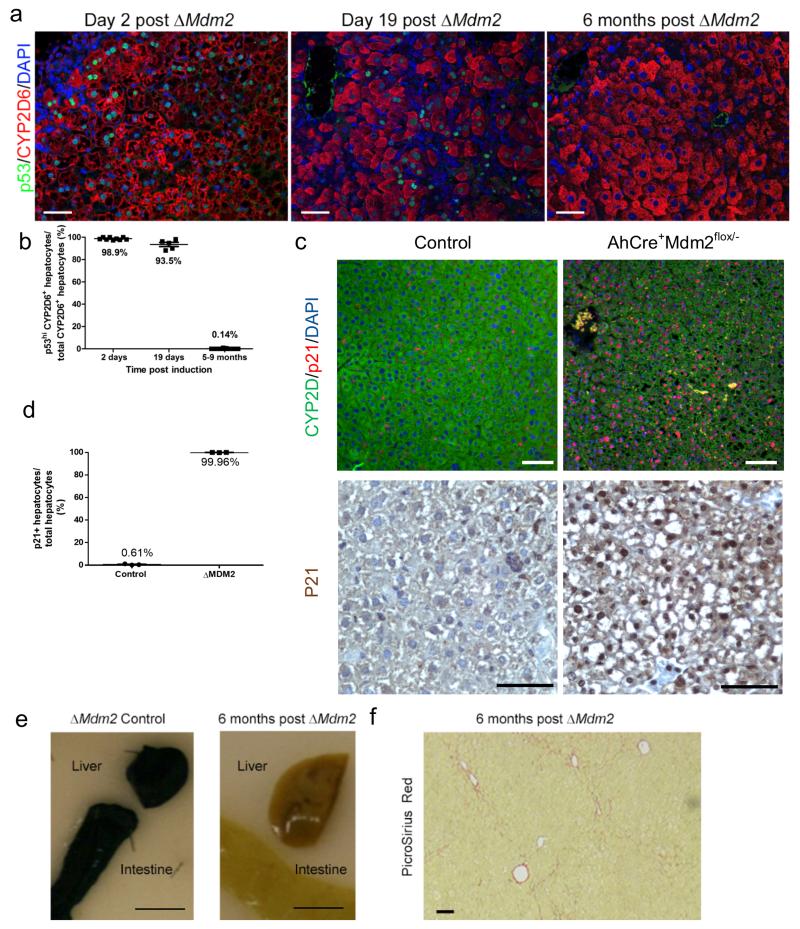
In AhCre+Mdm2flox/− mice following βNF administration, a large proportion developed liver damage then subsequently made full recoveries with dissipation of liver injury and the ductular reactions (Figure 3f) by 6 months. As the AhCre is highly efficient in the liver we used the presence of nuclear p53 in hepatocytes to ascertain the HPC mediated hepatocellular regeneration. As anticipated, two days following βNF we found the majority of hepatocytes expressed the p53 protein (98.9%) (Figure 4a and b). In addition to the high level of p53 in the liver, almost all hepatocytes (99.96%) express senescence marker p21 suggesting these cells enter either direct or indirect senescence 2 days following βNF (Figure 4c and d). 19 days following induction, p53 staining in hepatocytes was more heterogeneous, with 93.5% of hepatocytes still expressing p53 (Figure 4a and b). Six months following induction of Cre recombinase hepatocytes were largely p53 negative (only 0.14% of hepatocytes were positive). The sequential dilution of p53 positivity from the hepatocyte population reflects the progressive replacement of Mdm2-negative hepatocytes from the parenchyma by HPC derived (Mdm2 intact, p53 low) hepatocytes.
Figure 4. Loss of recombined ΔMdm2 hepatocytes over time during recovery.
(a) Immunohistochemistry for hepatocytes (CYP2D6, red), p53 (green) and DNA (DAPI, blue) in AhCre+Mdm2flox/−mice 2 days, 19 days and 6 months following βNF induction. White arrow denotes p53 low hepatocyte. (b) Quantification of p53 high, CYP2D6 positive hepatocytes 2 days, 19 days and 6 months following βNF induction (mean ± s.e.m, n = 9,5 and 11 mice for day 2, 19 and 5-9 months respectively). (c) p21 (red), CYP2D6 (green), DAPI (blue) immunohistochemistry (top panel), p21 iummunohistochemistry on AhCre+Mdm2flox/− mice 2 days following βNF induction compared to control (bottom panel). (d) Quantification of p21 expressing hepatocytes (bottom panel) on AhCre+Mdm2flox/− mice 2 days following βNF induction compared to control (mean ± s.e.m. n = 20 fields at ×200 magnification for each biological sample quantified, n = 3 mice per group). (e) Whole mount X-gal staining of intestine and liver from AhCre+Mdm2flox/−mice 2 days following ΔMdM2 mice and 6 months following ΔMdm2. Scale bars = 1cm (f) PicroSirius red (fibrillar collagen) staining in ΔMdm2 livers 6 months following induction. The representative images shown here are representative for 3-5 mice each group per experiment. Scale bars = 50μm.
To confirm that this loss of p53 positivity was not due to down-regulation of p53 by other mechanisms we used the AhCre+Mdm2flox/− crossed onto a silenced R26RLacZ reporter. Following recombination LacZ expression could be detected throughout the intestine and liver. Loss of reporter was seen over 6 months indicating that replacement of the recombined epithelium occurs from a non-recombined source (Figure 4e). Following recovery of AhCre+Mdm2flox/− mice no animals developed liver tumours or notable fibrosis (Figure 4f).
Endogenous HPCs are absolutely required for adult liver regeneration following Mdm2 loss
In our AhCre+Mdm2flox/− mouse model we wanted to evaluate the kinetics of cell replacement from p53 negative HPCs. Proliferating hepatocytes, positive for Ki67 were arranged in cord-like formations, with each cord of dividing hepatocytes being in contact with the distal tips of the ductular reaction (Figure 5a). This evidence, although indirect, suggested a role for HPCs in either supporting hepatocytes proliferation or differentiate into hepatocytes.
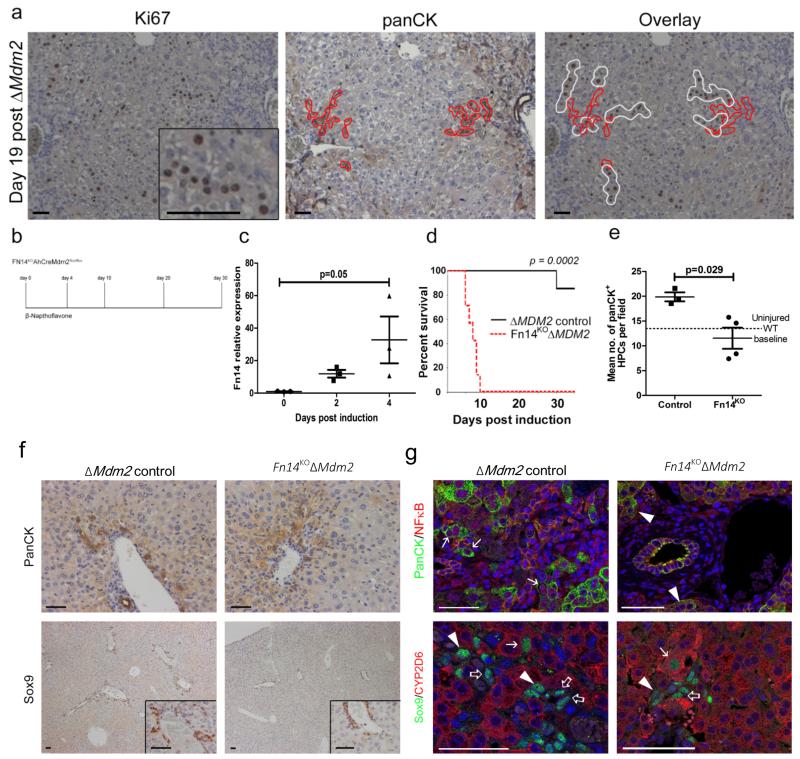
Figure 5. Fn14/TWEAK regulated HPCs are necessary for liver regeneration following hepatocyte Mdm2 deletion.
(a) Immunohistochemistry for proliferation (Ki67) demarcated in white and HPCs (panCK) demarcated in red 19 days following ΔMdm2 in AhCreMdm2flox/−. (b) Schematic representing the experimental time course using the Fn14KO AhCreMdM2flox/flox. (c) mRNA expression of Fn14 following ΔMdm2 induction in AhCre+Mdm2flox/flox (mean ± s.e.m, One-way ANOVA with Bonferroni correction. P = 0.05; n = 3 mice). (d) Kaplan–Meier survival curve of ΔMdm2 control versus Fn14KOΔMdm2 (Mantel-Cox test P = 0.0002, n = 7 mice each group). (e) Quantification of number of panCK+ HPCs in ΔMdm2 control versus Fn14KO ΔMdm2, dotted line denotes baseline HPC number in a WT uninjured liver (mean ± s.e.m, Mann Whitney test. P = 0.029; n = 3 vs. 4 mice). (f) Representative images of panCK+ and Sox9+ HPCs (insets high power) in the Fn14KO AhCre+ Mdm2flox/flox model with Fn14+ AhCre+ Mdm2flox/flox controls 4 days following 80 mg/kg βNF administration. (g) Active form nuclear NFκB p65 expressing panCK+ HPCs observed in Fn14+ mice (arrows) but not in HPCs from Fn14KO mice (arrowheads). Sox9+/CYP2D6+ co-staining association of Sox9+/CYP2D6low HPCs (open-arrows) with Sox9+/CYP2D6intermediate cells with intermediate morphology (arrowheads) and Sox9+/CYP2D6+ hepatocytes (arrows) in each experimental group. The representative images shown here are representative for 3-7 mice each group per experiment. Scale bars = 50 μm.
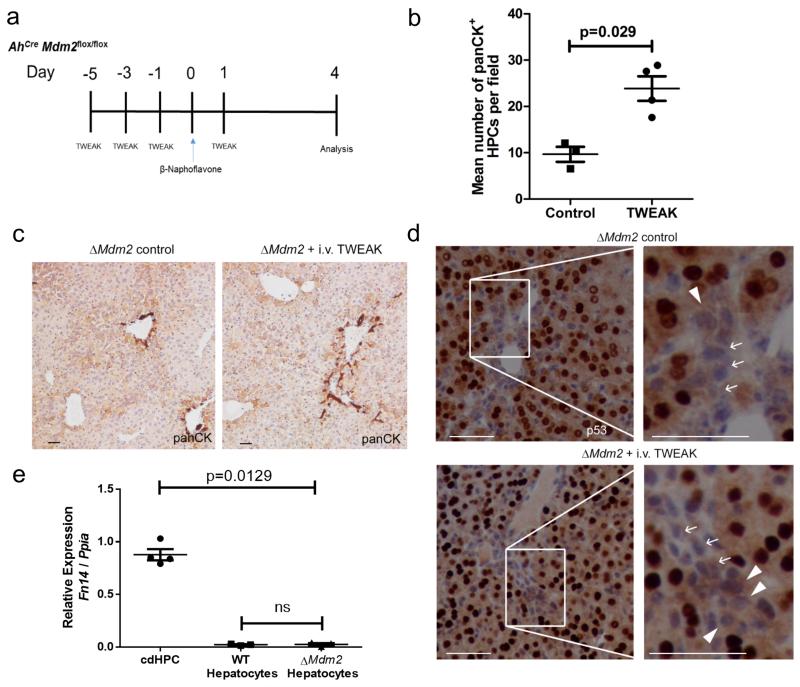
Fn14 has been identified as a critical receptor for mitogenic stimulation of HPCs31-34. To confirm the regeneration we see in the AhCre Mdm2 system is a direct result of HPC activation and differentiation, and not due to a small number of hepatocytes which escape Mdm2 recombination we crossed the AhCre+Mdm2flox/flox mouse onto an Fn14 null (Fn14KO) background34. We induced the AhCre+Mdm2flox/floxFn14KO mouse and tracked the fate of these animals over 30 days (Figure 5b). In our AhCre+Mdm2flox/flox model where Fn14 is intact we detect up-regulation of Fn14 mRNA following induction with βNF over four days (Figure 5c). Following Cre induction, Fn14KOΔMdm2 animals demonstrated a significantly reduced survival, and all had to be euthanized within 10 days (Figure 5d), unlike Fn14WTΔMdm2 mice in which 80% of animals survived to day 30. The induction of damage and repair on the background of Fn14 loss corresponded with failed ductular expansion (Figure 5e and f), indicating that in the AhCre+Mdm2flox/− mouse, HPCs are critical for hepatic regeneration. HPCs had a corresponding reduction in NFκB activity (Figure 5g), a downstream mediator of TWEAK-Fn14 signalling33. We treated ΔMdm2 mice (which are WT for the tweak receptor Fn14) throughout the onset of injury with 4 doses of intravenous TWEAK (Figure 6a). 4 days following activation of Cre, animals which had been given TWEAK had significantly increased numbers of ductular cells versus vehicle alone (Figure 6b and c). TWEAK treated ΔMdm2 mice also had a higher frequency of p53-negative hepatocytes adjacent to ductular cords (Figure 6d). To confirm the effect of TWEAK is on ducts instead of hepatocytes, we isolated hepatocytes from βNF-induced animals and did not observe upregulation of Fn14 in hepatocytes after injury (Figure 6e). These data suggest that following loss of Mdm2 in hepatocytes, large scale parenchymal injury prompts ductular expansion in a TWEAK/Fn14 dependent manner. This provides evidence of the beneficial effect of HPC to liver regeneration when hepatocyte senescence prevents normal hepatocyte regeneration.
Figure 6. TWEAK enhances ductular reaction through activation of HPCs.
(a) Schematic representing the experimental time course using the AhCreMdm2flox/flox mice that receive intravenous injection of TWEAK. (b) Quantification of number of panCK+ HPCs in ΔMdm2 control versus ΔMdm2 given recombinant TWEAK (mean ± s.e.m, Mann Whitney test. P = 0.029; n = 3-4 mice per group). (c) panCK staining in ΔMdm2 control mice versus ΔMdm2 mice given recombinant TWEAK. (d) p53 immunohistochemistry and analysis of portal tracts of both ΔMdm2 controls and mice treated with repeated injection of i.v. TWEAK, shows p53- cells with hepatocyte like morphology adjacent to portal tracts and areas of ductular expansion 4 days following induction (80mg/kg βNF). Arrows show p53- HPCs and arrowheads show p53- hepatocytes. (e) mRNA expression of Fn14 from isolated HPCs and, hepatocytes isolated from wild type mice, and hepatocytes isolated from ΔMdm2 mice (mean ± s.e.m, One-way ANOVA. P = 0.0129; n = 3 mice). The representative images shown here are representative for 3-4 mice. Scale bars = 50μm.
CD45−/CD31−/Ter119−/EpCAM+/CD24+/CD133+ marks a population of bipotent HPCs
The AhCre+Mdm2flox/flox model demonstrates the efficacy of adult HPCs to regenerate the adult liver. In human disease, cells analogous to the murine HPCs arise in response to biological or chemical damage24, 35 (not typically genetic alteration), so to assess whether ex vivo HPCs can also significantly repopulate the adult liver we used a chemical model of hepatocellular disease, the murine CDE diet, previously employed to trace the fate of HPCs in vivo11, 16, 17. Isolation of these HPCs by FACS has been variable8, 36, 37, therefore we used HPC markers described in the literature, in combination, to identify the most potent HPC sub-population.
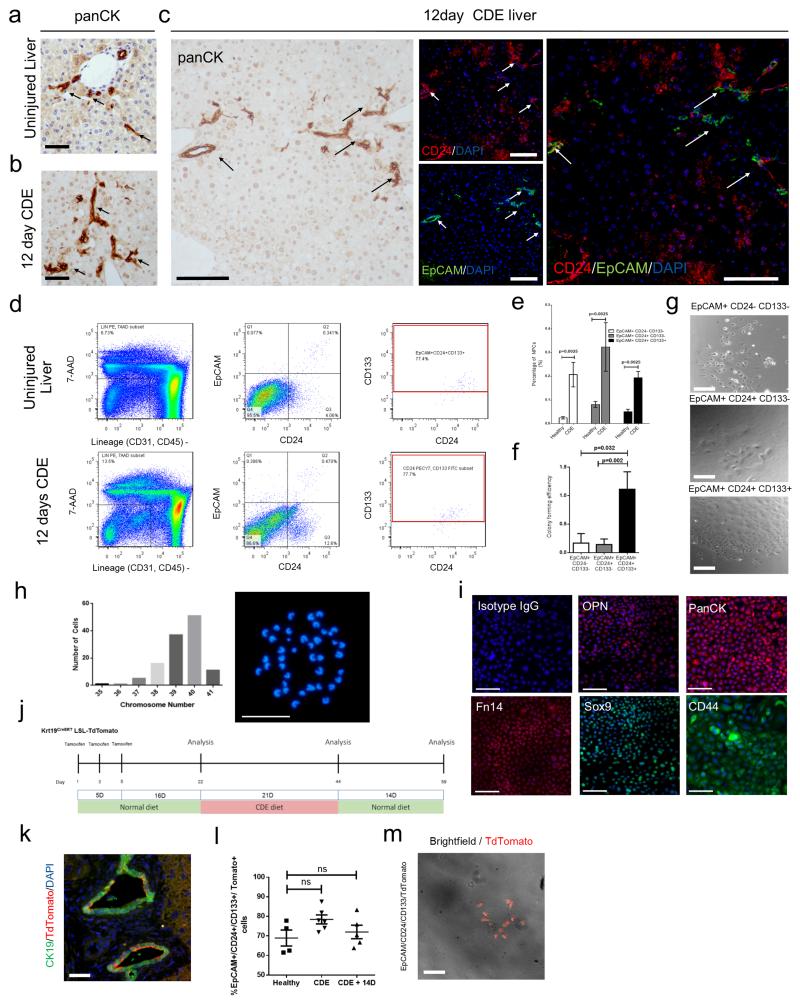
The CDE diet resulted in ductular reactions arising from the terminal ducts of the portal tracts compared to control diet (Figure 7a and b). Within the panCK expressing ductular reactions, we observe a subpopulation of HPCs that express the surface markers EpCAM and CD24; whilst EpCAM is very specific for ductular cells, CD24 labels both HPCs and a population of hepatocytes in vivo (Figure 7c, Supplementary figure 3d). To exclude these CD24 positive hepatocytes, and to ascertain whether these CD24+/EpCAM+ cells were bona fide HPCs we used FACS to isolate multiple candidate HPC populations. We negatively selected for haematopoietic (CD45), endothelial (CD31) and erythroid (Ter119) lineages then positively selected for EpCAM, CD24 and CD133 (Figure 7d), thereby generating three candidate HPC populations EpCAM+/CD24−/CD133−, EpCAM+/CD24+/CD133− and EpCAM+/CD24+/CD133+. Besides an increase in total NPC population, all three populations were enriched in the NPC fraction of livers from mice fed the CDE diet compared to the control diet (Figure 7e). These three populations were plated at clonal density to assess their colony forming potential. EpCAM+/CD24−/CD133− and EpCAM+/CD24+/CD133− populations formed infrequent, small colonies with mesenchymal characteristics, while the EpCAM+/CD24+/CD133+ population generated frequent colonies of packed epithelial cells, from hereon known as clonal density derived HPCs (cdHPCs) (Figure 7f and g).
Figure 7. In vitro expanded HPCs are genetically and phenotypically stable.
(a) Immunohistochemistry for HPCs (panCK) in uninjured liver and (b) mice treated with 12 days CDE diet. (c) Immunohistochemistry on serial sections for HPCs (panCK) and EpCAM (green), CD24 (red) and DAPI (blue). Arrows show EpCAM+ CD24+ HPCs. (d) FACS gating strategy to isolate 7AAD−/CD31−/CD45−/Ter119−/EpCAM+/CD24+/CD133+ HPCs from uninjured and CDE injured liver. The data shown here are representative images for 10 mice (e) Histogram representing the percentage representation of EpCAM+ CD24− CDE133−, EpCAM+ CD24+ CD133− and EpCAM+ CD24+ CD133+ in the NPC fraction of healthy or CDE treated livers (mean ± s.e.m, Mann-Whitney test. P = 0.0025; n = 4 mice each group). (f) Colony forming efficiency of EpCAM+ CD24− CDE133−, EpCAM+ CD24+ CD133− and EpCAM+ CD24+ CD133+ populations in vitro (mean ± s.e.m, One-way ANOVA. P = 0.032, and 0.002 respectively; n = 5 biological replicates per group).These data are representative of 3 individual experiments (g) Phase contrast images of the colonies formed by the EpCAM+ CD24− CD133−, EpCAM+ CD24+ CD133− and EpCAM+ CD24+ CD133+ populations. (h) Frequency of chromosome number in metaphase spreads from in vitro expanded HPCs, inset, a representative example of a normal karyotype from in vitro expanded cdHPCs. (i) Immunocytochemistry of HPC markers in in vitro expanded HPCs. (j) Schematic representation of the experiment to determine the origin of cdHPC. (k) Immunohistochemistry on healthy, tamoxifen induced Krt19CreERTLSLTdTomato mice for recombined HPC, CK19 (green), TdTomato (red), and DAPI (blue). (l) Percentage of EpCAM+ CD24+ CD133+ population expressing TdTomato in uninjured Krt19CreERTLSLTdTomato mice, Krt19CreERTLSLTdTomato mice treated with CDE diet, Krt19CreERTLSLTdTomato mice treated with CDE diet followed by 14 days of normal diet (mean ± s.e.m, One-way ANOVA. P >0.05; n = 5 mice). (m) Merged phase contrast and fluorescent image of isolated EpCAM+CD24+CD133+TdTomato+ cells from Krt19CreERTLSLTdTomato mice receiving CDE diet. Data are represented as mean ± s.e.m. n = ≥3 each group. in vitro data represents three independent experiments. Scale bars = 50μm.
The defined population of HPCs can be expanded in vitro
Over six weeks cdHPCs expand and proliferate in vitro. Historically expanding HPCs has been hampered by cells differentiating following replating; we therefore adapted a new method for passaging these cells where, rather than being passaged as a single cell suspension they are detached using highly diluted trypsin and lifted and replated as colonies, ensuring that cell-cell contact is maintained38. Using this method we found that the viability and phenotypically stability of passaged cdHPCs increased (Supplementary figure 4a). In our assay, only the cdHPC population can be expanded whilst maintaining an epithelial morphology, the other two populations failed to maintain any epithelial phenotype. The expanded cdHPC population can be maintained long-term in vitro (more than 30 passages and up to a year) and remains chromosomally stable having a normal karyotype (Figure 7h). The in vitro expanded cdHPCs maintained the co-expression of HPC markers EpCAM, CD24, and CD133 (Supplementary figure 4b). Expanded cdHPCs highly express HPCs related genes relative to housekeeping gene compared to hepatocytes (Supplementary figure 4c and d). Expanded cdHPCs maintained their HPC characteristics and were positive for a suite of markers (Sox9, CD44, Fn14, OPN1 and panCK) which have been used to define HPCs in vivo and the expression of these markers was maintained after 30 passages (Figure 7i, Supplementary figure 4e and f). We did not observe a significant change in the expression of most HPC related genes such as EpCAM, Ck19, Spp1, and Sox9, or Alb after long term passaging although Lgr5 expression increased (Supplementary figure 4e). After in vitro expansion, mean cell size reduced after 10 passages, however no significant changes in elongation assessed by cell roundness, or cell width to length ratio were seen (Supplementary figure 4g). We calculated that in our in vitro expansion system each cell has divided around 32 times at passage 10 (Supplementary figure 4h). cdHPCs did not express endothelial (CD31), haematopoietic (CD45) or mesenchymal (GFAP and desmin) markers (Supplementary figure 5a and b). To ascertain the bipotemtiality of cdHPCs we differentiated them towards biliary or hepatic lineages. Following 3-D culture of cdHPCs in Matrigel we observed cells forming tubules, which had a branched morphology and lumen (Supplementary figure 5c) and expressed the activated bile duct marker MIC1C3 and the biliary transcription factor HNF1β (Supplementary figure 5c). For hepatocyte differentiation cdHPCs were treated with recombinant WNT3A which has been shown in other systems to promote hepatocyte differentiation39. Following rWNT3A treatment clusters of cdHPCs increased in size and became hexagonal in shape, and increased glycogen storage by PAS staining (Supplementary figure 5d). Murine albumin increased at the transcript level and as protein in the culture medium (Supplementary figure 5e, upper histograms). Concurrent with this up-regulation we found loss of biliary and progenitor associated genes Krt19, Epcam, Dlk1, Afp and upregulation in hepatocyte transcription factor Hnf1a (Supplementary figure 5e, lower histograms). When we reanalysed this population for EpCAM and CD24 we identified three populations EpCAM−CD24lo, EpCAM−CD24Hi and EpCAM+CD24Hi. To further refine where in cdHPCs the most potent progenitor cell resides we investigated the self-renewal ability of these cdHPC populations using a secondary clone assay. EpCAM−CD24lo, EpCAM−CD24Hi and EpCAM+CD24Hi were sorted and plated at clonal density. EpCAM+ clones were capable of generating colonies; however this was significantly impaired in EpCAM− cdHPCs, regardless of CD24 expression status (Supplementary figure 6a-c).
To confirm the biliary/ductular origin of the cdHPCs, we isolated cdHPCs from Tamoxifen induced Krt19CreERTLSLtdTomato mice without liver injury , during CDE injury, and 2 weeks after CDE injury (Figure 7j). The recombination efficiency of Krt19 expressing cells after Tamoxifen administration in the Krt19CreERTLSLTomato mice was 62% (Figure 7k). Zero Tomato expression was observed in the hepatocytes in any animal (Supplementary figure 1a). Likewise the cdHPCs from Tamoxifen induced Krt19CreERTLSLTomato mice without liver injury were 70% Tomato+ demonstrating their ductular origin. This percentage of TdTomato expressing cdHPCs remain constant before, during CDE injury, and after recovery from CDE injury (Figure 7l and m), arguing against a significant non- ductular contribution to this population.
In vitro expanded HPCs can repopulate the liver after hepatocellular injury
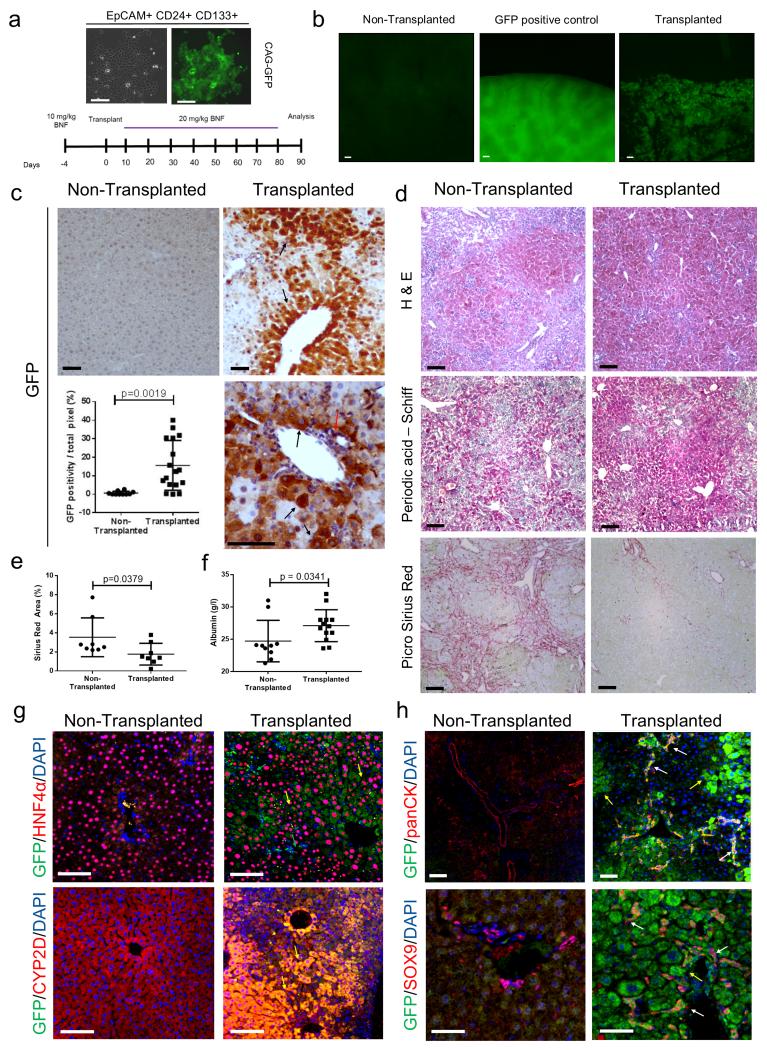
To ascertain the in vivo liver repopulating potency of cdHPCs we performed transplantation studies using cdHPCs. cdHPCs were transfected with a plasmid expressing CAG-GFP and stably transfected clones selected (Figure 8a). 5×106 GFP-cdHPCs were transplanted intrasplenically into the AhCre+Mdm2flox/flox mice which had previously received low dose of βNF (10 mg/Kg) to induce hepatocellular injury. Following transplantation, recipient AhCre+Mdm2flox/flox mice were given repeated doses of βNF (20 mg/Kg) every 10 days to progressively ablate endogenous Mdm2loxP hepatocytes (Figure 8a and Supplementary table 1). Three months following transplantation, large patches of GFP+ cells were observed in the transplanted liver compared to controls (Figure 8b). GFP positive cells in the parenchyma were large and morphologically consistent with hepatocytes (Figure 8c and Supplementary figure 7a). Transplanted animals had significantly improved liver histology with stronger glycogen staining (Figure 8d top and middle panel), less liver scarring, (Figure 8d bottom panel and Figure 8e) and increased serum albumin levels compared to controls (Figure 8f). The GFP expressing, cdHPC derived cells co-expressed the hepatocyte markers HNF4α and CYP2D6 and were found in patches adjacent to host GFP negative hepatocytes (Figure 8g and Supplementary figure 7b). Small GFP positive ductular cells were seen that expressed panCK and SOX9 (biliary markers), indicating bi-lineage differentiation of transplanted cdHPCs in vivo (Figure 8h). These GFP+ cells were found in both bile ducts and cords of ductular cells alongside host ductular cells (Supplementary figure 7b). cdHPCs hepatocytes were proliferating (Ki67+) in the recipient liver at 3 months following transplantation, whereas there was little proliferation in the endogenous hepatocytes of AhCre+Mdm2flox/flox recipient mice. Concurrently, more senescent cells in the non-transplanted group were observed with p21 staining (Supplementary figure 7c).
Figure 8. Relationship between activated HPCs and hepatocytes.
(a) A schematic representation of the AhCre+Mdm2flox/flox as a transplant recipient model of GFP expressing HPCs. Expanded HPCs stably transfected with a CAG-GFP expressing vector, left photomicrograph phase contrast, right GFP. (b) Low power epifluorescence of sham transplanted liver, ubiquitous CAG-GFP expressing liver and ΔMdm2 liver with CAG-GFP expressing transplanted in vitro expanded HPCs. (c) Immunohistochemistry for GFP in non-transplanted ΔMdm2 controls versus ΔMdm2 transplanted with CAG-GFP expressing HPCs. Black arrows denote GFP expressing hepatocytes and red arrow GFP expressing duct. Dot-plot shows quantification of GFP in transplanted versus non-transplanted liver (mean ± s.d, Student’s t-Test. P = 0,0019; n = 17 fields at ×200 magnification). (d) Histochemical analysis of non-transplanted AhCre+Mdm2flox/flox mice and cdHPC transplanted AhCre+Mdm2flox/flox mice. Hematoxylin and eosin (top panel), Periodic acid – Schiff (middle panel), PicroSirius Red (bottom panel). (e) PicroSirius Red area of non-transplanted AhCre+Mdm2flox/flox mice and cdHPC transplanted AhCre+Mdm2flox/flox mice (mean ± s.d, Mann-Whitney test. P = 0.0379; n = 8 mice each group). (f) Serum Albumin level of non-transplanted AhCre+Mdm2flox/flox mice and cdHPC transplanted AhCre+Mdm2flox/flox mice (mean ± s.d, Mann-Whitney test. P = 0.0341; n = 10 and 13 mice for non-transplanted group and transplanted group respectively). (g) Immunohistochemistry for GFP (green), hepatocytes (HNF4α or CYP2D6, red) and DNA (blue) in CAG-GFP HPC transplanted livers versus non-transplanted controls. (h) Immunohistochemistry for GFP (green), HPCs (panCK or Sox9, red) and DNA (blue) in non-transplanted controls versus CAG-GFP HPC transplanted animals. Data are represented as mean ± s.d. The data shown are representative of 3 different experiments with 8-10 mice each group. Scale bars = 50μm.
Discussion
The liver regenerates through hepatocytes proliferation following moderate liver injury, and hepatocyte cell therapy is a potential alternative to liver transplantation40, 41. Liver regeneration following chronic or severe liver injury is less well defined. HPCs are activated during chronic liver disease when hepatocyte proliferation is impaired15, 42, 43; however whether these HPCs regenerate parenchyma is controversial11, 21, 44, 45. Using targeted deletion of Mdm2 in mouse hepatocytes, we have demonstrated that HPC activation is sufficient to regenerate a large proportion of the liver parenchyma. In our model, upon Cre activation the majority of hepatocytes (98.9%) accumulate p53, inducing senescence. To address the possibility that the 1.1% of unrecombined hepatocytes may regenerate the liver, we investigated the degree of hepatocyte senescence by p21 staining. Although the proportion of p53 high hepatocytes was 98.9%, almost all hepatocytes (99.96%) were p21 positive, possibly reflecting either under-detection of p53 by immunohistochemistry or the occurrence of spreading senescence as previosly reported in vivo and in vitro 46, 47,48. The proliferation of p21 positive hepatocytes is irreversibly inhibited49 making it unlikely that p21 positive hepatocytes contribute to regeneration here. Furthermore, proliferating hepatocytes were located at the distal tips of the HPC containing ductular reactions following βNF induction, suggesting HPCs contribute to the formation of new p21 negative hepatocytes that are capable of proliferation.
To further investigate the importance of HPC activation, we used a model deficient in Fn14, a receptor for the HPC mitogen TWEAK33, 34. Mice with Fn14 knockout do not mount an appropriate ductular reaction, and have increased mortality. Administration of TWEAK enhanced the ductular reaction in AhCre+Mdm2flox/flox mice following liver injury. These findings suggest that TWEAK/Fn14 mediated ductular reaction are required to adequately regenerate liver parenchyma following such damage.
Huch and colleagues have shown that EpCAM+ cells isolated from human liver can be expanded as organoids in vitro50. Whether these cells can regenerate large amounts of liver parenchyma will be important for their development as a human cell therapy. Here, we demonstrate that large scale therapeutic liver repopulation can be achieved by transplanted HPCs.
CD45−/CD31−/Ter119−/EpCAM+/CD24+/CD133+ HPCs have high colony forming capacity and are of biliary origin. Using the the Krt19CreERTLSLTdTomato lineage tracing model we found the degree of hepatocyte formation from ductular cells in the CDE diet and recovery model is limited13, 21, 45; we speculate that this is due to the mild liver injury and lack of hepatocyte senescence, compared to advanced human liver disease51. This lack of hepatocyte senescence in the CDE model thereby allowing hepatocytes to regenerate themselves without recourse to a second tier of regeneration.
Although previous studies have isolated putative HPCs using FACS isolation, these techniques are based on single selection markers37,8 or did not completely exclude other NPC populations37,52. We investigated multiple potential HPC populations by using a highly stringent FACS gating strategy without the need of transgenic reporter. This is important as HPCs share markers with cholangiocytes and a defined selection strategy is required to reduce impurity if this is going to be translated into human studies. We have overcome the difficulty in expanding HPCs in vitro in monolayer conditions whilst maintaining their karyotypic and phenotypic stability. The CD45−/CD31−/Ter119−/EpCAM+/CD24+/CD133+ HPC population is extremely rare in healthy liver, making their isolation from healthy tissue logistically challenging. Furthermore, the liver repopulating capacity of these cells in a healthy liver is likely insignificant- previous lineage tracing studies have shown no contribution from HPCs to hepatocytes in healthy liver11,44 and hepatocytes are the primary source of regeneration in normal liver homeostasis3. Together our data suggests the existence of a facultative and functionally significant HPC population which does not contribute to homeostatic repair, but is activated in severe liver injury where hepatocyte senescence is widespread. We have used the term HPC throughout given the data confirming genuine progenitor characteristics of these cells within the ductular reaction, however in the absence of this functional evidence the term ductular cell may be more accurate.
To date the repopulation capacity of transplanted HPCs has been limited and their ability to improve the structure of chronically damaged liver unproven. Using repeated rounds of hepatocyte senescence and injury in the AhCre+Mdm2flox/flox mice combined wih HPC transplantation we found that the progeny of the transplanted HPCs can proliferate long-term and regenerate large areas of normal parenchyma. The transplanted HPCs demonstrated bi-lineage differentiation into large numbers of hepatocytes and cholangiocytes in vivo causing significant structural and functional improvement of the damaged liver, with reducced scarring and increase serum albumin in transplant recipients. Our characterisation of a defined population of HPCs indicates their biological significance and future therapeutic potential for the treatment of human liver disease.
Materials and Methods
Animal Models
Animals used in this study were all on a C57Bl/6 background aged within 8 to 12 weeks old at the start of the experiments, both male and female mice were used. Animals were housed in a specific pathogen free environment and kept under standard conditions with a 12 hour day/night cycle and access to food and water ad libitum. All animal experiments were carried out under procedural guidelines, severity protocols and with ethical permission from the Animal Welfare and Ethical Review Body (AWERB) and the Home Office (UK). Power calculations were not routinely performed, however animal numbers were chosen to reflect the expected magnitude of response taking into account the variability observed in previous experiments. AhCre+ mice were crossed with both Mdm2flox/flox, Mdm2flox/− and Mdm2flox/+ mice to generate AhCre+ Mdm2flox/flox, AhCre+ Mdm2flox/− mice and AhCre− Mdm2flox/flox and AhCre+ Mdm2flox/+ controls. AhCre+ Mdm2flox/flox or Mdm2+/+ mice were also crossed with Fn14KO (provided by Biogen Idec34). Genotyping was performed for AhCre using CCTGACTAGCATGGCGATAC (forward) and ATTGCCCCTGTTTCACTATC (reverse) primers and Mdm2 as previously described 28. AhCre was induced by intra-peritoneal (i.p.) injection of β-Napthoflavone (βNF, Sigma UK) at 20-80mg/kg, prepared previously under sterile conditions at 1% (w:v) in corn oil. K19Cre was induced by 3 individual i.p. injections of Tamoxifen (20mg/ml, Sigma UK) at the dose of 4mg. K19Cre animals received three weeks of normal diet after the last Tamoxifen injection before commencing the CDE diet regime.
BrdU (50 μg/g, Amersham, UK) was administered by i.p. injection. Choline Deficient Ethionine supplemented (CDE) protocols were as previously described6,31 . Recombinant murine TWEAK (R and D Systems) was delivered iv at 0.4μg on days -5,-3,-1 and 1 in sterile PBS as previously described32. Mice were sacrificed according to UK Home Office regulations. Unless livers were perfused, blood was harvested by cardiac puncture. Organs were harvested and stored following fixation in 10% formalin (in PBS) for 6hrs or methacarn (60% methanol 30% chloroform 10% glacial acetic acid) for 24hrs prior to embedding in paraffin blocks. Samples of liver were taken and snap frozen in an ethanol bath in dry ice in OCT media, RNAlater or directly for later analysis. Serum analysis used commercial kits according to manufacturer’s instructions for Alanine transaminase (ALT; Alpha Laboratories Ltd, UK), microalbumin (Olympus Diagnostics Lt, UK), Aspartate Aminotransferase (AST) and Alkaline Phosphatase (Alk Phos; both Randox Laboratories, UK).
Hepatocyte purification
Ex vivo primary cells were isolated using a modified retrograde perfusion technique53 consisting of: 1) Liver Perfusion Medium (Gibco) for 5 minutes; 2) Liver Digest Medium (Gibco) for 10 minutes. The liver was then excised and the capsule disrupted to yield a cell suspension which was collected in Liver Perfusion Medium (Gibco) and passed through a 100μm filter (BD Bioscences). Hepatocytes were pelleted by centrifugation at 135G for 1 minute separated them from a non-parenchymal cell (NPC) rich fraction and each resuspended in Williams E Medium (Gibco) with 5% FCS. The hepatocyte rich cell suspension was then purified as described previously53. Briefly cells were underlayered with a discontinuous Percoll gradient (1.06, 1.08 and 1.12g/ml Percoll in PBS). Cells were then spun at 750 G for 20 minutes at 20°C. The cell layer collected between the 1.08 and 1.12mg/ml Percoll layers were harvested and resuspended in Williams Medium. Cells were then purified a further 2 times using Percoll layers as previously and quantified prior to analysis. Routinely cells with hepatocyte morphology and expression of CYP2D6 (marker of hepatocellular differentiation) were obtained at over 99% purity (See Supplementary Fig 2, p<0.05).
HPC isolation
NPCs were retained from the supernatant following the initial 135G centrifugation step. Following red cell lysis (3 minutes in 160mM NH4Cl 10mM KHCO3 containing 0.01% EDTA), cells were re-suspended in Williams E Medium (Gibco) with 10% FCS and underlayered with an equal volume of 20% and 50% (v/v) Percoll (Sigma, UK) in PBS, respectively. Following centrifugation at 1400g for 20 minutes at 4°C, the HPC rich fraction lying between the 20% and 50% Percoll layers (enriched for non-parenchymal cells) was collected, washed twice and re-suspended in PBS with 2% FCS for FACS staining procedure. NPCs collected as described above were resuspended in PBS + 2% FCS prior to incubation with anti-mouse CD16/32 antibody (eBiosciences) for 10 minutes. After a wash with cold PBS, cells were resuspended in PBS + 2% FCS and were incubated with EpCAM-APC conjugated antibody (eBiosciences); CD24-PeCy7 conjugated antibody; CD133-FITC conjugated antibody CD45-PE conjugated antibody; CD31-PE conjugated; Ter119-PE conjugated antibody (Biolegend). Labelled cells were washed with PBS containing 2% FCS and analysed by FACS using a FACS-Aria II (BD) with 488nm and 640nm lasers using a 100 μm nozzle. Hematopoeitic cells (CD45+), erythroid cells (Ter119), and endothelial cells (CD31+) were excluded by gating. 10 μg/ml 7-Aminoactinomycin D (7-AAD) (Biolegend) was added immediately prior to sorting to exclude dead cells. Cells were analysed and sorted with FACS Aria II (BD). Trigger pulse width was used to exclude cell doublets from analysis and collection. Sorted cells were plated on rat tail collagen 1 (Sigma) coated plates supplemented with previously published HPC expansion medium 8. A step-by-step protocol describing the isolation and expansion of HPCs can be found at Nature Protocol Exchange (http://dx.doi.org/10.1038/protex.2015.051)54.
Colony forming assay
Cells were plated at clonal density (250 cells/cm2) on collagen coated plates with culture medium. The number of colonies comprised of more than 50 cells was counted two weeks after plating. Number of colonies formed / number of wells plated = Colony Forming Efficiency.
Expansion of HPCs
Sorted HPCs were cultured in HPC Expansion Medium: Williams’ Medium E (Gibco) containing 10% FCS, 17.6mM NaHCO3, 20mM HEPES pH7.5, 10mM Nicotinamide, 1mM Sodium Pyruvate, 1x Insulin, Transferrin, Selenium Solution (ITS) (Gibco), 100nM Dexamethasone, 0.2mM Ascorbic Acid, 14mM Glucose, 10ng/mL IL-6 (Peprotech), 10ng/mL HGF (Peprotech), 10ng/mL EGF (Sigma-Aldrich).
Cultured HPCs were washed with PBS. Cells were then incubated with diluted trypsin (0.25% Trypsin no EDTA, 20% Knockout Serum Replacement, 1mM CaCl2) at 37°C. Williams Medium E supplemented with 10% FCS were used to inhibit the reaction of trypsin. Cells were centrifuged at 300G for 5 minutes, supernatant was then discarded. Cells were resuspened in culture medium and replated on collagen coated plates.
Flow Cytometry analysis
Cells were rinsed with PBS and then incubated with Cell Dissociation Medium (GIBCO) for 20 minutes at 37°C to prevent excessive cleavage of desired epitopes for detection. Cells were then incubated with anti-mouse CD16/32 antibody (eBiosciences) at 1:50 dilution for 10 minutes on ice. After rinsing with PBS, cells were resuspened in PBS + 2% FCS and incubated with antibodies (indicated in List of Antibodies Table) for 1 hour at 4°C. Cells were then washed with PBS + 2% FCS and then resuspened for in PBS + 2% FCS for analysis or sorting. Stained samples were compared to unstained and isotype controls.
Immunohistochemistry
Three μm thick paraffin sections were stained for the published HPC markers: panCK, A6 kindly provided by Dr V. Factor (National Cancer Institute, Bethesda, MD), Sox9, CD24, Dlk1, EpCAM, and Fn14 or for the alternate antigens: BrdU, (AB6326 Abcam, UK), p53 (MS 104-P1, LabVision), Ki67, and CYP2D6 (Generous gift from Prof R Wolfe, University of Dundee); see Supplementary Table 2 for additional details. Cytospins and cells were fixed in cold methanol at 4°C for 5 minutes prior to staining Species isotype (Santa Cruz) staining controls were routinely performed. Detection was performed with DAB (DAKO) followed by counterstaining with Haematoxylin or alternately with Alexa 488, 555 or 650 (A21206, A21434A21436/S32355, and A21448 respectively; Invitrogen, UK) with a DAPI containing Vectasheld mounting media (Vector, UK). Apoptosis was assessed using the DeadEnd™ Colorimetric or Fluorometric (Alexa 488) TUNEL System (Promega #TB199) according to manufacturer's instructions.
Microscopy and cell counting
Images were obtained on a Zeiss Axiovert 200 microscope using a Zeiss Axiocam MR camera. Cell counts were performed manually on blinded slides and more than 20 consecutive non overlapping fields at ×200 or ×400 magnification. HPCs were defined as previously described and interlobular bile ducts were excluded from quantification32. Confocal image analysis was performed using a Leica SP5 system with the pinhole set to 1 airy unit. DAPI, Alexafluor 488, and 555 were detected using band paths of 415-480, 495-540 and 561-682nm for 405, 488 543 nm lasers respectively.
Cell Transplantation Assay
AhCre Mdm2flox/flox mice were used as recipients for transplantation assay. 10mg/kg of βNF was injected i.p. 4 days before transplant to induce liver injury, 5×10 GFP expressing cells were suspended in 200μL of PBS and injected intrasplenically after laparotomy. Transplantation control group receive 200μL PBS only. Recipient mice received intraperitoneal injections of 20mg/kg βNF every 10 days after transplantation to induce persistent liver injury. Mice were sacrificed and the liver was harvested 12 weeks after cell transplantation.
In Vitro Differentiation of Hepatic Progenitor Cells (HPCs)
100ng/mL of Murine Recombinant Wnt3a (R&D systems) and 1% of Dimethyl Sulfoxide (DMSO) were added to HPCs in combination with culture medium when cultured HPCs were 70% confluent. Differentiation process lasted for 6 days; medium was changed once in 3 days. Medium was kept for ELISA and cells were lysed for RNA isolation.
Formation of progenitor cell spheroids
HPCs were trypsinised into a single cell suspension by incubating with Cell Dissociation Medium (GIBCO) at 37°C for 15 minutes and resuspended in 50μL of undiluted Growth Factor Reduced Matrigel (BD) and plated on 24 well multiwell plates (Corning). Each 50μL of Matrigel that consist around 10,000 cells per spheroid was cultured. The plate was then left in the incubator at 37°C for 10 minutes for the Matrigel to set. Culture medium was then added to cover the Matrigel sphere. Spheres were cultured for 7 days and then analysed by immunocytochemistry.
Green Fluorescent Protein (GFP) transfection of progenitor cell line and derivation of stably transfected lines
Qiagen Effectene was used for stable transfection. 5×10 of in vitro expanded progenitor cells were plated overnight. 1μg of vector with a puromycin resistant CAG-GFP was used to stably transfect cells with GFP, kindly provided by Dr Sally Lowell (University of Edinburgh). Transfected cells were cultured for 3 days and GFP expression was detected with fluorescence microscopy. Around 10 percent of transfected cells expressed GFP after 4 days. The selection of stably transfected cells was carried with medium supplemented with 3 μg/ml puromycin for 14 days and colonies were picked with cloning rings and grown to generate clonally derived stably transfected lines. GFP expression of stably transfected lines were constantly monitored with fluorescent microscope.
Enzyme-linked Immunosorbent assay (ELISA) detection of Albumin
Conditioned medium from cultured cells was and stored at −80°C for further analysis. Stored medium was thawed at room temperature and tested for albumin concentration using Mouse Albumin ELISA Quantification Set following manufacturer’s protocol. Standard curve was generated using supplemented kits with different concentration of albumin. Absorbance at 450nm of was read using a spectrophotometer (SPECTROstar Omega).
Analysis of cell area, roundness, and ratio of width to length
Cell area, roundness, and ratio of width to length were analysed using the Operetta high content analysis system (PerkinElmer). Data analysis was performed using Columbus image data storage and analysis system.
Karyotyping of cells
Colcemid (Gibco) was added to the cultured cells for 18 hours. Cells were then trypsinised, washed with PBS and resuspended with 75mM KCL (Gibco) and incubated for 15 minutes at room temperature. Cells were then resuspended in fixatives (3:1 Methanol and Acetid Acid). Cells were washed and spread on glass slides. Slides were mounted with DAPI fluoromount (Southern Biotech). Number of chromosomes was counted using fluorescence microscopy.
Real time PCR and gene expression analysis
Genomic DNA (gDNA) was extracted from purified cell populations using whole liver using DNA Blood Mini Kit (Qiagen UK). Total RNA was extracted from 30-50mg tissue samples previously stored in RNAlater at −80°C or cultured cells, using a combination of TRIzol™ reagent (Invitrogen) and Qiagen RNeasy Mini system according to manufacturer’s instructions (Qiagen, UK). Reverse Transcription (including gDNA decontamination) and real time PCR was performed using reagents and primers (Quantifast and Quantitect respectively, Qiagen, UK) on a Roche Lightcycler 480. Mdm2flox integrity of gDNA was assessed using primers targeted to the floxed segment (Forward: ACGAGAAGCAGCAGCACATTG, Reverse: TTATAACCCACCACGCACAAGG) and a reference segment adjacent to Exon 3 (Forward: AACCTGGATTATCGCACAGTCG, Reverse: TCGCAGTGACCACTCTCTAATGC). cDNA was prepared from purified cell populations using Data were analysed using the LightCycler system following normalization to the housekeeping gene peptidylprolyl isomerase A (Ppia). All samples were run in triplicate.
Statistical analysis
Samples were randomized by a 'blinded' third party before being assessed separate 'blinded' assessor. Unblinding was performed immediately prior to final data analysis. Prism software (GraphPad Software, Inc) was used for all statistical analysis. Mean HPCs per ×200 magnification field from 30 fields for each mouse were compared. Data is presented as mean ± s.e.m. n refers to biological replicates. Normal distribution of data was determined using D’Agostino and Pearson omnibus normality test. For parametric data, data significance was analysed using a two-tailed unpaired Students t-test. In cases where more than two groups were being compared, then a one-way ANOVA was used. In instances where the n was too small to determine normal distribution or the data was non-parametric then a two-tailed Mann Whitney U-test was used. F tests were used to compare variances between groups.
Reproducibility of experiments
Routinely qPCR experiments were performed in technical triplicates when possible. For representative images whole liver lobes were examined histologically in multiple biological replicates. For figure 1b-f, k-l, 2a, b, d, e, I, j, Supplementary figures 2h, 3 a-c the images are representative for 30 mice in total over a 3-year period. For figure 2k, j, 4e, Supplementary figures 3a-d, h, the images are representative for 6 mice. For figure 3a-f, 4a, c, f, 5a, the images are representative for 40 mice in total over a 1-year period. For figure 5f, g, 6c, d the images are representative for 7 mice. For figure 7g, i, m, Supplementary figures 4a, f, 5b-d the images are representative for 5 separate clonal cultures. For figure 7h the image is representative of 50 cells. For figure 7 a-d, k, Supplementary figure 1b, c the images are representative for 10 mice. For figure 8 the images are representative for 23 mice. For Supplementary figures 2a-c, 4a the images are representative for 12 mice. For Supplementary figures 5a, 6a, 6b the images are representative for 7 separate clonal cultures.
In the cases of experiments in figures 1g-j, 2c, g, h, 5c, Supplementary figures 1a-c, 2a-h, 3e, 3f experiments were repeated twice. For figures 1g-j, 2c and Supplementary figures 2a-f these replicates consisted of repeated biological experiments with independent analysis, time points for analysis varied between experiments in 1h-j, 2c and Supplementary Figures 2e,f. For figures 2g, h and 5c and Supplementary Figures 2g, 3e, 3f, replicate experiments consisted of repeat analysis from biological samples from the level of RNA extraction. In the cases of experiments in figures 7e, f, 8a-h and Supplementary figures 4b, c, e, 5e, and 6c, flow cytometry and qPCR experiments were repeated for 3 times, transplantation experiments are shown as representative from 3 experiments.
Supplementary Material
Acknowledgements
The authors would like to thank Biogen Idec for supplying the Fn14KO mouse line. We would also like to thank Dr V Factor for kindly supplying the A6 antibody, and Professor Malcom Alison for proofreading the manuscript. T.B is funded by the Wellcome Trust and the Academy of Medical Sciences. W.Y Lu was supported by the University of Edinburgh, Charles Darwin Scholarship, Edinburgh Overseas Research Scholarship, and the UK Regenerative Medicine Platform. O.J.S is funded by Cancer Research UK and the European Research Council. S.J.F. is supported by the Sir Jules Thorn Charitable Trust, the Medical Research Council, and the UK Regenerative Medicine Platform.
Abbreviations
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- βNF
β-napthoflavone
- BrdU
Bromodeoxiuridine
- CDE
choline deficient ethionine supplemented
- GFP
Green fluorescent protein
- HPCs
Hepatic Progenitor Cells
- MDM2
Mouse double minute 2 homolog
- NPC
non-parenchymal cell
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labelling
Footnotes
Competing Fnancial Interests Statement: The authors have declared that no conflict of interest exists.
Reference List
- 1.Horslen SP, Fox IJ. Hepatocyte transplantation. Transplantation. 2004;77:1481–1486. doi: 10.1097/01.tp.0000113809.53415.c2. [DOI] [PubMed] [Google Scholar]
- 2.Horslen SP, et al. Isolated hepatocyte transplantation in an infant with a severe urea cycle disorder. Pediatrics. 2003;111:1262–1267. doi: 10.1542/peds.111.6.1262. [DOI] [PubMed] [Google Scholar]
- 3.Malato Y, et al. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. The Journal of clinical investigation. 2011;121:4850–4860. doi: 10.1172/JCI59261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erker L, et al. Therapeutic liver reconstitution with murine cells isolated long after death. Gastroenterology. 2010;139:1019–1029. doi: 10.1053/j.gastro.2010.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay DC. Cadaveric hepatocytes repopulate diseased livers: life after death. Gastroenterology. 2010;139:729–731. doi: 10.1053/j.gastro.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Boulter L, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nature medicine. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulter L, Lu WY, Forbes SJ. Differentiation of progenitors in the liver: a matter of local choice. The Journal of clinical investigation. 2013;123:1867–1873. doi: 10.1172/JCI66026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okabe M, et al. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- 9.Dorrell C, et al. Surface markers for the murine oval cell response. Hepatology. 2008;48:1282–1291. doi: 10.1002/hep.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nature genetics. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 11.Espanol-Suner R, et al. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology. 2012;143:1564–1575. e1567. doi: 10.1053/j.gastro.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jors S, et al. Lineage fate of ductular reactions in liver injury and carcinogenesis. The Journal of clinical investigation. 2015 doi: 10.1172/JCI78585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, et al. The origin and liver repopulating capacity of murine oval cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(Suppl 1):11881–11888. doi: 10.1073/pnas.1734199100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fickert P, et al. A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. The American journal of pathology. 2007;171:525–536. doi: 10.2353/ajpath.2007.061133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tirnitz-Parker JE, Tonkin JN, Knight B, Olynyk JK, Yeoh GC. Isolation, culture and immortalisation of hepatic oval cells from adult mice fed a choline-deficient, ethionine-supplemented diet. The international journal of biochemistry & cell biology. 2007;39:2226–2239. doi: 10.1016/j.biocel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Akhurst B, et al. A modified choline-deficient, ethionine-supplemented diet protocol effectively induces oval cells in mouse liver. Hepatology. 2001;34:519–522. doi: 10.1053/jhep.2001.26751. [DOI] [PubMed] [Google Scholar]
- 18.Fausto N. Liver regeneration. Journal of hepatology. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 19.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 20.Grompe M. Liver stem cells, where art thou? Cell stem cell. 2014;15:257–258. doi: 10.1016/j.stem.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Yanger K, et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell stem cell. 2014;15:340–349. doi: 10.1016/j.stem.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yimlamai D, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarlow BD, et al. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell stem cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouw AS, Clouston AD, Theise ND. Ductular reactions in human liver: diversity at the interface. Hepatology. 2011;54:1853–1863. doi: 10.1002/hep.24613. [DOI] [PubMed] [Google Scholar]
- 25.Golding M, et al. Oval cell differentiation into hepatocytes in the acetylaminofluorene-treated regenerating rat liver. Hepatology. 1995;22:1243–1253. doi: 10.1016/0270-9139(95)90635-5. [DOI] [PubMed] [Google Scholar]
- 26.Michalopoulos GK, Barua L, Bowen WC. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology. 2005;41:535–544. doi: 10.1002/hep.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ireland H, et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology. 2004;126:1236–1246. doi: 10.1053/j.gastro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Grier JD, Yan W, Lozano G. Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis. 2002;32:145–147. doi: 10.1002/gene.10066. [DOI] [PubMed] [Google Scholar]
- 29.Aravinthan A, et al. Hepatocyte expression of the senescence marker p21 is linked to fibrosis and an adverse liver-related outcome in alcohol-related liver disease. PloS one. 2013;8:e72904. doi: 10.1371/journal.pone.0072904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eischen CM, Boyd K. Decreased Mdm2 expression inhibits tumor development and extends survival independent of Arf and dependent on p53. PloS one. 2012;7:e46148. doi: 10.1371/journal.pone.0046148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenzini S, et al. Characterisation of a stereotypical cellular and extracellular adult liver progenitor cell niche in rodents and diseased human liver. Gut. 2010;59:645–654. doi: 10.1136/gut.2009.182345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird TG, et al. Bone marrow injection stimulates hepatic ductular reactions in the absence of injury via macrophage-mediated TWEAK signaling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6542–6547. doi: 10.1073/pnas.1302168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tirnitz-Parker JE, et al. Tumor necrosis factor-like weak inducer of apoptosis is a mitogen for liver progenitor cells. Hepatology. 2010;52:291–302. doi: 10.1002/hep.23663. [DOI] [PubMed] [Google Scholar]
- 34.Jakubowski A, et al. TWEAK induces liver progenitor cell proliferation. The Journal of clinical investigation. 2005;115:2330–2340. doi: 10.1172/JCI23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clouston AD, et al. Fibrosis correlates with a ductular reaction in hepatitis C: roles of impaired replication, progenitor cells and steatosis. Hepatology. 2005;41:809–818. doi: 10.1002/hep.20650. [DOI] [PubMed] [Google Scholar]
- 36.Rountree CB, et al. A CD133-expressing murine liver oval cell population with bilineage potential. Stem cells. 2007;25:2419–2429. doi: 10.1634/stemcells.2007-0176. [DOI] [PubMed] [Google Scholar]
- 37.Qiu Q, Hernandez JC, Dean AM, Rao PH, Darlington GJ. CD24-positive cells from normal adult mouse liver are hepatocyte progenitor cells. Stem cells and development. 2011;20:2177–2188. doi: 10.1089/scd.2010.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchiya A, et al. Long-term extensive expansion of mouse hepatic stem/progenitor cells in a novel serum-free culture system. Gastroenterology. 2005;128:2089–2104. doi: 10.1053/j.gastro.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 39.Hay DC, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overturf K, Al-Dhalimy M, Finegold M, Grompe M. The repopulation potential of hepatocyte populations differing in size and prior mitotic expansion. The American journal of pathology. 1999;155:2135–2143. doi: 10.1016/S0002-9440(10)65531-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overturf K, al-Dhalimy M, Ou CN, Finegold M, Grompe M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. The American journal of pathology. 1997;151:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- 42.Van Hul NK, Abarca-Quinones J, Sempoux C, Horsmans Y, Leclercq IA. Relation between liver progenitor cell expansion and extracellular matrix deposition in a CDE-induced murine model of chronic liver injury. Hepatology. 2009;49:1625–1635. doi: 10.1002/hep.22820. [DOI] [PubMed] [Google Scholar]
- 43.Fickert P, et al. The role of osteopontin and tumor necrosis factor alpha receptor-1 in xenobiotic-induced cholangitis and biliary fibrosis in mice. Laboratory investigation; a journal of technical methods and pathology. 2010;90:844–852. doi: 10.1038/labinvest.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodrigo-Torres D, et al. The biliary epithelium gives rise to liver progenitor cells. Hepatology. 2014;60:1367–1377. doi: 10.1002/hep.27078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell reports. 2014;8:933–939. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acosta JC, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nature cell biology. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson G, et al. A senescent cell bystander effect: senescence-induced senescence. Aging cell. 2012;11:345–349. doi: 10.1111/j.1474-9726.2012.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes & development. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H, et al. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes & development. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 50.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiemann SU, et al. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2002;16:935–942. doi: 10.1096/fj.01-0977com. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki A, et al. Flow cytometric isolation and clonal identification of self-renewing bipotent hepatic progenitor cells in adult mouse liver. Hepatology. 2008;48:1964–1978. doi: 10.1002/hep.22558. [DOI] [PubMed] [Google Scholar]
- 53.Goncalves LA, Vigario AM, Penha-Goncalves C. Improved isolation of murine hepatocytes for in vitro malaria liver stage studies. Malaria journal. 2007;6:169. doi: 10.1186/1475-2875-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu WY, et al. Isolation and expansion of the hepatic progenitor cell (HPC) population. Nat. Protoc. Exch. 2015 http://dx.doi.org/10.1038/protex.2015.051. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.