Abstract
The trade-off between survival and reproduction is fundamental to life history theory. Sexual selection is expected to favour a ‘live fast die young’ life history pattern in males due to increased risk of extrinsic mortality associated with obtaining mates. Sexual conflict may also drive a genetic trade-off between reproduction and lifespan in females. We found significant additive genetic variance in longevity independent of lifetime mating frequency, and in early life mating frequency. There was significant negative genetic covariance between these traits indicating that females from families characterized by high levels of multiple mating early in life die sooner than females that engage in less intense early life mating. Thus, despite heritable variation in both traits, their independent evolution is constrained by an evolutionary trade-off. Our findings indicate that, in addition to the well-known male-driven direct costs of mating on female lifespan (mediated by male harassment and harmful effects of seminal fluids), females with a genetic propensity to mate multiply live shorter lives. We discuss the potential role of sexual conflict in driving the evolutionary trade-off between reproduction and lifespan in Drosophila. More generally, our data show that, like males, females can exhibit a live fast die young life history strategy.
Senescence is defined as decreasing reproductive performance and increasing probability of death with age1. Theory proposes that senescence evolves as a result of decreased selection on genes expressed late in life as few individuals reach old age due to extrinsic mortality factors2,3,4,5. The Disposable Soma theory suggests that trade-offs result from the partitioning of finite resources between growth, maintenance and reproduction5. These trade-offs are important because they enable the accumulation of alleles with deleterious effects that are only expressed late in life (mutation accumulation2) and/or cause antagonistic pleiotropic effects on alleles, where genes that have positive effects on fitness early in life have deleterious effects on later survival (antagonistic pleiotropy3). Such alleles have the potential to generate genetic trade-offs among life history traits and constrain their independent evolution3,6.
The trade-off between survival and reproduction has long been recognised as a prominent feature of life history trajectories, and it is well established that reproduction reduces lifespan in many species7. Our understanding of the evolution of ageing, and how reproduction affects it has advanced greatly from research on Drosophila melanogaster. Numerous artificial selection experiments in this species provide evidence supporting a trade-off between early fecundity and late fecundity or survival. For example, artificial selection by Rose8 found higher rates of egg laying in late life than early life in females selected for extended longevity compared to control lines, and Partridge et al.9 found a decrease in fertility early in life in lines selected for longer life. Other studies have also found correlated responses to selection for late fecundity with increased lifespan in D. melanogaster10,11,12. The general finding of lifespan and fertility increasing in lines propagated from old adults, and pre-adult survival or fecundity of young adults showing a correlated decline with lifespan show that trade-offs are important in the evolution of ageing in this model system.
Sexual selection influences the costs of reproduction and the benefits of early versus late reproduction for each sex. Bateman’s principle predicts that males can increase their fitness by mating with many females, while females, due to a limitation on the number of eggs they can produce and their typically larger parental investment, are expected to maximise their reproductive success with only one or a few matings13,14. The dichotomy in reproductive investment between the sexes means that males typically allocate more resources to competition for matings than females14. Males are more likely to suffer increased mortality due to costly secondary sexual traits (costly in terms of energy expenditure and increased predation) and direct injury in combat competition to gain access to mates. Consequently, males are expected to pursue high risk ‘live fast, die young’ life history strategies that have potential to yield high fitness returns over short time periods15,16. Hence, it is predicted that selection will favour male reproductive strategies that sacrifice longevity for mating opportunities15,17. Indeed, empirical findings suggest that male mortality is higher than female mortality across a range of taxa18. In contrast, because females are limited by the time and energy requirements for offspring production, selection acting on females is expected to promote low risk, low wear and tear strategies with moderate rates of return over extended time periods16. In support of this hypothesis, male crickets selected for decreased lifespan increase early reproductive performance (calling), whereas female longevity was reduced without influencing their reproductive success19.
According to evolutionary theories of ageing, increased rates of extrinsic mortality should lead to accelerated rates of intrinsic mortality4, which reduces selection on late life performance20. This has led to the recognition that antagonistic coevolution between the sexes may promote an evolutionary trade-off between reproduction and lifespan. While evidence is mounting for diverse benefits of female multiple mating across species21,22,23, it is also well established that mating incurs a cost to females, and the archetypical example is D. melanogaster24,25,26,27,28,29,30,31. The increase in female mortality caused by sexually antagonistic adaptations in males should result in the accumulation of alleles with deleterious late life effects. Promislow32 suggests higher rates of sexual conflict lead to the evolution of higher rates of senescence. Maklakov et al.33 found the removal of sexual conflict via enforced monogamy in populations of the seed beetle Callosobruchus maculatus led to increased lifespan in virgin females from monogamous populations compared to polygamous populations. However, sexual conflict could also result in selection for increased somatic maintenance to repair the damage induced by toxic ejaculates and hence increase lifespan33,34. For example, Reznick, et al.35 found that higher extrinsic mortality mediated by predation in Poecilia reticulata guppies was associated with decelerated rates of intrinsic mortality. Promislow et al.36 also found that increased opportunity for sexual selection was genetically correlated with adult survivorship in D. melanogaster. Therefore, antagonistic coevolution could theoretically promote either accelerated or decelerated ageing.
An important prediction of the mutation accumulation and antagonistic pleiotropy theories of life history evolution is that the trade-off between reproduction and lifespan leads to negative genetic correlations. Implicit in this prediction is the assumption of the presence of additive genetic variation in which genes that alter reproduction early in life simultaneously alter survival in late life37. In this study we aimed to investigate the genetic trade-off between female longevity and early life mating frequency. However, quantification of genetic variation in lifespan is not straightforward because of the phenotypic cost of mating due to sexual conflict. Using a quantitative genetic design, we calculated quantitative genetic estimates for female lifespan while controlling for lifetime mating frequency, and genetic estimates for early life mating frequency while controlling for longevity in a natural population of D. melanogaster. We examined the relationship between early life mating frequency and lifetime mating rate to confirm that our measure of early life mating frequency was indicative of lifetime mating frequency. We then investigated the potential for an evolutionary trade-off between lifespan and early life mating by measuring the genetic covariance between the two traits.
Results
We found substantial phenotypic variation in longevity (mean ± SD = 40.460 days ± 10.361; range = 13–62). Raw sire family means of longevity are displayed in Fig. 1. There was a significant effect of total number of lifetime matings on longevity (Wald χ2 = 74.279 df = 1, p < 0.001), with longer lived females having a higher total number of matings in their life. There was a significant effect of start date (Wald χ2 = 10.439 df = 1, p = 0.001) but no significant effect of body size (Wald χ2 = 0.147, df = 1, p = 0.702) on longevity. Our genetic estimates for longevity after controlling for the number of lifetime matings showed considerable levels of additive genetic variation and narrow sense heritability (Table 1) and revealed significant variance among sires (χ2 = 17.385, df = 1, p < 0.001).
Figure 1. Raw sire family means of longevity.
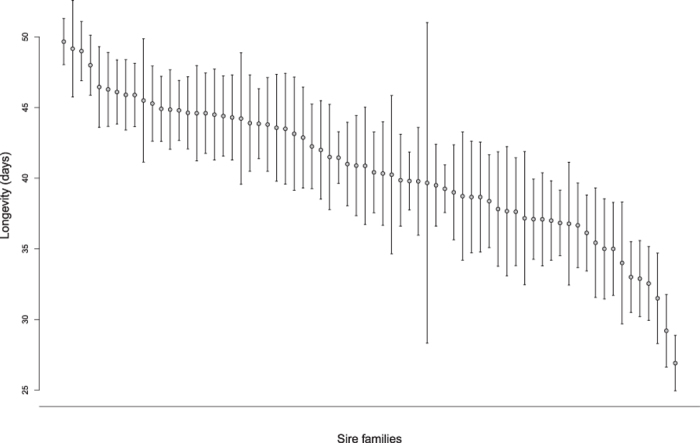
Table 1. Quantitative genetic parameters for longevity (after controlling for lifetime mating frequency) and early life mating frequency.
| N | Mean (SE) | n sires | n dams | VSire (SE) | VDam (SE) | VA (SE) | VP (SE) | VR (SE) | h2 (SE) | CVA(SE) | CVP(SE) | CVR (SE) | IA (SE) | PSire | PDam | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Longevity | 613 | 40.460 (0.420) | 70 | 197 | 12.602 (4.540) | 0 | 50.410 (18.161) | 96.393 (4.793) | 83.791 (4.867) | 0.523 (0.180) | 0.175 (0.034) | 0.243 (0.008) | 0.168 (0.029) | 0.031 (0.011) | <0.001* | 1 |
| Early life mating frequency | 613 | 2.085 (0.035) | 70 | 197 | 0.122 (0.039) | 0.002 (0.016) | 0.487 (0.156) | 0.644 (0.048) | 0.644 (0.048) | 0.634 (0.176) | 0.335 (0.059) | 0.420 (0.018) | 0.254 (0.049) | 0.112 (0.037) | <0.001* | 0.295 |
Number of offspring (N), trait means (longevity in days), number of sire (half-sib) and dam (full-sib) families (n), variance components for sires (VSire) and dams (VDam), additive genetic variation (VA), total phenotypic variation (Vp), residual variation (VR), narrow sense heritabilities (h2), mean-standardized additive genetic variances (Evolvabilities: CVA and IA), coefficient of phenotypic variation CVp, coefficient of residual variation CVR, and significance values for Sire and Dam effects (PSire and PDam). Standard errors (SE) are provided within brackets.
Early life mating frequency also showed considerable phenotypic variation (mean ± SD = 2.085 ± 0.88). Neither longevity (Wald χ2 = 0.810, df = 1, p = 0.368), body size (Wald χ2 = 2.070, df = 1, p = 0.150) or start date (Wald χ2 = 3.348, df = 1, p = 0.067) had a significant effect on the measure. Early life mating frequency displayed high narrow sense heritability and additive genetic variance (Table 1), and significant sire variance (χ2 = 23.77, df = 1, p < 0.001).
We found a significant negative genetic covariance between early life mating frequency and longevity (rg = −0.651, SE = 0.138). Figure 2 shows the relationship between sire family means of residual longevity and early life mating frequency. A significant positive correlation between early life and later life mating frequency was also found (rg = 0.692, SE = 0.171).
Figure 2. Sire family mean relationship between residual longevity and early life mating frequency.
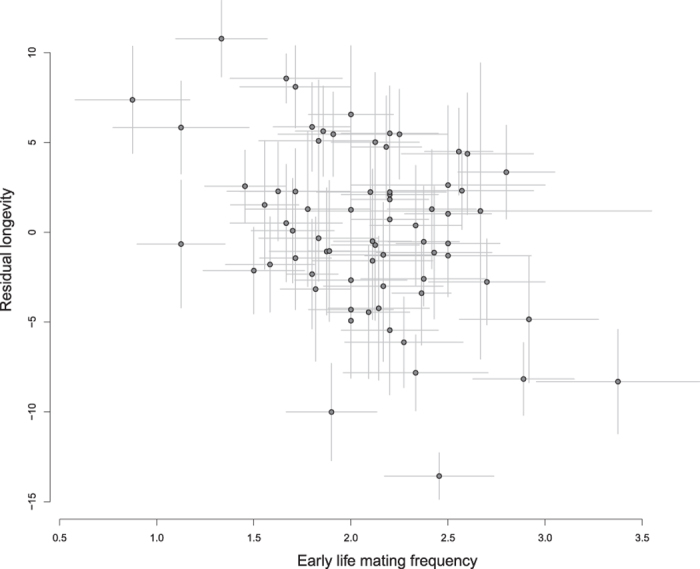
Residual longevity was obtained from a linear regression of longevity on lifetime mating frequency and start date. Points and error bars correspond to sire family means and standard errors, respectively.
Discussion
Our analyses of lifespan and early life mating frequency showed strong negative genetic covariance, indicative of an evolutionary trade-off. Numerous artificial selection experiments have selected for altered lifespan in this species and found correlated responses indicative of a trade-off between reproductive effort (e.g. fecundity, egg viability, competitive larval viability) and lifespan8,10,12 (for review see38). Our study has quantified the additive genetic basis of traits underlying the trade-off between lifespan and early life mating and found high levels of genetic variance in both traits. The genetic trade-off indicates that alleles that have a positive effect on early life mating also have a negative effect on longevity, as predicted by evolutionary theories of senescence. Additive genetic variation in these traits may in part be maintained by the opposing effects of their alleles on female fitness. Such alleles are likely to remain for longer periods at intermediate frequencies within a population, compared to alleles that have a positive effect on both traits whereby directional selection is expected to erode genetic variation37.
The negative genetic correlation between early life mating frequency and longevity is indicative of variation in female life history strategies. Male reproductive strategies are typically associated with elevated mortality risks and weaker selection for long lifespan compared to females, due to the high cost of bearing secondary sexual traits and higher extrinsic mortality16. For example, Robinson, et al.39 found a trade-off between annual breeding success and longevity in males, with larger horn size associated with reduced longevity in a population of Soay sheep. Lemaître, et al.40 also found increased rates of ageing in male red deer that controlled larger harems and rutted for longer periods which suggests males that invest more energy in reproduction early in life age at a faster rate. In females, Charmantier, et al.41 found a genetic trade-off between age at first reproduction and age at last reproduction in a population of free ranging mute swans (Cygnus olor), suggesting that increased early life performance trades off with earlier reproductive senescence. However, Bérubé et al.42 found no trade-off between female longevity and early life reproduction in two wild living populations of bighorn sheep (Ovis canadensis). We found that females from families characterized by high levels of multiple mating early in life died sooner than females from families that engaged in less intense early life mating. Our results thereby support the live fast die young trade-off in females. The positive correlation between early and late life mating suggests that our early life measure of mating frequency reflects overall lifetime mating strategy. Hence, variation in reproductive strategies within this population may constrain the evolution of lifespan.
The relationship between extrinsic mortality and lifespan is common to the main evolutionary explanations of senescence. Accordingly, it is now recognised that damage caused by sexual conflict may play an important role in the evolution of sex specific rates of mortality16. Importantly, our results show a genetic trade-off between early life mating and lifespan that is independent of the phenotypic effect of male seminal fluids on female lifespan. Therefore, in addition to the harmful effects of male seminal fluids on lifespan, our findings suggest that females with a genetic propensity to mate multiply live shorter lives. Sexual conflict in this species could contribute to the accumulation of alleles with deleterious effects in old age or accumulation of alleles that enhance early life fitness at the cost of late life fitness. It is possible that elevated rates of mortality in females due to toxic male seminal fluid proteins could drive a live fast die young strategy in females, due to weakened selection on somatic maintenance. The optimal strategy favoured by natural selection might be high rates of mating in early life because females would be unlikely to survive long enough in natural populations of flies to experience fitness costs associated with deleterious alleles expressed in late life. The intensity of female mating frequency may in part reflect a balance between the benefits to females of mating multiply and the cost of reduced lifespan. Our findings suggest that antagonistic coevolution not only imposes a phenotypic cost to mating but that it could potentially drive the evolution of lifespan of females in this species.
Methods
We used a full sib half sib breeding design to quantify genetic variation in female adult lifespan and lifetime mating frequency for 775 daughters distributed among 72 sire families and 198 dam families of D. melanogaster. Focal flies came from a laboratory population of sixth generation descendants of wild type D. melanogaster collected near Innisfail in Northern Queensland, Australia. To produce parents of focal females, grape agar plates were placed in the population cage for 4 h. The following day, we collected first instar larvae and transferred them to vials at a standard density of 50 larvae per vial. Vials contained 10 ml of sugar-maize medium. Offspring were collected 9–11 days later under CO2 anaesthesia within 8 h of eclosion and transferred to single sex vials. Males were kept at a density of 10 per vial and females at 5 per vial.
Parental generation matings were carried out when flies were 3–4 days old. Each male was mated to three virgin females to generate families of paternal half siblings and maternal full siblings. After mating, females were transferred to individual vials and moved to new vials every 48 h for four days. During peak eclosion, eight virgin female offspring (daughters) from each full sibling dam family were randomly collected; four were included in the lifetime mating frequency assay and four were frozen and later used to estimate full sibling dam family average female body size. Mating opportunities for daughters began at 3–5 days of age. Females were kept in individual vials and transferred to fresh food vials every week. Each female was given a mating opportunity with a sexually naïve male every Monday, Wednesday and Friday over her entire lifespan. Successful copulations were recorded and all pairs separated after 90 mins.
It is well known in this species that male seminal fluid proteins transferred during mating reduce female survival through their toxic effects25,43,44. It is also well established that male seminal fluid proteins mediate female remating27,43. Therefore, male effects are likely to introduce additional environmental variation to our genetic estimates of lifespan and female mating frequency. To address this problem we estimated the additive genetic variance of female mating frequency and lifespan by using standardized males as mating partners (see45 for rationale). Hence, to reduce male induced variation in female lifespan and female remating, for each mating opportunity, we standardised the male identity by randomly selecting naïve males from one of 10 isogenic lines. Each isogenic line had been generated through full sibling matings started with one founder pair taken from a replica of a LHM population46 (see below). We used 2–3 day old males for the mating trials. The standardization results in more precise estimates of polyandry by minimizing sampling variance induced by randomly selecting mates that differ in their effects on female survival and remating propensity.
Male isogenic lines were obtained through full sibling matings for 16 generations, followed by several generations of within line matings (approx. 15 individuals from each vial for each new generation). Full sibling matings were then reinstated for another 21 generations. Isogenic lines were then mass bred into population cages to allow collection of an adequate numbers of flies needed for mating trials. We standardized the identity of the male partner by randomly selecting males from the same isogenic line in each mating opportunity. Larvae were collected from population cages on grape agar plates and sexually naïve males collected at peak eclosion.
Statistical Analyses
To investigate the genetic basis of longevity and early life mating frequency, a subset (n = 613) was generated from the full dataset to include only females (daughters from the full sib half sib breeding design) that lived long enough to have at least six mating opportunities and that mated at least once. All females had a different number of mating opportunities throughout their life, depending on how long they lived. The rationale for subsetting the data to include a fixed number of mating opportunities (first six opportunities) was to control for the possible confounding effect of females with more mating opportunities having a lower lifetime remating proportion. Such a confounding effect may arise due to age related decline in mating rate, and increased probability of males failing to attempt courtship with females having had more mating opportunities. Furthermore, it is unlikely that females with a large number of mating opportunities will maintain high remating rates throughout all opportunities compared to females with only few mating opportunities. The subset also eliminated females that escaped or were accidentally killed. Thus, only females that had lived a ‘natural’ lifespan were included. The lme4 package47 implemented in R 3.03.348 was used to fit standard nested mixed models for a paternal half-sibling design. A Linear Mixed Model (LMM) on untransformed longevity was fitted, after residuals were tested for normal distribution, using the lmer function with sire and dam nested within sire as random effects. A LMM was fitted on log (x + 1) transformed number of matings accepted in the first six opportunities, hereafter referred to as early life mating frequency, using the lmer function with sire and dam nested within sire as random effects. All females were given the first opportunity to mate when they were 3–5 days old but for logistical reasons there was variation among sire and dam families in the date we started the assays. To control for potential temporal variation in longevity and early life mating frequency, we included assay start date as a fixed effect in both analyses. We also included total lifetime mating frequency as a fixed effect in our genetic analysis of longevity to control for the effect of mating frequency on lifespan. Longevity was included as a covariate in the genetic analysis of early life mating frequency. We also investigated the effect of body size on both traits. Significance of fixed effects was tested using Wald chi-square tests implemented in the Anova function of the car package49. Significance of the sire and dam variance components were determined using likelihood-ratio tests.
Genetic parameters for both traits were calculated using restricted maximum likelihood (REML) from LMMs using the lmer function. We performed the analyses on untransformed data because many genetic parameters (e.g. CVA and IA) cannot be used for comparative purposes if variance components are extracted when data are transformed50. Observational variance components were estimated from minimal models including only significant fixed effects. Narrow sense heritabilities (h2) were estimated from the ratio of additive genetic variance (VA: four times the sire variance component) to total phenotypic variance. Mean-standardized measures of evolvability were calculated, namely the coefficient of additive genetic variation (CVA), and IA50,51,52. CVp and CVr were also calculated as described in50. Standard errors for all quantitative genetic parameters were calculated by jackknifing across sire families53.
We used bivariate Animal Models in ASREML 354 to examine the genetic correlation between early life (number of matings accepted in opportunities 1–6) and later life mating frequency (number of matings accepted in opportunities 7- death). We also calculated the genetic correlation between longevity and early life mating frequency with bivariate animal models.
Additional Information
How to cite this article: Travers, L. M. et al. Live fast die young life history in females: evolutionary trade-off between early life mating and lifespan in female Drosophila melanogaster. Sci. Rep. 5, 15469; doi: 10.1038/srep15469 (2015).
Acknowledgments
We are grateful to Cameron Duggin, Leanda Mason, Carly Wilson and Nadia Sloan for their assistance throughout this study. We thank Joe Tomkins and Andreas Sutter for assistance with statistical analyses. We thank Gemma Fitzpatrick and Stephen Robinson for the maintenance of fly lines. We are also grateful to Jason Kennington for providing wild type flies for the experiment. This study was funded by the University of Western Australia and grants from the Australian Research Council to FGG (DP0985859) and LWS (DP110104594), and a grant from the Spanish Ministry of Economy to FGG (CGL2012-34685, co-funded by the European Regional Development Fund). FGG was also supported by the Spanish Ministry of Economy through the Ramon y Cajal program and the Spanish Severo Ochoa Program (SEV-2012-0262).
Footnotes
Author Contributions L.M.T., F.G.G. and L.W.S. conceived the study. L.M.T. conducted the experiment. L.M.T. and F.G.G. analysed the data. L.M.T. wrote the first draft of the manuscript, and all authors contributed to the final version.
References
- Rose M. R. Evolutionary biology of aging. Vol. 164 (Oxford University Press New York, 1991). [Google Scholar]
- Medawar P. B. Aging: An Unsolved Problem of Biology. (HK Lewis, 1952). [Google Scholar]
- Williams G. C. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 (1957). [Google Scholar]
- Partridge L. & Barton N. Evolution of aging: Testing the theory using Drosophila. Genetica 91, 89–98 (1993). [DOI] [PubMed] [Google Scholar]
- Kirkwood T. B. Evolution of ageing. Nature 270, 301–304 (1977). [DOI] [PubMed] [Google Scholar]
- Kirkwood T. B. L. & Rose M. R. Evolution of senescence: late survival sacrificed for reproduction. Philosophical Transactions of the Royal Society of London. Series B, Biological sciences. 332, 15–24 (1991). [DOI] [PubMed] [Google Scholar]
- Stearns S. C. The evolution of life histories. (Oxford University Press Oxford, 1992). [Google Scholar]
- Rose M. R. Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution 38, 1004–1010 (1984). [DOI] [PubMed] [Google Scholar]
- Partridge L., Prowse N. & Pignatelli P. Another set of responses and correlated responses to selection on age at reproduction in Drosophila melanogaster. Proceedings of the Royal Society of London B: Biological Sciences 266, 255–261 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. R. & Charlesworth B. Genetics of life history in Drosophila melanogaster II. Exploratory selection experiments. Genetics 97, 187–196 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckinbill L. S., Arking R., Clare M. J., Cirocco W. C. & Buck S. A. Selection for delayed senescence in Drosophila melanogaster. Evolution 38, 996–1003 (1984). [DOI] [PubMed] [Google Scholar]
- Partridge L. & Fowler K. Direct and correlated responses to selection on age at reproduction in Drosophila melanogaster. Evolution 46, 76–91 (1992). [DOI] [PubMed] [Google Scholar]
- Bateman A. J. Intra-sexual selection in Drosophila. Heredity 2, 349 (1948). [DOI] [PubMed] [Google Scholar]
- Trivers R. In Parental investment and sexual selection. Sexual Selection & the Descent of Man 136–179 (Aldine Publishing Company, 1972). [Google Scholar]
- Vinogradov A. E. Male reproductive strategy and decreased longevity. Acta biotheoretica 46, 157–160 (1998). [DOI] [PubMed] [Google Scholar]
- Bonduriansky R., Maklakov A., Zajitschek F. & Brooks R. Sexual selection, sexual conflict and the evolution of ageing and life span. Functional Ecology 22, 443–453, 10.1111/j.1365-2435.2008.01417.x (2008). [DOI] [Google Scholar]
- Carranza J. & Pérez-Barbería F. J. Sexual selection and senescence: male size‐dimorphic ungulates evolved relatively smaller molars than females. The American Naturalist 170, 370–380 (2007). [DOI] [PubMed] [Google Scholar]
- Finch C. Senescence, longevity, and the genome. (University of Chicago Press, Chicago, 1990). [Google Scholar]
- Hunt J., Jennions M. D., Spyrou N. & Brooks R. Artificial selection on male longevity influences age‐dependent reproductive effort in the Black field cricket Teleogryllus commodus. The American Naturalist 168, E72–E86 (2006). [DOI] [PubMed] [Google Scholar]
- Stearns S., Ackermann M., Doebeli M. & Kaiser M. Experimental evolution of aging, growth, and reproduction in fruitflies. Proceedings of the National Academy of Sciences 97, 3309–3313 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G. & Nilsson T. The evolution of polyandry: multiple mating and female fitness in insects. Animal Behaviour 60, 145–164 (2000). [DOI] [PubMed] [Google Scholar]
- Newcomer S. D., Zeh J. A. & Zeh D. W. Genetic benefits enhance the reproductive success of polyandrous females. Proceedings of the National Academy of Sciences 96, 10236–10241 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatyer R. A., Mautz B. S., Backwell P. R. Y. & Jennions M. D. Estimating genetic benefits of polyandry from experimental studies: a meta-analysis. Biological Reviews 87, 1–33, 10.1111/j.1469-185X.2011.00182.x (2012). [DOI] [PubMed] [Google Scholar]
- Fowler K. & Partridge L. A cost of mating in female fruitflies. Nature 338, 760–761 (1989). [Google Scholar]
- Chapman T., Liddle L. F., Kalb J. M., Wolfner M. F. & Partridge L. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature 373, 241–244 (1995). [DOI] [PubMed] [Google Scholar]
- Chapman T. Seminal fluid‐mediated fitness traits in Drosophila. Heredity 87, 511–521 (2001). [DOI] [PubMed] [Google Scholar]
- Wolfner M. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88, 85–93 (2002). [DOI] [PubMed] [Google Scholar]
- Rice W. R. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381, 232–234 (1996). [DOI] [PubMed] [Google Scholar]
- Holland B. & Rice W. R. Experimental removal of sexual selection reverses intersexual antagonistic coevolution and removes a reproductive load. Proceedings of the National Academy of Sciences of the United States of America 96, 5083–5088, 10.1073/pnas.96.9.5083 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S., Brown W. D. & Miller G. T. Evolution of female remating behaviour following experimental removal of sexual selection. Proceedings of the Royal Society of London. Series B: Biological Sciences 268, 557–563 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S. & Garcia–Gonzalez F. Harm to females increases with male body size in Drosophila melanogaster. Proceedings of the Royal Society of London. Series B: Biological Sciences 269, 1821–1828 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow D. Mate choice, sexual conflict, and evolution of senescence. Behavior genetics 33, 191–201 (2003). [DOI] [PubMed] [Google Scholar]
- Maklakov A. A., Fricke C. & Arnqvist G. Sexual selection affects lifespan and aging in the seed beetle. Aging Cell 6, 739–744, 10.1111/j.1474-9726.2007.00333.x (2007). [DOI] [PubMed] [Google Scholar]
- Williams P. D. & Day T. Antagonistic pleiotropy, mortality source interactions, and the evolutionary theory of senescence. Evolution 57, 1478–1488, 10.1111/j.0014-3820.2003.tb00356.x (2003). [DOI] [PubMed] [Google Scholar]
- Reznick D. N., Bryant M. J., Roff D., Ghalambor C. K. & Ghalambor D. E. Effect of extrinsic mortality on the evolution of senescence in guppies. Nature 431, 1095–1099 (2004). [DOI] [PubMed] [Google Scholar]
- Promislow D. E., Smith E. A. & Pearse L. Adult fitness consequences of sexual selection in Drosophila melanogaster. Proceedings of the National Academy of Sciences 95, 10687–10692 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick D. Costs of reproduction: an evaluation of the empirical evidence. Oikos 44, 257–267 (1985). [Google Scholar]
- Harshman L. G. Life span extension of Drosophila melanogaster: genetic and population studies. Population and Development Review 29, 99–126 (2003). [Google Scholar]
- Robinson M. R., Pilkington J. G., Clutton-Brock T. H., Pemberton J. M. & Kruuk L. E. Live fast, die young: trade‐offs between fitness components and sexually antagonistic selection on weaponry in Soay sheep. Evolution 60, 2168–2181 (2006). [PubMed] [Google Scholar]
- Lemaître J.-F., Gaillard J.-M., Pemberton J. M., Clutton-Brock T. H. & Nussey D. H. Early life expenditure in sexual competition is associated with increased reproductive senescence in male red deer. Proceedings of the Royal Society B: Biological Sciences 281, 10.1098/rspb.2014.0792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A., Perrins C., McCleery R. H. & Sheldon B. C. Quantitative genetics of age at reproduction in wild swans: support for antagonistic pleiotropy models of senescence. Proceedings of the National Academy of Sciences 103, 6587–6592 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérubé C. H., Festa-Bianchet M. & Jorgenson J. T. Individual differences, longevity, and reproductive senescence in Bighorn ewes. Ecology 80, 2555–2565, (1999). [Google Scholar]
- Wolfner M. F. Tokens of love: Functions and regulation of Drosophila male accessory gland products. Insect Biochemistry and Molecular Biology 27, 179–192, 10.1016/s0965-1748(96)00084-7 (1997). [DOI] [PubMed] [Google Scholar]
- Lung O. et al. The Drosophila melanogaster seminal fluid protein Acp62F is a protease inhibitor that is toxic upon ectopic expression. Genetics 160, 211–224 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez F. & Evans J. P. Fertilization success and the estimation of genetic variance in sperm competitiveness. Evolution 65, 746–756, 10.1111/j.1558-5646.2010.01127.x (2011). [DOI] [PubMed] [Google Scholar]
- Byrne P. G. & Rice W. Remating in Drosophila melanogaster: an examination of the trading-up and intrinsic male-quality hypotheses. Journal of Evolutionary Biology 18, 1324–1331 (2005). [DOI] [PubMed] [Google Scholar]
- Bates D., Maechler M. & Bolker B. lme4: Linear mixed-effects models using Eigen and S4. v. R package version 1.1-6. http://CRAN.R-project.org/package=lme4 (2014).
- R Core Team: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/. (2014).
- Fox J. & Weisberg S. An R Companion to Applied Regression. Second Edition, (Thousand Oaks CA, 2011). [Google Scholar]
- Garcia-Gonzalez F., Simmons L. W., Tomkins J. L., Kotiaho J. S. & Evans J. P. Comparing evolvabilities: common errors surrounding the calculation and use of coefficients of additive genetic variation. Evolution 66, 2341–2349 (2012). [DOI] [PubMed] [Google Scholar]
- Houle D. Comparing evolvability and variability of quantitative traits. Genetics 130, 195–204 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. F., Pélabon C. & Houle D. Heritability is not evolvability. Evolutionary Biology 38, 258–277 (2011). [Google Scholar]
- Roff D. A. An introduction to computer-intensive methods of data analysis in biology. (Cambridge University Press, 2006). [Google Scholar]
- ASReml user guide release 3.0 (VSN International Ltd, Hemel Hempstead, UK, 2009).


